Abstract
The enzymatic degradation of plant cell walls plays a central role in the carbon cycle and is of increasing environmental and industrial significance. The enzymes that catalyze this process include xylanases that degrade xylan, a β-1,4-xylose polymer that is decorated with various sugars. Although xylanases efficiently hydrolyze unsubstituted xylans, these enzymes are unable to access highly decorated forms of the polysaccharide, such as arabinoxylans that contain arabinofuranose decorations. Here, we show that a Clostridium thermocellum enzyme, designated CtXyl5A, hydrolyzes arabinoxylans but does not attack unsubstituted xylans. Analysis of the reaction products generated by CtXyl5A showed that all the oligosaccharides contain an O3 arabinose linked to the reducing end xylose. The crystal structure of the catalytic module (CtGH5) of CtXyl5A, appended to a family 6 noncatalytic carbohydrate-binding module (CtCBM6), showed that CtGH5 displays a canonical (α/β)8-barrel fold with the substrate binding cleft running along the surface of the protein. The catalytic apparatus is housed in the center of the cleft. Adjacent to the −1 subsite is a pocket that could accommodate an l-arabinofuranose-linked α-1,3 to the active site xylose, which is likely to function as a key specificity determinant. CtCBM6, which adopts a β-sandwich fold, recognizes the termini of xylo- and gluco-configured oligosaccharides, consistent with the pocket topology displayed by the ligand-binding site. In contrast to typical modular glycoside hydrolases, there is an extensive hydrophobic interface between CtGH5 and CtCBM6, and thus the two modules cannot function as independent entities.
Keywords: Carbohydrate-binding Protein, Enzyme Mutation, Enzyme Structure, Protein Structure, X-ray Crystallography, Glycoside Hydrolase
Introduction
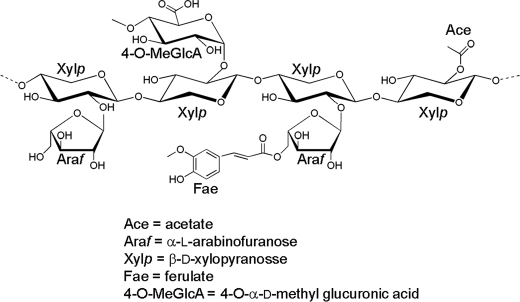
The plant cell wall, which is an important biological and industrial resource, primarily consists of interlocking polysaccharides (for review see Ref. 1). The biological conversion of the polysaccharides within the plant cell wall to their constituent monosaccharides is central to its biological and industrial exploitation (2, 3). An example of this chemical complexity is provided by xylan, which is the major hemicellulosic component of the wall. This polysaccharide includes a backbone of β-1,4-xylose residues in their pyranose configuration (Xylp), which are decorated at O2 with 4-O-methyl-d-glucuronic acid and at O2 and/or O3 with arabinofuranose (Araf) residues, whereas the polysaccharide can also be extensively acetylated. In addition, the Araf side chain decorations can also be esterified to ferulic acid that, in some species, provides a chemical link between hemicellulose and lignin (Fig. 1) (1). The precise structure of xylans varies between plant species, tissues, and during cellular differentiation (4).
FIGURE 1.
Schematic of xylan.
Reflecting the chemical complexity of plant structural polysaccharides, microbial plant cell wall-degrading microorganisms express a large number of enzymes, often in excess of 100 biocatalysts, that target specific linkages within these carbohydrate polymers (5–8). The majority of plant cell wall-degrading enzymes are glycoside hydrolases, although polysaccharide lyases and carbohydrate esterases also contribute to the catabolic process. These enzymes are grouped into families based on sequence and structural and catalytic conservation within the CAZy data base (9). As discussed in the accompanying article (48), many of these enzymes are appended to noncatalytic carbohydrate-binding modules (CBMs)4 that are also grouped into families on the CAZy data base. The xylan backbone is hydrolyzed by xylanases, the majority of which are located in glycoside hydrolase families 10 and 11, although they are also present in GH8 and GH30 (10, 11). The extensive decoration of the xylan backbone generally restricts the capacity of these enzymes to attack the polysaccharide prior to removal of the side chains (12).
Here, we report the biochemical properties and crystal structure of a GH5 enzyme that is appended to a family 6 CBM (CtCBM6). The enzyme (defined as CtXyl5A) is an arabinoxylan-specific xylanase that utilizes Araf decorations, appended to O3 of the Xylp bound at the active site, as an essential specificity determinant. The capacity of CtXyl5A to also accommodate arabinose side chains in all the other subsites (in addition to the active site) within the substrate binding cleft enables the enzyme to hydrolyze highly decorated arabinoxylans. The functional significance of the specificity of the arabinoxylanase, in the context of the plant cell wall degrading apparatus of the host bacterium, is discussed.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Components of CtXyl5A
DNA encoding CtGH5, CtGH5-CBM6, and CtCBM6 were amplified using primers, containing NheI and XhoI restriction sites, which are listed in supplemental Table S1. The amplified DNAs were cloned into NheI/XhoI-restricted pET21a such that the encoded recombinant proteins contained a C-terminal His6 tag. To express the Clostridium thermocellum proteins, Escherichia coli strain BL21(DE3), harboring appropriate recombinant plasmids, was cultured to mid-exponential phase in Luria broth at 37 °C, followed by the addition of isopropyl β-d-galactopyranoside at 1 mm to induce recombinant gene expression, and incubated for a further 5 h at 37 °C. The recombinant proteins were purified to >90% electrophoretic purity by immobilized metal ion affinity chromatography using TalonTM (Clontech), cobalt-based matrix, and elution with 100 mm imidazole, as described previously (13). When preparing the selenomethionine derivative of CtGH5-CBM6 for crystallography, the proteins were expressed in E. coli B834 (DE3), a methionine auxotroph, cultured in media comprising 1 liter of SelenoMet Medium BaseTM, 50 ml of SelenoMet Nutrient MixTM (Molecular Dimensions), and 4 ml of a 10 mg/ml solution of l-selenomethionine. Recombinant gene expression and protein purification were as described above except that all purification buffers were supplemented with 10 mm β-mercaptoethanol.
Mutagenesis
Site-directed mutagenesis was carried out using the PCR-based QuikChange method (Stratagene) deploying the primers listed in the supplemental Table S1.
Enzyme Assays
CtXyl5A and its derivatives were assayed for enzyme activity using the method of Miller (14) to detect the release of reducing sugar. The standard assay was carried out in 50 mm sodium phosphate buffer, pH 7.0, and the potential polysaccharide substrate was at 1 mg/ml. The reactions were initiated by the addition of enzyme up to 10 μm and incubated at 60 °C (unless otherwise stated) for up to 16 h. The identification of potential reaction products was also assessed by HPAEC using methodology described previously (15). The capacity of CtGH5 and CtGH5-CBM6 to hydrolyze xylooligosaccharides was assessed by HPAEC using 100 μm of oligosaccharide and 5 μm of protein.
Oligosaccharide Analysis
Rye arabinoxylan (5 g) was digested to completion (no further increase in reducing sugar and change in the HAEPC product profile) with 3 μm of CtXyl5A at 60 °C for 48 h. The oligosaccharide products were partially purified by size exclusion chromatography using a Bio-Gel P2 column as described previously (16). The structures of the oligosaccharides were analyzed by NMR, electrospray ionization mass spectrometry (ESI-MS), and HPAEC in combination with selective enzyme treatment. Partially methylated alditol acetate derivatives of the glycosyl residues of the oligosaccharides were prepared and analyzed by gas chromatography-electron impact mass spectrometry GC-EI-MS.
Preparation of the Partially Methylated Alditol Acetates
The mixture of oligosaccharides (∼500 μg) was per-O-methylated using the method of Ciucanu and Kerek (17). The per-O-methylated oligosaccharides were hydrolyzed with 2 n TFA, reduced, and acetylated to generate partially methylated alditol acetate derivatives (18).
