Abstract
Prodiginines are a class of red-pigmented natural products with immunosuppressant, anticancer, and antimalarial activities. Recent studies on prodiginine biosynthesis in Streptomyces coelicolor have elucidated the function of many enzymes within the pathway. However, the function of RedJ, which was predicted to be an editing thioesterase based on sequence similarity, is unknown. We report here the genetic, biochemical, and structural characterization of the redJ gene product. Deletion of redJ in S. coelicolor leads to a 75% decrease in prodiginine production, demonstrating its importance for prodiginine biosynthesis. RedJ exhibits thioesterase activity with selectivity for substrates having long acyl chains and lacking a β-carboxyl substituent. The thioesterase has 1000-fold greater catalytic efficiency with substrates linked to an acyl carrier protein (ACP) than with the corresponding CoA thioester substrates. Also, RedJ strongly discriminates against the streptomycete ACP of fatty acid biosynthesis in preference to RedQ, an ACP of the prodiginine pathway. The 2.12 Å resolution crystal structure of RedJ provides insights into the molecular basis for the observed substrate selectivity. A hydrophobic pocket in the active site chamber is positioned to bind long acyl chains, as suggested by a long-chain ligand from the crystallization solution bound in this pocket. The accessibility of the active site is controlled by the position of a highly flexible entrance flap. These data combined with previous studies of prodiginine biosynthesis in S. coelicolor support a novel role for RedJ in facilitating transfer of a dodecanoyl chain from one acyl carrier protein to another en route to the key biosynthetic intermediate 2-undecylpyrrole.
Keywords: Anticancer Drug, Bacterial Metabolism, Crystal Structure, Crystallography, Enzyme Kinetics, Gene Knockout, Immunosupressor, Protein Structure, Natural Product Biosynthesis
Introduction
Modular polyketide synthases (PKSs)5 and non-ribosomal peptide synthetases (NRPSs) are large multienzymes that catalyze the production of a wide range of biologically active compounds currently used as therapies for human diseases (1). Most PKSs and NRPSs are organized as assembly lines in which specific enzymatic domains catalyze elongation or modification of specific pathway intermediates. One key difference between these two types of assembly lines is that PKSs use acyl thioesters as substrates, whereas NRPSs use amino acids. Both PKS and NRPS pathway intermediates are tethered, via a thioester bond, to the phosphopantetheine (Ppant) arms of acyl/peptidyl carrier protein (ACP/PCP) domains throughout chain assembly. Thioester cleavage, most often catalyzed by a terminating or type I thioesterase (TE I), releases the fully assembled polyketide/peptide from the carrier domain.
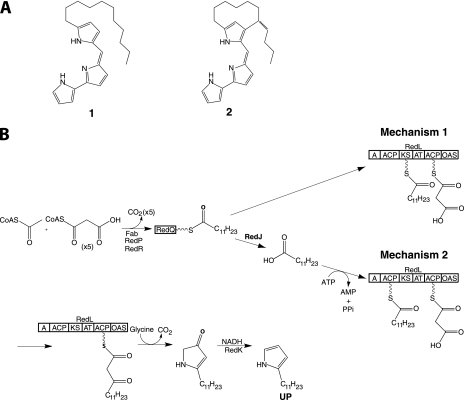
Prodiginine biosynthesis is directed by the red cluster in Streptomyces coelicolor and involves hybrid NRPS/PKS multienzymes employing an α-oxoamine synthase (OAS) in a highly unusual chain release mechanism to assemble undecylprodiginine (1) and its carbocyclic derivative streptorubin B (2) (Fig. 1A) (2–5). The therapeutic potential of prodiginines was demonstrated in prior studies in which analogues of undecylprodiginine exhibited immunosuppressant activity and analogues of streptorubin B exhibited anticancer activity (6, 7). Undecylprodiginine, 1, is assembled by RedH-mediated condensation of 2-undecylpyrrole (UP) and 4-methoxy-2,2′-bipyrrole-5-carboxaldehyde (5), which are biosynthesized by two distinct sets of enzymes encoded by genes within the red cluster of S. coelicolor (3, 8–11).
FIGURE 1.
Structures of prodiginines produced by S. coelicolor and proposed pathways for UP biosynthesis. A, structures of undecylprodiginine (1) and streptorubin B (2). B, biosynthesis of UP. Two possible mechanisms of dodecanoyl transfer from RedQ to RedL are shown. Fab, fatty acid biosynthesis enzyme; A, adenylation domain; AT, acyltransferase.
