Abstract
The enzymic degradation of plant cell walls plays a central role in the carbon cycle and is of increasing environmental and industrial significance. The catalytic modules of enzymes that catalyze this process are generally appended to noncatalytic carbohydrate-binding modules (CBMs). CBMs potentiate the rate of catalysis by bringing their cognate enzymes into intimate contact with the target substrate. A powerful plant cell wall-degrading system is the Clostridium thermocellum multienzyme complex, termed the “cellulosome.” Here, we identify a novel CBM (CtCBM62) within the large C. thermocellum cellulosomal protein Cthe_2193 (defined as CtXyl5A), which establishes a new CBM family. Phylogenetic analysis of CBM62 members indicates that a circular permutation occurred within the family. CtCBM62 binds to d-galactose and l-arabinopyranose in either anomeric configuration. The crystal structures of CtCBM62, in complex with oligosaccharides containing α- and β-galactose residues, show that the ligand-binding site in the β-sandwich protein is located in the loops that connect the two β-sheets. Specificity is conferred through numerous interactions with the axial O4 of the target sugars, a feature that distinguishes galactose and arabinose from the other major sugars located in plant cell walls. CtCBM62 displays tighter affinity for multivalent ligands compared with molecules containing single galactose residues, which is associated with precipitation of these complex carbohydrates. These avidity effects, which confer the targeting of polysaccharides, are mediated by calcium-dependent oligomerization of the CBM.
Keywords: Carbohydrate-binding Protein, Carbohydrate Complex, Carbohydrate Structure, Protein Structure, X-ray Crystallography
Introduction
Carbohydrate protein recognition plays a central role in biology, exemplified by microbe-mediated plant cell wall degradation. The release of sugars from plant cell walls is not only critical for the maintenance of the carbon cycle but is of increasing industrial and environmental significance through the development of second generation lignocellulose-based biofuels (1). The chemical and physical complexity of plant cell walls restricts their accessibility to enzyme attack and thus the recycling of photosynthetically fixed carbon is a relatively slow biological process.
Enzymes that catalyze plant cell wall degradation display complex molecular architectures. In addition to the catalytic module(s), these enzymes often contain one or more noncatalytic carbohydrate-binding modules (CBMs)4 (2). CBMs, by binding to their target ligand, bring the appended enzymes into intimate contact with its substrate thereby potentiating catalysis by reducing the accessibility problem (3–5). Carbohydrate-modifying enzymes and their component modules, which include CBMs, have been classified into sequence-based families in the CAZy database (6). Currently, there are 61 families of CBMs that recognize a variety of plant and mammalian glycans. In some families, exemplified by CBM1 and CBM10, ligand specificity is invariant; however, it is becoming increasingly apparent that carbohydrate recognition can be highly variable in different members of the same family. For example, CBM6 contains members that recognize xylan (β-1,4-xylose polymer), single cellulose (β-1,4-glucose polymer) chains, mixed linked β-1,4-β-1,3-glucans, laminarin (β-1,3-glucose polymer), and agarobiose (3,6-anhydro-l-galactose-d-galactose) (7–10).
Representative three-dimensional structures of around half the CBM families are available. The majority of these proteins display a β-sandwich fold in which the ligand-binding site for extended glycan chains is generally located on the β-sheet that includes the concave surface of the protein (for review see Ref. 2). For CBMs that recognize terminal mono- or disaccharides, the ligand-binding site is often located in the loops that connect the two β-sheets. Although there are examples of CBMs where the orientation of aromatic residues confers ligand specificity (12, 13), it is increasingly evident that polar interactions play a more important role in carbohydrate recognition (8, 9, 14). Recent reports have also revealed examples of CBMs that harness calcium in ligand recognition (8, 15, 16). CBMs are generally located in monomeric enzymes; thus cooperative (or avidity) effects occur rarely. Exceptions to this general rule are evident, however, in enzymes that contain multiple copies of CBMs that display the same specificity, where avidity effects between these modules has led to increased affinity for polysaccharides such as xylan (17–19). Given that oligomerization is a general feature of lectins (carbohydrate binding domains that are not components of enzymes), where avidity effects are common (20), it is surprising that similar macromolecular associations have not been observed in the CBM literature.
The multienzyme complex expressed by Clostridium thermocellum, referred to as the “cellulosome,” is one of the most powerful and intricate plant cell wall-degrading systems described to date (21, 22). The enzyme complex is anchored onto the plant cell wall by the noncatalytic scaffoldin protein, which contains a family 3 CBM that binds to crystalline cellulose (23). The catalytic subunits of the cellulosome also contain CBMs, which target β-glucans, xylans, uronic acids, and chitin (7, 16, 24–27), which direct the complex toward the target substrates for these enzymes. The extent to which the cellulosome can be directed toward the myriad of carbohydrate structures in plant cell wall, however, remains relatively unexplored, and indeed, numerous cellulosomal proteins contain modules of unknown function that could, potentially, comprise CBMs that display novel specificities. Furthermore, the quaternary structure of the cellulosome may provide a background for cooperativity between the CBMs within this protein complex, although such interactions have not been observed experimentally.
Here, we report the structure and biochemistry of a module, defined as CtCBM62, of the C. thermocellum cellulosomal protein Cthe_2193 (GenBankTM protein accession ABN53395.1; defined hereafter as CtXyl5A) that exhibits no significant sequence similarity to CBMs in the CAZy database. CtCBM62 displays a β-sandwich fold that binds to terminal α- and β-d-galactopyranose or l-arabinopyranose residues of complex polysaccharides. Ligand specificity is conferred primarily through extensive interactions with the axial O4 of galactose and arabinose, a distinctive feature of these two sugars. CtCBM62 also displays calcium-mediated oligomerization resulting in avidity effects that confer selectivity for polysaccharides rather than monovalent oligosaccharides.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Components of Cthe_2193
DNA encoding the following regions of CtXyl5A, GH5-CBM6-CBM13-Fn3-CtCBM62 Docl (mature CtXyl5A; see Fig. 1) and CtCBM62, were amplified using primers, containing NheI and XhoI restriction sites, listed in supplemental Table S1. The amplified DNAs were cloned into NheI/XhoI-restricted pET21a, such that the encoded recombinant proteins contain a C-terminal His6 tag. To express the two C. thermocellum proteins, Escherichia coli strain BL21(DE3), harboring appropriate recombinant plasmids, was cultured to mid-exponential phase in Luria broth at 37 °C followed by the addition of isopropyl β-d-galactopyranoside at 1 mm, to induce recombinant gene expression, and incubated for a further 5 h at 37 °C. The recombinant proteins were purified to >90% electrophoretic purity by immobilized metal ion affinity chromatography using TalonTM, a cobalt-based matrix, and eluted with 100 mm imidazole, as described previously (19). When preparing the selenomethionine derivative of CtCBM62 for crystallography, the protein was expressed in E. coli B834 (DE3), a methionine auxotroph, cultured in media comprising 1 liter of SelenoMet Medium BaseTM, 50 ml of SelenoMet Nutrient MixTM, and 4 ml of selenomethionine solution (10 mg/ml). Recombinant gene expression was as described above, as was protein purification, except that all buffers were supplemented with 10 mm β-mercaptoethanol.
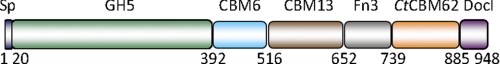
FIGURE 1.
Schematic of CtXyl5A. The abbreviated modules are as follows: Sp, signal peptide; DocI, type I dockerin modules.
