Abstract
Proteotoxicity resulting from accumulation of damaged/unwanted proteins contributes prominently to cellular aging and neurodegeneration. Proteasomal removal of these proteins upon covalent polyubiquitination is highly regulated. Recent reports proposed a role for autophagy in clearance of diffuse ubiquitinated proteins delivered by p62/SQSTM1. Here, we compared the turnover dynamics of endogenous ubiquitinated proteins by proteasomes and autophagy by assessing the effect of their inhibitors. Autophagy inhibitors bafilomycin A1, ammonium chloride, and 3-methyladenine failed to increase ubiquitinated protein levels. The proteasome inhibitor epoxomicin raised ubiquitinated protein levels at least 3-fold higher than the lysosomotropic agent chloroquine. These trends were observed in SK-N-SH cells under serum or serum-free conditions and in WT or Atg5−/− mouse embryonic fibroblasts (MEFs). Notably, chloroquine considerably inhibited proteasomes in SK-N-SH cells and MEFs. In these cells, elevation of p62/SQSTM1 was greater upon proteasome inhibition than with all autophagy inhibitors tested and was reduced in Atg5−/− MEFs. With epoxomicin, soluble p62/SQSTM1 associated with proteasomes and p62/SQSTM1 aggregates contained inactive proteasomes, ubiquitinated proteins, and autophagosomes. Prolonged autophagy inhibition (96 h) failed to elevate ubiquitinated proteins in rat cortical neurons, although epoxomicin did. Moreover, prolonged autophagy inhibition in cortical neurons markedly increased p62/SQSTM1, supporting its degradation mainly by autophagy and not by proteasomes. In conclusion, we clearly demonstrate that pharmacologic or genetic inhibition of autophagy fails to elevate ubiquitinated proteins unless the proteasome is affected. We also provide strong evidence that p62/SQSTM1 associates with proteasomes and that autophagy degrades p62/SQSTM1. Overall, the function of p62/SQSTM1 in the proteasomal pathway and autophagy requires further elucidation.
Keywords: Autophagy, Proteasome, Protein Degradation, Ubiquitin, Ubiquitination, p62/Sequestosome 1
Introduction
Chronic neurodegenerative disorders, such as Alzheimer and Parkinson diseases as well as amyotrophic lateral sclerosis, are a heterogeneous group of diseases characterized by selective loss of neurons in specific regions of the CNS. Despite their heterogeneity, they have similar features, including abnormal deposition of ubiquitinated protein aggregates in inclusion bodies within neurons in the respective affected areas of the CNS (reviewed in Ref. 1). The ubiquitinated protein aggregates are thought to result from dysfunction of the ubiquitin/proteasome pathway (UPP)2 or from structural changes in the protein substrates that prevent their degradation by the UPP (reviewed in Ref. 2). Emerging studies also implicate autophagy impairment in the formation of the ubiquitinated protein aggregates. Accordingly, two recent studies described prominent ubiquitin-positive aggregates in neurons of autophagy-deficient (Atg5−/− or Atg7−/−) mice (3, 4). Based on these results and on the finding that there was no apparent alteration in proteasome activity in the brains of autophagy-deficient mice (4), it was suggested that autophagy acts continuously to dispose of diffuse ubiquitinated proteins in a housekeeping role (3–5).
Although the UPP and autophagy were thought to work in parallel, recent investigations suggest a functional link between the two proteolytic pathways (reviewed in Ref. 6). The sequestosome 1/p62 (p62/SQSTM1) could play an important role in mediating the link between the two pathways. Because of its ability to interact with polyubiquitin chains, p62/SQSTM1 was suggested to be a receptor that binds and delivers polyubiquitinated proteins to the two proteolytic pathways (7). p62/SQSTM1 is a protein prone to aggregation and was first isolated in human tissues by Shin and co-workers (8). At its N terminus, p62/SQSTM1 has a Phox-BEM1 domain, which is a protein-protein interaction domain that assumes ubiquitin-like folding and can directly bind to proteasomes and other Phox-BEM1-containing proteins, including itself (9). Recently, p62/SQSTM1 was shown to also interact with LC3, a protein that is an autophagosomal marker (10). p62/SQSTM1 binds directly to LC3 via a 22-amino acid sequence, the LC3-interacting region (10, 11). By binding to polyubiquitinated proteins via its C-terminal ubiquitin-associated domain, to proteasomes via its N-terminal Phox-BEM1 domain, and to LC3-II via its LC3-interacting region domain, p62/SQSTM1 could direct polyubiquitinated proteins to the proteasome or to autophagy, when proteasomes are impaired or overwhelmed (10, 11). p62/SQSTM1 may thus be a candidate for the missing link between the UPP and autophagy.
Here, we compared the dynamics of the turnover of ubiquitinated proteins by proteasomes and autophagy by assessing the effect of pharmacologic inhibitors of each pathway on the accumulation of endogenous ubiquitinated proteins as well as on p62/SQSTM1. We conclusively demonstrate that autophagy impairment does not cause the accumulation of ubiquitinated proteins. The minimal accumulation of ubiquitinated proteins observed upon chloroquine treatment is due to weak proteasome inhibition by the lysosomotropic agent. Other autophagy inhibitors or genetic impairment of autophagy in Atg5−/− MEFs did not cause the accumulation of ubiquitinated proteins. Furthermore, it is clear that p62/SQSTM1 is associated with proteasomes and is degraded mainly by autophagy. The role of p62/SQSTM1 in both proteolytic pathways, i.e. proteasomes and autophagy, requires further elucidation.
EXPERIMENTAL PROCEDURES
Materials
The protease inhibitors used are as follows: chloroquine, bafilomycin A1, 3-methyladenine, and ammonium chloride (Sigma); epoxomicin (Peptides International Inc., Louisville, KY). The substrates used are as follows: Suc-LLVY-AMC (Bachem Bioscience Inc., King of Prussia, PA). The primary antibodies used are as follows: rabbit polyclonal anti-ubiquitinated proteins (1:1500, catalog no. Z0458, Dako North America, Carpinteria, CA); mouse monoclonal anti-ubiquitin antibody (for immunofluorescence detects mono- and polyubiquitinated proteins, 1:500, catalog no. BML-PW8810), anti-α4 (1:500, catalog no. PW8120), anti-Rpt6/S8 (1:1000, catalog no. PW9265), rabbit polyclonal anti-β5 (1:1000, catalog no. PW8895), in vitro synthesized Lys63-only and Lys48-only polyubiquitinated substrates (all from Enzo Life Sciences, Farmingdale, NY); mouse monoclonal anti-B-actin (1:10,000, catalog no. A-2228) and rabbit polyclonal anti-B-actin (1:10,000, catalog no. A-2066) both from Sigma; rabbit polyclonal anti-p62/SQSTM1 (1:1000, catalog no. PM045) and anti-Atg5 (1:1000, catalog no. PM050), and mouse monoclonal anti-Atg16 (1:1000, catalog no. M150–3) from MBL International Corp. (Woburn, MA). For the glycerol gradient fractionation the following were used: mouse monoclonal p62/SQSTM1 (Lck ligand) (1:500, catalog no. 610833, BD Transduction Laboratories, San Jose, CA); rabbit polyclonal anti-LC3 (1:1000, catalog no. Nb 100-2220, Novus Biological, Littleton, CO); secondary antibodies with HRP conjugate (1:10,000, Bio-Rad); and for immunofluorescence, Alexa-488 goat anti-mouse and Alexa-568 goat anti-rabbit (1:500, catalog no. A11029 and A11036, respectively, Molecular Probes, Carlsbad, CA).
Cells
Human neuroblastoma SK-N-SH cells were derived from peripheral tissue (12) and were obtained from ATCC. Under serum conditions, the cells were maintained at 37 °C and 5% CO2 in minimal essential media with Eagle's salts containing 2 mm l-glutamine, 1 mm sodium pyruvate, 0.4% minimal essential media vitamins, 0.4% minimal essential media nonessential amino acids, 100 units/ml penicillin, 100 μg/ml streptomycin, and 5% normal fetal bovine serum. Under serum-free conditions, the cells were maintained as in serum conditions except that the media lacked nonessential amino acids and fetal bovine serum.
Wild type and Atg5−/− mouse embryonic fibroblast cell lines (MEFs) were obtained from RIKEN BRC (Japan) and cultured at 37 °C and 5% CO2 in DMEM with 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% normal fetal bovine serum as described previously (13). The Atg5−/− cell line is deficient in Atg5, which is essential for autophagosome formation. Wild type and Atg5−/− MEFs were prepared from 13.5-day-old embryos and transformed with pEF321-T, an SV40 large T antigen expression vector to generate immortalized cell lines (13).
Rat cerebral cortical neuronal cultures were prepared from E18 embryos obtained from pregnant Sprague-Dawley females following the methods described previously (14). Cells were cultured at 37 °C and 5% CO2 in neurobasal media supplemented with B27 and 0.5 mm l-GlutaMAX. Cells were plated on 100-mm dishes precoated with 50 μg/ml poly-d-lysine and at a density of 6 million cells per dish. Experiments were carried out following 8 days in culture.
Cell Treatments
Cells were treated at 37 °C for different times with vehicle (0.5% DMSO) or different concentrations of the protease inhibitors listed above. Drugs were added dropwise directly into the medium with a gentle swirl of the culture plate. At the end of the incubation, all cultures were washed twice with phosphate-buffered saline (PBS) and processed for the different assays as described below. Cell washes removed unattached cells; therefore, subsequent assays were performed on adherent cells only.
Cell Viability
Cell survival was assessed with the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay as described previously (15).
Western Blotting
After treatment, cells were rinsed twice with PBS and harvested by gently scraping into hot (100 °C) SDS buffer (0.01 m Tris-EDTA, pH 7.5, and 1% SDS) to make sure all intracellular proteins were included. Samples were subjected to a 5-min boil at 100 °C followed by a brief sonication. After determination of the protein concentration with the bicinchoninic acid assay kit (Pierce), the following was added to each sample: β-mercaptoethanol (358 mm), bromphenol blue (0.005%), glycerol (20%), and SDS (4%) in stacking gel buffer (0.1 m Tris-Cl, pH 6.8). Following SDS-PAGE on 8 or 10% polyacrylamide gels, proteins were transferred to an Immobilon-P membrane (Millipore, Bedford, MA). The membranes were probed with the respective antibodies, and antigens were visualized by a standard chemiluminescent horseradish peroxidase method with the ECL reagent. Semi-quantification of protein detection was done by image analysis with the ImageJ program (National Institutes of Health, //rsb.info.nih.gov). Relative intensity (no units) is the ratio between the value for each protein and the value for the respective loading control.