Recent genetic studies have suggested a plausible pathway for the biosynthesis of UP (Fig. 1B) (3). RedP and RedR, which are homologues of fatty acid biosynthetic enzymes FabH (3-ketoacyl-ACP synthase III) and FabF (3-ketoacyl-ACP synthase II), respectively, are proposed to synthesize a dodecanoyl thioester tethered to an ACP (RedQ) in concert with the primary metabolic ketoreductase, dehydratase, and enoylreductase enzymes of fatty acid biosynthesis (3, 10, 11). The dodecanoyl group is then transferred to RedL, which contains an adenylation domain (A domain), two ACPs, an acyltransferase, a ketosynthase (KS), and an OAS (3, 11). The KS domain is proposed to catalyze 2-carbon elongation of a dodecanoyl thioester by decarboxylative condensation with a malonyl thioester attached to the C-terminal ACP domain of RedL. The OAS domain catalyzes condensation of the resulting 14-carbon β-ketothioester with glycine to form a β-keto-α-amino acid, followed by decarboxylation, cyclization, and dehydration to form 2-undecylpyrrolin-4-one. Finally, RedK reduces the keto group in 2-undecylpyrrolin-4-one and dehydrates the corresponding alcohol to yield UP (3).
Two mechanisms for transfer of the dodecanoyl group from RedQ to RedL during UP biosynthesis are possible. Mechanism 1 involves direct transacylation from dodecanoyl-RedQ to the active site Cys of the RedL KS domain (Fig. 1B). However, Mo et al. (3) reported that for a redR deletion mutant, in which dodecanoyl-RedQ and thus UP formation was inhibited, UP production could be restored by feeding dodecanoic acid but not its corresponding N-acetylcysteamine thioester. These results are consistent with transfer mechanism 2, in which dodecanoyl-RedQ is hydrolyzed to form dodecanoic acid, which is activated by adenylation and transferred to the N-terminal ACP domain of RedL by its A domain (Fig. 1B). The RedL A domain lacks the conserved aspartate residue of NRPS A domains that adenylate amino acids and has sequence similarity to long-chain fatty acid-AMP ligases, which adenylate the carboxyl group of long-chain fatty acids, activating them for transfer to ACPs (11, 12).
Transfer mechanism 2 requires a specific thioesterase to release dodecanoic acid by hydrolysis of dodecanoyl-RedQ. One candidate thioesterase is RedJ, which has high sequence similarity to editing, or type II, thioesterases (TE II), such as RifR (38% sequence identity) from the rifamycin pathway in Amycolatopsis mediterranei (11, 13). Based on sequence similarity and genetic studies, previous reports have assigned RedJ an editing function similar to TE IIs from other natural product biosynthetic pathways (11, 14). TE IIs are thought to relieve stalled assembly lines by removing non-productive intermediates from carrier protein domains (13, 15–18). Such nonproductive intermediates may arise from premature decarboxylation by a KS domain (16, 17) or incorrect priming of carrier protein domains (15, 18). Consistent with the proposed editing function, previously characterized TE IIs exhibit low substrate selectivity, hydrolyze a range of substrates from a range of ACPs, and have low activity (13, 16, 17).
Crystal and NMR solution structures of chain-terminating TE Is (19–24) and TE IIs (13, 25) from PKS and NRPS pathways demonstrate structural variability among TEs. All PKS and NRPS TEs adopt an α/β-hydrolase fold and contain a serine-histidine-aspartate catalytic triad. In addition to the α/β-hydrolase core, the TEs have a helical lid domain positioned above the active site. For PKS TE Is, the lid domain has a fixed position in which it maintains the structure of an active site tunnel (19, 20). However, for TE IIs and NRPS TE Is, the lid region is flexible based on crystal and NMR solution structures that captured distinct conformations and movement of the lid (13, 21–23, 25). Lid movement has been proposed to regulate access of substrates to the active site, to recognize specific ACPs, and to change the size and shape of the substrate chamber (13, 21–23, 25). Furthermore, TE quaternary structures vary. TE Is from PKS pathways are dimers, whereas TE Is from NRPS pathways and TE IIs are monomeric.
Here we report structural, biochemical, and genetic characterization of RedJ from the prodiginine biosynthetic pathway of S. coelicolor. The data demonstrate selectivity of RedJ for dodecanoyl-ACP substrates and support a novel role for RedJ in transfer mechanism 2 (Fig. 1B). In addition, eight independent views of RedJ in two crystal structures provide snapshots of lid motion and insights about the mechanism of substrate selectivity and access to the active site.