Mutagenesis
Site-directed mutagenesis was carried out using the PCR-based QuikChange method (Stratagene) deploying the primers listed in supplemental Table S1.
Binding Assays
Affinity gel electrophoresis was used to screen for the binding of CtCBM62 to polysaccharides, following the method of Ref. 28. The proteins were subjected to nondenaturing PAGE, in the presence of 5 mm CaCl2, deploying parallel gels containing no ligand and the target polysaccharide at 100 μg/ml, respectively. The gels were also loaded with BSA, which acts as a nonbinding negative control. After electrophoresis, the gels were stained with Coomassie Blue, and proteins that bound to the polysaccharide displayed reduced electrophoretic migration in the presence of the complex carbohydrate. The binding of CtCBM62 to its ligands was quantified by isothermal titration calorimetry (ITC), as described previously (25). Titrations were carried out in 50 mm Na-HEPES buffer, pH 7.5, containing 5 mm CaCl2 (or 5 mm EDTA) at 25 °C. The reaction cell contained protein at 145 μm, while the syringe contained either the oligosaccharide at 10 mm or polysaccharide at 3–5 mg/ml. The titrations were analyzed using Microcal Origin version 7.0 software to derive n, Ka, and ΔH values, although ΔS was calculated using the standard thermodynamic equation, RTlnKa = ΔG = ΔH − TΔS.
Analytical Ultracentrifugation
Sedimentation velocity experiments were performed using an Optima XLA analytical ultracentrifuge (Beckman Coulter) with an An60 Ti four-hole rotor. Purified CtCBM62 (see above) was dialyzed into either CaCl2 buffer (50 mm Na-HEPES, pH 7.5, 150 mm NaCl, 10 mm CaCl2) or EDTA buffer (50 mm Na-HEPES, pH 7.5, 150 mm NaCl, 10 mm EDTA) using a Microdialyzer System 100 (Pierce). In each experiment, 400 μl of 1 mg/ml CtCBM62 and 410 μl of reference buffer were loaded into 12-mm double-sector Epon centerpieces equipped with quartz windows and equilibrated for ∼1 h at 20 °C. Experiments were performed at a rotor speed of 50,000 rpm and a wavelength of 280 nm. Data were collected over 8.5-h periods using a radial step size of 0.003 cm. The partial specific volume (v̄) of 0.7211 was calculated from the amino acid sequence. The buffer viscosity of 0.00892 centipoise and density of 1.0042 g/ml for the CaCl2 buffer or 1.005 g/ml for the EDTA buffer were calculated using the program SEDNTERP. All data analysis for sedimentation velocity experiments were performed using the program SEDFIT, version 12.1b. Continuous sedimentation coefficient distribution c(s) analyses were restrained by Marquardt-Levenberg regularization at a confidence interval p = 0.68 with uniform prior knowledge (29). The base line, meniscus, frictional coefficient, systematic time invariant, and radial invariant noise were fitted. The final fit c(Pδ)(s) implemented prior knowledge of discrete species for determining the size distribution (30). The r.m.s.d. values for all reported experiments were 0.008 or less.
Crystallography
CtCBM62 was crystallized using the hanging drop vapor diffusion technique at 20 °C with an equal volume (1 μl) of protein and reservoir solution. The native and selenomethionine (in the presence of 5 mm DTT) forms of apo-CtCBM62 were crystallized in 20% (w/v) PEG 3350, 0.2 m trisodium citrate, pH 8.3 (PEG/IonTM 1 screen condition 46; Hampton Research) at 150 mg/ml. CtCBM62 in complex with 10 mm xyloglucan oligosaccharides was crystallized in the same condition described above at a protein concentration of 191 mg/ml. CtCBM62, in complex with 10 mm 16-α-GalMan3, was crystallized in 0.5 m ammonium sulfate in 0.1 m Na-HEPES, pH 7.4, containing 30% 2-methyl-2,4-pentanediol (condition 59 of the PACT screen) also with protein at 191 mg/ml. The CtCBM62 crystals grew over 4–5 days, after which 25 μl of mother liquor, including 30% (v/v) glycerol, was added as the cryoprotectant before being flash frozen in liquid nitrogen.
All the crystals of CtCBM62 were in space group F432 with unit cell dimensions of a = b = c = 192.2 Å, with one protein molecule in the asymmetric unit. The structure of apo-CtCBM62 was solved by SIRAS, exploiting both the anomalous scattering from the seleniums and the isomorphous differences between the selenomethionine derivative and the native diffraction data. After processing the diffraction data in XDS (31) and SCALA (32), the unmerged intensities were input to the HKL2MAP (33) interface to SHELX (34), and the heavy atom substructure was solved in SHELXD. The SHELXE solvent flattened map was of sufficient quality to build manually, in COOT (35), the CtCBM62 molecule. The ligand-bound structures of CtCBM62 were determined by molecular replacement with AMORE (36) using the apo structure as the search model. All structures were refined to convergence using REFMAC5 (37) with manual corrections being applied in COOT (35). The data collection, phasing, and refinement statistics are displayed in Table 1, and the PDB codes for the protein structures are as follows: 2YFU, 2YFZ, and 2YG0.
TABLE 1.
Diffraction data collection and refinement statistics of CtCBM62
Values in parentheses refer to the highest resolution shell.
| CtCBM62 SeMet | CtCBM62 native | CtCBM62/xyloglucan oligosaccharide | CtCBM62/GM3 | |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength | 1.54 | 1.54 | 1.073 | 1.073 |
| Space group cell dimensions | F432 | F432 | F432 | F432 |
| a, b, c | a = b = c = 191.75 Å | a = b = c = 192.16 Å | a = b = c = 191.82 Å | a = b = c = 191.72 Å |
| Resolution | 47.94–2.43 Å | 48.04–1.65 Å | 44–1.80 Å | 47.93–1.80 Å |
| (2.56–2.43 Å) | (1.74–1.65 Å) | (1.90–1.80 Å) | (1.90–1.80 Å) | |
| Rmerge | 0.113 (0.453) | 0.066 (0.406) | 0.113 (0.537) | 0.081 (0.479) |
| I/σI | 47.5 (12.1) | 33.7 (7.1) | 18.2 (4.6) | 21.9 (5.4) |
| Completeness | 99.9% (99.1%) | 100.0% (99.8%) | 100.0% (100.0%) | 99.8% (100.0%) |
| Redundancy | 53.7 (43.3) | 15.7 (13.2) | 10.5 (10.6) | 10.4 (10.5) |
| Anomalous completeness | 99.7% (98.1%) | |||
| Anomalous redundancy | 29.7 (23.2) | |||
| Refinement | ||||
| No. of reflections | – | 37038 (5272) | 21020 (2996) | 28437 (4098) |
| Rwork/Rfree | – | 0.1512 (0.1624) | 0.1637 (0.1803) | 0.1621 (0.1787) |
| No. of atoms | ||||
| Protein | – | 1058 | 1061 | 1053 |
| Ligand/Ion | – | 13 | 64 | 30 |
| Water | – | 224 | 136 | 190 |
| B-factor | ||||
| Protein | – | 13 | 12 | 15 |
| Ligand/Ion | – | 19 | 47 | 26 |
| Water | – | 28 | 24 | 29 |
| Root mean square deviations | ||||
| Bond lengths | 0.015 Å | 0.014 Å | 0.014 Å | |
| Bond angles | 1.42° | 1.29° | 1.69° | |
| PDB codes | 2YFU | 2YFZ | 2YG0 | |
RESULTS
CtCBM62 Is a Component of the C. thermocellum Cellulosomal Protein CtXyl5A
An initial analysis of the C. thermocellum genome was performed as described in Cantarel et al. (6). The analysis identified 72 genes encoding modular proteins with elements corresponding to CAZy families of catalytic modules and CBMs, which, based on the presence of type I dockerins, were components of the cellulosome. The functions of many of these proteins have been demonstrated biochemically or inferred from sequence similarity to enzymes with known activities. There remain, however, several cellulosomal subunits that contain modules with no sequence similarity with proteins of known function. The open reading frame identified as locus Cthe_2193 (defined hereafter as CtXyl5A) is one such protein. The protein is likely to be secreted because it contains an N-terminal 20-residue signal peptide. The mature polypeptide includes 928 amino acids and consists of six modules, which are displayed in Fig. 1. In addition to the type I dockerin, the protein contains two CBMs from families 6 and 13, a fibronectin domain, and a GH5 module, which is likely to be the catalytic component of the protein (see the accompanying paper, which describes the biochemical properties of the full-length enzyme (38)). A particularly noteworthy feature of the open reading frame is the sequence extending from 740 to 878, defined hereafter as CtCBM62, which displays no significant sequence similarity to proteins in the CAZy database. To investigate the function of CtCBM62, a recombinant form of this protein module was expressed in soluble form in E. coli and purified to electrophoretic homogeneity.