Peptidase Activity of the 20 S Proteasome
Total cell lysates were prepared on ice by homogenization in 0.01 m Tris-EDTA, pH 7.5, buffer. The lysates were cleared by a 15-min centrifugation (19,000 × g) at 4 °C. The cleared samples were normalized for protein concentration determined with the bicinchoninic acid assay kit (Pierce). The chymotrypsin-like activity was assayed colorimetrically in 25 μg of protein/sample with the substrate Suc-LLVY-AMC (400 μm in DMSO) after 24-h incubations at 37 °C as described previously (16). Values obtained were in the linear range of the reaction (data not shown).
Glycerol Density Gradient Centrifugation
Cells were harvested in 25 mm Tris-HCl, pH 7.5, 2 mm ATP, and 1 mm DTT. Following homogenization and sonication, the lysates were centrifuged (19,000 × g for 15 min) at 4 °C. The cleared supernatants (2 mg of protein/sample) were subjected to centrifugation (83,000 × g for 24 h) at 4 °C in a Beckman SW41 rotor in a 10–40% glycerol gradient (fractions 13 to 1) made in the same lysis buffer. Following centrifugation, 13 fractions (800 μl each) were collected and analyzed. Aliquots (50 μl) of each fraction were assayed for chymotrypsin-like activity with the substrate Suc-LLVY-AMC colorimetrically after 24-h incubations at 37 °C as described previously (16).
Proteins were precipitated with acetone from 700 μl of each fraction and subjected to Western blot analysis (10% gels). The membranes were probed with the respective antibodies, and antigens were visualized by a standard chemiluminescent horseradish peroxidase method with the ECL reagent.
In-gel Proteasome Activity and Detection
Upon treatment with vehicle (DMSO) or the respective drugs, cells were washed twice with PBS and were harvested with the following buffer A: 50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 5 mm ATP (grade 1; Sigma), 1 mm DTT and 10% glycerol, which preserves 26 S proteasome assembly (17). Following homogenization and centrifugation (19,000 × g for 15-min) at 4 °C, the protein concentration of the cleared supernatants was determined with the Bradford assay (Bio-Rad) and normalized with buffer A. The cleared supernatants (80 μg of protein/lane for proteasome activity and 40 μg of protein/lane for Western blotting) were resolved by nondenaturing PAGE using a modification of the method described previously (18). We used a gel consisting of three layers of equal amounts, from the bottom up, of 5, 4, and 3% polyacrylamide with RhinohideTM polyacrylamide strengthener (Molecular Probes). Bromphenol blue was added to the protein samples prior to loading. Nondenaturing minigels were run at 150 V for either 3 h or 90 min. The gels were then incubated on a rocker for 10 min at 37 °C with 15 ml of 400 μm Suc-LLVY-AMC in buffer B (buffer A modified to contain 1 mm ATP). Proteasome bands were visualized upon exposure to UV light (360 nm) and were photographed with a NIKON Cool Pix 8700 camera with a 3-4219 fluorescent green filter (Peca Products, Inc.). Proteins on the native gels were transferred (110 mA) for 2 h onto PVDF membranes. Western blot analyses were then carried out sequentially for detection of the 20 S and 26 S proteasomes with anti-β5 and anti-Rpt6/S8 subunit antibodies. The anti-β5 antibody reacts with a core particle subunit and therefore detects both the 26 S and 20 S proteasomes. The anti-Rpt6/S8 antibody reacts with a regulatory particle subunit, thus only detecting 26 S proteasomes. Antigens were visualized by a chemiluminescent horseradish peroxidase method with the ECL reagent. Aliquots of the samples were also boiled for 5 min in Laemmli buffer and loaded onto 10% gels (40 μg of protein/lane) for Western blot analysis following SDS-PAGE.
Filter Trap Assay
Cells were washed twice with PBS and harvested with RIPA buffer (20 mm Tris-HCl, pH 7.5, 137 mm NaCl, 1 mm EGTA, 10% glycerol (v/v), 1 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, 1 mm β-glycerophosphate, 2.5 mm sodium pyrophosphate, 50 mm sodium fluoride, 1% Nonidet P-40, and protease inhibitor mixture). Following homogenization, lysates were centrifuged at 4 °C (for 15 min at 19,000 × g). Cell pellets were resuspended by sonication in harvesting buffer containing 2% SDS; 100 μg of protein/sample were trapped by filtration through a pre-wet Trans-Blot nitrocellulose membrane 0.2 μm adapted to a 96-well dot blot apparatus (Bio-Rad) as described previously (19). Because of the 0.2-μm pore size of this membrane, only aggregated proteins are retained, although the soluble ones pass through the pores of the membrane. To detect aggregates, the membranes were probed with the anti-p62/SQSTM1 or the anti-ubiquitinated protein antibodies.
In Situ Proximity Ligation Assay (PLA)
After treatment, SK-N-SH cells were washed twice with PBS and fixed in 4% paraformaldehyde for 15 min at room temperature. The PLA was performed as described previously (20) and with the Duolink PLA detection kit 613 from Axxora, LLC, San Diego. Slides were mounted with Vectashield medium containing DAPI (Vector Laboratories, Inc., Burlingame, CA). Cell staining was visualized with an UltraViewVoX spinning disk confocal microscope (PerkinElmer Life Sciences).
Immunofluorescence Analysis
After treatment, SK-N-SH cells were fixed for 30 min in 4% paraformaldehyde and post-fixed for 2 min in ice-cold methanol, followed by blocking and permeabilization in 2.5% BSA, 5% normal goat serum, and 0.3% Triton X-100 for 1 h, and co-incubated overnight at 4 °C with the respective antibodies. Slides were mounted with ProLong® Gold antifade mounting reagent with DAPI (Invitrogen). Cell staining was visualized with an Axio Imager M2 microscope (Carl Zeiss Micro Imaging, Inc. Thornwood, NY).
Statistical Analysis
statistical significance was estimated using one-way analysis of variance (Tukey-Kramer multiple comparison test) with the Instat 2.0, Graphpad software (San Diego).
RESULTS
Chloroquine, Ammonium Chloride, and Bafilomycin A1 as Autophagy Inhibitors
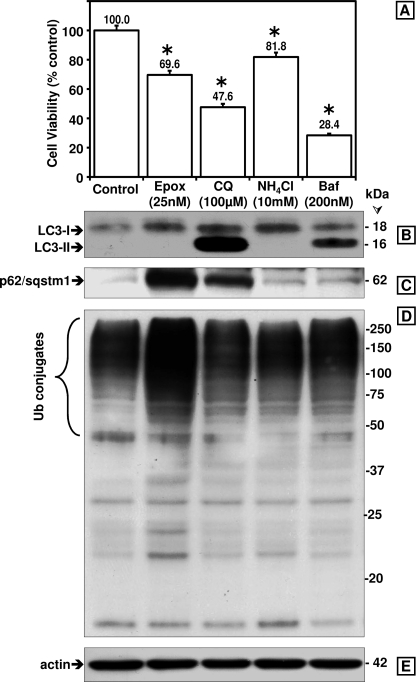
To investigate the link between autophagy and degradation of ubiquitinated proteins, SK-N-SH cells were treated with three different autophagy inhibitors, namely chloroquine (CQ, 100 μm), ammonium chloride (NH4Cl, 10 mm), and bafilomycin A1 (200 nm) at concentrations reported in the literature to inhibit autophagy. We compared the effects of the three drugs on cell viability and autophagy inhibition. In parallel studies, we treated cells with epoxomicin (Epx, 25 nm), an irreversible proteasome inhibitor. Fig. 1A shows that at the concentrations tested bafilomycin A1 was the most toxic of the drugs tested.
FIGURE 1.
Comparison between the effects of proteasome and autophagy inhibitors on cytotoxicity as well as on LC3-I and LC3-II, p62/SQSTM1, and ubiquitinated protein levels in SK-N-SH cells. A, cell viability was assessed with the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. Data represent the mean ± S.E. from at least eight determinations. The viability for each condition was compared with the viability of cells treated with vehicle only (control, 100%). The asterisk identifies the values that are significantly different (p < 0.001) from the control. Western blot analyses (8 and 10% gels) were used to detect LC3-I and LC3-II (B), p62/SQSTM1 (C), ubiquitinated (Ub) proteins (D), and actin as loading control (E) in total extracts of human SK-N-SH neuroblastoma cells (40 μg of protein/lane). Cells were treated with the different inhibitors for 24 h. Molecular mass markers in kDa are shown on the right. Similar results were obtained in duplicate experiments. Epx, epoxomicin; CQ, chloroquine; NH4Cl, ammonium chloride; Baf, bafilomycin A1; Ub, ubiquitin.
Autophagy inhibition was assessed by accumulation of the autophagosome marker LC3-II, which is accepted as an overall indicator of autophagy impairment. LC3-II levels were most prominent after chloroquine treatment (123-fold increase) as indicated by the strong LC3-II band on the Western blots (Fig. 1B). In bafilomycin A1-treated cells, LC3-II levels (32-fold increase) were lower than in chloroquine-treated cells, and ammonium chloride and epoxomicin treatment failed to induce LC3-II accumulation (Fig. 1B). These data show that among the three autophagy inhibitors tested, chloroquine most effectively raised LC3-II levels. Proteasome inhibition by epoxomicin had no inhibitory effect on autophagy.
Epoxomicin and to a Lesser Extent Chloroquine Increased p62/SQSTM1 and Ubiquitinated Protein Levels but the Two Other Autophagy Inhibitors Did Not
p62/SQSTM1 was suggested to be specifically degraded by autophagy (10, 21), and its levels were proposed to increase in response to autophagy inhibition (22). We compared how the four proteolytic inhibitors listed above affected p62/SQSTM1 as well as ubiquitinated protein levels. It is clear that from the four drugs tested, epoxomicin most effectively increased p62/SQSTM1 (9-fold, Fig. 1C) and ubiquitinated protein (3-fold, Fig. 1D) levels in SK-N-SH cells. Chloroquine caused a detectable increase in p62/SQSTM1 (6-fold), albeit to a lesser extent than epoxomicin. Ammonium chloride and bafilomycin A did not increase p62/SQSTM1 levels. Another autophagy inhibitor, 3-MA, failed to increase the levels of ubiquitinated proteins, p62/SQSTM1 and LC3-II (supplemental Fig. 1). Upon treatment with chloroquine, we observed a 1.7-fold increase in ubiquitinated proteins. Actin levels were not increased by any of the treatments (Fig. 1E).