GC-EI-MS Analysis
Partially methylated alditol acetate derivatives were analyzed with a Hewlett-Packard 5890 gas chromatograph-mass spectrometer. The partially methylated alditol acetates were separated with an SP 2330 column (30 m × 0.25 mm, 0.25-μm film thickness, Supelco) using the following temperature gradient: 80 °C for 2 min, 80–170 °C at 30 °C/min, 170–240 °C at 4 °C/min, and 240 °C held for 20 min. Samples were ionized by electrons impact at 70 eV.
Preparation of Per-O-methylated Oligoglycosyl Alditols
The sample (∼500 μg) was reduced with sodium borohydride to generate oligoglycosyl alditols, which were per-O-methylated as described previously (19).
MALDI-TOF Mass Spectrometry (MALDI-TOF-MS)
Positive ion MALDI-TOF mass spectra were recorded using an Applied Biosystems Voyager-DE biospectrometry workstation. Samples (1 μl of a mg/ml solution) were mixed with an equal volume of matrix solution (0.1 m 2,5-dihydroxybenzoic acid and 0.03 m 1-hydroxyisoquinoline in aqueous 50% MeCN) and dried on MALDI target plate. Typically, spectra from 200 laser shots were summed to generate a mass spectrum.
ESI-MS
The multiple stage ESI mass spectra were recorded in a Thermo Scientific LTQ XL ion trap mass spectrometer. Per-O-methylated oligoglycosyl alditols in methanol were diluted with 50% acetonitrile/water containing 0.1% TFA. Samples were infused through a fused silica capillary (150 μm inner diameter × 363 μm outer diameter × ∼60 cm, Thermo Finnigan) into the source at flow rate of 3 μl/min using the syringe pump provided with the instrument. The electrospray source was operated at a voltage of 5.0 kV, and the capillary heater was set to 275 °C. All the experiments were performed in the positive-ion mode.
NMR Spectroscopy
Oligosaccharides (∼2 mg) were dissolved in D2O (0.5 ml, 99.9%; Cambridge Isotope Laboratories). 1H NMR spectra were recorded with Varian Inova NMR spectrometer operating at 500 MHz at 298 K. All two-dimensional spectra were recorded using standard Varian pulse programs.
Isothermal Titration Calorimetry
The binding of CtCBM6 to ligands was quantified by isothermal titration calorimetry (ITC), as described previously (20). Titrations were carried out in 50 mm Na-HEPES buffer, pH 7.5, containing 5 mm CaCl2 at 25 °C. The reaction cell contained protein at 145 μm, and the syringe contained the monosaccharide or oligosaccharide at 5–15 mm, and polysaccharide, when used as the titrant, was at 3–5 mg/ml. The titrations were analyzed using Microcal Origin version 7.0 software to derive, n, Ka, and ΔH values, and ΔS was calculated using the standard thermodynamic equation, RTlnKa = ΔG = ΔH − TΔS.
Crystallography
Proteins were crystallized using the hanging drop vapor technique at 20 °C with an equal volume (1 μl) of protein and reservoir solution. Native (10 mg/ml) and selenomethionine (3 mg/ml) CtGH5-CBM6 were crystallized in 16–24% PEG 3000, 150 mm sodium citrate, pH 5.5. A CtGH5-CBM6 construct containing two additional methionines, W391M/W397M, was produced to facilitate structure solution by selenomethionine single wavelength anomalous x-ray scattering. Crystals were cryoprotected by the inclusion of 25% glycerol in the crystallization solution and flash-frozen in liquid nitrogen. Diffraction data were collected at ID14.4 ESRF, Grenoble, France, at the selenium K absorption edge to enable structure solution by single wavelength anomalous x-ray scattering. The diffraction data were processed in MOSFLM (21) and SCALA (22), and the heavy atom substructure was solved using SHELXCDE (23) as part of CCP4i, and an initial model was built in Arp/wArp (24), which was completed manually in COOT (25). The complete initial model was used to determine the structure of the wild type protein by molecular replacement and refined at higher resolution from data collected at the Diamond Light Source, UK. The crystal of the reported structure had been soaked in 20 mm “Fraction 1” in an attempt to obtain a structure of the enzyme in complex with carbohydrate, although no sugar molecules, other than glycerol, were observed in the electron density.
All structures were refined to convergence using REFMAC5 (26) with manual corrections being applied in COOT (25). The data collection, phasing, and refinement statistics are displayed in supplemental Table S2, and the PDB code for the protein structure is 2y8k.
RESULTS
Expression and Purification of CtXyl5A
To investigate the function of the CtGH5 and CtCBM6 components of CtXyl5A, the modules were expressed as either individual entities or covalently linked. Although CtCBM6 and CtGH5-CBM6 were expressed in soluble form at high levels in E. coli, CtGH5 was predominantly insoluble, and only a small amount of soluble protein was generated in the enteric bacterium. All three proteins were purified by immobilized metal ion affinity chromatography to electrophoretic homogeneity.
CtXyl5A Is an Arabinoxylanase
Screening the capacity of CtXyl5A to hydrolyze plant structural polysaccharides revealed that the enzyme was able to degrade rye and wheat arabinoxylan, displayed limited activity against oat spelt xylan but was unable to act on glucuronoxylan, birch, or beech xylan (Table 1). The enzyme displayed no activity against a range of mannans, pectins, galactans, arabinans, and β-glucans (data not shown). The individual kinetic constants of CtXyl5A against rye and wheat arabinoxylan could not be determined as the Km value was greater than the maximum concentration of soluble substrate; however, the catalytic efficiency of the enzyme was similar for both rye and wheat arabinoxylan. The high Km value may reflect weak affinity for the substrate, or the glycosidic bonds targeted by CtXyl5A occur rarely in the arabinoxylan substrates. The enzyme displayed trace activity against xylohexaose with a kcat/Km value estimated to be <101 min−1 m−1. These data indicate that CtXyl5A hydrolyzes arabinoxylans but does not act on xylans that contain few arabinofuranose side chains. This is in sharp contrast to typical xylanases, located mainly in GH10 and GH11, which display a preference for the poorly decorated xylans from birch and beech (12). These data show that CtXyl5A displays specificity for arabinoxylans and as such is defined as an arabinoxylanase, an activity not previously reported.
TABLE 1.
Catalytic activity of CtXyl5A and its variants
The enzymes were assayed at 60 °C in 50 mm sodium phosphate buffer, pH 7.0, containing substrate at a concentration of 1 mg ml−1. The reaction was monitored by the release of reducing sugar (14). The catalytic rate could be used to determine kcat/Km values as the substrate concentration was ≪ Km (the rate of reaction was directly proportion to substrate concentration up to 2 mg ml−1). Note that CtXyl5A is the full-length enzyme, and CtGH5-CBM6 and CtGH5 are derivatives of the enzymes containing the catalytic module appended to the CBM6 and the catalytic module, respectively.
| Proteins |
kcat/Km |
||
|---|---|---|---|
| Rye arabinoxylan | Wheat arabinoxylan | Oat spelt xylan | |
| min−1mg−1ml | |||
| CtXyl5A | 1322 ± 357 | 808 ± 76 | 15 ± 1.2 |
| CtXyl5A + 2 mm CaCl2 | 2271 ± 274 | 1658 ± 56 | NDa |
| CtXyl5A + 5 mm EDTA | 858 ± 102 | 343 ± 32 | ND |
| CtGH5-CBM6 | 1012 ± 83 | 652 ± 21 | 14 ± 2.2357 |
| W424A (CtGH5-CBM6) | 1656 ± 173 | 728 ± 127 | ND |
| F478A (CtGH5-CBM6) | 983 ± 52 | 713 ± 66 | ND |
| E279A (CtGH5-CBM6) | NAb | NA ± 357 | ND |
| E171A (CtGH5-CBM6) | NA | NA | ND |
| CtGH5 | 1.1 ± 0.45 | 0.6 ± 0.07 | NA |
a ND means not determined.
b NA means no activity detected.