Identification of a Novel CBM in CtXyl5A
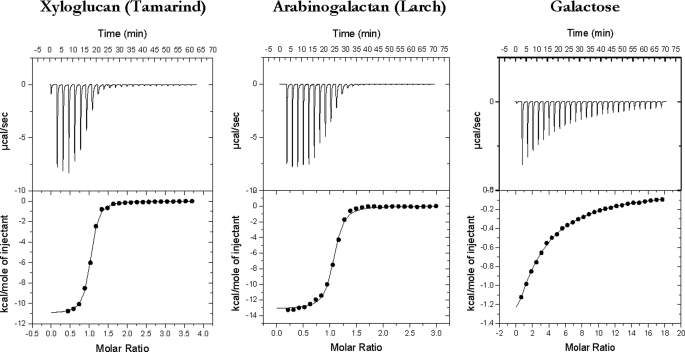
Biochemical analysis of CtCBM62 failed to reveal catalytic activity against an extensive range of plant structural polysaccharides, including cellulose and various hemicellulose and pectin polymers (data not shown). To explore whether CtCBM62 fulfills a noncatalytic carbohydrate binding function, the capacity of the protein to bind to a range of polysaccharides was assessed by affinity gel electrophoresis. The data, reported in Table 2, show that the protein module binds to xyloglucan, arabinogalactan, and galactomannan, but does not recognize the other polymers evaluated, including the arabinoxylans that are hydrolyzed by CtXyl5A (38). The protein module therefore includes a functional CBM and represents the founding member of a new family designated CBM62. To investigate the specificity of CtCBM62 in more detail, the capacity of the protein to bind to galactomannan- and xyloglucan-derived oligosaccharides and monosaccharides was assessed by ITC. The data, reported in Table 3, with example titrations displayed in Fig. 2, show that CtCBM62 does not bind to mannooligosaaccharides or cellulooligosaccharides, the backbone structures of galactomannan and xyloglucan, respectively, or the repeating XXXG (X is a backbone glucose decorated α1,6 with Xyl and G represents unsubstituted glucose) motif of xyloglucan (39). The protein, however, does bind to 16-α-d-GalMan3, 3646-α-d-Gal2Man5, and a mixture of XLXG, XLLG, and XXLG, where L is X in which the xylose is decorated with a β1,2-linked d-galactose residue. The stoichiometry of binding was unity for galactose residues (in the oligosaccharides) indicating that CtCBM62 has a single ligand-binding site. The thermodynamic data show that the binding of CtCBM62 to all its ligands is enthalpically driven, whereas the change in entropy makes a negative contribution to overall affinity, as observed for the majority of CBMs studied to date (14, 16, 27). These data indicate that CtCBM62 binds to galactomannan, arabinogalactan, and xyloglucan, which are all decorated with galactose residues, through recognition of the hexose sugar. This view was confirmed by the observation that the protein binds to galactose and arabinose, in which O4 is axial, but not to sugars such as xylose, mannose, and glucose, where O4 is equatorial. Indeed, the similar affinity of CtCBM62 for 16-α-d-GalMan3 and galactose, and the lack of significant binding to XXXG, suggests that the backbone components of mannan and xyloglucan do not make significant interactions with the protein. It should be noted, however, that the affinity of the protein for Gal and 16-α-GalMan3 is 2 orders of magnitude lower than for galactomannan, whereas the Ka values for the xyloglucan oligosaccharides are also significantly lower than for the corresponding polysaccharide (Table 3). As mature CtXyl5A (which includes all the GH5-CBM6-CBM13-Fn3-CBM62-Doc1 domains) also exhibits much higher affinity for xyloglucan and carob galactomannan than galactose (Table 3), CtCBM62 displays its polysaccharide targeting function in the full-length enzyme. To summarize, the capacity of CtCBM62 to bind to xyloglucan and galactomannan is conferred by its recognition of either α- or β-d-galactose.
TABLE 2.
Polysaccharide specificity of CtCBM62 determined by affinity polyacrylamide gel electrophoresis
| Liganda | Bindingb |
|---|---|
| Arabinogalactan | ++ |
| Oat-spelt xylan | − |
| Wheat arabinoxylan | − |
| Laminarin | − |
| Lichenan (Icelandic moss) | − |
| Xyloglucan (Tamarind) | +++ |
| Glucomannan (Konjac) | − |
| Galactomannan (Gal:Man, 21:79) (Carob) | ++ |
| Galactomannan (Gal:Man, 38:40) (Guar) | +++ |
| Pullulan | − |
| Galactan (Potato) | − |
| Pustulan | − |
| Rhamnogalacturonan (Soybean) | − |
| Hydroxyethylcellulose | − |
| Glucuronoxylan | − |
| Arabinan (debranched) | − |
| Pectic galactan (Potato) | − |
| Polygalacturonic acid | − |
| Rhamnogalacturonan I (Potato) | − |
a Ligands were screened at a concentration of 0.1 mg/ml.
b The following symbols are used: −, no detectable binding; ++, significant binding; +++, strong binding +++.
TABLE 3.