To further investigate the cross-talk between the UPP and autophagy, we decided to focus our studies on chloroquine and epoxomicin, because ammonium chloride and bafilomycin A1 failed to increase p62/SQSTM1 and ubiquitinated protein levels, and the latter inhibitor was highly cytotoxic.
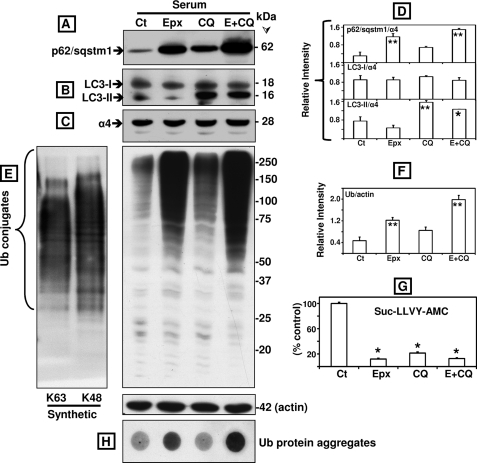
We compared the effects of inhibiting each pathway alone with inhibiting both pathways together (Fig. 2). Similar results were obtained in cells maintained under serum or serum-free conditions (the latter not shown). p62/SQSTM1 levels were raised 4-fold over control by epoxomicin (p < 0.01) but only 2-fold by chloroquine (p > 0.05, not significant) (Fig. 2, A and D). In cells treated with both inhibitors, p62/SQSTM1 levels were higher than in cells treated with epoxomicin alone, but the difference was not statistically significant (p > 0.05). As expected, LC3-II levels were increased only in cells treated with chloroquine as proteasome inhibition does not block autophagy function. Chloroquine alone raised LC3-II levels 2-fold (p < 0.01, Fig. 2, B and D). LC3-II levels were similar in cells treated with chloroquine alone or in combination with epoxomicin (p > 0.05). The levels of LC3-I and the α4 subunit of the 20 S proteasome did not show any significant changes (Fig. 2, B–D). Together, these data clearly demonstrate that proteasome inhibition most efficiently increases p62/SQSTM1 levels when compared with autophagy impairment, the latter ascertained by accumulation of LC3-II in cells treated with chloroquine.
FIGURE 2.
Comparison between the effects of epoxomicin (Epx or E, 25 nm), chloroquine (100 μm), and the two combined (E+CQ) on p62/SQSTM1, LC3-I and LC3-II, ubiquitinated proteins, protein aggregates, and proteasome activity in SK-N-SH cells maintained in serum conditions. Western blot analyses were used to detect p62/SQSTM1 (A), LC3-I and LC3-II (B), the proteasome subunit α4 (C), and ubiquitinated proteins as well as actin as loading control (E) in total extracts of human SK-N-SH neuroblastoma cells (40 μg of protein/lane). In vitro synthesized Lys63-only and Lys48-only polyubiquitinated substrates (5 μg of protein/lane) were also loaded in the 1st two lanes in E. Molecular mass markers in kDa are shown on the right. Cells were treated with each inhibitor by itself or in combination for 24 h. The levels of p62/SQSTM1, LC3-I and LC3-II (D), and ubiquitinated proteins (F) were semi-quantified by densitometry. Data represent the relative intensity for all proteins. Values represent mean ± S.D. from at least duplicate experiments. The asterisks identify values that are significantly different (*, p at least <0.05; **, p < 0.01) from control (Ct). The chymotrypsin-like activity of the proteasome (G) was measured in cleared supernatants obtained from total cell homogenates (25 μg of protein/sample). The peptidase activity was assayed colorimetrically after 24-h incubations at 37 °C with Suc-LLVY-AMC. Data represent the mean ± S.E. from three experiments. The asterisks identify values that are significantly different (*, p at least <0.001) from control conditions. Aggregates of ubiquitinated (Ub) proteins were assessed with the filter trap assay (H).
Both Epoxomicin and Chloroquine Inhibit Suc-LLVY-AMC Hydrolysis
Inhibition of lysosomal function was recently shown to reduce proteasome activity (23), and therefore, we compared the effect of epoxomicin and chloroquine on the cleavage of Suc-LLVY-AMC, a short substrate used to measure proteasome activity. Similar results were obtained under serum and serum-free conditions (the latter not shown). As shown in Fig. 2G, Suc-LLVY-AMC hydrolysis measured in total lysates was blocked by at least 80% in cells treated with epoxomicin or chloroquine alone or combined.
Despite a similar decline in Suc-LLVY-AMC hydrolysis observed in cells treated with each or both drugs, the levels of ubiquitinated proteins were quite different. Treatment with epoxomicin caused an ∼3-fold rise in ubiquitinated protein levels (p < 0.01) compared with a much smaller increase (1.8-fold, p > 0.05) in cells treated with chloroquine (Fig. 2, E and F). Combined treatment with epoxomicin and chloroquine enhanced ubiquitinated protein levels by 4-fold, which was significantly different from the values observed under epoxomicin alone (p < 0.01, Fig. 2, E and F). Ubiquitin protein aggregates were detected with the filter trap assay in cells treated with epoxomicin by itself or in combination with chloroquine (Fig. 2H). Actin levels were not changed upon proteasome and/or autophagy inhibition (Fig. 2E).
These data clearly demonstrate that measuring Suc-LLVY-AMC hydrolysis using total cell lysates does not accurately reflect proteasome activity. This is not surprising, because it is well established that the substrate Suc-LLVY-AMC is cleaved not only by the proteasome (24) but also by other chymotrypsin-like proteases as well as by calpains (25). In addition, it is clear that the overall levels of ubiquitinated proteins in chloroquine-treated cells are very low. This includes Lys48- and Lys63-linked chains as both are detected with the polyubiquitin antibody from DAKO (Carpinteria, CA) used in these studies (Fig. 2E, left panel). This is relevant to autophagy because Lys63-linked chains were postulated to selectively facilitate the clearance of ubiquitinated proteins via the autophagic pathway (26).
Chloroquine Inhibits Proteasome Activity albeit to a Lesser Extent than Epoxomicin
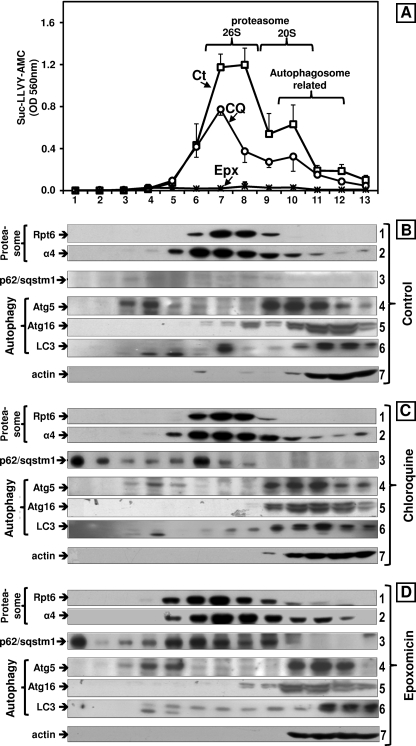
The decline in Suc-LLVY-AMC hydrolysis together with the accumulation of ubiquitinated proteins observed in cells treated with chloroquine could reflect proteasome inhibition by the lysosomotropic agent. To test this hypothesis, total extracts from cells treated with chloroquine under serum or serum-free conditions (the latter not shown) were fractionated by glycerol density gradient centrifugation. We also evaluated control and epoxomicin-treated cells. Fractions were analyzed for Suc-LLVY-AMC hydrolysis, which reflects the chymotrypsin-like activity.
Compared with controls, the chymotrypsin-like activity of chloroquine-treated cells was significantly reduced (Fig. 3A) in the fractions corresponding to the elution of both forms of the proteasome as follows: fractions 7–8 (peak for 26 S) and fractions 9–10 (20 S); compare chloroquine treatment (circles) with control (squares). Almost no Suc-LLVY-AMC hydrolysis was detected in cells treated with epoxomicin (Fig. 3A, crosses close to the x axis).
FIGURE 3.
Effects of epoxomicin (25 nm) and chloroquine (100 μm) on the sedimentation velocity of proteasomes and autophagy-related proteins in SK-N-SH cells maintained in serum conditions. Total lysates (2 mg of protein/sample) were fractionated by glycerol density gradient centrifugation (10–40% glycerol corresponding to fractions 13 to 1). A, aliquots (50 μl) of each fraction obtained from control (Ct, squares), chloroquine- (CQ, circles), and epoxomicin (Epx, crosses)-treated cells under serum conditions were assayed for chymotrypsin-like activity with Suc-LLVY-AMC. Data represent the mean ± S.E. from three experiments. B–D, immunoblot analyses of each fraction probed with antibodies that react with the proteasome (α4, core particle; Rpt6/S8, 19 S regulatory particle), with autophagy-related proteins (Atg5, Atg16, and LC3) and with p62/SQSTM1. Proteins were precipitated with acetone from 700 μl of each fraction. The fractions were obtained from control (B), chloroquine (C), and epoxomicin-treated (D) cells. Similar results were obtained in triplicate experiments and under serum and serum-free conditions. The numbers on the right designate each row.
To confirm the proteasome elution pattern, aliquots from each fraction were subjected to Western blot analysis with the anti-α4 antibody that reacts with a subunit of the 20 S core particle and with the anti-Rpt6/S8 antibody that reacts with an ATPase subunit of the 19 S particle (Fig. 3, B–D, rows 1 and 2). Immunodetection of the α4 and Rpt6/S8 subunits confirmed a similar elution pattern under all treatment conditions. From these experiments, we can conclude that a 24-h treatment with 100 μm chloroquine inhibits both the 20 S and 26 S forms of the proteasome, although not as effectively as 25 nm epoxomicin.
p62/SQSTM1 Associates with 26 S Proteasomes and Not with Individual Autophagy-related Proteins, including LC3, upon Fractionation of Total Cells Lysates by Glycerol Gradient Centrifugation
Western blot analyses of the glycerol gradient fractions were probed for p62/SQSTM1 and for three individual autophagy-related proteins Atg5, Atg16, and LC3. As expected, under control conditions no p62/SQSTM1 was detected (Fig. 3B, row 3). The three individual autophagy-related proteins co-eluted with the 20 S proteasome and in lighter fractions (Fig. 3B, rows 4–6). A similar elution pattern was observed in cells treated with chloroquine (Fig. 3C, rows 4–6) and epoxomicin (Fig. 3D, rows 4–6). As a loading control, we also probed the blots for actin (Fig. 3, B–D, row 7).