Characterization of the Reaction Products Generated by CtXyl5A from Arabinoxylan
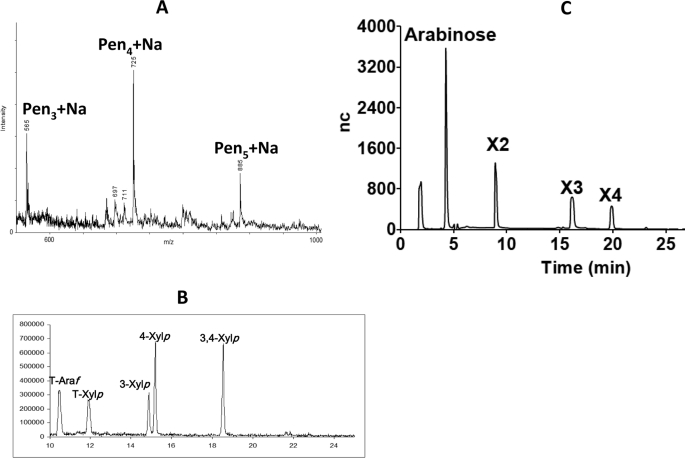
To explore the substrate specificity of CtXyl5A in more detail, the reaction products generated by treating rye arabinoxylan with the enzyme were partially purified by size exclusion chromatography to remove high molecular weight polymers. The fractions containing the majority of the products were pooled (designated henceforth as fraction 1). Fraction 1 was reduced and per-O-methylated, and the products were analyzed by MALDI-TOF-MS. The data revealed that the major reaction products were pentose-containing oligosaccharides with degrees of polymerization (DPs) of 3 (m/z 565), 4 (m/z 725), and 5 (m/z 885), respectively (Fig. 2A). Partially methylated alditol acetate derivatives were then prepared from per-O-methylated fraction 1 and analyzed by GC-EI-MS (Fig. 2B). This semi-quantitative analysis revealed terminal Araf (methylated at O2, O3, and O5), terminal Xylp (methylated at O2, O3, and O4), 3-linked Xylp, 4-linked Xylp, and 3,4-linked Xylp. No Xylp residues decorated at O2 or at both O2 and O3 were observed. These data indicate that the oligosaccharides consist of a backbone of (1→4)-linked Xylp residues decorated with Araf side chains at O3 of internal or reducing Xylp residues (3,4-linked Xylp), or at O3 of nonreducing terminal Xylp residues (3-linked Xylp). Fraction 1 was also treated with CjAbf51A, an arabinofuranosidase that releases Araf residues from O2 or O3 of singly branched Xylp residues in the xylan backbone (27). HPAEC analysis of the CjAbf51A digestion products revealed the presence of arabinose, xylobiose, xylotriose, and xylotetraose (Fig. 2C), indicating that the predominant CtXyl5A products are xylooligosaccharides in which at least one of the Xylp residues bear a mono-Araf side chain. By contrast, GH10 and GH11 xylanases generate predominantly xylose and xylobiose from wheat arabinoxylan, reflecting a preference for undecorated regions of the polysaccharide (12).
FIGURE 2.
Analysis of the reaction products generated by CtXyl5A from arabinoxylan. Rye arabinoxylan was incubated with CtXyl5A until the reaction was complete, and the products were purified by size exclusion chromatography. Fraction 1 contained the most abundant oligosaccharides. A shows the MALDI-TOF MS analysis of permethylated and NaBH4-reduced oligosaccharides in fraction 1. Molecules that contained exclusively pentaose sugars are labeled Pen with the DP in subscript. B shows GC-EI-MS analysis of fraction 1. All the hydroxyls of T-Araf and T-Xylp are methylated, whereas 4-Xylp 3,4-Xylp, and 3-Xylp signify the positions of the hydroxyls that are not methylated and were thus involved in a linkage prior to TFA cleavage. C shows HPAEC analysis of fraction 1 treated with the arabinofuranosidase CjAbf51A. Peaks X2, X3, and X4 co-migrate with xylobiose, xylotriose, and xylotetraose. nc, nanocoulomb.
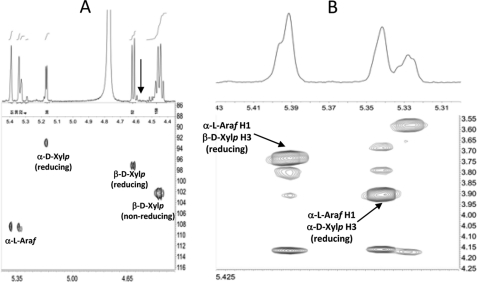
The oligosaccharides in fraction 1 were analyzed by several two-dimensional NMR methods, including gCOSY, HSQC, TOCSY, and ROESY. These analyses provided scalar and dipolar correlations that allowed the resonances of the most abundant spin systems to be assigned to specific sugar residues (supplemental Table S3; for a more detailed description of this approach, see, for example, Refs. 19, 28, 29). Upfield shifts typical of reducing residues (19, 28, 29) were observed for two C1 resonances (δ 92.4 and 96.6) in the HSQC spectrum of the CtXyl5A-generated oligosaccharides (Fig. 3A). In combination with other two-dimensional NMR data, this allowed these two resonances to be assigned to α-Xylp and β-Xylp residues at the reducing end of the oligosaccharides. However, the exact 1H and 13C shifts of these reducing residues indicate that they are structurally distinct from the unbranched (4-linked) sugars at the reducing end of oligosaccharides, generated by more typical endoxylanases (19, 28, 29). The data reveal the presence of an Araf side chain at O3 (along with a β-Xylp at O4) of the reducing Xylp residues of the CtXyl5A-generated oligosaccharides. For example, the C3 resonances of the reducing α-Xylp and β-Xylp units exhibit diagnostic downfield glycosylation shifts (δC 77.7 and 77.8), relative to the corresponding unbranched reducing residues produced by more typical endoxylanases (δC 71.2 and 73.8). Furthermore, the ROESY spectrum of fraction 1 (Fig. 3B) revealed strong dipolar interactions between the two most intense α-Araf H1 resonances (δH 5.342 and 5.391) and the reducing α-Xylp and β-Xylp H3 resonances (δH 3.906 and 3.736, respectively), indicating that most of the α-Araf residues are linked to O3 of reducing Xylp moieties. The identification of branched, reducing Xylp residues in fraction 1 is consistent with the detection of 3,4-linked Xylp residues in the partially methylated derivatives (Fig. 2B). Resonances corresponding to unbranched 4-linked β-Xylp residues at the reducing end of the oligosaccharides (e.g. H1 at δ 4.584, see Fig. 3A) were not detectable in the NMR spectra. Integration of the Xylp and Araf H1 resonances in the one-dimensional spectrum of the CtXyl5A-generated oligosaccharides (Fig. 3A) allowed the following quantitative conclusions to be drawn; the oligosaccharides have an average backbone DP of 2.76 and an average overall DP of 4.04; >99% of the oligosaccharides have an α-l-Araf side chain on O3 of the reducing Xylp residue; ∼30% of the oligosaccharides have a second α-l-Araf side chain.
FIGURE 3.
NMR analysis of the oligosaccharides generated by CtXyl5A. A depicts a partial HSQC spectrum of fraction 1 showing upfield shifts of reducing Xylp residues. The arrow indicates the barely detectable H1 resonance of (unbranched) reducing 4-linked residues. B is a partial ROESY spectrum of fraction 1 showing interglycosidic dipolar contacts between the Araf H1 and the reducing Xylp H3 resonances.