Affinity and thermodynamic parameters of CtCBM62 binding to polysaccharides, oligosaccharides and monosaccharides
| Ligand | Ka × 104 | ΔG | ΔH | TΔS | na |
|---|---|---|---|---|---|
| m−1 | kcal mol−1 | kcal mol−1 | kcal mol−1 | ||
| Galactomannan (Carob) | 56.4 ± 3.1 | −7.85 ± 0.03 | −11.23 ± 0.06 | −3.38 ± 0.03 | 1.00 ± 0.003 |
| 63,64-Di-α-d-galactosyl mannopentaose | 2.21 ± 0.72 | −4.56 ± 1.14 | −8.69 ± 1.6 | −4.13 ± 0.46 | 0.992 ± 0.15 |
| 61-α-d-Galactosyl mannotriose | 0.29 ± 0.02 | −4.33 ± 0.03 | −7.04 ± 0.75 | −2.71 ± 0.72 | 1.07 ± 0.89 |
| Galactose | 0.32 ± 0.003 | −4.79 ± 0.01 | −8.59 ± 0.17 | −3.8 ± 0.16 | 1.07 ± 0.02 |
| Mannopentaose | NBb | ||||
| Xyloglucan (Tamarind) | 177 ± 17.7 | −8.53 ± 0.06 | −11.63 ± 0.07 | −3.1 ± 0.01 | 1.01 ± 0.003 |
| XLXLG | 2.01 ± 0.20 | −5.87 ± 0.06 | −10.74 ± 0.46 | −4.87 ± 0.40 | 1.08 ± 0.036 |
| XXXG | <0.01 | ||||
| Arabinogalactan (Larch) | 84.1 ± 8.4 | −8.09 ± 0.06 | −13.16 ± 0.10 | −5.07 ± 0.04 | 1.04 ± 0.005 |
| Arabinan (debranched) | 9.42 ± 0.49 | −6.79 ± 0.03 | −10.84 ± 0.10 | −4.05 ± 0.07 | 1.02 ± 0.007 |
| Arabinose | 0.11 ± 0.005 | −4.17 ± 0.02 | −9.10 ± 0.20 | −4.93 ± 0.18 | 0.99 ± 0.22 |
| Xylan (oat spelt) | <0.01 | ||||
| Cellohexaose | NB | ||||
| Cellotetraose | NB | ||||
| Xylose | NB |
a The ITC data were fitted to a single site binding model for all ligands. For polysaccharide ligands, in which the molar concentration of binding sites is unknown, the n value was iteratively fitted as close as possible to one, by adjusting the molar concentration of the ligand.
b NB, no binding.
FIGURE 2.
Representative ITC data of CtCBM62 binding to soluble ligands. The ligand (3 mg/ml xyloglucan, 5 mg/ml arabinogalactan, 10 mm arabinose) in the syringe was titrated into CtCBM52 (145 μm) in the cell. The top half of each panel shows the raw ITC heats; the bottom half shows the integrated peak areas fitted using a one single binding model by MicroCal Origin software. ITC was carried out in 50 mm Na-HEPES, pH 7.5, containing 5 mm CaCl2, at 25 °C.
CtCBM62 Displays Avidity Effects
The tighter binding of CtCBM62 to polysaccharides, compared with monovalent oligosaccharides and monosaccharides, is likely mediated through avidity effects, which occurs when a protein with multiple binding sites interacts with a multivalent ligand, such as a polysaccharide. Indeed, avidity effects are invariably associated with cross-linking and thus precipitation of the ligand-protein complex (18), which occurs when CtCBM62 binds to xyloglucan, galactomannan, and arabinogalactan (data not shown).
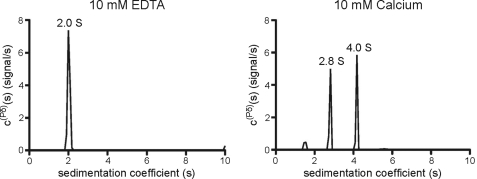
To explore the potential role of calcium (which plays a key role in cellulosome function (40)) in the avidity effects displayed by CtCBM62, the capacity of the protein to bind to xyloglucan and galactose was assessed in the presence of EDTA and calcium, respectively. The data, displayed in Table 4, show that for the simple monovalent ligand galactose, neither the addition of calcium nor the chelating agent EDTA influenced affinity for these carbohydrates. By contrast, CtCBM62 binds to the galactose-containing polysaccharide xyloglucan >100-fold more tightly in the presence of calcium than with EDTA. The data described above indicate that calcium enhances affinity for multivalent ligands but does not influence the recognition of carbohydrates containing a single galactose residue. The most likely explanation for the role of calcium is through its capacity to mediate oligomerization of CtCBM62, which would result in enhanced affinity for multivalent ligands through avidity effects. To explore this possibility, CtCBM62 was subjected to size exclusion chromatography, using a Sephadex S100 column. In the presence of EDTA, CtCBM62 was eluted in a volume of 89 ml. Replacing the chelating agent with calcium reduced the elution volume to 74 ml (data not shown). Based on the calibration of the column with proteins of known molecular weights, CtCBM62 migrated as a monomer in the presence of EDTA and a dimer when calcium was included in the elution buffer. Calcium-mediated oligomerization of CtCBM62 was confirmed using analytical ultracentrifugation. In the presence of EDTA, CtCBM62 sediments as a monomer with a sedimentation coefficient of 2.0 S. Calcium induces CtCBM62 to form a dimer at 4.2 S that is in dynamic equilibrium with the monomer, causing a shift to 2.8 S (Fig. 3). These data indicate that the capacity of calcium to enhance the affinity of CtCBM62 for multivalent ligands is mediated by metal ion-dependent oligomerization of the protein, which leads to avidity effects.
TABLE 4.
The influence of calcium on the affinity of CtCBM62 for xyloglucan and galactose
| Ligand | Ka × 104 | ΔG | ΔH | TΔS | na |
|---|---|---|---|---|---|
| m−1 | kcal mol−1 | kcal mol−1 | kcal mol−1 | ||
| Xyloglucan + calciumb | 129 ± 14.3 | −8.35 ± 0.07 | −11.4 ± 0.11 | −3.14 ± 0.18 | 1.00 ± 0.004 |
| Xyloglucan − calciumc | 0.95 ± 0.02 | −5.43 ± 0.01 | −11.5 ± 0.15 | −6.08 ± 0.16 | 1.08 ± 0.01 |
| Xyloglucan + EDTAd | 0.59 ± 0.05 | −5.15 ± 0.06 | −12.7 ± 1.68 | −7.53 ± 1.64 | 1.04 ± 0.08 |
| Galactose + calciumb | 0.41 ± 0.003 | −3.58 ± 0.01 | −9.58 ± 0.12 | −6.0 ± 0.18 | 1.02 ± 0.02 |
| Galactose + EDTAb | 0.38 ± 0.013 | −3.32 ± 0.01 | −9.40 ± 0.17 | −6.08 ± 0.16 | 1.12 ± 0.11 |
a The ITC data were fitted to a single site binding model for all ligands. For polysaccharide ligands in which the molar concentration of binding sites is unknown, the n value was iteratively fitted to as close as possible to one, by adjusting the molar concentration of the ligand.
b CtCBM62 was dialyzed in the presence of 2 mm CaCl2, and the metal was added to the ITC experiment.
c CtCBM62 was dialyzed in buffer lacking CaCl2, and ITC was carried out in the absence of calcium.
d CtCBMX was dialyzed into buffer lacking CaCl2, and the ITC was performed in the presence of 10 mm EDTA.
FIGURE 3.
Analytical ultracentrifugation of CtCBM62. Sedimentation velocity centrifugation was carried out as described under “Experimental Procedures.” Freshly prepared protein was dialyzed against 4000 volumes of 20 mm Na-HEPES buffer, pH 7.0, containing 10 mm CaCl2 or 10 mm EDTA. CtCBM62 was centrifuged at a concentration of 1 mg/ml at 20 °C.