In accordance with our previous results (Figs. 1 and 2), significantly lower levels of p62/SQSTM1 were detected in cells treated with chloroquine (Fig. 3C, row 3) than with epoxomicin (Fig. 3D, row 3). Most of the p62/SQSTM1 co-eluted with the 26 S proteasome, except for a “putative” aggregated form detected in the heaviest fraction (fraction 1) observed in cells treated with chloroquine or epoxomicin. These results clearly demonstrate that “soluble” p62/SQSTM1 has a higher affinity for 26 S proteasomes than for the autophagy protein LC3.
Corroboration That Chloroquine Inhibits the Proteasome
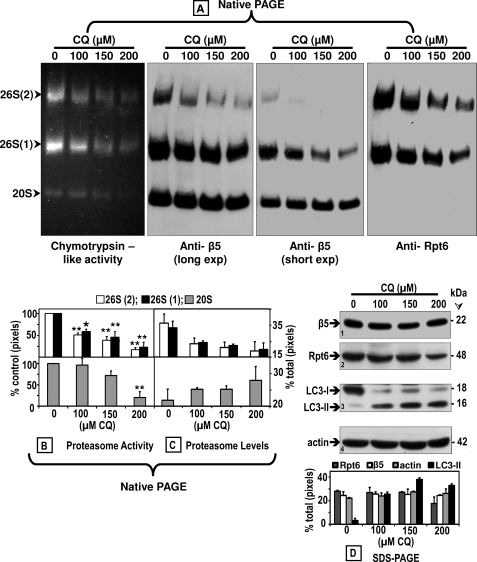
To confirm that chloroquine inhibits the proteasome, we assessed its activity with the in-gel assay using the Suc-LLVY-AMC substrate as described under “Experimental Procedures.” The in-gel assay for measuring proteasome activity has the advantage of differentiating between the two forms of the 26 S proteasome (one capped and two capped) as well as the 20 S proteasome. Cells were treated for 24 h without (0, control) or with three concentrations of chloroquine (100, 150, or 200 μm). An equal amount of protein (80 μg) from the cleared cell lysates was loaded onto each lane of the native gels.
These studies clearly demonstrate a concentration-dependent decline in proteasome activity (Fig. 4A, left panel). Semi-quantification of the bands detected in Fig. 4A show a significant (mostly p < 0.01) decline in proteasome activity (Fig. 4B) in both 26 S forms (upper graph: two capped, white bars; one capped, black bars) and in 20 S proteasomes (bottom graph).
FIGURE 4.
Chloroquine inhibits the proteasome activity and affects its assembly. Crude extracts were prepared from control (0) SK-N-SH cells or cells treated with increasing CQ concentrations for 24 h. Cleared lysates (80 μg/sample) were subjected to nondenaturing gel electrophoresis. A, proteasomal chymotrypsin-like activity was measured with Suc-LLVY-AMC by the in-gel assay (left panel). 26 S and 20 S proteasomes were detected by immunoblotting with an anti-β5 antibody, a subunit of the core proteasome particle (20 S) (two middle panels (long and short exposures (exp))). The 26 S holoenzyme was further identified with an antibody that reacts with the Rpt6/S8 (ATPase) subunit of the 19 S regulatory particle (right panel). Proteasomal 26 S (two capped and one capped) and 20 S forms are indicated by arrowheads on the left. Activity (B) and immunoblot (C) bands on the nondenaturing (native) gel were semi-quantified by densitometry. Data represent total pixels compared with control (no chloroquine) and means and S.E. from three experiments. B, asterisks identify values that are significantly different (*, p at least <0.05; **, p < 0.01) from control (Ct). In parallel experiments (D), cells were harvested for SDS-PAGE followed by Western blot analyses (40 μg of protein/lane) to detect proteasome subunits β5 and Rpt6/S8, autophagy proteins LC3-I and LC3-II, and actin (loading control) after treatment with increasing chloroquine concentrations for 24 h. The level of each protein band was semi-quantified by densitometry (graph). Data represent total pixels and means ± S.D. from duplicate experiments. Molecular mass markers in kDa are shown on the right. The small numbers within the immunoblots designate each row.
Immunoblot analyses of the native gels with the anti-β5 (Fig. 4A, two middle panels) as well as with the anti-Rpt6/S8 (Fig. 4A, right panel) antibodies revealed a substantial concentration-dependent decline in the levels of 26 S proteasomes (Fig. 4C, semi-quantification). On the contrary, the levels of 20 S proteasome increased slightly (Fig. 4A, two middle panels; Fig. 4C, semi-quantification). The total levels of β5 subunit did not vary although Rpt6/S8 was slightly decreased by treatment with the highest (200 μm) chloroquine concentration (Fig. 4D, rows 1 and 2). These findings suggest that the decrease in proteasome activity observed in cells treated with chloroquine is linked to a parallel decline in 26 S proteasome assembly. As expected, LC3-II was detected only in cells treated with chloroquine (Fig. 4D, row 3). Actin levels were unaltered (Fig. 4D, row 4).
Proteasomal Association with p62/SQSTM1 upon Epoxomicin or Chloroquine Treatment
p62/SQSTM1 levels are almost undetectable under basal conditions. To increase p62/SQSTM1 levels, we treated cells with epoxomicin or chloroquine to investigate the association of p62/SQSTM1 with proteasomes and/or LC3 as an indicator for autophagosomes. Not surprisingly, proteasome inhibition assessed by the in-gel assay was much stronger in cells treated with 25 nm epoxomicin than with 100 μm chloroquine (Fig. 5A, left panel). Immunoblot analyses of the native gels with the anti-β5 (Fig. 5A, middle panel) as well as with the anti-Rpt6/S8 (Fig. 5A, right panel) antibodies revealed a decrease in 26 S proteasomes and a slight increase in 20 S proteasomes upon treatment with chloroquine. In cells treated with epoxomicin, such changes were not obvious because, besides the conventional 26 S and 20 S proteasome bands, additional bands were detected with both antibodies (Fig. 5A). These additional bands most likely correspond to increased amounts of proteasome precursor complexes and intermediates shown to occur in cells upon proteasome inhibition (27).
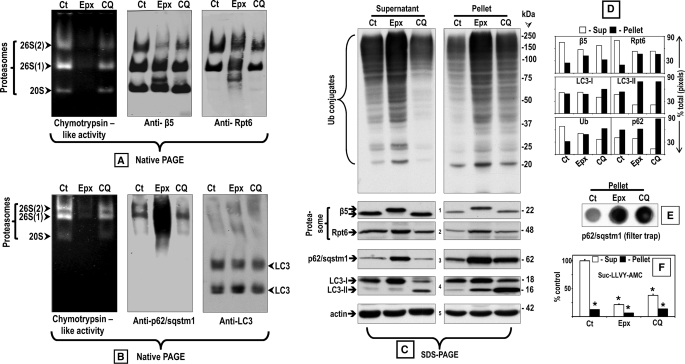
FIGURE 5.
Comparison between the effects of epoxomicin (25 nm) and chloroquine (100 μm) on p62/SQSTM1 and its association with proteasomes and LC3 in supernatant and pellet fractions. Crude extracts were prepared from SK-N-SH cells control (Ct) or treated with each inhibitor for 24 h. Cleared lysates (80 μg/sample) were subjected to nondenaturing gel electrophoresis. A and B, proteasomal chymotrypsin-like activity was measured with Suc-LLVY-AMC by the in-gel assay (left panels). A, 26 S and 20 S proteasomes were detected by immunoblotting with an anti-β5 antibody, a subunit of the core proteasome particle (20 S) (middle panel), and with the anti-Rpt6/S8 antibody, an ATPase subunit of the 19 S regulatory particle (right panel). Proteasomal 26 S (two capped and one capped) and 20 S forms are indicated by arrowheads on the left. B, p62/SQSTM1 (middle panel) and LC3 (right panel) were detected by immunoblotting with the respective antibodies. C, aliquots of the supernatant (cleared lysate) and pellet fractions were run on SDS-PAGE followed by immunoblotting with the anti-ubiquitinated proteins antibody as well as the same antibodies listed in A and B. Molecular mass markers in kDa are shown on the right. The small numbers next to the immunoblots designate each row. The level of the proteins in each immunoblot was semi-quantified by densitometry (D). Data represent % of total pixels (total = supernatant + pellet). The pellet fractions were also subjected to the filter trap assay to detect protein aggregates (E). The chymotrypsin-like activity of the supernatant (Sup) and pellet fractions was measured with Suc-LLVY-AMC (F). The asterisks identify values that are significantly different (*, p at least <0.001) from the control supernatant.
To identify LC3 that is associated with autophagosomes, native gels were run for a much shorter time (90-min instead of 3 h) just until the dye front reached the bottom of the gels (Fig. 5B). The in-gel assay revealed proteasome activity only (Fig. 5B, left panel), but Western blotting with the anti-LC3 antibody detected LC3 indicative of autophagosomes on the bottom half of the gel (Fig. 5B, right panel). Under native conditions, there is no separation between LC3-I and LC3-II; only “total” LC3 is detected in two bands that could represent different autophagosome populations, just like three proteasome bands are observed under native conditions. The levels of total LC3 do not change, probably because often when LC3-I decreases LC3-II increases. In epoxomicin-treated cells, which are the conditions in our experiments under which p62/SQSTM1 levels are the highest, p62/SQSTM1 staining was observed exclusively in association with the proteasome (Fig. 5B, middle panel). In chloroquine-treated cells, almost no p62/SQSTM1 was associated with proteasomes or LC3-associated with autophagosomes (Fig. 5B, middle panel).
To determine the fate of ubiquitinated proteins and p62/SQSTM1 upon chloroquine treatment, cells were harvested in the same buffer used for running native gels. For these native gels, cells are harvested in a buffer that preserves 26 S proteasome assembly (see “Experimental Procedures”), and after homogenization, two fractions are generated upon centrifugation as follows: the cleared supernatant, which is run on the native gel, and a pellet containing insoluble particles that are usually discarded. For SDS-PAGE, both fractions, the cleared supernatant and the pellet, were analyzed followed by Western blotting to detect ubiquitinated proteins, p62/SQSTM1, proteasomes (anti-β5 and anti-Rpt6/S8), and autophagosomes (anti-LC3) (Fig. 5, C, semi-quantification in D). For loading control, the blots were probed for actin (Fig. 5C, row 5). In addition, aggregates in the pellet fractions were assessed by the filter trap assay (Fig. 5E), and Suc-LLVY-AMC hydrolysis was measured in the supernatant and pellet fractions (Fig. 5F).