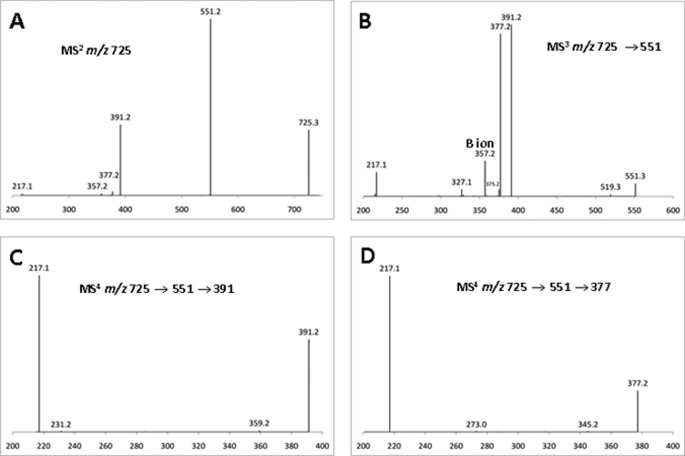
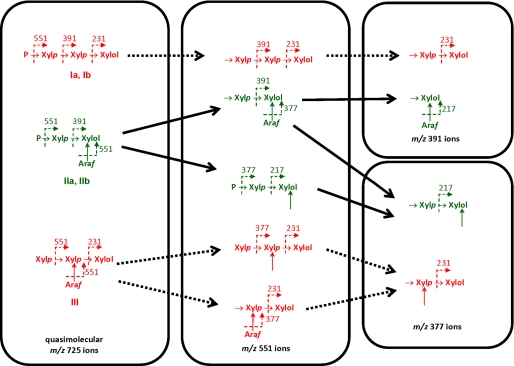
To analyze fraction 1 by ESI-MSn, the oligosaccharides in this sample were treated with NaBH4, and the resulting oligoglycosyl alditols were methylated prior to fragmentation. This procedure imparts a distinctive mass label to the newly formed alditol end of the oligosaccharide, facilitating ESI-MSn analysis (19). The data, examples of which are shown in Fig. 4, provided unambiguous evidence supporting the presence of branched reducing residues in the oligosaccharides in fraction 1. This conclusion is exemplified by the analysis of the possible tetrasaccharides in fraction 1. Thus, based on the structure of the polysaccharide substrate, linkage, and NMR analysis of fraction 1, only five different tetrasaccharide structures (Ia, Ib, IIa, IIb, and III) are theoretically possible (Fig. 5). The ESI-MSn analysis provided information regarding the topology of the oligomers but did not define the stereochemistry (identity) of the individual pentose residues. Therefore, the terminal pentose residues at the nonreducing end of the main chain in structures Ia, Ib, IIa, IIb, displayed in Fig. 5, are indicated by the letter P (as the sugar can be either Araf or Xylp residues). However, in Fig. 5, nonterminal backbone residues, and sugars attached to branched backbone units (backbone sugars that are linked at O4 and O3 to other sugars), are known to be Xylp and Araf, respectively. Thus, structure I could be (Araf)-Xylp-Xylp-Xylol (Ia) or Xylp-Xylp-Xylp-Xylol (Ib) in which Araf is an arabinose decoration appended to the following xylose residue, whereas Xylol is the alditol form of the xylose at the reducing end. Structure II could be Araf-Xylp-(Araf)-Xylol (IIa) or Xylp-Xylp-(Araf)-Xylol (IIb), and III is Xylp-(Araf)-Xylp-Xylol. The quasimolecular (M + Na+) ion at m/z 725, corresponding to these DP4 structures was selected for MS2 (Fig. 4A). The fragmentation pattern is dominated by y ions (19, 30), which contain the alditol end of the oligomer. The y ion (m/z 551) generated by loss of a single terminal pentosyl residue was selected as the precursor for MS3 fragmentation (Fig. 4B). Comparison of this MS3 spectrum (Fig. 4, A and B) to the theoretical fragmentation pattern for all possible m/z 551 ions (Fig. 5) indicates that structures I and III are not present, as these would fragment to form ions at m/z 231, which were not observed. This was confirmed by MS4 analysis (Fig. 4, C and D), in which MS3 fragment ions at m/z 391 and 377 were selected as precursors. Here, the extremely low abundance of ions at m/z 231 confirms the absence of significant amounts of structures I and III (Fig. 5). However, all ions predicted for structure II were observed, notably the high abundance ion at m/z 217, which consists of the alditol residue with two unmethylated hydroxyl groups that were exposed by cleavage of glycosidic bonds during MS2 and MS3 (Fig. 4).
FIGURE 4.
ESI-MS of the tetrasaccharides in fraction 1. The tetrasaccharides in fraction 1, which contains the most abundant products, were analyzed by ESI-MSn. A shows the fragmentation of the m/z 725 ion, which includes the tetrasaccharides. B shows the fragmentation of the m/z 551 ion derived from the m/z 725 ion in A. C and D depict the fragmentation pattern of the m/z 391 and m/z 377 ions, respectively, derived from the m/z 551 ion generated in B. The masses of y ions are indicated unless otherwise stated.
FIGURE 5.
Structure of the tetrasaccharides generated by CtXyl5A. Based on the data displayed in Fig. 4, the structures of the tetrasaccharides in fraction 1 were identified. The sugars labeled P can be Araf or Xylp. The data showed that the oligosaccharide ions colored green were present, and those colored red were not evident. The solid arrows between oligosaccharides show the conversion of one oligosaccharide into another, through ESI-MS fragmentation. Dotted arrows between oligosaccharides identify theoretical ESI-MS-mediated oligosaccharide conversions that did not occur in these analyses. The dotted arrow between sugar linkages within the oligosaccharides shows the fragmentation site and the ion identified. Arrows pointing at sugars (but do not link two sugars together) identify hydroxyl groups that were not methylated as they composed a glycosidic linkage in a parental ion. Xylol is the reducing end xylose that has been reduced to its alditol form by NaBH4.
When the DP5 oligoglycosyl alditols in fraction 1 were analyzed by MSn, virtually all of the alditol moieties were branched (supplemental Figs. S1 and S2). ESI-MSn data for the DP5 oligoglycosyl alditols also provide further insight into the extent to which Araf side chains can decorate the xylooligosaccharides produced by CtXyl5A. Notably, MS4 of the m/z 537 ion (derived from the alditol pentasaccharide) generates an m/z 363 y ion that yields an m/z 217 ion at MS5. As shown in the schematic of supplemental Fig. S2, these species can only be generated if the xylosyl alditol and the adjacent Xylp are both branched. The detection of a m/z 377 ion at MS3, however, demonstrates that the structure Xylp-Xylp-(Araf)-Xylol is also present. Fragmentation of DP3 oligoglycosyl alditols yields an m/z 217 y ion at MS3, although only trace amounts of a m/z 231 ion were evident (supplemental Figs. S3 and S4). This again demonstrates that the xylosyl alditol contains a branch, and thus the structure of the trisaccharide is predicted to be Xylp-(Araf)-Xylol.
Binding of CtXyl5A to Arabinoxylan
The terminal reaction products produced by endo-acting glycoside hydrolases reflects an iterative process in which the products from initial hydrolytic reactions serve as substrates in subsequent rounds of catalysis. Analysis of the structure of the terminal reaction products (which are unable to be further hydrolyzed) provides insight into the possible modes of substrate binding to both the negative and positive subsites (see below). The subsite nomenclature of glycoside hydrolases was defined previously by Davies et al. (31). Briefly, the scissile bond is positioned between subsites −1 and +1, and subsites that extend toward the nonreducing and reducing ends of the substrate are assigned increasing negative and positive numbers, respectively. The Xylp residues at the reducing and the nonreducing end of the oligosaccharide products are derived from substrate bound at the −1 and +1 subsites, respectively. As ∼99% of the reducing end Xylp residues contain an O3 Araf branch, it is evident that the arabinose decoration of the xylose bound at the −1 subsite is a key specificity determinant of the enzyme. The detection of terminal Xylp (in which O2, O3, and O4 are methylated) and 3-linked Xylp residues, both of which occur at the nonreducing end of the oligosaccharide backbone, indicates that a Xylp with an Araf side chain at O3 can be accommodated in the +1 subsite of CtXyl5A, but a side chain in this position is not a specificity determinant. As both (Araf)-Xylp-(Araf)-Xylol and Xylp-Xylp-(Araf)-Xylol were identified in the tetrasaccharide, an O3-Araf side chain is present on some, but not all, of the Xylp residues bound in the −2 subsite. Thus, although an O3-Araf side chain can be accommodated at the −2 subsite, the arabinose decoration does not define enzyme specificity. The identification of Xylp-(Araf)-Xylp-(Araf)-Xylol in the pentasaccharide reaction products not only confirms that Araf can be present at the −1 and −2 subsites, but it also demonstrates that the +2 and +3 (if it exists) subsites can accommodate Xylp residues bearing arabinose side chains. It should be noted, however, that Xylp-(Araf)-Xylp-(Araf)-Xylp is a potential substrate for the enzyme (binding from subsites −2 to +1), suggesting that this molecule is only hydrolyzed very slowly by the enzyme, possibly because it is unable to access the +2 subsites. This is consistent with the absence of Xylp or (Araf)-Xylp in the reaction products; xylose or decorated xylose can only be generated if the substrate is hydrolyzed when it occupies only +1 of the positive subsites of the enzyme. Thus, to summarize, subsites −2 to +2 of CtXyl5A can accommodate Xylp residues that contain an O3-Araf side chain; however, only at the −1 subsite does the arabinose decoration act as an essential specificity determinant.
