Abstract
Predatory marine cone snails (genus Conus) utilize complex venoms mainly composed of small peptide toxins that target voltage- and ligand-gated ion channels in their prey. Although the venoms of a number of cone snail species have been intensively profiled and functionally characterized, nothing is known about the initiation of venom expression at an early developmental stage. Here, we report on the expression of venom mRNA in embryos of Conus victoriae and the identification of novel α- and O-conotoxin sequences. Embryonic toxin mRNA expression is initiated well before differentiation of the venom gland, the organ of venom biosynthesis. Structural and functional studies revealed that the embryonic α-conotoxins exhibit the same basic three-dimensional structure as the most abundant adult toxin but significantly differ in their neurological targets. Based on these findings, we postulate that the venom repertoire of cone snails undergoes ontogenetic changes most likely reflecting differences in the biotic interactions of these animals with their prey, predators, or competitors. To our knowledge, this is the first study to show toxin mRNA transcripts in embryos, a finding that extends our understanding of the early onset of venom expression in animals and may suggest alternative functions of peptide toxins during development.
Keywords: Embryo, Mass Spectrometry (MS), Neurotoxin, NMR, Toxins, Toxin Ontogeny, Toxin Structure Function
Introduction
Cone snails of the genus Conus are predatory marine gastropods that utilize venom to capture prey. Conus venoms mainly consist of small disulfide-rich peptides commonly referred to as conotoxins or conopeptides. Each of the ∼700 Conus species synthesizes its own characteristic repertoire of toxic peptides. It has been estimated that the toxin repertoire of cone snails comprises >100,000 different bioactive compounds with various neurological targets (1). Remarkably, this vast library of bioactive peptides has been generated by a relatively small number of gene superfamilies (2, 3). Conotoxins are translated as precursor proteins with an N-terminal signal sequence, an intermediate pro-region followed by the mature toxin at the C terminus. Comparisons between the different gene superfamilies revealed high conservation within the primary amino acid sequence for the signal and pro-sequence, whereas the mature toxin region exhibits hypermutation between a conserved disulfide scaffold (2). The venom repertoire of cone snails is further extended through the addition of post-translational modifications that increase toxin potency (4, 5) and aid in stabilizing the three-dimensional structure of the molecule (6, 7). Such is the diversification of conotoxins that venom profiles differ significantly among individuals of the same species (8–10). The exact mechanism underlying the accelerated evolution of conotoxins is not yet understood, but it has been suggested that rapid genetic divergence is driven by the various interactions between the toxins and the snail's biotic environment. Besides their function in predation, anecdotal evidence points to the role for conotoxins in deterring predators and competitors (2, 11). Given the diversity of disulfide-rich peptides in Conus, other functions, such as regulation of social behavior as observed in other molluscs (12, 13), may exist in this genus.
Very little is known about the onset of venom synthesis in cone snails. Female cone snails typically lay their eggs in egg capsules attached to a benthic substrate. Prehatching time, hatching size, and total prejuvenile development greatly vary between different Conus species and depend to a large extent on the presence or absence of a feeding larval stage (14–17). The few studies addressing feeding behavior of juvenile snails indicate that predation can occur a few days after larval metamorphosis (18), suggesting initialization of venom biosynthesis at an earlier developmental stage. Although juveniles of Conus textile were shown to feed on the same prey as adults (18), specimens of Conus magus exhibited a change in prey type from polychaetes to fish as they matured (19). Whether the venom composition changed during this transition was not determined.
Among the most extensively studied cone snail toxins are the α-conotoxins, known antagonists of the nicotinic acetylcholine receptors (nAChRs)4 (20–22). Recent studies have also identified N-type calcium channels as another neuronal target for a number of α-conotoxins (23, 24). α-Conotoxins inhibit these channels via activation of the GABAB receptor (24) and target these receptors and ion channels with exquisite selectivity. Target specificity not only varies between toxins from different Conus species but, remarkably, even between peptides isolated from the same individual (25). It is likely that the expression profile of α-conotoxins exhibiting different target specificity reflects an adaptation to the biotic environment.
In this study, we demonstrate that prior to hatching, embryos of Conus victoriae are capable of expressing venom mRNA. Interestingly, embryonic α- and O-conotoxin sequences differ significantly from adult toxin transcripts. Embryonic α-conotoxins were chosen for further characterization. Although the novel embryonic α-conotoxin Vc1.2 shares the same three-dimensional structure of the previously reported adult toxin Vc1.1, these peptides exhibit different affinities for the GABAB receptor/N-type Ca2+ channels and distinct subtype selectivities for the nAChR. Despite thorough electrophysiological investigations, the target receptor for a second embryonic toxin, Vc1.3, could not be determined, suggesting a novel target for this particular venom species. It appears that cone snails at different developmental stages differ in the relative abundance of their bioactive peptides potentially reflecting their particular ontogenetic stage.
EXPERIMENTAL PROCEDURES
Specimen Collection and Histological Preparation
Specimens of C. victoriae were collected from Broome, Western Australia, maintained in flow-through seawater tanks at 24 °C, and fed every 2 weeks with live specimens of Austrocochlea spp. Approximately 4 weeks post-collection, two specimens of C. victoriae laid egg capsules, each containing between 20 and 50 eggs derived from two independent matings. For histological preparations, adult snails were transferred to seawater containing 2% MgCl2 for 4 h followed by overnight fixation in 4% paraformaldehyde/phosphate-buffered saline (PBS). Specimens were washed in water for 15 min, decalcified for 5 h in 5% formic acid, and stored in 70% ethanol until further processing. Embryos from two specimens of C. victoriae were removed from their egg capsules 18 days after egg deposition and fixed in 4% paraformaldehyde/PBS. Embryos were sequentially washed in PBS and embedded in 2% agarose/PBS preheated to 60 °C. Once set, the agarose blocks were stored in 70% ethanol until further processing. Adult snails and embryos were processed, sectioned (7 μm), and stained with Mallory's trichrome stain (26) following routine histological procedures.
Conotoxin cDNA Isolation and Sequencing
Venom ducts were dissected and embryos collected from two adult specimens of C. victoriae 18 days after egg deposition, immediately snap-frozen in liquid nitrogen, and stored at −80 °C. The two sets of embryos represent the progeny of two independent mating events. Frozen embryos and venom duct tissues were ground under liquid nitrogen. Total RNA was extracted using TRIzol reagent (Invitrogen), and DNase I was treated with Turbo DNase (Ambion). RNA extraction and DNase treatment were performed according to the manufacturer's instructions. Total RNA concentrations were determined using a spectrophotometer, and RNA integrity was verified by gel electrophoresis. cDNA was reverse-transcribed from 720 ng of DNase-treated RNA using the transcriptor high fidelity cDNA synthesis kit (Roche Applied Science). Primary reverse transcription PCRs (RT-PCR) were performed in volumes of 30 μl containing 2 μl of cDNA (60 ng), 0.3 μl of TITANIUM TaqDNA polymerase (Clontech), 1× Advantage 2 PCR buffer (Clontech), 200 μm of each deoxynucleotide triphosphate (dNTPs), and 0.2 μm of forward and reverse oligonucleotides (supplemental Table 1). PCR cycle conditions were 1 cycle at 94 °C for 3 min and 30 cycles at 94 °C, 54 °C for 30 s and 72 °C for 30 s, and then 72 °C for 10 min. To rule out false amplification of genomic DNA, a negative control was performed using a reverse transcription reaction from which the enzyme reverse transcriptase was excluded. Nested PCRs were performed as described above except 2 μl of the 1:5 diluted primary PCR was used as DNA template, and oligonucleotides were replaced with 0.2 μm of nested oligonucleotides (supplemental Table 1), and the annealing temperature was reduced to 43 °C for 30 s. All PCR amplicons were analyzed by gel electrophoresis, cloned into pGEM-T plasmid vectors (Promega), and subsequently sequenced as described previously (27). All sequences analyzed in this study were deposited in GenBankTM (National Center for Biotechnology Information, National Library of Medicine, Bethesda). Nucleotide sequences were translated into the predicted amino acid residues, and comparative alignments of the protein and nucleotide sequences were performed using MAFFT E-INS-i sequence alignment by means of local pairwise alignment information (28). The putative signal peptides were predicted using SignalP software (29).