Under control conditions, ubiquitinated proteins and proteasomes were detected mostly in the soluble fraction (Fig. 5, C, upper panel as well as rows 1 and 2, and D). LC3-I was distributed evenly between the two fractions (Fig. 5, C, row 4 and D). LC3-II and p62/SQSTM1 levels were very low in both fractions (Fig. 5, C and D).
In cells treated with chloroquine, ubiquitinated proteins were more abundant in the pellet fraction (Fig. 5, C, upper panel, and D). Proteasome levels were still higher in the soluble fraction (Fig. 5, C, rows 1 and 2, and D). LC3-I was slightly more abundant in the pellet (Fig. 5, C, row 4, and D). Most of LC3-II and p62/SQSTM1 were found in the pellet (Fig. 5, C, rows 4 and 3, D).
In epoxomicin-treated cells, ubiquitinated proteins, proteasomes, and LC3-I were evenly distributed between the two fractions (Fig. 5, C, upper panel as well as rows 1, 2, and 4, and D). Notably, the β5 subunit displayed a higher molecular weight, possibly because epoxomicin (Mr = 554.7) is covalently bound to it. LC3-I levels were higher than in controls supporting the view that LC3-I is a proteasome substrate (28). p62/SQSTM1 was 1.6-fold more abundant in the pellet than in the supernatant (Fig. 5, C, row 3, and D). LC3-II was almost undetectable in the supernatant, but low levels were present in the pellet (Fig. 5C, row 4).
In chloroquine- and in epoxomicin-treated cells, the pellet fraction contains aggregates with p62/SQSTM1 (Fig. 5E). Under all conditions, proteasome activity assessed with Suc-LLVY-AMC was very low in the pellet fraction (Fig. 5F).
Overall, these findings establish that when the proteasome is inhibited by epoxomicin and p62/SQSTM1 levels are high, p62/SQSTM1 is distributed among two fractions as follows: the soluble (supernatant) fraction was found to be associated with proteasomes and the aggregated (pellet) fraction that also contains inactive proteasomes and autophagosome proteins. Upon autophagy inhibition induced by chloroquine, most of the p62/SQSTM1 co-localizes with autophagosomes in the pellet fraction.
Validation That p62/SQSTM1 Is Co-localized with the Proteasome
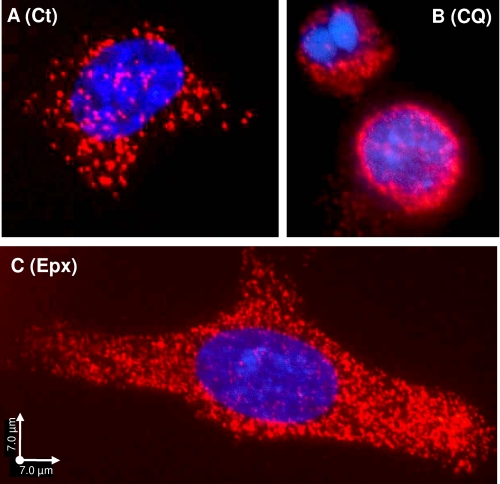
We used a PLA for detecting in situ whether p62/SQSTM1 is co-localized with the proteasome. In this assay, a pair of oligonucleotide-labeled secondary antibodies (PLA probes) generates an individual fluorescent signal when bound to the primary antibodies in close proximity (20, 29, 30). The PLA allows for the in situ detection of individual endogenous proteins that are in close proximity within cells. We found that p62/SQSTM1 and the proteasome co-localize in situ even under basal conditions (Ct, control), when p62/SQSTM1 levels are low (Fig. 6A). In cells treated with CQ (100 μm), the co-localization signal was mostly perinuclear (Fig. 6B). In the epoxomicin-treated cells (25 nm), the signal was distributed throughout the cell (Fig. 6C). The same pattern was obtained with the anti-Rpt6/S8 (Fig. 6) or anti-α4 (data not shown) proteasome antibodies. Under lower magnification (data not shown), most cells in each condition exhibited a signal pattern similar to the one shown in each figure panel. Omission of each or both of the primary antibodies produced no signal (data not shown). These results further substantiate that these two molecules, i.e. p62/SQSTM1 and proteasomes, are closely located within cells.
FIGURE 6.
Protein interaction between p62/SQSTM1 and proteasomes detected by the PLA. SK-N-SH cells control (A, Ct) or treated for 24 h with 100 μm chloroquine (B, CQ) or 25 nm epoxomicin (C, Epx) were co-incubated with a proteasome antibody (Rpt6/S8) and a p62/SQSTM1 antibody. Nuclear staining, DAPI, and PLA signal, Texas Red. Scale bars, 7 μm.
Protein Aggregation Induced by Epoxomicin and Chloroquine
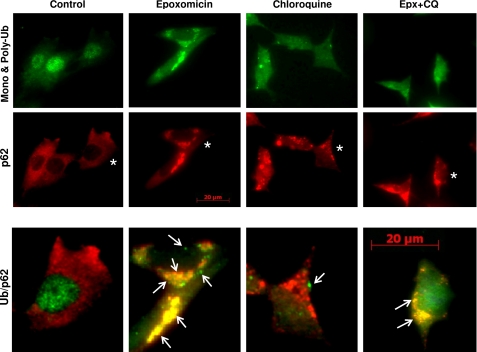
We assessed the effect of the two proteolytic inhibitors alone or in combination on protein aggregation with antibodies that detect mono- and polyubiquitinated proteins (Fig. 7, ubiquitin, green) and p62/SQSTM1 (Fig. 7, red). Upon epoxomicin treatment for 24 h, abundant protein aggregates were observed containing both ubiquitinated proteins and p62/SQSTM1, with few aggregates staining for ubiquitin alone (Fig. 7, merged images on right panels). Chloroquine treatment caused the appearance of protein aggregates containing mostly p62/SQSTM1 with a few containing ubiquitinated proteins as well. Cells treated with epoxomicin and chloroquine showed a pattern similar to those treated with epoxomicin alone. These data strongly indicate that autophagy did not clear the protein aggregates formed in cells treated with epoxomicin alone and that few ubiquitin protein aggregates form upon autophagy impairment.
FIGURE 7.
Effects of epoxomicin and chloroquine on p62/SQSTM1 and ubiquitin-protein aggregates. Double immunofluorescence staining of SK-N-SH cells control or treated with 25 nm epoxomicin or 100 μm chloroquine alone or combined (Epx + CQ) is shown. p62/SQSTM1 (p62) aggregates were visualized with the anti-p62/SQSTM1 antibody (red), and mono- and polyubiquitinated proteins were visualized with the anti-ubiquitin antibody (green). Enlarged merged (ubiquitin /p62) images of cells marked with an asterisk and treated under the different conditions are shown in the bottom panels. Scale bar, 20 μm. Arrows point to ubiquitin-protein aggregates.
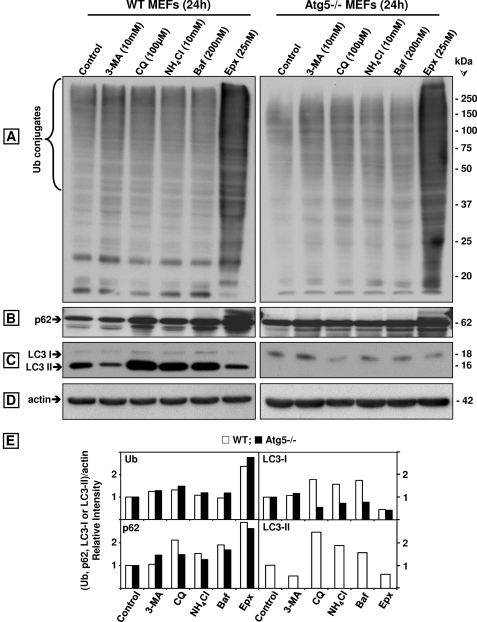
Chloroquine Inhibits Proteasome Activity in Wild Type and Atg5−/− MEFs
To establish if proteasome inhibition by chloroquine was observed in other cell lines besides the human neuroblastoma SK-N-SH cells, we tested the effects of the lysosomotropic agent on wild type (WT) and autophagy-deficient (Atg5−/−) MEFs. Contrary to what was observed with SK-N-SH cells (Fig. 2G), chloroquine at 100 and 200 μm failed to inhibit Suc-LLVY-AMC hydrolysis measured with total cells lysates obtained from WT (white bars) and Atg5−/− (black bars) MEFs (Fig. 8A). In addition, inhibition of Suc-LLVY-AMC hydrolysis by epoxomicin was found to be significantly less effective in MEFs (Fig. 8A) than in SK-N-SH cells (Fig. 2G). The proteasome inhibitor at 25 nm reduced Suc-LLVY-AMC hydrolysis in SK-N-SH cells by almost 90% (Fig. 2G), although even at a concentration as high as 150 nm only 23% inhibition was observed in WT (white bars) and Atg5−/− (black bars) MEFs (Fig. 8A).
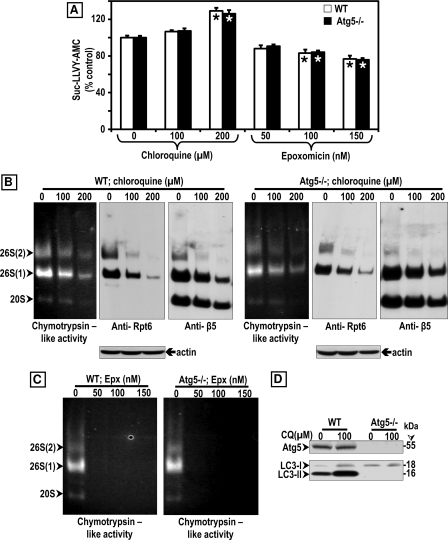
FIGURE 8.