CtCBM6 Specificity
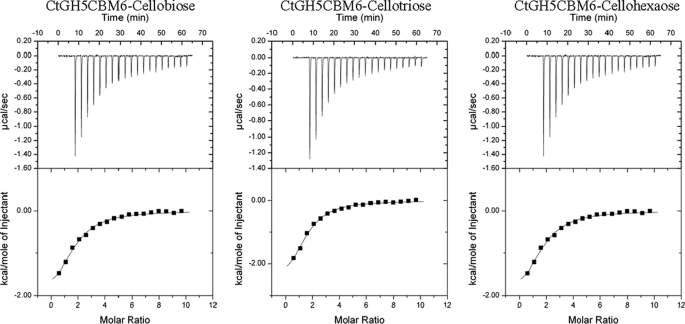
To investigate whether CtCBM6 is a functional CBM, the capacity of CtGH5-CBM6 to bind to various carbohydrates was assessed by ITC. The data showed that CtGH5-CBM6 bound to cellohexaose and cellobiose with similar affinity (Table 2; example titrations are shown in Fig. 6). By contrast, binding to glucose was too low to quantify. The protein also displayed affinity for the reaction products generated by CtXyl5A and for undecorated xylooligosaccharides. The protein did not appear to bind to various xylans or to β-1,3-β-1,4-glucans. This indicates that CtGH5-CBM6 recognizes the terminal region of these polysaccharides, as the concentration of ligand available to the protein in these polymers, which have DPs >300, would be very low, and thus binding would not be detected. It is possible that the catalytic module, rather than CBM6, mediates binding to the xylo- and cello-oligosaccharides. To test this hypothesis, the ligand binding profile of variants of CtGH5-CBM6, in which either Trp-424 or Phe-478 had been substituted with Ala, was assessed. As discussed below, these two aromatic residues are highly conserved in the CBM6 family and includes the primary binding site in this protein family (32). Both CtGH5-CBM6:W424A and CtGH5-CBM6:F478A, although catalytically active (Table 1), displayed no binding to the xylan- and cellulose-derived oligosaccharides (Table 2). It is therefore evident that the CBM6 component of CtGH5-CBM6 mediates the observed binding to oligosaccharides.
TABLE 2.
Binding of CtXyl5A derivatives to polysaccharides and oligosaccharides
The binding of derivatives of CtXyl5A to ligands was measured by ITC. The protein was at the 145 μm in the cell and polysaccharide (3–5 mg/ml) or oligosaccharide (5–15 mm) was in the syringe. ITC was carried out in 50 mm Na/HEPES buffer, pH 7.5, at 25 °C. The concentration of the oligosaccharides generated by the digestion of wheat arabinoxylan (WAX) was fitted to give an n value close to 1.
| CtXyl5A derivative | Ligand | Ka × 103 | ΔG | ΔH | TΔS | n |
|---|---|---|---|---|---|---|
| m−1 | kcal mol−1 | kcal mol−1 | kcal mol−1 | |||
| CtGH5-CBM6 | Cellobiose | 13.7 ± 1.0 | −5.6 | −3.8 ± 0.11 | 1.8 | 1.03 ± 0.13 |
| CtGH5-CBM6 | Cellotriose | 17.4 ± 3.8 | −5.8 | −4.7 ± 0.75 | 1.1 | 1.02 ± 0.13 |
| CtGH5-CBM6 | Cellohexaose | 21.0 ± 4.9 | −5.9 | −3.2 ± 0.44 | 2.7 | 1.12 ± 0.12 |
| CtGH5-CBM6 | Xylotetraose | 3.4 ± 0.2 | −4.8 | −1.2 ± 0.13 | 3.6 | 1.24 ± 0.15 |
| CtGH5-CBM6 | Xylotriose | 2.5 ± 0.6 | −4.6 | −1.3 ± 0.03 | 3.3 | 1.26 ± 0.13 |
| CtGH5-CBM6 | Xylobiose | 2.4 ± 0.4 | −4.6 | −1.2 ± 0.06 | 3.4 | 1.1 ± 0.10 |
| CtGH5-CBM6 | WAXa treated with CtXyl5A | 9.1 ± 2.1 | −5.4 | −2.4 ± 0.5 | 3.0 | 1.01 ± 0.12 |
| CtGH5-CBM6 | WAX treated with CtXyl5A and CjAbF51Abb | 15.1 ± 2.3 | −5.7 | −4.9 ± 0.45 | 0.8 | 1.08 ± 0.06 |
| CtGH5-CBM6:E279A | Cellohexaose | 13.4 ± 2.1 | −5.6 | −4.2 ± 0.98 | 1.4 | 1.12 ± 0.14 |
| CtGH5-CBM6:E279A | WAX treated with CjXyn10Ac | 14.2 ± 1.2 | −5.7 | −3.6 ± 0.83 | 2.1 | 1.12 ± 0.12 |
| CtGH5CBM6E279A | No binding to WAX | |||||
| CtGH5-CBM6:W424A | No binding to cellohexaose, xylotetraose, WAX treated with CjXyn10A, CtXyl5A, or both CtXyl5A and CjAbf51A | |||||
| CtGH5-CBM6:F478A | No binding to cellohexaose, WAX treated with CjXyn10A, CtXyl5A, or both CtXyl5A and CjAbf51A | |||||
| CtCBM6 | No binding to cellohexaose, xylotetraose, xylotriose, xylobiose, WAX treated with CtXyn5A or CjXyn10A | |||||
a WAX means wheat arabinoxylan.
b CjXyn10A indicates GH10 xylanase from Cellvibrio japonicas.
c CjAbf51A indicates GH51 arabinofuranosidase from C. japonicus.
FIGURE 6.
Representative ITC data of CtGH5-CBM6 to oligosaccharides. The ligands (10 mm arabinose) in the syringe was titrated into CtGH5-CBM6 (100 μm) in the cell. The top half of each panel shows the raw ITC heats; the bottom half of each panel shows the integrated peak areas fitted using a single binding model by MicroCal Origin software. ITC was carried out in 50 mm Na-HEPES, pH 7.5, at 37 °C.
Crystal Structure of CtGH5-CBM6
The structure of CtGH5-CBM6 was solved by selenomethionine single wavelength anomalous x-ray scattering, and the resulting structure was used as a starting model for refinement against native data extending to 1.5 Å resolution (supplemental Table S2) (PDB code 2y8k). The polypeptide chain is visible from Ser-37 to Ile-516.
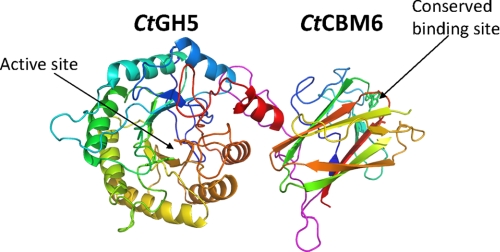
CtGH5
As expected, the N-terminal CtGH5 module displays a (β/α)8 barrel architecture, although α-helix 8 points away from the barrel and toward CtCBM6 module (discussed below) (Fig. 7). GH5 enzymes are members of clan GH-A in which the two catalytic residues are invariant glutamates presented at the end of β-strands 4 and 7 (33, 34). From the structure of CtGH5-CBM6, the catalytic acid-base is likely to be Glu-171 (end of β-strand 4) and the catalytic nucleophile Glu-279 (end of β-strand 7). The catalytic role of these two residues is confirmed by the observation that the mutants E171A and E279A are inactive (Table 1). A narrow V-shaped cleft, ∼25 Å in length, extends along the full length of the protein and sits over the top of the β-barrel. The dimensions of the cleft, in the center of which is the catalytic apparatus, suggest that the protein contains ∼5 subsites extending from −3 to +2.