Peptide Synthesis
The embryonic peptides Vc1.2 and Vc1.3 were synthesized using Fmoc (N-(9-fluorenyl)methoxycarbonyl) solid-phase peptide chemistry (Liberty peptide synthesizer, CEM Corp.). Peptides were cleaved from the solid-phase resin with TFA/H2O/triisopropylsilane/3,6-dioxa-1,8-octane-dithiol (90:2.5:2.5:5) for 2 h. The crude peptides were isolated by ether precipitation, dissolved 30:70, v/v, in ACN/H2O, lyophilized, and purified on a C18 column (5-μm particle size, dimensions are 15 cm × 4.6 mm, Discovery C18 column, Supelco Inc.) using an Agilent 1200 HPLC system (Agilent) with a linear gradient from 10 to 60% buffer B (99% ACN, 0.1% TFA; buffer A, 0.1% TFA) over 30 min. The purified linear peptides were oxidized in 100 mm NH4HCO3 in water/ACN (v/v, 90:10%) at a concentration of 0.5 mg/ml for 16 h at room temperature. Oxidized peptides were isolated to a final purity of >94% on a C18 column (5-μm particle size, dimensions: 9.4 cm × 2.5 mm, Eclipse C18 column, Agilent) using an Agilent 1100 HPLC system with the gradient described above. The identities of the fully oxidized peptides were confirmed by high resolution mass spectrometry (6510 Q-TOF LC/MS mass spectrometer, Agilent).
Electrophysiological Studies on Embryonic Peptides
RNA preparation, oocyte preparation, and expression of nAChR subunits in Xenopus oocytes were performed as described previously (30). Briefly, plasmids with cDNA encoding the rat α3, α4, α9, α10, β2, and β4 nAChR and human α7 subunits were subcloned into the oocyte expression vector pNKS2 and were used for mRNA preparation using mMESSAGE mMACHINE kit (Ambion Inc.). All oocytes were injected with 5 ng of cRNA and then kept at 18 °C in ND96 buffer (96 mm NaCl, 2 mm KCl, 1 mm CaCl2, 1 mm MgCl2, and 5 mm HEPES, pH 7.4) supplemented with 50 mg/liter gentamycin and 5 mm pyruvic acid 2–5 days before recording. Membrane currents were recorded from Xenopus oocytes using an automated work station with eight channels in parallel, including drug delivery and on-line analysis (OpusXpress 6000A work station; Molecular Devices Inc.) and a two-electrode virtual ground voltage clamp circuit with a GeneClamp 500B amplifier (Molecular Devices). Both the voltage recording and current injecting electrodes were pulled from borosilicate glass (GC150T-15, Harvard Apparatus Ltd.) and had resistances of 0.2–1.5 megohms when filled with 3 m KCl. All recordings were conducted at room temperature (20–23 °C) using a bath solution of ND96 as described above. During recordings, the oocytes were perfused continuously at a rate of 1.5 ml/min, with 300-s incubation times for the conotoxin. Acetylcholine (100 mm for α7, 30 mm for all other nAChR subtypes) was applied for 2 s at 5 ml/min, with 300-s washout periods between applications. Cells were voltage-clamped at a holding potential of −80 mV. Data were sampled at 500 Hz and filtered at 50 Hz. Peak current amplitude was measured before and following incubation of the peptide (31).
Concentration-response curves for antagonists were fitted by unweighted nonlinear regression to the logistic equation, Ex = Emax XnH/(XnH + IC50nH), where Ex is the response; X is the antagonist concentration; Emax is the maximal response, nH is the slope factor, and IC50 is the concentration of antagonist that inhibits the agonist response by 50%. All electrophysiological data were pooled (n = 4–8 for each data point) and represent the means ± S.E. of the fit. Computation was carried out using SigmaPlot 11.0 (Systat Software).
Dorsal root ganglion (DRG) neurons were enzymatically dissociated from ganglia of 7–14-day-old Wistar rats according to standard protocols as described previously (24). The external recording solution contained 150 mm tetraethylammonium chloride, 2 mm BaCl2, 10 mm d-glucose, 10 mm HEPES, pH 7.3–7.4. Patch recording electrodes were filled with an internal solution containing 140 mm CsCl, 1 mm MgCl2, 5 mm MgATP, 0.1 mm NaGTP, 5 mm 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid-Cs4, 10 mm HEPES, pH 7.3, with CsOH and had resistances of 1.0–2.5 megohms. Membrane currents were recorded using the whole-cell configuration of the patch clamp technique with an Axopatch 200B amplifier (Molecular Devices). A voltage protocol using step depolarizations from −80 to −10 mV was applied when examining high voltage-activated (HVA) calcium channel currents with Ba2+ as the charge carrier. Test potentials 150 ms in duration were applied every 20 s. Leak and capacitative currents were subtracted using a −P/4 pulse protocol. Membrane currents were acquired by a computer using pClamp 9.2 software (Molecular Devices), filtered at 2 kHz, and sampled at 8 kHz by the Digidata 1322A (Molecular Devices). Sampled data were stored digitally on a computer for further analysis.
NMR Spectroscopy and Structure Calculations
NMR experiments were performed at 25 °C on a 1.5 mm Vc1.2 sample in 95% H2O, 5% 2H2O, pH 3.6. Two-dimensional homonuclear TOCSY spectra, with a spin-lock time of 70 ms, and double quantum filtered correlated spectroscopy spectra were acquired on a DRX-600 spectrometer equipped with a triple resonance probe. Two-dimensional NOESY spectra with a mixing time of 250 ms, 15N HSQC, and 13C HSQC spectra were recorded on an Avance-800 spectrometer equipped with a TCI cryoprobe. Spectra were processed using TOPSPIN version 1.3 (Bruker Biospin Pty. Ltd.) and analyzed using XEASY (32). Backbone and side chain 1H, 13C, and 15N chemical shifts were assigned. NOEs were assigned automatically using CYANA 2.1 (33, 34). The f and y angle constraints were predicted using TALOS+ (35) based on chemical shifts and were used in structure calculations when the predictions were consistent with an analysis of 3JHNHa coupling constants based on double quantum filtered-COSY spectra.
A family of 200 structures was calculated using Xplor-NIH (36) using standard simulation annealing scripts. The 80 lowest energy structures were then subject to energy minimization in water; during this process, a box of water with a periodic boundary of 18.856 Å was built around the peptide structure, and the ensemble was energy-minimized on the basis of NOE and dihedral constraints and the geometry of the bonds, angles, and impropers (37). From this set of structures, a final family of 20 lowest energy structures was chosen for analysis using PROCHK-NMR (38) and MOLMOL (39). The final structures had no experimental distance violations greater than 0.2 Å or dihedral angle violations greater the 5°. The final structures and the associated structural constraints have been deposited in the BioMagResBank (40) under accession number 20126.
Protein Extraction and Mass Spectrometric Analyses
Venom ducts were dissected from four snails, and crude venom was manually squeezed from the ducts and air-dried. Approximately 1 mg of venom was reconstituted in 1 ml of ice-cold 30% ACN, 0.3% trifluoroacetic acid (TFA) followed by sonication for 10 min on ice. The extracts were centrifuged at 13,000 × g for 20 min; the supernatants were lyophilized and reconstituted in 500 μl of ultrapure water. Because of the complexity of the venom samples, extracts were separated by reversed-phase HPLC on a micropreparative C18 column (3.5 μm particle size, dimensions: 2.1 × 100 mm, X-Bridge, Waters) prior to mass spectrometric analysis using a linear gradient from 10 to 60% buffer B (95% ACN, 0.1% TFA; buffer A, 0.1% TFA) over 70 min. Reversed-phase venom fractions were individually analyzed on a MALDI-TOF mass spectrometer (QSTAR Pulsar, positive reflector mode, AB SCIEX). In addition to MALDI-TOF MS, venom extracts were analyzed by electrospray ionization-MS/MS. Samples were loaded onto a C18 reversed-phase column (ProtoCol nano column, particle size 300 Å and 3 μm, dimensions 75 μm × 100 mm, SGE Analytical Sciences) and analyzed using a Hybrid Quadrupole-TOF LC/MS/MS mass spectrometer (QSTAR Elite, AB SCIEX). Solvent A contained 0.1% formic acid, and solvent B consisted of 95% ACN, 0.1% formic acid. Separation was performed with a solvent B gradient of 5–60% over 90 min, followed by 60–80% B over 10 min. Acquired data were analyzed manually using Analyst QS software (version 2.0, AB SCIEX) accounting for the presence of various post-translationally modified peptide precursors (e.g. sulfation of tyrosines, γ-carboxylation of glutamate, hydroxylation of proline, and C-terminal amidation).