Crystal Structure of CtCBM62
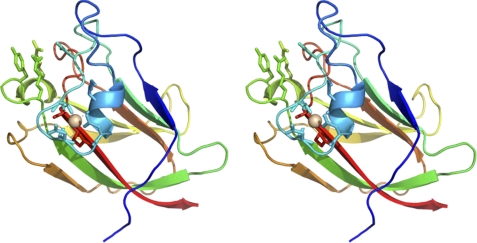
To explore the mechanism by which CtCBM62 binds to both α- or β-d-galactose, the crystal structure of the protein module was determined. In the conditions used to obtained crystals of the protein, no calcium was added, and thus the CBM crystallized in its monomeric form. The final structure corresponds to residues 739–878 of full-length CtXyl5A. CtCBM62 displays a canonical β-sandwich fold (2) comprising two β-sheets containing five antiparallel β-strands on the concave face (β-strands 1, 2, 4, 5, and 7) and three antiparallel β-strands on the convex face (β-strands 3, 6, and 8) (Fig. 4). The protein contains a bound metal located at the beginning of the loop connecting α-1 helix to the β-2 strand which, based on its B-factor in comparison to neighboring atoms, its octahedral coordination, the bond distances, its satisfactory refinement with no residual positive or negative difference density, and its interaction with only oxygen ligands, has been modeled as calcium. This structural calcium is conserved in many CBM families that display a β-sandwich fold (2). CtCBM62 displays greatest structural similarity to MvCBM32 (PDB code 1EUT (41) r.m.s.d. of 2.7 Å over 131 Cα atoms), CpCBM32 (PDB code 2J1A (42) r.m.s.d. of 2.4 Å over 126 Cα atoms), and SpCBM47 (PDB code 2J1R (43) r.m.s.d. of 2.3 Å over 125 Cα atoms). These proteins all display <14% sequence identity with CtCBM62.
FIGURE 4.
Crystal structure of CtCBM62 and the location residues that interact with galactose and calcium. Stereo figure of the overall structure of CtCBM62 displayed as a protein schematic, continuously color ramped from N to C terminus, from blue to red. The bound calcium is drawn as a light pink sphere, with coordinating amino acids drawn as sticks, with the H-bonds between them as dashed lines. The carbohydrate-binding amino acids are also drawn as sticks, and the same view is used for the other structural figures.
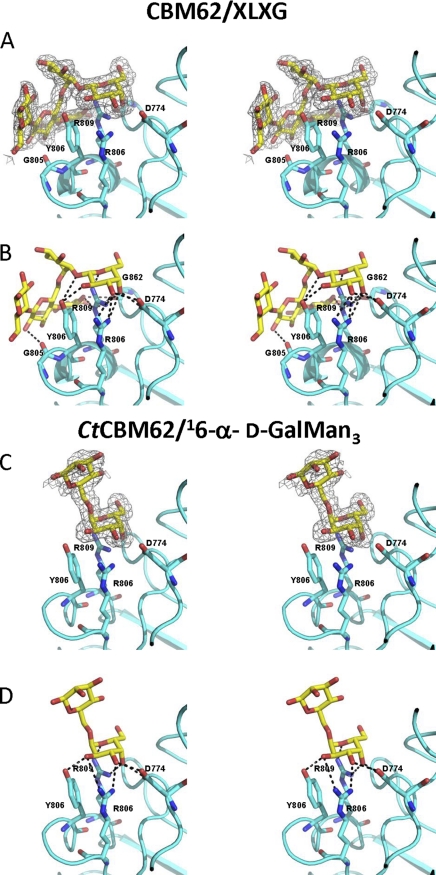
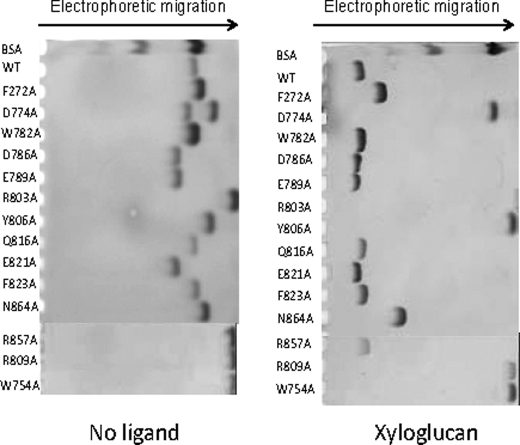
Inspection of the surface of the protein reveals a shallow pocket formed by the loops connecting β-1 and α-1, α-1 and β-2, β-2 and α-2, although α-2 also contributes to the pocket (Fig. 5), a topology consistent with the specificity of the protein for the terminal sugars of complex polysaccharides. The crystal structure of CtCBM62 in complex with 16-α-d-GalMan3 or XLXG confirms that the pocket includes the ligand-binding site and provides insight into how the protein recognizes the galactosyl residue, which makes identical interactions with the protein in both ligand complexes. Thus, the α-face of the pyranose ring of the bound galactose makes extensive hydrophobic contacts with Trp-754, which is aligned parallel to the sugar ring. Such hydrophobic interactions are a generic feature of CBM ligand recognition (12–14, 19, 44). The bound galactose also makes several direct hydrogen bonds with the protein; O1 and O2 make polar contacts with the OH of Tyr-806, whereas O3 forms hydrogen bonds with Nη1 and Nη2 of Arg-803 and Oδ2 of Asp-774. The O4 of galactose makes numerous potential polar interactions with CtCBM62; the hydroxyl is within hydrogen bonding distance of Oδ1 and Oδ2 of Asp-774 and Nη1 of Arg-803 and Arg-809. Finally, the endocyclic oxygen of the galactose makes a polar contact with Nη2 of Arg-809. The extensive interactions made by CtCBM62 with the axial O4 galactose explains why the protein displays tight specificity for this sugar; an equatorial O4, evident in glucose, mannose, and xylose, would not make polar contacts with the protein and, indeed, is predicted to make steric clashes with the side chain of Asp-774. Affinity gel electrophoresis (Fig. 6) and ITC (data not shown) showed that the replacement by alanine of all these residues resulted in the complete loss in ligand binding, confirming the role of these amino acids in galactose recognition. By contrast, mutation of residues on the concave surface of the protein, W782A, D786A, E789A, Q816A, E821A, F823A, and R856A (Fig. 6) did not appear to cause a reduction in affinity for xyloglucan. Similar observations were made with galactomannan and arabinogalactan (data not shown). These data confirm that the sugar-binding site in CtCBM62 is located in the loops connecting the two β-sheets and not on the concave surface of the protein.
FIGURE 5.
Structure of CtCBM62 in complex with xyloglucan and galactomannan-derived oligosaccharides. A and C display the stereo figure of the carbohydrate-binding site of CtCBM62, with unbiased Fo-Fc electron density displayed as a mesh and key amino acids and the carbohydrates drawn as sticks. The protein backbone is shown for reference. In the XLXG complex, the electron density is contoured at 3σ, 0.14 e−/Å3, and for GM3, the map is also at 3σ, e−/Å3. B and D display the same view as A and C, respectively, of the carbohydrate-binding site of CtCBM62, with hydrogen bonds drawn as dashed lines and key amino acids labeled. In both B and D only the amino acids that make hydrogen bonds with the ligand are shown.
FIGURE 6.
Affinity gel electrophoresis of wide type and mutants of CtCBM62 against xyloglucan. Wide type and mutants of CtCBM62 were electrophoresed on nondenaturing polyacrylamide gels containing no ligand or 1 mg/ml xyloglucan. BSA was used as a nonpolysaccharide binding control.
Significantly, CtCBM62 does not make polar contact with the O6 of galactose suggesting that the protein may interact with l-arabinopyranose in which O4 is also axial. Data presented in Table 3 confirm that CtCBM62 does bind to arabinopyranose, albeit with lower affinity than galactose. This may reflect the strain imposed by forcing the sugar ring into its pyranose conformation.