FIGURE 7.
Crystal structure of CtGH5-CBM6. In the protein schematic of CtGH5-CBM6, both modules are color ramped from the N terminus (blue) to C terminus (red). The loop connecting the two modules is colored magenta. The two catalytic residues (Glu-171 and Glu-279) in CtGH5 and the two aromatic amino acids that are conserved in the ligand-binding site of family 6 CBMs (Trp-424 and Phe-478) are shown in stick format. The figure, and the other structural figures, was drawn with PyMol (DeLano Scientific).
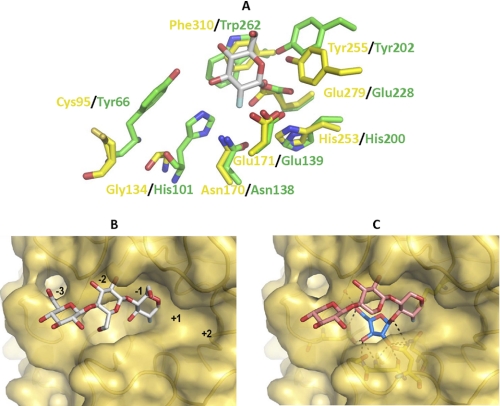
An analysis of structural homologues of the CtGH5 component of CtGH5-CBM6 by the DaliLite webserver identified a large number of GH5 and clan GH-A enzymes that displayed significant structural similarity to CtGH5. The Pseudoalteromonas haloplanktis cellulase Cel5G (PDB 1tvn), with a root mean square deviation of 2.8 Å over 253 Cα atoms and a Z-score of 24.1, and the Bacillus agaradhaerens cellulase BaCel5A (PDB 1qi2), with a root mean square deviation of 2.9 Å over 254 Cα atoms and a Z-score of 23.6, are representative and close structural homologues. The critical −1 subsite, where the transition state is formed, is similar in the arabinoxylanase and the GH5 cellulases. In addition to the two catalytic glutamates, CtGH5 contains several key residues that have been identified as “strictly conserved” in family GH5 enzymes (35). These residues in the CtGH5 module, which superimpose with amino acids in the active site of BaCel5A (the cellulase residues are shown in parentheses), are as follows: Asn-170 (Asn-138), Glu-171 (Glu-139), Tyr-255 (Tyr-202), Glu-279 (Glu-228), and Phe-310 (Trp-262) (Fig. 8A). The catalytic acid base, Glu-171, makes hydrogen bonds with Asn-139 and His-253, and these interactions likely contribute to both the position and ionization state of this critical amino acid. Asn-170 is highly conserved in clan GH-A glycoside hydrolases and plays an important role in transition state stabilization by making a hydrogen bond with the O2 of the sugar at the −1 subsite (36). The position of the catalytic nucleophile, Glu-279, is stabilized through a hydrogen bond with Tyr-255, whereas Phe-310, based upon comparison with other related hydrolases, is likely to form the sugar-binding hydrophobic platform in subsite −1.
FIGURE 8.
Superimposition of CtGH5 and the cellulase BaCel5A. A shows the superimposition of the residues in the active site (−1 subsite) of BaCel5A (PDB 1qi2; colored green), which interact with the substrate, with the equivalent amino acids (colored yellow) in CtGH5. B shows the solvent-accessible surface of CtGH5 in which 2-deoxy-2-fluorocellotriose, derived from BaCel5A, has been superimposed. C depicts a model of xylotriose, containing Araf appended to O3 of Xylp-1, bound to CtGH5. The tetrasaccharide ligand is modeled on the superimposed structure of 2-deoxy-2-fluorocellotriose and the glycerol and water molecules in the putative arabinose binding pocket. A and B, bound ligand is colored silver (carbons), and the Xylp and Araf residues in C are colored salmon pink and blue (carbons), respectively.
Despite numerous attempts, no structure of CtGH5-CBM6 in complex with its substrate or reaction products has been obtained, in part due to the preference of this protein to crystallize with the N-terminal residues of a symmetry-related molecule positioned in the substrate binding cleft, and because co-crystallization experiments did not yield diffracting crystals. Consequently, it is difficult to define precisely the structural basis for the unusual substrate specificity displayed by the arabinoxylanase. Superimposing BaCel5A in complex with 2-deoxy-2-fluorocellotriose with CtGH5 provides some insight into the specificity displayed by the arabinoxylanase. As discussed above, the catalytic apparatus, the residues that interact with O2 and the endocyclic oxygen of the −1 sugar, and the hydrophobic platform are conserved in CtGH5 (Fig. 8A). It is evident, however, that the arabinoxylanase lacks the residues that in other GH5 enzymes hydrogen bond with O3 of the active site sugar. For instance, His-101 and Tyr-66 in BaCel5A hydrogen bond with O3 of the −1 Glc, whereas the equivalent residues in CtGH5 are Gly-134 and Cys-95, respectively (Fig. 8A). Indeed, in the −1 subsite of the arabinoxylanase, there is a large pocket around the O3 of the superimposed Glc that could accommodate a sugar decoration such as Araf (Fig. 8B). The pocket contains a tyrosine (Tyr-92) that may make hydrophobic interactions with the arabinose and contains several polar residues, Glu-68, Asn-135, Asn-139, and Asn-170, that could make polar contacts with the sugar. Based on the presence of glycerol and water molecules within this region of the enzyme, an Araf molecule was modeled into the pocket (Fig. 8C).
CtCBM6
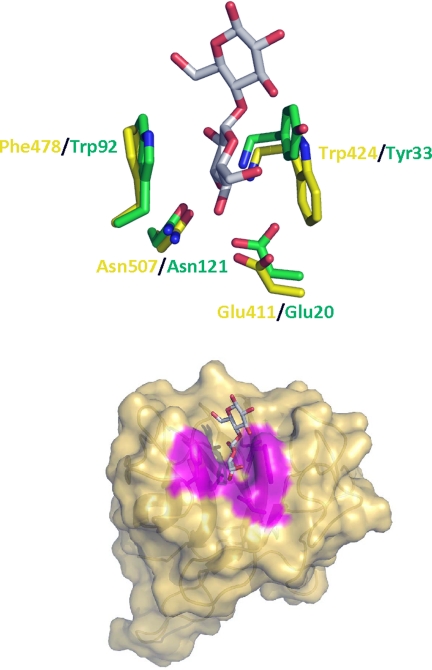
The structure of the CtCBM6 module displays a β-sandwich fold typical of other family CBM6 members (Fig. 7) (32, 37, 38). The twisted pair of β-sheets, which can be viewed as forming an extended barrel, consist of five and four anti-parallel β-strands, respectively. The structure of CtCBM6 shows strong similarity with numerous CBM6 members. The closest homologue is the CBM6 module (designated CmCBM6) from the Cellvibrio mixtus lichenase CmLic5A (PDB 1uz0; root mean square deviation 1.5 Å over 123 Cα atoms and a Z-score of 18.1). The major binding site in the CBM6 family is in the loops connecting the two β-sheets. This region, referred to as site A (32, 37), may comprise a pocket if terminal sugars are recognized (39) or a cleft for the binding of internal regions of polysaccharides (32). A central feature of site A is a pair of aromatic residues, which bind to the α- and β-face, respectively, of the terminal sugar (or central sugar in the case of xylan-binding modules) and an asparagine, located at the base of the site that makes critical hydrogen bonds with O2, O3, or O4. Specificity is conferred by additional polar and hydrophobic interactions (37). Site A in CtCBM6 displays a pocket-like topology and contains all the key ligand binding residues present in CmCBM6 (Fig. 9) (40). The pair of aromatic residues in CmCBM6, Trp-92 and Tyr-33, which straddle the nonreducing terminal sugar, correspond to Phe-478 and Trp-424, respectively, in CtCBM6. Furthermore, Glu-20 and Asn-121 in CmCBM6, which make polar contacts with O3 and O4 of the nonreducing terminal sugar in cello- and xylooligosaccharides, superimpose with Glu-411 and Asn-507, respectively, in CtCBM6. Finally, the amide nitrogen of Tyr-33 in CmCBM6 makes a polar contact with O2 and O3 of the terminal sugar, a contact that is likely to be replicated by that of Trp-424 in CtCBM6. The structural conservation between site A in CtCBM6 and CmCBM6 is consistent with the similar ligand specificities displayed by this binding site in the two proteins (Table 2) (39). Thus, both proteins bind to xylo- and gluco-configured oligosaccharides but do not display affinity for the corresponding polysaccharides. Thus, the structural similarity between CmCBM6 and CtCBM6 is consistent with the view that the Clostridium module targets the terminal regions of oligosaccharides. In CmCBM6 cellooligosaccharides can bind to site A in both orientations, consistent with the targeting of O1/O4, O2, and O3, but not the endocyclic oxygen or O6, which would adopt different positions in the two orientations. Therefore, it is highly likely that CtCBM6 will also bind to xylo- and cellooligosaccharides in both orientations. Given that the key interactions with the ligand at site A is with the terminal sugar, it is perhaps surprising that CtCBM6 does not display measurable binding to xylose or glucose. It is possible that the entropic cost of locking the sugar into a pyranose ring conformation may contribute to the weak binding, although it is also possible that the protein makes indirect, water-mediated interactions to the penultimate sugar in the oligosaccharides, as observed in CmCBM6-ligand complexes (40).