RESULTS
Identification of Novel Conotoxin Transcripts in C. victoriae Embryos
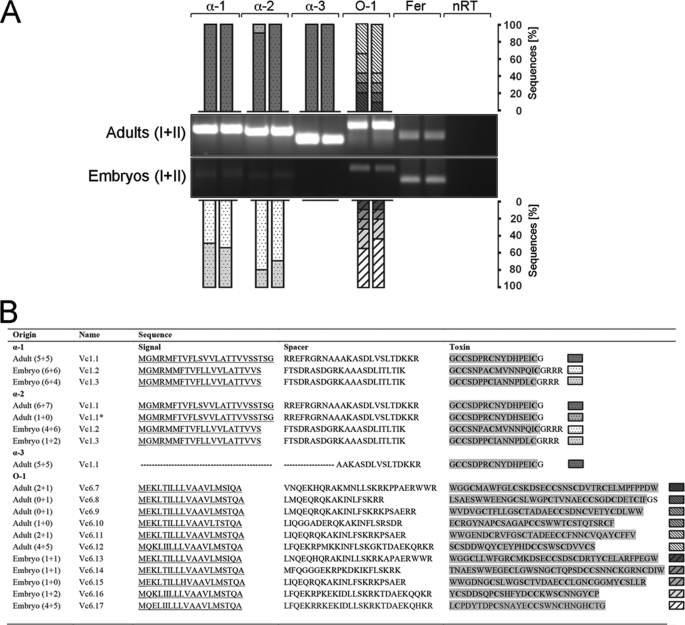
RT-PCR using universal α- (41–43) and O-conotoxin (44) oligonucleotides led to the discovery of a number of novel toxin mRNA transcripts expressed in embryos and adults of C. victoriae and confirmed the presence of α-conotoxin Vc1.1 mRNA in the venom duct of adult specimens (Fig. 1) (43). All novel sequences were named according to the nomenclature previously used for C. victoriae venom peptides, where the first two letters indicate the species, the following number (1 or 6) represents the toxin family (α or O), and the last number implies the order of toxin discovery (e.g. Vc1.2 is the second α-conotoxin identified for C. victoriae) (45).
FIGURE 1.
A, universal α- and O-conotoxin RT-PCR showing toxin mRNA expression in embryos (n = 2, each pooled from ∼50 embryos) and adult specimens (n = 2) of C. victoriae. 110 ng of cDNA was amplified using three α- (α-1, α-2, and α-3) and one O-conotoxin (O-1) oligonucleotide pair. Ferritin (Fer) served as a reference gene. No reverse transcriptase controls (nRT) were performed to rule out contamination by genomic DNA. RT-PCRs were subsequently cloned and subjected to nucleotide sequencing. Bars represent the number of clones identified per RT-PCR expressed as percentages. DNA ladder: 1 kb plus (Invitrogen). *, analogue of Vc1.1 carrying one amino acid substitution. B, conotoxin sequences identified by RT-PCR showing the predicted signal peptide sequences (underlined), the spacer region, and the mature toxin region highlighted in gray. The number of clones sequenced per replicate (n = 2) is given in parentheses (43). GenBankTM accession numbers are as follows: Vc1.2, GU046308; Vc1.3, GU046309; Vc6.7, JF433900; Vc6.8, JF433901; Vc6.9, JF433902; Vc6.10, JF433903; Vc6.11, JF433904; Vc6.12, JF433905; Vc6.13, JF433906; Vc6.14, JF433907; Vc6.15, JF433908; Vc6.16, JF433909; and Vc6.1, JF433910.
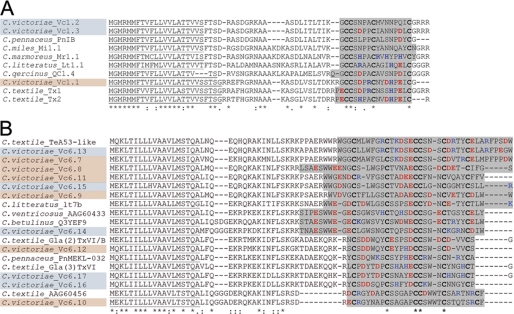
A total of five adult-specific and six embryo-specific O-superfamily toxins were identified, with no sequence overlap between these two life stages (Fig. 1B). Interestingly, the O-conotoxin expression pattern was nearly identical between the two sets of embryos tested with the exception of Vc6.15 which was only identified in one set of embryos (Fig. 1B). The most abundant transcript was Vc6.12 (47% of cloned sequences) in adults and Vc6.17 (54%) in embryos. O-superfamily toxins exhibit a characteristic pattern of 6 cysteines (C-C-CC-C-C) that form three disulfide bonds. Members of this family are known modulators of voltage-sensitive calcium, potassium, and sodium channels (44, 46). Alignment of the novel O-conotoxins with other members of the O-superfamily revealed high sequence similarity to the O2 gene superfamily (Fig. 2B), peptides with an as yet unidentified target receptor (44, 47).
FIGURE 2.
Comparative alignment of novel embryonic α- (A) and O-conotoxins (B) isolated from C. victoriae with toxin sequences from other cone snail species. Alignment was performed using MAFFT E-INS-i sequence alignment by means of local pairwise alignment information (28). Sequences obtained from C. victoriae embryos and adults are highlighted blue and orange, respectively. Predicted protein signal sequences are underlined (SignalP). Conserved cysteine residues are shown in boldface, and the predicted mature toxin regions are highlighted gray (50). Basic and acidic amino acids are shown in blue and red, respectively. Dashes denote gaps. Amino acid conservations are denoted by an asterisk, and colons and periods represent a high and low degree of similarity, respectively.
Universal α-conotoxin RT-PCR identified two novel transcripts in the embryos (Vc1.2 and Vc1.3; Fig. 1) and confirmed the presence of Vc1.1 in the adults (43). Interestingly, utilizing a number of different universal α-conotoxin oligonucleotides (α-1, α-2, and α-3; Fig. 1) did not lead to the identification of additional sequences suggesting that these toxins represent the most abundant α-conotoxin transcripts. In the adult, all but one clone (Vc1.1*) represented Vc1.1 (Fig. 1B). This finding is consistent with previous studies addressing α-conotoxin expression in C. victoriae (43, 45). The two cDNA transcripts identified from the embryos were almost equally represented with 55% for Vc1.2 and 45% for Vc1.3 using the α-1 oligonucleotide and slightly higher percentages for Vc1.2 (75%) when using the α-2 oligonucleotide (Fig. 1). The frequency of sequences obtained by RT-PCR screening generally indicates relative abundances of mRNA transcripts (48, 49). Thus, based on the number of clones obtained by primary RT-PCR analyses, relative abundances of toxin mRNAs varied greatly between the two life stages tested. The presence of such a limited yet distinct number of α-conotoxin transcripts was intriguing and was therefore further investigated.
Embryonic and Adult α-Conotoxin Repertoire of C. victoriae
To further investigate α-conotoxin expression in adult versus embryos, toxin-specific oligonucleotides were designed for Vc1.1 and Vc1.2 (supplemental Table 1). Sequence similarities among the three transcripts precluded the design of specific PCR oligonucleotides for Vc1.3. Nested PCR can be utilized to detect low abundant transcripts and was performed on primary α-conotoxin PCRs using internal toxin-specific oligonucleotides. Amplicons were successfully generated demonstrating that embryos express mRNA encoding the adult toxin Vc1.1 and that adults possess mRNA for the embryonic toxin Vc1.2 (supplemental Fig. 1).
Sequence Alignment of Novel Embryonic Toxins
Alignment of the novel embryonic toxin peptides with available α-conotoxin sequences revealed high similarity of the novel peptides to α-conotoxins sharing the conserved 4/7 cysteine pattern with 4 residues between C1 and C2, and 7 residues between C3 and C4 (Fig. 2). The predicted signal sequences and pro-regions share 100% identity although differences in the mature toxin regions are apparent. Based on observations made for other conotoxins, further proteolytic C-terminal cleavage and subsequent amidation of -CGRRR- to -C-NH2- are likely to occur (50). Interestingly, protein alignment revealed the highest similarity of the pro-region and the mature toxin region between the embryonic peptides and α-conotoxin PnIB from Conus pennaceus, a potent inhibitor of the α7 subtype of the nAChR (51), although the adult toxin Vc1.1 is a known antagonist of the α9α10 subtype (52). To further elucidate potential differences in the neuronal target, electrophysiology was performed.