Comparison between the effects of CQ and Epx on proteasome activity in WT and Atg5−/− MEF. A, chymotrypsin-like activity was measured in cleared supernatants obtained from total cell homogenates (25 μg of protein/sample). The peptidase activity was assayed colorimetrically after 24-h incubations at 37 °C with Suc-LLVY-AMC. Data represent the mean ± S.E. from eight determinations. The asterisks identify values that are significantly different (*, p at least <0.01) from control (0). B and C, crude extracts were prepared from WT and Atg5−/− MEFs control (0) or treated with increasing chloroquine (B) or epoxomicin (C) concentrations for 24 h. Cleared lysates (80 μg/sample) were subjected to nondenaturing gel electrophoresis. The proteasomal chymotrypsin-like activity was measured with Suc-LLVY-AMC by the in-gel assay (left panels). 26 S and 20 S proteasomes were detected by immunoblotting with the anti-Rpt6/S8 antibody (for 26 S, middle panels) and the anti-β5 antibody (for 26 S and 20 S, right panels). Proteasomal 26 S (two capped and one capped) and 20 S forms are indicated by arrowheads on the left. In parallel experiments (D), cells were harvested for SDS-PAGE followed by Western blot analyses (40 μg of protein/lane) to detect the autophagy proteins Atg5, LC3-I, and LC3-II under control (0) and CQ treatment for 24 h. Molecular mass markers in kDa are shown on the right.
Based on these surprising results, we decided to assess proteasome activity in MEFs by the in-gel assay (Fig. 8, B and C). Just like in SK-N-SH cells (Fig. 4A), it is clear that chloroquine inhibits proteasome activity and impairs its assembly in a concentration-dependent manner in WT (Fig. 8B, three left panels) and Atg5−/− (Fig. 8B, three right panels) MEFs. For loading control, the blots were probed for actin (Fig. 8B). When assessed by the in-gel assay, epoxomicin effectively inhibited proteasome activity in WT (Fig. 8C, left panel) and Atg5−/− (Fig. 8C, right panel) MEFs. These findings support the view that chloroquine inhibits proteasome activity in cell lines other than SK-N-SH cells, although it is significantly less potent than epoxomicin. In addition, MEFs contain other proteases besides the proteasome that effectively cleave the substrate Suc-LLVY-AMC, and their activity was not reduced by chloroquine or epoxomicin. The nature of these proteases remains to be established. That the Atg5−/− is indeed defective in autophagy is demonstrated in Fig. 8D, as upon chloroquine treatment LC3-II was only detected in WT and not in the Atg5−/− MEFs.
No Increases in Ubiquitinated Proteins and p62/SQSTM1 Are Observed in Atg5−/− MEFs unless They Are Treated with Epoxomicin
Genetically impairing autophagy as in Atg5−/− MEFs did not cause an increase in ubiquitinated proteins or p62/SQSTM1. As shown in Fig. 9, WT (white bars) and Atg5−/− (black bars) MEFs treated with each of the four autophagy inhibitors tested exhibited similar levels of ubiquitinated proteins as controls (Fig. 9, A and E). As expected, the levels of ubiquitinated proteins in epoxomicin-treated MEFs were the highest (Fig. 9, A and E) even when compared with MEFs treated with chloroquine concentrations as high as 200 μm (supplemental Fig. 2). All autophagy inhibitors except for 3-MA elevated p62/SQSTM1 levels in WT but not in Atg5−/− MEFs, and epoxomicin raised p62/SQSTM1 to higher levels than any other treatment (Fig. 9, B and E). LC3-II was detected in WT but not in Atg5−/− MEFs (Fig. 9, C and E), and actin was unaltered under all conditions (Fig. 9D).
FIGURE 9.
Comparison of the effects of the autophagy inhibitors 3-MA, CQ, NH4Cl, and bafilomycin A1 (Baf) with the proteasome inhibitor Epx on ubiquitinated (Ub) proteins, p62/SQSTM1, LC3-I, and LC3-II in wild type (WT) and Atg5−/− MEFs. Western blot analyses (8 or 10% gels) to detect ubiquitinated proteins (A), p62/SQSTM1 (p62, B), LC3-I and LC3-II (C), and actin (D, loading control) in total extracts (40 μg of protein/lane) of MEFs treated with each inhibitor for 24 h. Molecular mass markers in kDa are shown on the right. The protein levels were semi-quantified by densitometry (E). Data represent relative intensity obtained from at least four cell culture dishes per condition.
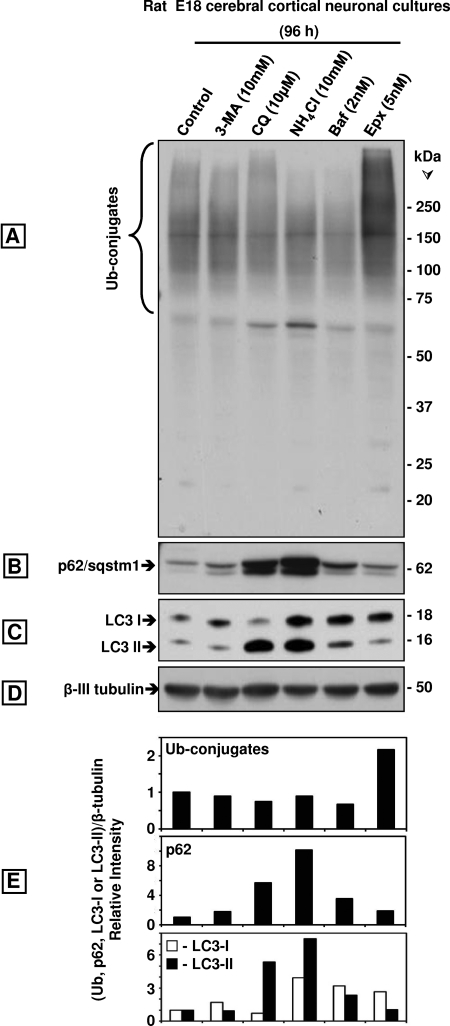
Prolonged Incubations with Four Autophagy Inhibitors Failed to Increase Ubiquitinated Protein Levels but Raised p62/SQSTM1 in Rat Cerebral Cortical Neuronal Cultures
We also investigated the effect of pharmacologically inhibiting autophagy with four different drugs on rat E18 cerebral cortical neuronal cultures. The four autophagy inhibitors failed to increase ubiquitinated protein levels upon 96 h of incubation (Fig. 10, A and E). We used lower drug concentrations than with SK-N-SH cells or MEFs because we incubated the cells for a longer time, and the higher concentrations were cytotoxic. That autophagy was inhibited is confirmed by the increase in LC3-II observed in cells treated with all autophagy inhibitors except for 3-MA, as the latter prevents autophagosome formation (Fig. 10, C and E). As expected, epoxomicin clearly increased the levels of ubiquitinated proteins at concentrations as low as 5 nm for 96 h. Treatments for shorter time points (24 and 48 h) showed a similar trend (supplemental Fig. 3). The levels of p62/SQSTM1 increased markedly upon 96 h of treatment with the autophagy inhibitors but not with epoxomicin (Fig. 10, B and E). In epoxomicin-treated cells, p62/SQSTM1 levels were higher by 24 h of treatment but then declined in the later time points (supplemental Fig. 3). No changes were detected in β-tubulin (Fig. 10C).
FIGURE 10.
Comparison of the effects of the autophagy inhibitors 3-MA, CQ, NH4Cl, and bafilomycin A1 (Baf) with the proteasome inhibitor Epx on ubiquitinated (Ub) proteins, p62/SQSTM1, LC3-I, and LC3-II in rat E18 cerebral cortical neuronal cultures treated with each inhibitor for 96 h. Western blot analyses (8 or 10% gels) to detect ubiquitinated proteins (A), p62/SQSTM1 (p62, B), LC3-I, and LC3-II (C) and β-tubulin (D, loading control) in total extracts of the neuronal cultures (40 μg of protein/lane). Molecular mass markers in kDa are shown on the right. The protein levels were semi-quantified by densitometry (E). Data represent relative intensity obtained from at least three cell culture dishes per condition.
DISCUSSION
In this study, we demonstrate that autophagy impairment per se fails to cause accumulation of ubiquitinated proteins. We compared the effect of the proteasome inhibitor epoxomicin with four autophagy inhibitors as follows: chloroquine, ammonium chloride, bafilomycin A1, and 3-methyladenine. Chloroquine and ammonium chloride are weak bases that become protonated and accumulate inside lysosomes raising the pH values, thus inactivating lysosomal hydrolases (31). The change in pH also causes accumulation of autophagosomes in cells by inhibiting the fusion of autophagosomes with lysosomes (32). Chloroquine is a more effective inhibitor than ammonium chloride thus requiring a 100-fold lower concentration (33). In some studies, concentrations of ammonium chloride as high as 100 mm are required to observe LC3-II accumulation (34). It is thus not surprising that we did not observe LC3-II accumulation with 10 mm ammonium chloride. Bafilomycin A1 is a potent and specific inhibitor of vacuolar H+-ATPase and prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes (35). We also tested the effect of 3-MA, which inhibits phosphatidylinositol 3-OH kinase (PI3K) and blocks autophagosome formation; thus, LC3-II cannot be detected (36). Except for chloroquine in SK-N-SH cells, none of the autophagy inhibitors induced the accumulation of ubiquitinated proteins. The same trend was observed in SK-N-SH cells maintained under serum or serum-free conditions, in rat primary cortical neuronal cultures, as well as in WT and Atg5−/− MEFs, the latter exhibiting genetically impaired autophagy. Together, these results clearly indicate that autophagy inhibition by itself, via pharmacologic or genetic means, does not lead to the accumulation of ubiquitinated proteins.
The increase in ubiquitinated proteins in epoxomicin-treated SK-N-SH cells was considerably higher than in cells treated with chloroquine. As Lys63-linked ubiquitination was suggested to selectively facilitate clearance by autophagy (26), we investigated if the DAKO antibody used in our studies had a similar affinity for Lys48- and Lys63-ubiquitinated substrates. We confirmed that the DAKO antibody detects both types of ubiquitin chains. Thus, the low levels of ubiquitinated proteins detected upon chloroquine treatment were not caused by a lack of reactivity of the antibody with Lys63-ubiquitinated proteins.
We considered if the limited accumulation of ubiquitinated proteins observed under chloroquine treatment could be due to proteasomal inhibition, so we compared the effect of chloroquine and epoxomicin on proteasome activity. The lysosomotropic agent inhibited 26 S and 20 S proteasomal activities assessed in total lysates as well as by glycerol density fractionation and the in-gel assay, albeit to a lesser extent than epoxomicin. The decrease in 26 S proteasome activity was partially due to its disassembly and was observed in SK-N-SH cells as well as in WT and Atg5−/− MEFs. No overall changes in the levels of individual proteasome subunits were observed upon chloroquine treatment. Another study with human neuroblastoma cells confirmed that chloroquine inhibited proteasome activity measured in total cell lysates (23). Notably, chloroquine was shown to inhibit 20 S proteasomes by binding to regions between the α and β subunits 20 Å away from the proteolytic sites (37), supporting the view that chloroquine belongs to a new class of proteasome inhibitors that do not bind to its active sites.