FIGURE 9.
Superimposition of CtCBM6 and CmCBM6. A shows the superimposition of the residues in the ligand-binding site of CmCBM6 (PDB 1uz0; colored green) with the equivalent amino acids (colored yellow) in CtCBM6. B show the solvent-accessible surface of CtCBM6 in complex with cellotriose (superimposed from CmCBM6). Amino acids whose side chains are predicted to contribute to ligand recognition are colored magenta. In both panels ligand is shown in silver (carbon) stick representation.
Linker Connecting CtGH5 with CtCBM6
CtCBM6 is connected to CtGH5 by a sequence extending from residues Gly-336 to Thr-373. This linker, which adopts a stable conformation based on its B-factor, makes numerous internal polar contacts and forms hydrogen bonds with β-strand 3 and the loop connecting β-strands 3 and 4 of CtCBM6 and α-helices 7 and 8 of CtGH5. Furthermore, the C-terminal region of α-helix 8 and the internal region of α-helix 7 make hydrogen bonds with β-strands 3 and 7 of CtCBM6. The polar contacts between the two modules are augmented by a large number of apolar interactions mediated by the linker sequence. The resultant burial of a significant hydrophobic surface, at the interface between CtGH5 and CtCBM6, likely explains why these two modules (or domains) do not fold independently, as occurs in other glycoside hydrolases that contain catalytic modules and CBMs (41). This view is consistent with the observation that CtCBM6, when expressed as a discrete entity (Thr-373 to Ile-516), does not bind to cellohexaose or xylohexaose, and CtGH5 (Asn-32 to Thr-373) exhibits very low catalytic activity and is considerably more thermolabile than CtGH5-CBM6 (supplemental Fig. S5).
DISCUSSION
This study reveals a C. thermocellum protein that displays arabinoxylanase activity, an activity not previously reported. The vast majority of xylanases are derived from GH10 and GH11 and target the β-1,4-d-xylose polymeric backbone. These enzymes do not generally distinguish between different xylans, although highly decorated forms of the polysaccharide, such as rye arabinoxylan, are poorly degraded as steric constraints restrict enzyme access (12). Indeed, the only other examples of xylanases that utilize side chains as essential specificity determinants are glucuronoxylan-specific enzymes from GH30. These enzymes make critical interactions with the 4-O-methylglucuronic acid (linked α-1,2 to the xylan backbone) that decorates the xylose at the −2 subsite (42). CtXyl5A is highly unusual in that its essential Araf decoration is attached to the xylose positioned in the active site. The only other example of an active site side chain specificity determinant is the α-1,6-Xylp that decorates the −1 Glc in the xyloglucan cellobiohydrolase, OXG-RCBH, from Geotrichum sp. (43).
The function of CtXyl5A within the context of C. thermocellum, which has the genetic capacity to recruit 72 different enzymes into the cellulosomes (44), including seven GH10 and GH11 xylanases, is intriguing. It is likely that the GH10 and GH11 enzymes target xylans that are sparsely decorated with arabinose side chains. By contrast, CtXyl5A most likely hydrolyzes xylans where tandem Xylp residues contain Araf decorations. The recognition of the termini of xylo- and gluco-configured polymers by CtCBM6 suggests that the arabinoxylanase is targeted to regions of the plant cell wall that is undergoing degradation and is therefore accessible to enzyme attack. Although the primary function of CBMs is to bring their cognate enzymes into close contact with appropriate substrates (45), there is increasing evidence that a subset of these modules, from CBM families 6, 9, and 35, target the termini of polysaccharides and thus may play a similar function to CtCBM6 (37, 46, 47). In conclusion, CtXyl5A displays a specificity that is complementary to endoxylanases from GH10, GH11, and GH30. As such the enzyme will make a contribution to the toolbox of biocatalysts required to degrade plant cell walls to their constituent sugars, which can then be used in the biofuel and bioprocessing industries.
Supplementary Material
Acknowledgments
We thank the European Synchrotron Radiation Facility, Diamond Light Source, and the staff at both these facilities for help with data collection. We also thank the Department of Energy Office Center for Plant and Microbial Complex Carbohydrates (supported by Grant DE-FG02-93ER20097) for supporting critical infrastructure and analytical instrumentation required for this research.
This work was supported by Grants PTDC/BIAPRO/69732/2006 from Fundação para a Ciência e Tecnologia (Portugal) and SFRH/BD/16731/2004 (to M. A. S. C.), Bioenergy Science Center, United States Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the Department of Energy Office of Science, and Department of Energy Grant DE-FG02-96ER20220.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Tables 1–3.
The atomic coordinates and structure factors (code 2y8k) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CBM
- carbohydrate-binding module
- PDB
- Protein Data Bank
- DP
- degrees of polymerization
- EI
- electron ionization
- ESI
- electrospray ionization
- ITC
- isothermal titration calorimetry
- HPAEC
- high performance anion exchange chromatography.