Determination of the Neuronal Target Receptor
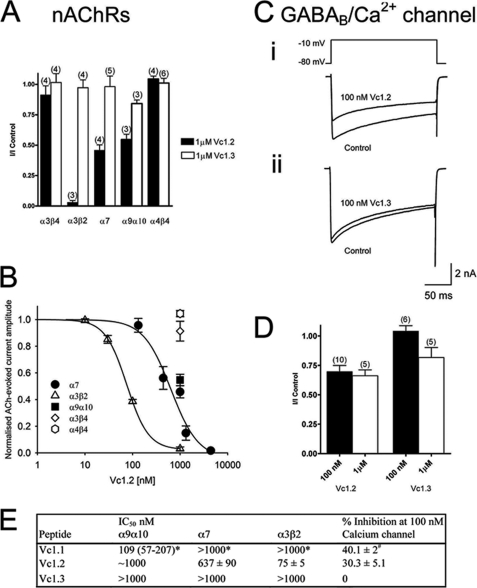
The effects of Vc1.2 and Vc1.3 were examined on ACh-evoked currents mediated by various nAChR subtypes expressed in Xenopus oocytes. ACh (30–100 mm) was applied at 5-min intervals, and the peak amplitude of the corresponding membrane current was assessed. The peptide was applied 5 min prior to co-application of ACh plus peptide. Vc1.2 (1 mm) completely inhibited ACh-evoked currents mediated by α3β2 nAChRs and inhibited the ACh-evoked current amplitude mediated by α7 and α9α10 nAChRs by 54 ± 5 and 46 ± 4% (n = 3–5), respectively (Fig. 3A). In contrast, Vc1.2 did not inhibit the α3β4, α4β2, or α4β4 nAChR subtypes (n = 3–5) (Fig. 3A). Vc1.3 (1 mm) exhibited no significant activity at any of the neuronal nAChR subtypes (Fig. 3A). Concentration-response curves obtained for Vc1.2 displayed the following order of selectivity and corresponding IC50 values: α3β2 (75 ± 5 nm) > α7 (637 ± 90 nm) > α9α10 (∼1 mm) (Fig. 3, B and E). These data indicate that Vc1.2 selectively and potently targets the α3β2 and to a lesser extent the α7 nAChR subtype. The embryonic toxin Vc1.2 therefore exhibits a distinct nAChR selectivity to the adult toxin Vc.1.1 (Fig. 3E).
FIGURE 3.
Vc1.2 and Vc1. 3 inhibition of nAChRs expressed in Xenopus oocytes and Ca2+ channel currents in rat DRG neurons. A, bar graph of the relative inhibition of nAChRs expressed in Xenopus oocytes by 1 μm Vc1.2 and Vc1.3. Vc1.2 (1 μm) completely inhibited α3β2 nAChRs and inhibited α7 and α9α10 by 54 and 45%, respectively. Vc1.3 was inactive at all neuronal nAChR subtypes. B, concentration-response curves obtained for Vc1.2 inhibition of nAChR subtypes. Vc1.2 was most active at α3β2 nAChRs with an IC50 of 75 ± 5 nm (▴; n = 3), 637 ± 90 nm for α7 (●; n = 4–6), and ∼1 μm for α9α10 (■; n = 3). C, superimposed depolarization-activated whole-cell Ba2+ currents elicited by voltage steps from a holding potential of −80 to −10 mV in the absence (control) and presence of 100 nm Vc1.2 (panel i) and 100 nm Vc1.3 (panel ii), respectively. D, bar graph of the relative inhibition of HVA Ca2+ channel currents in rat DRG neurons by 100 nm and 1 μm Vc1.2 and Vc1.3. Numbers in parentheses reflect number of cells. E, comparison of the IC50 values of Vc1.1, Vc1.2, and Vc1.3 at different nAChR subtypes and percentage inhibition of calcium currents in isolated DRG neurons. Values determined in this study represent mean ± S.E. * indicates mean plus 95% confidence interval (54), and # represents mean ± S.E. (24).
Vc1.1 has previously been shown to inhibit N-type (CaV2.2) Ca2+ channel currents via activation of the G protein-coupled GABAB receptor in sensory neurons (24). Therefore, the activity of Vc1.2 and Vc1.3 was also examined on depolarization-activated whole-cell Ba2+ currents in rat DRG neurons. Application of Vc1.2 inhibited HVA Ca2+ channel currents by 30.3% at 100 nm (n = 9) (Fig. 3, C–E). Vc1.3 was not active at 100 nm and inhibited the Ba2+ current amplitude by <20% at 1 μm (n = 7) (Fig. 3, C–E). Both embryonic peptides Vc1.2 and Vc1.3 exhibit a reduced potency for GABAB receptor-mediated inhibition of HVA Ca2+ channel currents compared with Vc1.1 (Fig. 3E).
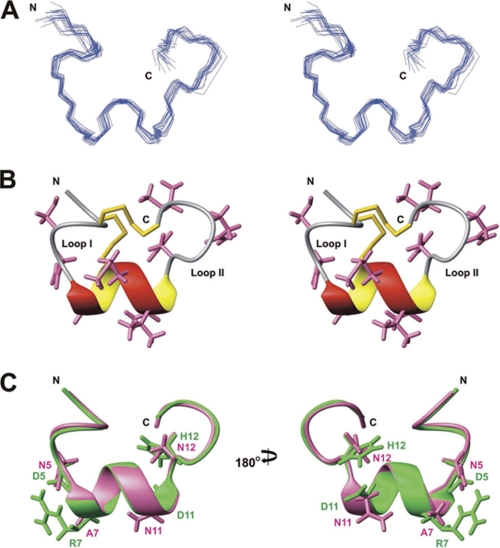
Structure Determination of Vc1.2
To investigate whether subtype selectivity for the nAChR and affinity toward the GABAB receptor arise from a difference in the three-dimensional structures of the peptides, the solution structure of Vc1.2 was determined using nuclear magnetic resonance (NMR) spectroscopy. As can be seen in the one-dimensional 1H NMR spectra recorded at different pH values and temperatures (supplemental Fig. 2), the backbone amide proton peak of Cys2 was visible at pH 3.2 but not at pH 5.5 because of the exchange of solvent water; other peaks did not shift appreciably over the pH range 3.2–5.5, consistent with the fact that Vc1.2 does not contain charged residues and indicating that the structure is maintained over this pH range. A summary of experimental constraints and structural statistics for Vc1.2 is given in Table 1. The final 20 structures (Fig. 4A) fit well with experimentally derived distance and angle constraints and are well defined over the entire length of the polypeptide. The closest-to-average structure of Vc1.2 (Fig. 4B) is characterized by an α-helix (residues 6–11), as was also seen in other α-conotoxins with the same loop I and loop II lengths, such as Vc1.1 (30) and PnIA (53). The N-terminal residues 2–4 also appear to form a 310 helix-like turn structure. The trans orientations of the peptide bond preceding both Pro6 and Pro3 were established by the intense Hα–Hδ nuclear Overhauser effects (NOEs) between the prolines and their preceding residues. Superposition of the backbone heavy atoms (N, Cα, and C′) of the final ensemble of 20 Vc1.2 structures with those of Vc1.1 (30) gave average group root-mean-square deviation values of 0.66 Å, a value no larger than the root-mean-square deviation within the Vc1.2 family, indicating that the backbone structures are highly conserved in these α-conotoxins. Therefore, the different subtype specificities of Vc1.2 and Vc1.1 for nAChR binding must arise from the specific amino acid side chain differences in these toxins. Amino acid residues affecting the affinity of Vc1.1 for the α9α10 subtype were determined by scanning mutagenesis (54). Vc1.2 differs from Vc1.1 in only four of those residues (Asp5, Arg7, Asp11, and His12, see Figs. 4C and 5). Based on high sequence similarities between Vc1.2 and Vc1.3, it is anticipated that the two embryonic toxins share the same structure. The additional Pro7 in Vc1.3 is unlikely to affect the α-helical structure as demonstrated by comparing the solution structure of Vc1.2 with that of PnIA, an α-conotoxin with Pro6 and Pro7 in the first loop (supplemental Fig. 3). Therefore, differences in their ability to mediate GABAB receptor/N-type Ca2+ channel inhibition are likely to arise from specific side chain differences (Pro7, Ile9, Ala10, and Leu15 in Vc1.3, see Fig. 5). Likewise, Vc1.3 differs from Vc1.2 in only three side chains important for α3β2 and α7 binding (Asn5, Ile9, and Leu15 in Vc1.3 (51, 55)) indicating that changes in these side chains may abolish binding to these nAChR subtypes (Fig. 5).
TABLE 1.