It is likely that an ∼50% proteasome inhibition as observed upon chloroquine treatment only causes a weak accumulation of ubiquitinated proteins, because under homeostatic conditions, there is a surplus of proteasome activity in cells. Only when most of the proteasome activity is inhibited, as in the case of epoxomicin treatment, the accumulation of ubiquitinated proteins is detectable. A similar phenomenon was observed in yeast and flies. Yeast cells continue to grow when 70–80% of their proteasome activity is inhibited by peptide aldehydes or β-lactone (38) thus confirming that under homeostatic conditions the capacity of the proteasome in yeast exceeds the required activity. Thus, a 20–30% proteasome capacity is sufficient for yeast cell survival and growth under homeostatic conditions. Similarly, feeding young flies sublethal concentrations of the proteasome inhibitor PSI had no apparent effect on survival or the levels of polyubiquitinated proteins (39). However, in old flies the disassembled state of the 26 S proteasome severely diminishes its activity, therefore rendering old flies exceptionally sensitive to proteasome inhibitors.
Besides proteasomal dysfunction observed with the pharmacologic inhibition of autophagy, proteasome impairment was also detected in autophagy-deficient transgenic mice resulting from genetically inactivated autophagy. Accordingly, reduced proteasome activity was observed in brain cortical extracts of cathepsin D-deficient mice (40). However, in transgenic mice lacking Atg7 (Atg7−/−) exclusively in neurons, no apparent proteasomal dysfunction was detected in the brain (4). Both studies report ubiquitin-positive neuronal inclusions despite the observed incidence (40) or absence (4) of proteasome impairment. The discrepancy in the outcome of proteasome activity reported in these two studies could be due to different genetic approaches. Although in the first study cathepsin D deficiency was targeted to all cells (41), in the second study, atg7 knock-out was specific to neurons in the CNS (4). In the latter study, it is possible that proteasome activity was impaired in the neurons exhibiting ubiquitin-positive neuronal inclusions but not in the ubiquitin-negative neurons or glia. Because total brain proteasome activity was assessed in the Atg7−/− mice, the observed rise in glia numbers could cause a parallel rise in proteasome activity thus concealing any potential decrease occurring in neurons with high levels of ubiquitinated proteins.
In conclusion, it is clear that the limited accumulation of ubiquitinated proteins observed under autophagy impairment results from weak proteasome inhibition by the lysosomotropic agents themselves or by proteasome overload. Thus, caution needs to be exercised when using autophagy-incompetent cells to conclude that autophagy acts continuously in a housekeeping role to dispose of diffuse ubiquitinated proteins, because in these cells the turnover of proteasomal substrates is compromised.
Epoxomicin and chloroquine affected MEFs differently from SK-N-SH cells. Both drugs inhibited proteasome activity assessed by the in-gel assay in MEFs and SK-N-SH cells. Chloroquine impaired autophagy in WT MEFs as indicated by LC3-II accumulation; the latter was not detected in Atg5−/− MEFs confirming that they are deficient in autophagy. However, hydrolysis of the chymotrypsin-like substrate Suc-LLVY-AMC by total MEF lysates was insensitive to chloroquine inhibition and at least 4-fold less sensitive to epoxomicin inhibition, when compared with SK-N-SH cells. Because Suc-LLVY-AMC is cleaved not only by the proteasome (24) but also by other chymotrypsin-like proteases as well as by calpains (25), we postulate that MEFs have enzymes that cleave this substrate and that are insensitive to epoxomicin and/or chloroquine. These enzymes are present in WT as well as in Atg5−/− MEFs. Notably, a recent paper identified an Atg5/Atg7-independent autophagy in MEFs (42, 43). This alternative form of autophagy was detected in several types of mammalian embryonic organs such as brain, liver, and heart. It plays a role in mitochondrial clearing during erythrocyte maturation and is inhibited by bafilomycin A1, but its sensitivity to chloroquine was not tested.
We demonstrate that p62/SQSTM1 is clearly associated with the 26 S proteasome. Under proteasome inhibition, there is a robust increase in p62/SQSTM1 levels in comparison with autophagy inhibition, which raises p62/SQSTM1 levels to a much lower level in SK-N-SH cells. Furthermore, proteasome inhibition in these cells leads to a clear association between endogenous soluble p62/SQSTM1 and proteasomes, established by three different methods (glycerol gradient fractionation, in-gel assay, and proximity ligation assay). We showed this association without having to overexpress any proteins, a process that perturbs the natural interactome balance (44).
p62/SQSTM1 was also found with inactive proteasomes in protein aggregates upon proteasome inhibition. In addition, we show by immunofluorescence analysis that p62/SQSTM1 is co-localized with ubiquitin protein aggregates in cells treated with epoxomicin alone or in combination with chloroquine. Several studies support our findings that proteasome inhibition alone causes protein aggregation (45). Some of these aggregates seem to be cleared by autophagy, because a recent study reports that no aggregates were observed in cells treated with proteasome inhibitors alone, although ubiquitinated proteins were still detected by Western blotting (46). These findings could indicate that aggregates can be cleared by autophagy but diffuse ubiquitinated proteins cannot.
Upon autophagy inhibition, we found p62/SQSTM1 with LC3-II in protein aggregates (immunoblotting) and in large dots with no ubiquitinated proteins (immunofluorescence). These dots could reflect lysosomal dilation and autophagosome-autophagosome fusion caused by chloroquine blocking autophagosomes from fusing with lysosomes (47, 48). p62/SQSTM1 seems to play an important role in aggregate clearance by autophagy. We previously demonstrated that preventing p62/SQSTM1 up-regulation by RNAi abolishes the aggregation but not the accumulation of diffuse ubiquitinated proteins or cell cytotoxicity induced by a product of inflammation (49). These findings were corroborated by other studies demonstrating that a reduction in p62/SQSTM1 significantly inhibited the recruitment of LC3 to autophagosomes increasing cell death induced by mutant huntingtin (22) and suppressed the appearance of ubiquitin-protein aggregates in hepatocytes and neurons of autophagy-deficient mice (21). Because p62/SQSTM1 binds to polyubiquitinated proteins, excess levels of this shuttling factor observed under autophagy impairment could compromise substrate delivery to the proteasome (50). However, in our studies with primary neuronal cultures, prolonged (96 h) autophagy inhibition with four different agents resulted in a drastic elevation of endogenous p62/SQSTM1 without a corresponding accumulation of ubiquitinated proteins. It is likely that elevation of p62/SQSTM1 levels by certain conditions is caused by induction of its transcription, although other conditions inhibit its degradation. That autophagy could play a more important role than the proteasome in p62/SQSTM1 degradation is supported by the marked elevation of p62/SQSTM1 in MEFs and primary neurons, with all agents that inhibit autophagy (except for 3-MA) and its reversal in the Atg5−/− MEFs. The role of both types of p62/SQSTM1 regulation, i.e. its up-regulation or degradation, in protein clearance and aggregation needs further characterization.
p62/SQSTM1 is a scaffold protein that encodes multiple binding domains for proteins involved in the UPP, autophagy, signaling such as through NFκB, extrinsic apoptotic pathway, and tumorigenesis (reviewed in Ref. 51). By mediating/interacting with so many different pathways, p62/SQSTM1 seems to play a critical role in cellular life and death decisions.
Our studies do not rule out that chaperone-mediated autophagy (CMA) is activated when proteasomes are blocked. CMA is independent of vesicle formation or membrane invagination, and thus does not require LC3-II, and it also has its own delivery system not mediated by p62/SQSTM1 (reviewed in Refs. 52, 53). CMA clients are directly targeted to lysosomes for degradation by the chaperone Hsc70, which recognizes the motif KFERQ present in ∼30% of cytoplasmic proteins. The relation between CMA activation and proteasome impairment remains to be established.
In conclusion, our studies convincingly demonstrate that autophagy impairment does not cause accumulation of ubiquitinated proteins unless the proteasome is affected. Low levels of ubiquitinated proteins detected upon autophagy impairment result from weak proteasome inhibition by lysosomotropic agents or by impaired flux through the proteasome because of substrate excess. Finally, great care needs to be exercised when attempting to infer that p62/SQSTM1 is a selective autophagy cargo receptor because it interacts with many client molecules, including proteasomes thus playing a function in a variety of pathways, including the UPP and autophagy. The roles of p62/SQSTM1 in cellular function require further investigation.
Supplementary Material
Acknowledgments
We thank Dr. N. Mizushima (Dept. of Physiology and Cell Biology, Tokyo Medical and Dental University, Tokyo, Japan) for the wild type and Atg5−/− MEF immortalized cell lines. The MEFs were provided by the RIKEN BRC through the National Bio-Resource Project of the MEXT, Japan. We also thank Dr. T. Schmidt-Glenewinkel (Dept. of Biological Sciences, Hunter College of City University of New York) for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants NS41073 (SNRP) NINDS (to M. E. F.-P.), AG028847 from NIA (to M. E. F.-P.), UL-RR024996 from NCRR/CTSC (TL1 training award to N. M.), and RR03037 from NIGMS/RCMI (infrastructure to Hunter College, City University of New York).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- UPP
- ubiquitin/proteasome pathway
- CQ
- chloroquine
- Epx
- epoxomicin
- LC3
- light chain 3
- 3-MA
- 3-methyladenine
- MEF
- mouse embryonic fibroblast
- PLA
- proximity ligation assay
- Rpt
- regulatory particle triple-A ATPase
- Suc
- succinyl
- AMC
- 7-amino-4-methylcoumarin
- CMA
- chaperone-mediated autophagy.