EXPERIMENTAL PROCEDURES
Plasmids
The redJ coding sequence located at 19,170–20,012 bp of cosmid SC3F7 from the S. coelicolor ordered genomic cosmid library was PCR-amplified using the forward primer 5′-CATATGTCGCCCGCTGACCTGCTC-3′ introducing an NdeI site (boldface type) and the reverse primer 5′-AAGCTTTCAGAATGTCCATGTTGCTTC-3′ introducing a HindIII site (boldface type). The resulting PCR product was digested with NdeI and HindIII and ligated into the corresponding sites of the pET-28a(+) expression vector to provide pMMA1. For crystallization, a plasmid, pRedJT, was used to produce a truncated RedJ, RedJT (5-residue N-terminal truncation and 19-residue C-terminal truncation). The redJT construct was PCR-amplified under standard conditions from pMMA1 and ligated into pMCSG7 to give pRedJT (26). The forward primer was 5′-TACTTCCAATCCAATGCCCTGCTCTCCCAGCGTTCC-3′, and the reverse primer was 5′-TTATCCACTTCCAATGCTAGAGTTCGGTGCCCAGGTG-3′. Normal type indicates the sequences complementary to DNA encoding the protein, and boldface type indicates overhangs used for ligation-independent cloning.
The redQ coding sequence was amplified from SC3F7 cosmid using the forward primer 5′-GGCCCGCGCATATGAGCACCACCTACGA-3′ (introducing an NdeI site, indicated in boldface type) and the reverse primer, 5′-ATGCAGGATCCTCATGACGCGGTGGCCG-3′ (introducing a BamHI site, indicated in boldface type). The resulting PCR product was digested with NdeI and BamHI and ligated into the corresponding sites in pET15b to provide the redQ expression plasmid, pSJM4.
pMMA1 derivatives were generated using the QuikChange XLII site-direct mutagenesis kit (Agilent Technologies) to provide expression plasmids for RedJ containing L150N, V158T, L162N, L187N, and I215N substitutions. Mutagenic primers 5′-CGCGCCGACCACACCAATTCCGACACCGG-3′ and 5′-CAGGGCGGTGTCGGAATTGGTGTGGTCGGCGCG-3′ provided the expression plasmid pMMA1L150N, primers 5′-CGCCCTGCGCGAGACCATCCGGGACCTG-3′ and 5′-CAGGTCCCGGATGGTCTCGCGCAGGGCG-3′ provided pMMA1V158T, primers 5′-GAGGTCATCCGGGACAATGGCGGCCTCGACGAC-3′ and 5′-GTCGTCGAGGCCGCCATTGTCCCGGATGACCTC-3′ provided pMMA1L162N, primers 5′-GCTCCGCGCCGACAATCGCGCCTGCGAGC-3′ and 5′-GCTCGCAGGCGCGATTGTCGGCGCGGAGC-3′ provided pMMA1L187N, and primers 5′-GCCGCCGACCCCAACGCCACCCC-3′ and 5′-GGGGTGGCGTTGGGGTCGGCGGC-3′ provided pMMA1I215N (mutations are underlined). A pRedJT derivate with a S107A substitution was generated using the same method. Mutagenic primers 5′-CCTGTTCGGGCACGCCATGGGCGCCCTG-3′ and 5′-CAGGGCGCCCATGGCGTGCCCGAACAGG-3′ provided pRedJT_S107A. All constructs were confirmed by sequencing.
Protein Expression
Plasmids pMMA1, pMMA1L150N, pMMA1V158T, pMMA1L162N, pMMA1L187N, pMMA1I215N, and pSJM4 were used to transform Escherichia coli BL21(DE3) cells. The resulting transformants were grown in LB medium at 37 °C containing either 50 μg/ml kanamycin for pMMA1 or 100 μg/ml apramycin for pSJM4, to an A600 = 0.5–0.6, equilibrated to 22–25 °C, induced with 0.1 mm isopropyl β-d-thiogalactopyranoside (IPTG), and incubated for ∼18 h. Plasmids pRedJT and pRedJT_S107A were similarly used to transform E. coli BL21(DE3) cells, and the transformants were cultured at 37 °C to A600 = 0.8–1 in 0.5 liters of TB (6 g of tryptone, 12 g of yeast extract, 1.16 g of KH2PO4, 6.25 g of K2HPO4, 20 ml of glycerol) containing 0.1 mg/ml ampicillin. Cultures were incubated for 1 h at 20 °C, induced with 0.2 mm IPTG, and allowed to express for ∼18 h.
Selenomethionyl (SeMet) His6-RedJT was produced using a protocol from Guerrero et al. (27). Briefly, a colony of E. coli BL21(DE3) cells bearing pRedJT was cultured for 18 h at 37 °C in TB. This culture was centrifuged, and the pellet was resuspended to an A600 = 0.4 in a minimal medium containing 50 μg/ml dl-selenomethionine. The cells were cultured at 37 °C to an A600 = 0.6, incubated at 20 °C for 1 h, induced with 1 mm IPTG, and allowed to express at 20 °C for 18 h. In all cases, cells were harvested by centrifugation at 5670 × g for 25 min at 4 °C, and cell pellets were frozen at −20 °C.