Structural statistics for Vc1.2
| No. of distance constraints | 202 |
| Intra-residue (i = j) | 105 |
| Sequential (|i − j| = 1) | 67 |
| Short (1< |i −j| < 6) | 29 |
| Long | 1 |
| No. of dihedral constraints | 16 |
| Energy (kcal/mol)a | |
| ENOE | 4.1 ± 0.6 |
| Deviations from ideal geometryb | |
| Bonds | 0.0038 ± 0.0002 Å |
| Angles | 0.6980 ± 0.0184° |
| Impropers | 0.5730 ± 0.0273° |
| Mean global R.M.S.D. (Å)c | |
| Backbone heavy atoms (N, Ca, C′) | 0.66 ± 0.19 |
| All heavy atoms | 1.02 ± 0.21 |
| Ramachandran plotd | |
| Most favored | 89.6% |
| Allowed | 10.4% |
| Additionally allowed | 0% |
| Disallowed | 0% |
a The values for ENOE are calculated from a square well potential with force constants of 50 kcal mol−1 Å2.
b The values for the bonds, angles, and impropers show the deviations from ideal values based on perfect stereochemistry.
c Mean pairwise root-mean-square deviation (R.M.S.D.) over all residues calculated in MOLMOL.
d Data are as determined by the program PROCHECK-NMR for all residues except Gly and Pro.
FIGURE 4.
Solution structure of Vc1.2. A, stereo view of the family of 20 final structures of Vc1.2, superimposed over the backbone heavy atoms. B, stereo ribbon view of the closest-to-average structure of Vc1.2 highlighting the α-helix and two disulfide bonds. Side chains are shown except for prolines. C, comparison of the solution structures of Vc1.2 (pink) and Vc1.1 (30) (Protein Data Bank code 2H8S, green). Structures are superimposed over the backbone heavy atoms. Side chains of Vc1.1 residues important for α9α10 nAChR binding (30) and equivalent residues in Vc1.2 are shown.
FIGURE 5.
Alignment of α-conotoxins from C. victoriae highlighting residues important for biological function. Differences in sequence between the embryonic toxins Vc1.2 and Vc1.3 to the adult toxin Vc1.1 are shown in green. Amino acid residues likely to have caused a loss in activity toward the α9α10 neuronal nicotinic subtype (54), the N-type calcium channels,3 and the α7 subtype (55) are highlighted in pink and yellow and depicted with a frame, respectively. The disulfide bonds between Cys1–Cys3 and Cys2–Cys4 are indicated by black lines. The backbone loops formed by this disulfide connectivity are shown below the sequences. All three C termini are likely to be amidated (#), a common modification in conotoxins.
Comparative Anatomy of the Embryonic and Adult Foregut
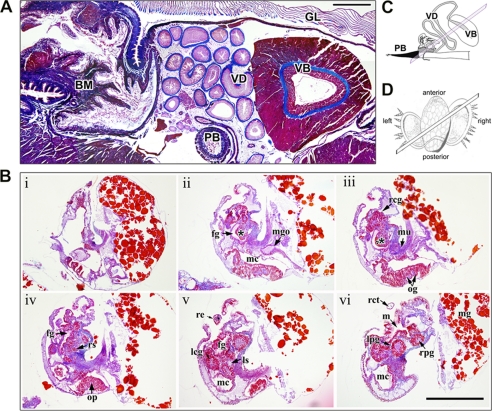
Regions of the foregut important for venom biosynthesis, transport, and delivery were identified in adult specimens of C. victoriae (Fig. 6A). The venom apparatus consists of a long convoluted venom duct for toxin biosynthesis, a muscular venom bulb believed to be involved in venom transport (56), and harpoon-like radula teeth responsible for injecting the venom into the prey (Fig. 6, A and C) (57). Toxin biosynthesis and potential post-translational modifications take place in the columnar epithelial layer of the venom duct (58). Following biosynthesis, the venom is packed into ovoid-shaped granules that are densely packed into larger circular vesicles upon secretion from the epithelial cells (58). Histological examination of the embryos revealed that at the time when embryos of C. victoriae were sampled and fixed for sectioning, they did not possess the characteristic features of a functional venom apparatus. Although the radula sac could be resolved in histological sections (Fig. 6B, panel iv), the proboscis and venom duct were not yet differentiated. The venom duct in Conus is believed to develop from an outpocketing of the mid-esophageal wall (15); however, this differentiation could not be observed in any of the embryonic specimens examined.
FIGURE 6.
Histological preparations of the adult venom apparatus and embryos of C. victoriae. Sections were Mallory-stained and cut at 7 μm thickness. A, cross-section through the venom apparatus of C. victoriae showing the venom duct (VD), the venom bulb (VB), the proboscis (PB), the gill (GL), and the buccal mass (BM). Scale bar, 200 μm. B, serial sections (panels i–vi) through an embryo depicting foregut (fg), left cerebral ganglion (lcg), left pedal ganglion (lpg), left statocyst (ls), mouth (m), mantle cavity (mc), midgut (mg), midgut opening (mgo), muscle (mu), osphradial ganglia (og), osphradium (op), right cerebral ganglion (rcg), right cephalic tent (rct), right eye (re), radula sack (rs), right pedal ganglion (rpg), style sac (ss). The incipient venom gland is marked with an asterisk. Scale bar, 200 μm. C, schematic of the venom apparatus showing orientation of section shown in A. D, drawing of the larvae showing orientation of serial sections shown in B.
Mass Spectrometric Analysis of Venom Peptide Preparations
Liquid chromatography coupled with mass spectrometry (LC/MS) revealed a complex composition of C. victoriae venom (supplemental Fig. 4A). Although the presence of multiple analogues of Vc1.1 containing hydroxyproline and/or γ-carboxyglutamate was confirmed in this complex mixture (supplemental Fig. 4B (45, 59)), novel embryonic α-conotoxins were not detected in the adult's venom despite extensive and targeted LC-MS/MS analysis for various candidate venom peptide precursors. This finding indicates that although adults express embryonic toxin mRNAs, minimal or no translation into bioactive peptides takes place. Alternatively, the translated peptides may have been present in the venom but could not be detected using LC/MS due to unanticipated post-translational modifications. Analyses of LC/MS data obtained for C. victoriae venom anticipated disulfide bond formation, C-terminal amidation, hydroxylation of prolines, and γ-carboxylation of glutamate as well as differential C- and N-terminal cleavage. Electrospray ionization-MS/MS on the hybrid quadrupole-TOF mass spectrometer (QSTAR Elite, AB SCIEX) is highly sensitive allowing for the detection of peptides in the sub-femtomole range. Thus, failure to identify Vc1.2 and Vc1.3 in the venom of C. victoriae is unlikely to reflect the sensitivity of the detection method used.
DISCUSSION
Molecular sequencing revealed venom mRNA expression in embryos of C. victoriae and led to the identification of five novel O- and two α-conotoxin transcripts as well as confirmed the presence of mRNA encoding Vc1.1, a pharmacologically active peptide identified previously in adult specimens of C. victoriae (43). Thus, targeting different developmental stages proved to be a powerful technique for the discovery of novel bioactive peptides that are masked in the adult by the presence of highly abundant transcripts. Testing the embryonic α-conotoxins against different subtypes of the neuronal nicotinic receptor revealed that the embryonic toxin peptides had different target specificities. The embryonic peptide Vc1.2 exhibited high affinity toward the α3β2 and α7 nAChR subtype but lower activity toward α9α10, the preferred receptor subtype for Vc1.1 (52).
Little is known about the presence or distribution of the nAChRs in invertebrates (60). In mammals, the α7 subtype is among the most abundant nicotinic receptors (61). With an unusually high permeability for calcium ions, this subtype regulates many calcium-dependent events throughout the central and peripheral nervous system (61). In contrast, in mice, the expression of the α3β2 subtype is restricted to the habenulointerpeduncular tract in the brain (62). Similarly, expression of the α9α10 nicotinic receptor is restricted to the cochlear hair cells, peripheral blood lymphocytes (63), skin keratinocytes (64, 65), and dorsal root ganglia (66, 67) where co-expression with α7 has been observed (68). Given this subtype-specific expression pattern of the nAChRs, conotoxins that selectively antagonize different nicotinic subtypes are likely to exhibit distinct biological functions.