In both ligand complexes, components of the oligosaccharides, in addition to the terminal galactose, are evident. Indeed the successive Gal-β-Xyl and Xyl-α-Glc linkages in XLXG curve the xyloglucan oligosaccharide into a shape that is perfectly accommodated by the surface of CtCBM62. These topological features may contribute to the slightly higher affinity of the protein for XLXG, compared with galactose; the increase in affinity may reflect polar interactions between the Glc backbone and residues (Gly-805 and Gly-862; Fig. 5) on the surface of the protein. To accommodate the Glc or Man backbone of XLXG and 16-α-d-GalMan3, respectively, the side chain of Gln-809 is twisted by 114° which, although imposing an energetic cost, enables both Oϵ1 and Nϵ2 of this residue to make polar contacts with the backbone of at least xyloglucan. By contrast, the Man backbone of 16-α-d-GalMan3 did not make direct polar interactions with CtCBM62.
As discussed above, CtCBM62 appears to undergo calcium-mediated oligomerization leading to increased affinity for polysaccharides through avidity effects. Inspection of the structure of CtCBM62 reveals a calcium-binding site that is conserved in several other CBM families that display a jelly roll fold (CBM4, CBM6, CBM22, and CBM29 (2)). The calcium ion makes coordinate bonds with the O of Lys-763, Oδ1 of Asp-766, O of Asp-766, the backbone O, and Oγ of Thr-771, O of Ala-868, and Oϵ1 of Glu-869, Fig. 4, and as such it fulfills the octahedral geometry typical of many calcium-binding sites. The calcium-binding loop contributes to a crystal packing interface with a symmetry-equivalent molecule in an adjacent asymmetric unit. The physicochemical characteristics of this particular interface are not greatly dissimilar from other potential dyad interactions in the crystal. The D766A, T771A, E869A, D766A/E869A and D766A/T771A/E869A mutants of CtCBM62 (in which the calcium-binding site has been disrupted by mutating, in various combinations, the three residues whose side chains make polar contacts with the metal) displayed calcium-dependent high affinity for xyloglucan, supplemental Table S2. Thus, the calcium-binding loop does not appear to contribute to the oligomerization of the protein in solution. Attempts to map the dimer-binding site by replacing all the surface Asp and Glu residues with alanine (mutations listed in supplemental Table S2) failed to influence the avidity effects displayed by the protein. Thus, currently, the site of CtCBM62 oligomerization remains unclear.
CtCBM62 Defines a New CBM Family
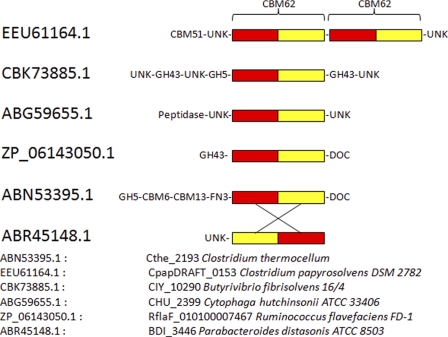
CtCBM62 was subjected to BLAST analysis to identify other proteins that are related to the CBM. The analysis revealed that CtCBM62 displayed very distant similarity to a CBM32 module, comprising residues 2086–2229 in the Caldicellulosiruptor kronotskyensis protein Calkro_0121 (BLAST score of 7e−4), with significant sequence identity restricted to the N-terminal 17 residues of the two protein modules. No sequence similarity was identified between CtCBM62 and any other CAZy protein, including the remaining 1000 CBM32 modules. Thus. CtCBM62 represents the founding member of a new CBM family, defined as CBM62. The BLAST search, however, identified five non-CAZy protein modules that display >42% sequence identity with CtCBM62 with e values <10−23 (supplemental Fig. S2). These proteins are therefore likely to be members of family CBM62. Two of these CBM62 modules are components of large extracytoplasmic proteins of unknown function, although the other three contain members of glycoside hydrolase families. The residues that comprise the ligand-binding site in CtCBM62 are highly conserved in the five proteins suggesting that they may also bind to galactose (and arabinopyranose). Indeed, one of these proteins, C7IBP5, is a member of family GH98, where endo-β-galactosidase is the only reported activity. In addition to the five close homologs, there are 40 proteins that also show sequence similarity with CtCBM62 with e values <10−5. The limited sequence similarity is generally restricted to the C-terminal portion of CtCBM62. Closer analysis, however, showed that noncontiguous regions of CtCBM62 display extensive sequence similarity with these proteins. When a rearranged linear sequence of one of these proteins, a conserved hypothetical protein from Parbacteroides distasonis (GenBankTM accession number ABR45148), in which the N- and C-terminal regions are permuted at position 69 (the N-terminal Phe-466 and C-terminal Asn-600 residues are ligated together and cleavage of the protein occurs between Asn-535 and Asp-536), is compared with CtCBM62, then sequence similarity with the apparent “noncontiguous homologs” extends across the whole sequence of the CBM. Thus, the proteins display noncontiguous stretches of homology with a score of 3e−7. If CtCBM62 is compared with the rearranged Parbacteroides protein, then sequence similarity is contiguous with an amino acid identity of ∼40% and a score of e−17 (Fig. 7 and supplemental Fig. S2). The sequence similarity of noncontiguous regions of these proteins is strongly indicative of a “circular permutation.” In this process, genetic events result in the ligation of the terminal residues of the protein, which is subsequently cleaved to yield a new N and C terminus (45, 46). The cleavage or ligation event likely occurred in CBM62 members at a position equivalent to the peptide bond linking Gly-797 and Tyr-798 in CtCBM62.
FIGURE 7.
Circular permutation in the CBM62 family. BLAST analysis of CtCBM62, corresponding to residues 739–885 of CtXyl5A, identified five proteins that displayed >42% sequence identity, which are identified by their GenBankTM accession numbers. ABR45148.1 is an example of a group of 40 proteins that display noncontiguous similarity to CtCBM62. Extensive co-linear identity to CtCBM62 is evident after ABR45148.1 has been subject to a circular permutation. This genetic event is indicated by the crossover lines, leading to the ligation of the N and C terminus of the encoded protein and cleavage at the interface of the red and yellow sequence, resulting in the generation of a new N and C terminus. The sequence alignment of CtCBM62 (ABN53395) and ABR45148 before and after circular permutation is shown in supplemental Fig. S2.
The vast majority of CBM families display a β-sandwich fold in which the strand order and a structural calcium are highly conserved features (2). It is therefore likely that these families arose from a common progenitor sequence. As the β-strand order in CtCBM62 is similar to other CBM families (e.g. CBM4, -6, -15, -22, -29, -35, and -36), it is unlikely that the protein module arose through a circular permutation event. It would therefore appear that a cleavage and ligation event occurred in an ancestral member of CBM62, which resulted in the large number of proteins that display noncontiguous sequence similarity to CtCBM62. It should be noted that alignment of these noncontiguous sequences with (rearranged) CtCBM62 shows that the key ligand-binding residues are highly conserved, suggesting that these proteins may also target terminal galactose residues. In general, proteins that have been subjected to circular permutation adopt a β-barrel or β-sandwich fold in which the N and C termini are in close proximity. Circular permutation events have been observed in a range of enzymes, including those that modify carbohydrates (45, 46). As CBMs generally display a β-sandwich fold in which the two termini are in close proximity (2), these proteins are candidates for permutation events. Indeed, a recent report has shown that CBM36 and CBM60 display highly conserved ligand-binding sites derived from different regions of the respective proteins, which likely reflect a circular permutation event (22). The evolutionary rationale (if any) for circular permutation events is not entirely clear. Intuitively, such events, which retain overall three-dimensional structure, may introduce subtle structural changes that alter the function of the protein. In view of the circular permutation event within CBM62, we suggest the division of the family into two subfamilies. The two subfamilies display contiguous sequence similarity to CtCBM62 or are related to the protein by a circular permutation event, respectively.