REFERENCES
- 1. Mayer R. J. (2003) Drug News Perspect. 16, 103–108 [DOI] [PubMed] [Google Scholar]
- 2. Ciechanover A. (2005) Cell Death Differ. 12, 1178–1190 [DOI] [PubMed] [Google Scholar]
- 3. Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. (2006) Nature 441, 885–889 [DOI] [PubMed] [Google Scholar]
- 4. Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. (2006) Nature 441, 880–884 [DOI] [PubMed] [Google Scholar]
- 5. Klionsky D. J. (2006) Nature 441, 819–820 [DOI] [PubMed] [Google Scholar]
- 6. Koga H., Kaushik S., Cuervo A. M. (2011) Ageing Res. Rev. 10, 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shin J. (1998) Arch. Pharm. Res. 21, 629–633 [DOI] [PubMed] [Google Scholar]
- 8. Park I., Chung J., Walsh C. T., Yun Y., Strominger J. L., Shin J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 12338–12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wooten M. W., Hu X., Babu J. R., Seibenhener M. L., Geetha T., Paine M. G., Wooten M. C. (2006) J. Biomed. Biotechnol. 2006, 62079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 11. Seibenhener M. L., Geetha T., Wooten M. W. (2007) FEBS. Lett. 581, 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biedler J. L., Roffler-Tarlov S., Schachner M., Freedman L. S. (1978) Cancer Res. 38, 3751–3757 [PubMed] [Google Scholar]
- 13. Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 14. Biederer T., Scheiffele P. (2007) Nat. Protoc. 2, 670–676 [DOI] [PubMed] [Google Scholar]
- 15. Mosmann T. (1983) J. Immunol. Methods 65, 55–63 [DOI] [PubMed] [Google Scholar]
- 16. Wilk S., Orlowski M. (1983) J. Neurochem. 40, 842–849 [DOI] [PubMed] [Google Scholar]
- 17. Elsasser S., Schmidt M., Finley D. (2005) Methods Enzymol. 398, 353–363 [DOI] [PubMed] [Google Scholar]
- 18. Ogburn K. D., Figueiredo-Pereira M. E. (2006) J. Biol. Chem. 281, 23274–23284 [DOI] [PubMed] [Google Scholar]
- 19. Wanker E. E., Scherzinger E., Heiser V., Sittler A., Eickhoff H., Lehrach H. (1999) Methods Enzymol. 309, 375–386 [DOI] [PubMed] [Google Scholar]
- 20. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., Landegren U. (2006) Nat. Methods. 3, 995–1000 [DOI] [PubMed] [Google Scholar]
- 21. Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 22. Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qiao L., Zhang J. (2009) Neurosci. Lett. 456, 15–19 [DOI] [PubMed] [Google Scholar]
- 24. Stein R. L., Melandri F., Dick L. (1996) Biochemistry 35, 3899–3908 [DOI] [PubMed] [Google Scholar]
- 25. Sasaki T., Kikuchi T., Yumoto N., Yoshimura N., Murachi T. (1984) J. Biol. Chem. 259, 12489–12494 [PubMed] [Google Scholar]
- 26. Tan J. M., Wong E. S., Kirkpatrick D. S., Pletnikova O., Ko H. S., Tay S. P., Ho M. W., Troncoso J., Gygi S. P., Lee M. K., Dawson V. L., Dawson T. M., Lim K. L. (2008) Hum. Mol. Genet. 17, 431–439 [DOI] [PubMed] [Google Scholar]
- 27. Meiners S., Heyken D., Weller A., Ludwig A., Stangl K., Kloetzel P. M., Krüger E. (2003) J. Biol. Chem. 278, 21517–21525 [DOI] [PubMed] [Google Scholar]
- 28. Gao Z., Gammoh N., Wong P. M., Erdjument-Bromage H., Tempst P., Jiang X. (2010) Autophagy 6, 126–137 [DOI] [PubMed] [Google Scholar]
- 29. Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gústafsdóttir S. M., Ostman A., Landegren U. (2002) Nat. Biotechnol. 20, 473–477 [DOI] [PubMed] [Google Scholar]
- 30. Weibrecht I., Leuchowius K. J., Clausson C. M., Conze T., Jarvius M., Howell W. M., Kamali-Moghaddam M., Söderberg O. (2010) Expert Rev. Proteomics 7, 401–409 [DOI] [PubMed] [Google Scholar]
- 31. Wibo M., Poole B. (1974) J. Cell Biol. 63, 430–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paludan C., Schmid D., Landthaler M., Vockerodt M., Kube D., Tuschl T., Münz C. (2005) Science 307, 593–596 [DOI] [PubMed] [Google Scholar]
- 33. Poole B., Ohkuma S. (1981) J. Cell Biol. 90, 665–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Underwood B. R., Imarisio S., Fleming A., Rose C., Krishna G., Heard P., Quick M., Korolchuk V. I., Renna M., Sarkar S., García-Arencibia M., O'Kane C. J., Murphy M. P., Rubinsztein D. C. (2010) Hum. Mol. Genet. 19, 3413–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamamoto A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., Tashiro Y. (1998) Cell Struct. Funct. 23, 33–42 [DOI] [PubMed] [Google Scholar]
- 36. Seglen P. O., Gordon P. B. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 1889–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sprangers R., Li X., Mao X., Rubinstein J. L., Schimmer A. D., Kay L. E. (2008) Biochemistry 47, 6727–6734 [DOI] [PubMed] [Google Scholar]
- 38. Lee D. H., Goldberg A. L. (1996) J. Biol. Chem. 271, 27280–27284 [DOI] [PubMed] [Google Scholar]
- 39. Vernace V. A., Arnaud L., Schmidt-Glenewinkel T., Figueiredo-Pereira M. E. (2007) FASEB J. 21, 2672–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qiao L., Hamamichi S., Caldwell K. A., Caldwell G. A., Yacoubian T. A., Wilson S., Xie Z. L., Speake L. D., Parks R., Crabtree D., Liang Q., Crimmins S., Schneider L., Uchiyama Y., Iwatsubo T., Zhou Y., Peng L., Lu Y., Standaert D. G., Walls K. C., Shacka J. J., Roth K. A., Zhang J. (2008) Mol. Brain 1, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saftig P., Hetman M., Schmahl W., Weber K., Heine L., Mossmann H., Köster A., Hess B., Evers M., von Figura K. (1995) EMBO J. 14, 3599–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., Shimizu S. (2009) Nature 461, 654–658 [DOI] [PubMed] [Google Scholar]
- 43. Shimizu S., Arakawa S., Nishida Y. (2010) Autophagy 6, 290–291 [DOI] [PubMed] [Google Scholar]
- 44. Fredriksson S. (2009) Nat. Methods 6, i–ii (Application Notes) [Google Scholar]
- 45. Johnston J. A., Ward C. L., Kopito R. R. (1998) J. Cell Biol. 143, 1883–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu K., Dunner K., Jr., McConkey D. J. (2010) Oncogene 29, 451–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., Bamber B. A., Bassham D. C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J. S., Bredesen D. E., Brodsky J. L., Brumell J. H., Brunk U. T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L. S., Choi A., Chu C. T., Chung J., Clarke P. G., Clark R. S., Clarke S. G., Clavé C., Cleveland J. L., Codogno P., Colombo M. I., Coto-Montes A., Cregg J. M., Cuervo A. M., Debnath J., Demarchi F., Dennis P. B., Dennis P. A., Deretic V., Devenish R. J., Di Sano F., Dice J. F., Difiglia M., Dinesh-Kumar S., Distelhorst C. W., Djavaheri-Mergny M., Dorsey F. C., Dröge W., Dron M., Dunn W. A., Jr., Duszenko M., Eissa N. T., Elazar Z., Esclatine A., Eskelinen E. L., Fésüs L., Finley K. D., Fuentes J. M., Fueyo J., Fujisaki K., Galliot B., Gao F. B., Gewirtz D. A., Gibson S. B., Gohla A., Goldberg A. L., Gonzalez R., González-Estévez C., Gorski S., Gottlieb R. A., Häussinger D., He Y. W., Heidenreich K., Hill J. A., Høyer-Hansen M., Hu X., Huang W. P., Iwasaki A., Jäättelä M., Jackson W. T., Jiang X., Jin S., Johansen T., Jung J. U., Kadowaki M., Kang C., Kelekar A., Kessel D. H., Kiel J. A., Kim H. P., Kimchi A., Kinsella T. J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovács A. L., Kroemer G., Kuan C. Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H. Y., Lenardo M. J., Levine B., Lieberman A., Lim K. L., Lin F. C., Liou W., Liu L. F., Lopez-Berestein G., López-Otín C., Lu B., Macleod K. F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A. J., Meléndez A., Michels P., Miotto G., Mistiaen W. P., Mizushima N., Mograbi B., Monastyrska I., Moore M. N., Moreira P. I., Moriyasu Y., Motyl T., Münz C., Murphy L. O., Naqvi N. I., Neufeld T. P., Nishino I., Nixon R. A., Noda T., Nürnberg B., Ogawa M., Oleinick N. L., Olsen L. J., Ozpolat B., Paglin S., Palmer G. E., Papassideri I., Parkes M., Perlmutter D. H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F., Rohrer B., Rubinsztein D. C., Ryan K. M., Sadoshima J., Sakagami H., Sakai Y., Sandri M., Sasakawa C., Sass M., Schneider C., Seglen P. O., Seleverstov O., Settleman J., Shacka J. J., Shapiro I. M., Sibirny A., Silva-Zacarin E. C., Simon H. U., Simone C., Simonsen A., Smith M. A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P. E., Subauste C. S., Sugimoto S., Sulzer D., Suzuki T., Swanson M. S., Tabas I., Takeshita F., Talbot N. J., Tallóczy Z., Tanaka K., Tanaka K., Tanida I., Taylor G. S., Taylor J. P., Terman A., Tettamanti G., Thompson C. B., Thumm M., Tolkovsky A. M., Tooze S. A., Truant R., Tumanovska L. V., Uchiyama Y., Ueno T., Uzcátegui N. L., van der Klei I., Vaquero E. C., Vellai T., Vogel M. W., Wang H. G., Webster P., Wiley J. W., Xi Z., Xiao G., Yahalom J., Yang J. M., Yap G., Yin X. M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R. L. (2008) Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yoon Y. H., Cho K. S., Hwang J. J., Lee S. J., Choi J. A., Koh J. Y. (2010) Invest. Ophthalmol. Vis. Sci. 51, 6030–6037 [DOI] [PubMed] [Google Scholar]
- 49. Wang Z., Figueiredo-Pereira M. E. (2005) Mol. Cell. Neurosci. 29, 222–231 [DOI] [PubMed] [Google Scholar]
- 50. Korolchuk V. I., Mansilla A., Menzies F. M., Rubinsztein D. C. (2009) Mol. Cell 33, 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Moscat J., Diaz-Meco M. T. (2009) Cell 137, 1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cuervo A. M. (2010) Trends Endocrinol. Metab. 21, 142–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kettern N., Dreiseidler M., Tawo R., Höhfeld J. (2010) Biol. Chem. 391, 481–489 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.