The structures of Vc1.1 and Vc1.2 were almost identical, and changes in their target specificity were mediated by substitutions of a small number of amino acid side chains, although the disulfide scaffold was conserved (25, 55). This extraordinary ability to generate peptides with novel neuronal activities but equal structural stability has enabled cone snails to quickly adapt to changes in their biotic environment and rapidly diversify. Combining our findings on the structure-activity relationships of Vc1.2 and Vc1.3 with data obtained from scanning mutagenesis studies on Vc1.1 (54),5 residues causing a shift in target specificity can now be predicted. Amino acid substitutions affecting Asp5 to Arg7 and Asp11 to Ile15 caused a significant decrease in the affinity of Vc1.1 toward the α9α10 subtype (54). Because only four of these amino acids differ between Vc1.1 and Vc1.2, substitutions of one or more of these residues are likely to have caused the shift in subtype specificity. Interestingly, in Vc1.1, three of these side chains are charged, and one is an imidazolium ring. In Vc1.2, all are replaced by uncharged residues. This charge loss may contribute to the decreased affinity of Vc1.2 for the α9α10 subtype. Unlike Vc1.1 and Vc1.2, Vc1.3 did not inhibit GABAB receptor/N-type Ca2+ channels. Vc1.3 differs from the other two peptides in five positions, four of which are known to be important sites for GABAB receptor recognition.5 Likewise, specific differences in amino acids between Vc1.3 and Vc1.2 must contribute to a loss in activity toward the α3β2 and α7 subtypes (51, 55).
This poses the following questions. Why do these toxins have a different biological target than the most abundant α-conotoxin in the adult snail? Why do cone snail embryos express toxin-encoding mRNAs in the first place? Histological investigations demonstrated that at the time of sampling, the cells of the mid-esophagus had not yet formed the esophageal ventral groove that will later develop into the venom gland. However, it is possible that the cells of the incipient venom gland had begun to hypertrophy and produce mRNA transcripts of toxin genes prior to tissue differentiation. Embryos were harvested approximately 2 weeks before hatching occurred. Unfortunately, hatched stages could not be recovered. Morphological studies of embryos of Conus anemone showed that immediately prior to hatching the esophageal diverticulum was filled with secretory granules (15) likely to contain conotoxins (58). Oocytes of some marine organisms such as sea urchins and starfish store maternal mRNAs enabling rapid biosynthesis of vital proteins in the developing embryo (69–71). Given that C. anemone embryos synthesize venom granules in the incipient venom duct tissue, toxin mRNA transcripts identified in C. victoriae embryos are not likely to be of maternal origin.
Although we were unable to detect transcripts at the protein level, it is unlikely that the embryonic toxins solely represent silent transcripts. Electrophysiological investigations demonstrated that the embryonic toxin Vc1.2 is active in its mature state and combined with the presence of a variety of different O-superfamily toxins, it indicates that embryos express functionally active peptides.
Juveniles of C. pennaceus and Conus mediterraneus have been reported to feed on small gastropods shortly after hatching (17, 72). Based on these findings, venom mRNA expression in cone snail embryos could represent preparation of the venom machinery for a predatory lifestyle (Fig. 7B). C. victoriae is therefore likely to hatch from the egg capsule as a short lived nonfeeding larva or a juvenile. Similarly, mRNA encoding chymotrypsin-like preproprotease, a highly expressed protein in the intestine of the adult gastropod Halotis rufescens, was detected in amebocyte of the digestive tissue of embryos well before metamorphosis and gut morphogenesis (73). When the relationship between the morphogenesis and appearance of secretory components were studied in embryos of the viper Vipera palaestinae, neurotoxins and venom-specific enzymes were detected by immunohistochemistry together with secretion of granules into the lumen of the venom gland days before hatching (74). As proposed for cone snails, it can be hypothesized that snake embryos synthesize venom to prime for a predatory lifestyle. Juveniles of C. magus experienced an age-related change in prey type from polychaetes to fish (19). It is now well understood that the venom composition of fish-hunting cone snails is different from that of mollusc and worm hunters (2). Adult C. victoriae are molluscivorous (mollusc-eating). The feeding behavior of juvenile C. victoriae has not been investigated, but differences in toxin expression between the embryos and the adults may indicate that similar lifestyle changes occur in C. victoriae. Differences in the relative abundances of conotoxins may therefore indicate that the venom composition undergoes ontogenetic changes as observed in other venomous animals (75). Behavioral studies on newly hatched juveniles and investigations on relative abundances of conotoxins at different developmental stages are needed to further support this notion.
FIGURE 7.
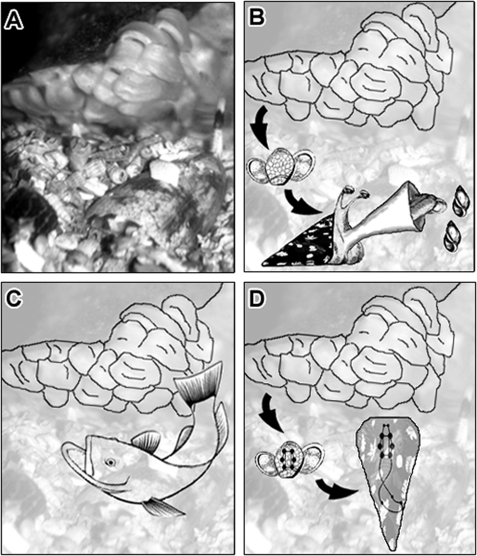
Potential usage of toxins in cone snail embryos. A, C. victoriae guarding egg capsules. Cone snail embryos may express toxin mRNA transcripts as preparation for a predatory life style (B) or to deter predators (C), or toxin peptides may be involved in other biological pathways such as neuronal signaling (D).
Cone snail embryos and newly hatched juveniles may synthesize venom for defense rather than predation (Fig. 7C). Injection of the venom into the snail's prey causes immediate paralysis (76), and thus compounds that are utilized for hunting can also serve for defense. Anecdotal evidence for the usage of venom in defense by adult cone snails is emerging (11). Synthesis of defensive or deterrent compounds is a common phenomenon in embryos and larvae of many marine organisms (77–80). Defensive compounds include glycosides (79), alkaloids (81), cyclic peptides (82), halogenated phenols (80), and terpenes (77). Interestingly, comparisons between adult and larval deterrent profiles revealed that the same compounds are utilized by different life stages, but concentrations can vary extensively (78, 80, 82). In cone snails, full development of the venom apparatus occurs during or shortly after hatching (16, 17, 72). It can be hypothesized that juveniles of C. victoriae inject the novel embryonic toxins into their prey and potential predators/competitors. Consequently, changes in the venom repertoire may reflect differences in the type of predators and competitors with which these animals interact.
It is now well understood that most toxins are proteins that have originally been recruited from ancestral body proteins through gene duplication and subsequent mutation and/or deletion events (83). The three-dimensional scaffold of the newly generated toxin multigene family is generally preserved, although the remaining residues diversify to generate molecules with novel biological activities (83). For example, the snake three-finger neurotoxins are derivations of endogenous neuropeptides similar to a family of proteins found in humans, the SLUR proteins (84). SLUR proteins therefore belong to a group of toxin-like proteins with nontoxin endogenous activities. For example, SLURP-1 is a disulfide-rich endogenous ligand of the α7 nicotinic receptor subtype (85) and is expressed in a variety of different tissue types, including skin, gums, stomach, and the esophagus (86). The bee ω-conotoxin-like protein 1 (OCLP1) is another example of a toxin-like peptide that potentially represents an ancestral toxin protein (87). OCLP1 exhibits the characteristic disulfide scaffold of cone snail ω-conotoxins and is highly expressed in the bee brain where it has been suggested to modulate voltage-gated Ca2+ channel activity (87).
As whole embryos were taken for molecular sequencing, toxin transcripts might have originated from tissues other than the incipient venom gland. The embryonic peptides could therefore represent toxin-like compounds that function as endogenous neuronal modulators in the developing snail embryo (Fig. 7D). Sequencing and phylogenetic analysis of toxins and toxin-like peptides from the venom gland and tissues not involved in venom biosynthesis could be revealing in this context.
In summary, this study has identified novel α- and O-toxin peptides in embryos of the cone snail C. victoriae. Embryonic α-conotoxins differ significantly in their biological function from the most abundant α-conotoxin in the adult, although the three-dimensional structure is conserved. We suggest that the venom of cone snails undergoes ontogenetic variations and that the early onset of venom expression in embryos most likely represents preparation for predation and/or defense, although a role in endogenous processes cannot be ruled out. Future analyses of embryos from additional mating events will be informative in this context. Behavioral studies and further characterization of the venom composition in embryos and adult snails will provide insights into the mechanisms underlying the generation of biodiversity in Conus.