DISCUSSION
This study reveals a novel CBM family that recognizes galactose-containing polysaccharides, which is located in an enzyme that displays arabinoxylanase activity (the catalytic module is described in the accompanying paper (38)), and is a component of the C. thermocellum cellulosome. The β-sandwich fold displayed by CtCBM62 is typical of the majority of CBM families (2). Although historically the ligand-binding site of CBMs is associated with the concave surface of these proteins, this appears to apply only to proteins that recognize the internal regions of polysaccharides. The location of the CtCBM62 ligand-binding site in the loops that connect the β-sheets is an example of an increasingly common feature of CBMs that recognize terminal sugars (8, 16, 22, 47).
An intriguing feature of CtXyl5A is that the enzyme contains a CBM that binds terminal d-galactose or l-arabinopyranose residues of polysaccharides, a specificity mediated by numerous interactions with the axial O4, which distinguishes these two sugars from other neutral carbohydrates, notably mannose, xylose, and glucose, which are widely distributed in plant structural polysaccharides (48). In general, plant cell wall hydrolases contain CBMs that target the substrate recognized by the enzyme and/or crystalline cellulose (2). Although most arabinoxylans contain arabinofuranose and 4-O-methyl-d-glucuronic acid side chains, it is apparent that these polysaccharides are very heterogeneous molecules, and eucalyptus contains arabinoxylans that are also decorated with d-galactose (49). It is therefore possible that the primary substrate for CtXyl5A is an arabinoxylan that also contains sugar side chains (d-galactose) recognized by CtCBM62. Such a targeting strategy has resonance with the observation that a CBM32 targets LacNAc (Gal-β1,4-GlcNAc), but the appended enzymes functions as an N-acetyl-β-hexosaminidase (42). Recent studies, however, have shown that CBMs may direct their appended enzymes to their target substrates by binding to a polysaccharide that is in close proximity with the substrate. Examples include xylanases and pectinases appended to cellulose-specific CBMs (50). It is therefore possible that arabinoxylans are in close association with xyloglucans, galactomannans, and/or arabinogalactans in certain cell walls, and by targeting the β- and α-d-galactose or l-arabinopyranose residues, CtCBM62 brings the catalytic module of CtXyl5A into close proximity with its substrate.
The observations that CtCBM62 binds more tightly to complex polysaccharides than to monovalent ligands and binding is associated with the formation of an insoluble polysaccharide lattice are classic features of avidity effects (18, 20). The rationale for the increase in Ka relates to the decrease in the frequency with which the protein will completely dissociate from its complex ligand, compared with monovalent carbohydrates. Examples of such avidity effects have been observed previously in enzymes containing multiple CBMs that recognize the same complex ligand (17, 18). By contrast, avidity effects through the oligomerization of a discrete enzyme-located CBM, as observed here for CtCBM62, have not been reported previously, although such events are a very common feature of plant and animal lectins (20).
Analysis of the structure of derivatives of CtXyl5A showed that only CtCBM62 was capable of dimerization (data not shown). The biological significance of calcium-induced dimerization of CtCBM62, within the context of full-length CtXyl5A, is particularly intriguing when one considers that the module is derived from a protein that is a component of the cellulosome. The cellulosome assembles through the interaction of the dockerin module, present on the enzymes, with one of the nine cohesins located on the noncatalytic scaffoldin protein, CipA (for review see Refs. 21, 40). It is possible that calcium-induced dimerization of CtCBM62 recruits two molecules of CtXyl5A onto the same cellulosome molecule. The oligomerization, however, may also lead to cellulosome cross-linking (if the dockerins in the dimer bind to different molecules of CipA), and thus contribute to the formation of polycellulosomes, which are formed on the surface of the bacterium (11).
In conclusion, this report reveals a novel CBM that represents the founding member of family 62. CtCBM62 binds to the terminal d-galactose and l-arabinopyranose residues in polysaccharides, specificities not previously observed in CBMs that are components of the C. thermocellum cellulosome. The CBM uniquely mediates protein dimerization, which confers polysaccharide, rather than oligosaccharide, targeting through avidity effects. The catalytic module appended to CtCBM62 is an arabinoxylanase (described in the accompanying paper (38)), suggesting that the enzyme targets xylans that contain a highly complex repertoire of side chains.
Supplementary Material
This work was supported by Grants PTDC/BIAPRO/69732/2006 from Fundação para a Ciência e Tecnologia (Portugal) and SFRH/BD/16731/2004 (to M. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
The atomic coordinates and structure factors (codes 2YFU, 2YFZ, and 2YG0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) ABN53395.1.
- CBM
- carbohydrate-binding module
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1. Ragauskas A. J., Williams C. K., Davison B. H., Britovsek G., Cairney J., Eckert C. A., Frederick W. J., Jr., Hallett J. P., Leak D. J., Liotta C. L., Mielenz J. R., Murphy R., Templer R., Tschaplinski T. (2006) Science 311, 484–489 [DOI] [PubMed] [Google Scholar]
- 2. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Black G. W., Rixon J. E., Clarke J. H., Hazlewood G. P., Ferreira L. M., Bolam D. N., Gilbert H. J. (1997) J. Biotechnol. 57, 59–69 [DOI] [PubMed] [Google Scholar]
- 4. Bolam D. N., Ciruela A., McQueen-Mason S., Simpson P., Williamson M. P., Rixon J. E., Boraston A., Hazlewood G. P., Gilbert H. J. (1998) Biochem. J. 331, 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ferreira L. M., Durrant A. J., Hall J., Hazlewood G. P., Gilbert H. J. (1990) Biochem. J. 269, 261–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Czjzek M., Bolam D. N., Mosbah A., Allouch J., Fontes C. M., Ferreira L. M., Bornet O., Zamboni V., Darbon H., Smith N. L., Black G. W., Henrissat B., Gilbert H. J. (2001) J. Biol. Chem. 276, 48580–48587 [DOI] [PubMed] [Google Scholar]
- 8. Henshaw J., Horne-Bitschy A., van Bueren A. L., Money V. A., Bolam D. N., Czjzek M., Ekborg N. A., Weiner R. M., Hutcheson S. W., Davies G. J., Boraston A. B., Gilbert H. J. (2006) J. Biol. Chem. 281, 17099–17107 [DOI] [PubMed] [Google Scholar]