REFERENCES
- 1. Brett C. T., Waldren K. (1996) Physiology and Biochemistry of Plant Cell Walls. Topics in Plant Functional Biology, Chapman and Hall Ltd., London [Google Scholar]
- 2. Himmel M. E., Bayer E. A. (2009) Curr. Opin. Biotechnol. 20, 316–317 [DOI] [PubMed] [Google Scholar]
- 3. Ragauskas A. J., Williams C. K., Davison B. H., Britovsek G., Cairney J., Eckert C. A., Frederick W. J., Jr., Hallett J. P., Leak D. J., Liotta C. L., Mielenz J. R., Murphy R., Templer R., Tschaplinski T. (2006) Science 311, 484–489 [DOI] [PubMed] [Google Scholar]
- 4. Ebringerova A., Hromadkova Z., Heinze T. (2005) in Polysaccharides 1: Structure, Characterization and Use (Heinze T. ed) pp. 1–67, Springer-Verlag, Berlin [Google Scholar]
- 5. DeBoy R. T., Mongodin E. F., Fouts D. E., Tailford L. E., Khouri H., Emerson J. B., Mohamoud Y., Watkins K., Henrissat B., Gilbert H. J., Nelson K. E. (2008) J. Bacteriol. 190, 5455–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weiner R. M., Taylor L. E., 2nd, Henrissat B., Hauser L., Land M., Coutinho P. M., Rancurel C., Saunders E. H., Longmire A. G., Zhang H., Bayer E. A., Gilbert H. J., Larimer F., Zhulin I. B., Ekborg N. A., Lamed R., Richardson P. M., Borovok I., Hutcheson S. (2008) PLoS Genet. 4, e1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berg Miller M. E., Antonopoulos D. A., Rincon M. T., Band M., Bari A., Akraiko T., Hernandez A., Thimmapuram J., Henrissat B., Coutinho P. M., Borovok I., Jindou S., Lamed R., Flint H. J., Bayer E. A., White B. A. (2009) PLoS One 4, e6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu J., Bjursell M. K., Himrod J., Deng S., Carmichael L. K., Chiang H. C., Hooper L. V., Gordon J. I. (2003) Science 299, 2074–2076 [DOI] [PubMed] [Google Scholar]
- 9. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gilbert H. J. (2010) Plant Physiol. 153, 444–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gilbert H. J., Stålbrand H., Brumer H. (2008) Curr. Opin. Plant Biol. 11, 338–348 [DOI] [PubMed] [Google Scholar]
- 12. Pell G., Taylor E. J., Gloster T. M., Turkenburg J. P., Fontes C. M., Ferreira L. M., Nagy T., Clark S. J., Davies G. J., Gilbert H. J. (2004) J. Biol. Chem. 279, 9597–9605 [DOI] [PubMed] [Google Scholar]
- 13. Charnock S. J., Bolam D. N., Nurizzo D., Szabó L., McKie V. A., Gilbert H. J., Davies G. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14077–14082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miller G. L. (1959) Anal. Chem. 31, 426–428 [Google Scholar]
- 15. Charnock S. J., Lakey J. H., Virden R., Hughes N., Sinnott M. L., Hazlewood G. P., Pickersgill R., Gilbert H. J. (1997) J. Biol. Chem. 272, 2942–2951 [DOI] [PubMed] [Google Scholar]
- 16. Proctor M. R., Taylor E. J., Nurizzo D., Turkenburg J. P., Lloyd R. M., Vardakou M., Davies G. J., Gilbert H. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2697–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ciucanu I., Kerek F. (1984) Carbohydr. Res. 131, 209–217 [Google Scholar]
- 18. Carpita N. C., Shea E. M. (1989) in Analysis of Carbohydrates by GLC and MS (Biermann C. J., McGinnis G. D. eds) pp. 157–216, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 19. Mazumder K., York W. S. (2010) Carbohydr. Res. 345, 2183–2193 [DOI] [PubMed] [Google Scholar]
- 20. Charnock S. J., Bolam D. N., Turkenburg J. P., Gilbert H. J., Ferreira L. M., Davies G. J., Fontes C. M. (2000) Biochemistry 39, 5013–5021 [DOI] [PubMed] [Google Scholar]
- 21. Leslie A. G. W. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, No. 26 [Google Scholar]
- 22. Evans P. (1993)CCP4 Daresbury Study Weekend: Data Collection and Processing, DL/SCI/R34, pp. 114–122, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 23. Sheldrick G. M. (1990) Acta Crystallogr. A 46, 467–473 [Google Scholar]
- 24. Lamzin V. S., Wilson K. S. (1993) Acta Crystallogr. D Biol. Crystallogr. 49, 129–147 [DOI] [PubMed] [Google Scholar]
- 25. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 26. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 27. Beylot M. H., McKie V. A., Voragen A. G., Doeswijk-Voragen C. H., Gilbert H. J. (2001) Biochem. J. 358, 607–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gruppen H., Hoffmann R. A., Kormelink F. J., Voragen A. G., Kamerling J. P., Vliegenthart J. F. (1992) Carbohydr. Res. 233, 45–64 [DOI] [PubMed] [Google Scholar]
- 29. Hoffmann R. A., Leeflang B. R., de Barse M. M., Kamerling J. P., Vliegenthart J. F. (1991) Carbohydr. Res. 221, 63–81 [DOI] [PubMed] [Google Scholar]
- 30. Domon B., Costello C. E. (1988) Biochemistry 27, 1534–1543 [DOI] [PubMed] [Google Scholar]
- 31. Davies G. J., Wilson K. S., Henrissat B. (1997) Biochem. J. 321, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Czjzek M., Bolam D. N., Mosbah A., Allouch J., Fontes C. M., Ferreira L. M., Bornet O., Zamboni V., Darbon H., Smith N. L., Black G. W., Henrissat B., Gilbert H. J. (2001) J. Biol. Chem. 276, 48580–48587 [DOI] [PubMed] [Google Scholar]
- 33. Henrissat B., Callebaut I., Fabrega S., Lehn P., Mornon J. P., Davies G. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7090–7094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jenkins J., Lo Leggio L., Harris G., Pickersgill R. (1995) FEBS Lett. 362, 281–285 [DOI] [PubMed] [Google Scholar]
- 35. Hilge M., Gloor S. M., Rypniewski W., Sauer O., Heightman T. D., Zimmermann W., Winterhalter K., Piontek K. (1998) Structure 6, 1433–1444 [DOI] [PubMed] [Google Scholar]
- 36. Williams S. J., Notenboom V., Wicki J., Rose D. R., Withers S. G. (2000) J. Am. Chem. Soc. 122, 4229–4230 [Google Scholar]
- 37. Abbott D. W., Ficko-Blean E., van Bueren A. L., Rogowski A., Cartmell A., Coutinho P. M., Henrissat B., Gilbert H. J., Boraston A. B. (2009) Biochemistry 48, 10395–10404 [DOI] [PubMed] [Google Scholar]
- 38. van Bueren A. L., Morland C., Gilbert H. J., Boraston A. B. (2005) J. Biol. Chem. 280, 530–537 [DOI] [PubMed] [Google Scholar]
- 39. Henshaw J. L., Bolam D. N., Pires V. M., Czjzek M., Henrissat B., Ferreira L. M., Fontes C. M., Gilbert H. J. (2004) J. Biol. Chem. 279, 21552–21559 [DOI] [PubMed] [Google Scholar]
- 40. Pires V. M., Henshaw J. L., Prates J. A., Bolam D. N., Ferreira L. M., Fontes C. M., Henrissat B., Planas A., Gilbert H. J., Czjzek M. (2004) J. Biol. Chem. 279, 21560–21568 [DOI] [PubMed] [Google Scholar]
- 41. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vrsanská M., Kolenová K., Puchart V., Biely P. (2007) FEBS J. 274, 1666–1677 [DOI] [PubMed] [Google Scholar]
- 43. Yaoi K., Kondo H., Hiyoshi A., Noro N., Sugimoto H., Tsuda S., Mitsuishi Y., Miyazaki K. (2007) J. Mol. Biol. 370, 53–62 [DOI] [PubMed] [Google Scholar]
- 44. Fontes C. M., Gilbert H. J. (2010) Annu. Rev. Biochem. 79, 655–681 [DOI] [PubMed] [Google Scholar]
- 45. Bolam D. N., Ciruela A., McQueen-Mason S., Simpson P., Williamson M. P., Rixon J. E., Boraston A., Hazlewood G. P., Gilbert H. J. (1998) Biochem. J. 331, 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boraston A. B., Creagh A. L., Alam M. M., Kormos J. M., Tomme P., Haynes C. A., Warren R. A., Kilburn D. G. (2001) Biochemistry 40, 6240–6247 [DOI] [PubMed] [Google Scholar]
- 47. Montanier C., van Bueren A. L., Dumon C., Flint J. E., Correia M. A., Prates J. A., Firbank S. J., Lewis R. J., Grondin G. G., Ghinet M. G., Gloster T. M., Herve C., Knox J. P., Talbot B. G., Turkenburg J. P., Kerovuo J., Brzezinski R., Fontes C. M., Davies G. J., Boraston A. B., Gilbert H. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3065–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Montanier C. Y., Correia M. A. S., Flint J. E., Zhu Y., McKee L. S., Prates J. A. M., Polizzi S. J., Coutinho P. M., Lewis R. J., Henrissat B., Fontes C. M. G. A., Gilbert H. J. (2011) J. Biol. Chem. 286, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.