Supplementary Material
Acknowledgments
We thank Bruce Abaloz and Dr. Neil Young for help with histological preparation, Dr. Bruce Livett for reviewing the manuscript, John Ahern for the maintenance of specimens, and Dr. Robyn Bradbury and Johan Paz for assistance in collection of specimens.
Note Added in Proof
Since our manuscript was accepted, we have learned of some earlier studies that have shown embryonic toxin expression in sea anemones (88, 89) and Hydra embryos (90).
This work was supported in part by Australian Research Council Grant DP110101331 (to A. W. P. and N. A. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4 and Table 1.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) GU046308, GU046309, JF433900, JF433901, JF433902, JF433903, JF433904, JF433905, JF433906, JF433907, JF433908, JF433909, JF433910.
D. J. Adams and B. Callaghan, unpublished data.
- nAChR
- nicotinic acetylcholine receptor
- ACN
- acetonitrile
- HVA
- high voltage-activated
- DRG
- dorsal root ganglion
- ACh
- acetylcholine.
REFERENCES
- 1. Terlau H., Olivera B. M. (2004) Physiol. Rev. 84, 41–68 [DOI] [PubMed] [Google Scholar]
- 2. Olivera B. M. (2006) J. Biol. Chem. 281, 31173–31177 [DOI] [PubMed] [Google Scholar]
- 3. Norton R. S., Olivera B. M. (2006) Toxicon 48, 780–798 [DOI] [PubMed] [Google Scholar]
- 4. Pisarewicz K., Mora D., Pflueger F. C., Fields G. B., Marí F. (2005) J. Am. Chem. Soc. 127, 6207–6215 [DOI] [PubMed] [Google Scholar]
- 5. Lopez-Vera E., Walewska A., Skalicky J. J., Olivera B. M., Bulaj G. (2008) Biochemistry 47, 1741–1751 [DOI] [PubMed] [Google Scholar]
- 6. Craig A. G., Park M., Fischer W. H., Kang J., Compain P., Piller F. (2001) Toxicon 39, 809–815 [DOI] [PubMed] [Google Scholar]
- 7. Loughnan M. L., Nicke A., Jones A., Adams D. J., Alewood P. F., Lewis R. J. (2004) J. Med. Chem. 47, 1234–1241 [DOI] [PubMed] [Google Scholar]
- 8. Jakubowski J. A., Kelley W. P., Sweedler J. V., Gilly W. F., Schulz J. R. (2005) J. Exp. Biol. 208, 2873–2883 [DOI] [PubMed] [Google Scholar]
- 9. Davis J. M., Boswell B. A., Bächinger H. P. (1989) J. Biol. Chem. 264, 8956–8962 [PubMed] [Google Scholar]
- 10. Jones A., Bingham J. P., Gehrmann J., Bond T., Loughnan M., Atkins A., Lewis R. J., Alewood P. F. (1996) Rapid Commun. Mass Spectrom. 10, 138–143 [DOI] [PubMed] [Google Scholar]
- 11. Olivera B. M. (2002) Annu. Rev. Ecol. System 33, 25–47 [Google Scholar]
- 12. Cummins S. F., Nuurai P., Nagle G. T., Degnan B. M. (2010) Peptides 31, 394–401 [DOI] [PubMed] [Google Scholar]
- 13. Vreugdenhil E., Jackson J. F., Bouwmeester T., Smit A. B., Van Minnen J., Van Heerikhuizen H., Klootwijk J., Joosse J. (1988) J. Neurosci. 8, 4184–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kohn A. J., Perron F. E. (1994) J. Marine Biol. Assoc. UK. 74, 744 [Google Scholar]
- 15. Ball A. D. (2002) Bolletino. Malacologico. 38, 51–78 [Google Scholar]
- 16. Ball A. D. (1999) in The Seagrass Flora and Fauna of Rottnest Island, Western Australia (Wells D. I., Wells F. E. eds) pp. 137–162, Western Australian Museum, Perth, Western Australian [Google Scholar]
- 17. Perron F. E. (1981) Mar. Biol. 61, 215–220 [Google Scholar]
- 18. Perron F. E. (1980) J. Exp. Marine Biol. Ecol. 42, 27–38 [Google Scholar]
- 19. Nybakken J., Perron F. E. (1988) Mar. Biol. 98, 239–242 [Google Scholar]
- 20. Cartier G. E., Yoshikami D., Gray W. R., Luo S., Olivera B. M., McIntosh J. M. (1996) J. Biol. Chem. 271, 7522–7528 [DOI] [PubMed] [Google Scholar]
- 21. Loughnan M., Bond T., Atkins A., Cuevas J., Adams D. J., Broxton N. M., Livett B. G., Down J. G., Jones A., Alewood P. F., Lewis R. J. (1998) J. Biol. Chem. 273, 15667–15674 [DOI] [PubMed] [Google Scholar]
- 22. Johnson D. S., Martinez J., Elgoyhen A. B., Heinemann S. F., McIntosh J. M. (1995) Mol. Pharmacol. 48, 194–199 [PubMed] [Google Scholar]
- 23. Callaghan B., Adams D. J. (2010) Channels 4, 51–54 [DOI] [PubMed] [Google Scholar]
- 24. Callaghan B., Haythornthwaite A., Berecki G., Clark R. J., Craik D. J., Adams D. J. (2008) J. Neurosci. 28, 10943–10951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Whiteaker P., Christensen S., Yoshikami D., Dowell C., Watkins M., Gulyas J., Rivier J., Olivera B. M., McIntosh J. M. (2007) Biochemistry 46, 6628–6638 [DOI] [PubMed] [Google Scholar]
- 26. Pantin C. F. (1946) Notes on Microscopical Technique for Zoologists, Cambridge University Press, Cambridge [Google Scholar]
- 27. Safavi-Hemami H., Bulaj G., Olivera B. M., Williamson N. A., Purcell A. W. (2010) J. Biol. Chem. 285, 12735–12746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Katoh K., Kuma K., Toh H., Miyata T. (2005) Nucleic Acids Res. 20, 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 30. Clark R. J., Fischer H., Nevin S. T., Adams D. J., Craik D. J. (2006) J. Biol. Chem. 281, 23254–23263 [DOI] [PubMed] [Google Scholar]
- 31. Nevin S. T., Clark R. J., Klimis H., Christie M. J., Craik D. J., Adams D. J. (2007) Mol. Pharmacol. 72, 1406–1410 [DOI] [PubMed] [Google Scholar]
- 32. Bartels C., Xia T. H., Billeter M., Guntert P., Wuthrich K. (1995) J. Biomol. NMR 6, 1–10 [DOI] [PubMed] [Google Scholar]
- 33. Güntert P., Mumenthaler C., Wüthrich K. (1997) J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 34. Herrmann T., Güntert P., Wüthrich K. (2002) J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 35. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 37. Linge J. P., Williams M. A., Spronk C. A., Bonvin A. M., Nilges M. (2003) Proteins 50, 496–506 [DOI] [PubMed] [Google Scholar]
- 38. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 39. Koradi R., Billeter M., Wuthrich K. (1996) J. Mol. Graph. 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 40. Seavey B. R., Farr E. A., Westler W. M., Markley J. L. (1991) J. Biomol. NMR 1, 217–236 [DOI] [PubMed] [Google Scholar]
- 41. López-Vera E., Aguilar M. B., Schiavon E., Marinzi C., Ortiz E., Restano Cassulini R., Batista C. V., Possani L. D., Heimer de la Cotera E. P., Peri F., Becerril B., Wanke E. (2007) FEBS J. 274, 3972–3985 [DOI] [PubMed] [Google Scholar]
- 42. Luo S., Zhangsun D., Zhang B., Quan Y., Wu Y. (2006) J. Pept. Sci. 12, 693–704 [DOI] [PubMed] [Google Scholar]
- 43. Sandall D. W., Satkunanathan N., Keays D. A., Polidano M. A., Liping X., Pham V., Down J. G., Khalil Z., Livett B. G., Gayler K. R. (2003) Biochemistry 42, 6904–6911 [DOI] [PubMed] [Google Scholar]
- 44. Zhangsun D., Luo S., Wu Y., Zhu X., Hu Y., Xie L. (2006) Chem. Biol. Drug Des. 68, 256–265 [DOI] [PubMed] [Google Scholar]