- 9. Henshaw J. L., Bolam D. N., Pires V. M., Czjzek M., Henrissat B., Ferreira L. M., Fontes C. M., Gilbert H. J. (2004) J. Biol. Chem. 279, 21552–21559 [DOI] [PubMed] [Google Scholar]
- 10. van Bueren A. L., Morland C., Gilbert H. J., Boraston A. B. (2005) J. Biol. Chem. 280, 530–537 [DOI] [PubMed] [Google Scholar]
- 11. Mayer F., Coughlan M. P., Mori Y., Ljungdahl L. G. (1987) Appl. Environ. Microbiol. 53, 2785–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raghothama S., Simpson P. J., Szabó L., Nagy T., Gilbert H. J., Williamson M. P. (2000) Biochemistry 39, 978–984 [DOI] [PubMed] [Google Scholar]
- 13. Simpson P. J., Xie H., Bolam D. N., Gilbert H. J., Williamson M. P. (2000) J. Biol. Chem. 275, 41137–41142 [DOI] [PubMed] [Google Scholar]
- 14. Pell G., Williamson M. P., Walters C., Du H., Gilbert H. J., Bolam D. N. (2003) Biochemistry 42, 9316–9323 [DOI] [PubMed] [Google Scholar]
- 15. Jamal-Talabani S., Boraston A. B., Turkenburg J. P., Tarbouriech N., Ducros V. M., Davies G. J. (2004) Structure 12, 1177–1187 [DOI] [PubMed] [Google Scholar]
- 16. Montanier C., van Bueren A. L., Dumon C., Flint J. E., Correia M. A., Prates J. A., Firbank S. J., Lewis R. J., Grondin G. G., Ghinet M. G., Gloster T. M., Herve C., Knox J. P., Talbot B. G., Turkenburg J. P., Kerovuo J., Brzezinski R., Fontes C. M., Davies G. J., Boraston A. B., Gilbert H. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3065–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bolam D. N., Xie H., White P., Simpson P. J., Hancock S. M., Williamson M. P., Gilbert H. J. (2001) Biochemistry 40, 2468–2477 [DOI] [PubMed] [Google Scholar]
- 18. Boraston A. B., McLean B. W., Chen G., Li A., Warren R. A., Kilburn D. G. (2002) Mol. Microbiol. 43, 187–194 [DOI] [PubMed] [Google Scholar]
- 19. Charnock S. J., Bolam D. N., Nurizzo D., Szabó L., McKie V. A., Gilbert H. J., Davies G. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14077–14082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vijayan M., Chandra N. (1999) Curr. Opin. Struct. Biol. 9, 707–714 [DOI] [PubMed] [Google Scholar]
- 21. Bayer E. A., Belaich J. P., Shoham Y., Lamed R. (2004) Annu. Rev. Microbiol. 58, 521–554 [DOI] [PubMed] [Google Scholar]
- 22. Correia M. A., Abbott D. W., Gloster T. M., Fernandes V. O., Prates J. A., Montanier C., Dumon C., Williamson M. P., Tunnicliffe R. B., Liu Z., Flint J. E., Davies G. J., Henrissat B., Coutinho P. M., Fontes C. M., Gilbert H. J. (2010) Biochemistry 49, 6193–6205 [DOI] [PubMed] [Google Scholar]
- 23. Tormo J., Lamed R., Chirino A. J., Morag E., Bayer E. A., Shoham Y., Steitz T. A. (1996) EMBO J. 15, 5739–5751 [PMC free article] [PubMed] [Google Scholar]
- 24. Carvalho A. L., Goyal A., Prates J. A., Bolam D. N., Gilbert H. J., Pires V. M., Ferreira L. M., Planas A., Romão M. J., Fontes C. M. (2004) J. Biol. Chem. 279, 34785–34793 [DOI] [PubMed] [Google Scholar]
- 25. Charnock S. J., Bolam D. N., Turkenburg J. P., Gilbert H. J., Ferreira L. M., Davies G. J., Fontes C. M. (2000) Biochemistry 39, 5013–5021 [DOI] [PubMed] [Google Scholar]
- 26. Dvortsov I. A., Lunina N. A., Chekanovskaya L. A., Schwarz W. H., Zverlov V. V., Velikodvorskaya G. A. (2009) Microbiology 155, 2442–2449 [DOI] [PubMed] [Google Scholar]
- 27. Najmudin S., Guerreiro C. I., Carvalho A. L., Prates J. A., Correia M. A., Alves V. D., Ferreira L. M., Romão M. J., Gilbert H. J., Bolam D. N., Fontes C. M. (2006) J. Biol. Chem. 281, 8815–8828 [DOI] [PubMed] [Google Scholar]
- 28. Freelove A. C., Bolam D. N., White P., Hazlewood G. P., Gilbert H. J. (2001) J. Biol. Chem. 276, 43010–43017 [DOI] [PubMed] [Google Scholar]
- 29. Brown P. H., Schuck P. (2006) Biophys. J. 90, 4651–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brown P. H., Balbo A., Schuck P. (2007) Biomacromolecules 8, 2011–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kabsch W. (2010) Acta Crystallogr. D66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Evans P. (1993) CCP4 Daresbury Study Weekend: Data Collection and Processing, DL/SCI/R34, pp. 114–122, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 33. Schneider T. R., Sheldrick G. M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 34. Sheldrick G. M. (1990) Acta Crystallogr. A 46, 467–473 [Google Scholar]
- 35. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 36. Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 37. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 38. Correia A. S., Mazumder K., Bras J. L., Firbank S. J., Zhu Y., Lewis R. J., York W. S., Fontes C. M., Gilbert H. J. (2011) J. Biol. Chem. 286, ******** [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gloster T. M., Ibatullin F. M., Macauley K., Eklöf J. M., Roberts S., Turkenburg J. P., Bjørnvad M. E., Jørgensen P. L., Danielsen S., Johansen K. S., Borchert T. V., Wilson K. S., Brumer H., Davies G. J. (2007) J. Biol. Chem. 282, 19177–19189 [DOI] [PubMed] [Google Scholar]
- 40. Fontes C. M., Gilbert H. J. (2010) Annu. Rev. Biochem. 79, 655–681 [DOI] [PubMed] [Google Scholar]
- 41. Gaskell A., Crennell S., Taylor G. (1995) Structure 3, 1197–1205 [DOI] [PubMed] [Google Scholar]
- 42. Ficko-Blean E., Boraston A. B. (2006) J. Biol. Chem. 281, 37748–37757 [DOI] [PubMed] [Google Scholar]
- 43. Boraston A. B., Wang D., Burke R. D. (2006) J. Biol. Chem. 281, 35263–35271 [DOI] [PubMed] [Google Scholar]
- 44. Nagy T., Simpson P., Williamson M. P., Hazlewood G. P., Gilbert H. J., Orosz L. (1998) FEBS Lett. 429, 312–316 [DOI] [PubMed] [Google Scholar]
- 45. Lindqvist Y., Schneider G. (1997) Curr. Opin. Struct. Biol. 7, 422–427 [DOI] [PubMed] [Google Scholar]
- 46. Lo W. C., Lee C. C., Lee C. Y., Lyu P. C. (2009) Nucleic Acids Res. 37, D328–D332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abbott D. W., Ficko-Blean E., van Bueren A. L., Rogowski A., Cartmell A., Coutinho P. M., Henrissat B., Gilbert H. J., Boraston A. B. (2009) Biochemistry 48, 10395–10404 [DOI] [PubMed] [Google Scholar]
- 48. Brett C. T., Waldren K. (1996) Physiology and Biochemistry of Plant Cell Walls. Topics in Plant Functional Biology, Chapman and Hall Ltd., London [Google Scholar]
- 49. Shatalov A. A., Evtuguin D. V., Pascoal Neto C. (1999) Carbohydr. Res. 320, 93–99 [DOI] [PubMed] [Google Scholar]
- 50. Hervé C., Rogowski A., Blake A. W., Marcus S. E., Gilbert H. J., Knox J. P. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 15293–15298 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.