- 45. Jakubowski J. A., Keays D. A., Kelley W. P., Sandall D. W., Bingham J. P., Livett B. G., Gayler K. R., Sweedler J. V. (2004) J. Mass Spectrom. 39, 548–557 [DOI] [PubMed] [Google Scholar]
- 46. Heinemann S. H., Leipold E. (2007) Cell. Mol. Life Sci. 64, 1329–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Halai R., Craik D. J. (2009) Nat. Prod. Rep. 26, 526–536 [DOI] [PubMed] [Google Scholar]
- 48. Gatton M. L., Peters J. M., Gresty K., Fowler E. V., Chen N., Cheng Q. (2006) Am. J. Trop. Med. Hyg. 75, 212–218 [PMC free article] [PubMed] [Google Scholar]
- 49. Stock D. W., Buchanan A. V., Zhao Z., Weiss K. M. (1996) Genomics 37, 234–237 [DOI] [PubMed] [Google Scholar]
- 50. Santos A. D., McIntosh J. M., Hillyard D. R., Cruz L. J., Olivera B. M. (2004) J. Biol. Chem. 279, 17596–17606 [DOI] [PubMed] [Google Scholar]
- 51. Luo S., Nguyen T. A., Cartier G. E., Olivera B. M., Yoshikami D., McIntosh J. M. (1999) Biochemistry 38, 14542–14548 [DOI] [PubMed] [Google Scholar]
- 52. Vincler M., Wittenauer S., Parker R., Ellison M., Olivera B. M., McIntosh J. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17880–17884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hu S. H., Gehrmann J., Guddat L. W., Alewood P. F., Craik D. J., Martin J. L. (1996) Structure 4, 417–423 [DOI] [PubMed] [Google Scholar]
- 54. Halai R., Clark R. J., Nevin S. T., Jensen J. E., Adams D. J., Craik D. J. (2009) J. Biol. Chem. 284, 20275–20284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Millard E. L., Daly N. L., Craik D. J. (2004) Eur. J. Biochem. 27, 2320–2326 [DOI] [PubMed] [Google Scholar]
- 56. Le Gall F., Favreau P., Richard G., Letourneux Y., Molgó J. (1999) Toxicon 37, 985–998 [DOI] [PubMed] [Google Scholar]
- 57. Kohn A. J., Nybakken J. W., Van Mol J. J. (1972) Science 176, 49–51 [DOI] [PubMed] [Google Scholar]
- 58. Marshall J., Kelley W. P., Rubakhin S. S., Bingham J. P., Sweedler J. V., Gilly W. F. (2002) Biol. Bull. 203, 27–41 [DOI] [PubMed] [Google Scholar]
- 59. Townsend A., Livett B. G., Bingham J. P., Truong H. T., Karas J. A., O'Donnell P., Williamson N. A., Purcell A. W., Scanlon D. (2009) Int. J. Pept. Res. Ther. 15, 195–203 [Google Scholar]
- 60. Jones A. K., Brown L. A., Sattelle D. B. (2007) Invert. Neurosci. 7, 67–73 [DOI] [PubMed] [Google Scholar]
- 61. Sargent P. B. (1993) Annu. Rev. Neurosci. 16, 403–443 [DOI] [PubMed] [Google Scholar]
- 62. Drago J., McColl C. D., Horne M. K., Finkelstein D. I., Ross S. A. (2003) Cell. Mol. Life Sci. 60, 1267–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Peng H., Ferris R. L., Matthews T., Hiel H., Lopez-Albaitero A., Lustig L. R. (2004) Life Sci. 76, 263–280 [DOI] [PubMed] [Google Scholar]
- 64. Kurzen H., Berger H., Jager C., Hartschuh W., Maas-Szabowski N. (2005) Exp. Dermatol. 14, 155 [Google Scholar]
- 65. Arredondo J., Nguyen V. T., Chernyavsky A. I., Bercovich D., Orr-Urtreger A., Kummer W., Lips K., Vetter D. E., Grando S. A. (2002) J. Cell Biol. 159, 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Haberberger R. V., Bernardini N., Kress M., Hartmann P., Lips K. S., Kummer W. (2004) Autonomic Neurosci. 113, 32–42 [DOI] [PubMed] [Google Scholar]
- 67. Lips K. S., Pfeil U., Kummer W. (2002) Neuroscience 115, 1–5 [DOI] [PubMed] [Google Scholar]
- 68. Hone A. J., Whiteaker P., Christensen S., Xiao Y., Meyer E. L., McIntosh J. M. (2009) J. Neurochem. 111, 80–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Brandhorst B. P. (1985) Dev. Biol. 1, 525–527 [DOI] [PubMed] [Google Scholar]
- 70. Nash M. A., Kozak S. E., Angerer L. M., Angerer R. C., Schatten H., Schatten G., Marzluff W. F. (1987) J. Cell Biol. 104, 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Howell A. M., Cool D., Hewitt J., Ydenberg B., Smith M. J., Honda B. M. (1987) J. Mol. Evol. 25, 29–36 [Google Scholar]
- 72. Franc A. (1943) Faculty of Science. Ph.D. thesis, pp. 1–158, University of Algeria [Google Scholar]
- 73. Degnan B. M., Groppe J. C., Morse D. E. (1995) Roux's Arch. Dev. Biol. 205, 97–101 [DOI] [PubMed] [Google Scholar]
- 74. Fein A., Bdolah A., Kochva E. (1971) Dev. Biol. 24, 520–532 [DOI] [PubMed] [Google Scholar]
- 75. Escoubas P., Corzo G., Whiteley B. J., Célérier M. L., Nakajima T. (2002) Rapid Commun. Mass Spectrom. 16, 403–413 [DOI] [PubMed] [Google Scholar]
- 76. Olivera B. M. (1997) Mol. Biol. Cell. 8, 2101–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Coll J. C., Leone P. A., Bowden B. F., Carroll A. R., König G. M., Heaton A., de Nys R., Maida M., Aliño P. M., Willis R. H., Babcock R. C., Florian Z., Clayton M. N., Miller R. L., Alderslade P. N. (1995) Mar. Biol. 123, 1432–1793 [Google Scholar]
- 78. Slattery M., Hines G. A., Starmer J., Paul V. J. (1999) Coral Reefs 18, 75–84 [Google Scholar]
- 79. Howden M. E., Lucas J., McDuff M., Salathe R. (1974) in Crown-of-Thorns Starfish: Seminar Proceedings, September 6, 1974, Government Publishing Service, Canberra, Brisbane, Australia [Google Scholar]
- 80. Cowart J. D., Fielman K. T., Woodin S. A., Lincoln D. E. (2000) Mar. Biol. 136, 993–1002 [Google Scholar]
- 81. Lindquist N., Fenical W. (1991) Experientia 47, 504–506 [Google Scholar]
- 82. Lindquist N., Hay M. E., Fenical W. (1992) Ecol. Monogr. 62, 547–568 [Google Scholar]
- 83. Fry B. G., Roelants K., Champagne D. E., Scheib H., Tyndall J. D., King G. F., Nevalainen T. J., Norman J. A., Lewis R. J., Norton R. S., Renjifo C., de la Vega R. C. (2009) Annu. Rev. Genomics Hum. Genet. 10, 483–511 [DOI] [PubMed] [Google Scholar]
- 84. Fry B. G. (2005) Genome Res. 15, 403–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Faghih R., Gfesser G. A., Gopalakrishnan M. (2007) Recent Pat. CNS Drug Discov. 2, 99–106 [DOI] [PubMed] [Google Scholar]
- 86. Mastrangeli R., Donini S., Kelton C. A., He C., Bressan A., Milazzo F., Ciolli V., Borrelli F., Martelli F., Biffoni M., Serlupi-Crescenzi O., Serani S., Micangeli E., El Tayar N., Vaccaro R., Renda T., Lisciani R., Rossi M., Papoian R. (2003) Eur. J. Endocrinol. 13, 560–570 [PubMed] [Google Scholar]
- 87. Kaplan N., Morpurgo N., Linial M. (2007) J. Mol. Biol. 369, 553–566 [DOI] [PubMed] [Google Scholar]
- 88. Moran Y., Weinberger H., Reitzel A. M., Sullivan J. C., Kahn R., Gordon D., Finnerty J. R., Gurevitz M. (2008) J. Mol. Biol. 380, 437–434 [DOI] [PubMed] [Google Scholar]
- 89. Moran Y., Weinberger H., Lazarus N., Gur M., Kahn R., Gordon D., Gurevitz M. (2009) J. Mol. Evol. 69, 115–124 [DOI] [PubMed] [Google Scholar]
- 90. Genikhovich G., Kürn U., Hemmrich G., Bosch T. C. (2006) Dev. Biol. 289, 466–481 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.