Protein Purification
For apo-RedQ purification, the appropriate E. coli cell pellets were suspended in 10–15 ml of buffer A (50 mm NaH2PO4 pH 8.0, 300 mm NaCl, 1 mm DTT, 10% glycerol) containing 10 mm imidazole. Cells were lysed by sonication (5 × 10 s with 50-s intervals), and the cell debris was pelleted by centrifugation (20,000 × g, 30 min, 4 °C). The cell lysate was filtered through a 0.45-μm syringe filter and then shaken with 1 ml of nickel-NTA resin (Qiagen) for 1 h at 4 °C and transferred to a column. The nickel-NTA resin was washed with 5 column volumes of buffer A with 10 mm imidazole, followed by 5 column volumes of buffer A containing 30 mm imidazole. The N-terminal polyhistidine-tagged protein was eluted using buffer A with 300 mm imidazole. Apo-RedQ was further purified using a Superdex 75 10/300 (GE Healthcare) column (50 mm NaH2PO4, pH 7.2, 1 mm tris(2-carboxyethyl)phosphine (TCEP)). Fractions containing apo-RedQ (confirmed by ESI-LC-MS) were pooled, exchanged with a storage buffer (50 mm NaH2PO4 pH 7.2, 1 mm TCEP, 1 mm EDTA, 20% glycerol), concentrated using a microcon YM-10 (Millipore), and stored in aliquots at −80 °C.
For RedJT and pRedJT_S107A purification, cell pellets were resuspended in buffer B (50 mm Tris, pH 7.5, 300 mm NaCl, 10% glycerol) with 0.1 mg/ml lysozyme and lysed by sonication. Cell lysates were centrifuged at 34,540 × g for 45 min. The supernatant was passed through a 0.45-μm filter and loaded onto a 5-ml His trap column (GE Healthcare). Proteins were eluted using a gradient of 15–300 mm imidazole in buffer B over 10 column volumes. For removal of the His6 tag from RedJT, pooled fractions from the His column were incubated for 4 h at room temperature with tobacco etch virus protease (30:1 molar ratio of protein to protease) and 2 mm dithiothreitol (DTT), dialyzed overnight in buffer B, and loaded onto a His trap column to separate untagged RedJ from His-tagged tobacco etch virus protease. Untagged RedJ was collected in the flow-through. After a final gel filtration step (HiPrep 16/60 Sephacryl S100 HR equilibrated with buffer B containing 2 mm DTT), the protein was concentrated to 8 mg/ml, flash-frozen in liquid nitrogen, and stored at −80 °C. RedJ variants produced from pMMA1 were purified by a similar protocol, but the His6 tag was not removed.
ACP Acylation
Sfp from Bacillus subtilis was used for the conversion of apo-ACP to corresponding acyl-ACPs as described previously (28). Briefly, each reaction mixture contained 50 μm apo-ACP, 150 μm acyl-CoA, 0.5 mm TCEP, 2.5 mm MgCl2, and 1–2 μm Sfp in sodium/potassium phosphate buffer, pH 6.0, and was incubated for 1–16 h at 37 °C. Formation of the acyl-ACP was monitored using ESI-LC-MS. A Microcon YM-3 filter (Millipore) was used to remove the excess acyl-CoA substrate, exchange the buffer for 50 mm phosphate buffer, and concentrate the acyl-ACP product.
Acetyl-FabC (ACP) was generated by enzymatic decarboxylation of malonyl-FabC using E. coli β-ketoacyl acyl carrier protein synthase III (FabH) as described previously (29). Briefly, malonyl-FabC was incubated with 10 nm FabH, and decarboxylation was monitored using ESI-LC-MS as described below. The resulting acetyl-FabC was concentrated and further purified using a Superdex 75 10/300 column (GE Healthcare) equilibrated with 50 mm NaH2PO4, pH 7.2, 1 mm TCEP buffer and stored as aliquots at −80 °C.
HPLC and LC-MS Analyses
Samples for LC-MS and LC-MS/MS analysis were injected via autosampler onto a Discovery (3 μm, 15 cm × 2.1 mm) (Supelco) reverse-phase column operated at a flow rate of 200 μl/min. The outflow was directed into the mass spectrometer. A gradient elution method was used employing Solvent A (99% H2O, 1% acetonitrile, 0.05% TFA) for 5 min, a gradient to 100% solvent B (1% H2O, 99% acetonitrile, 0.05% TFA) over 25 min, and 100% solvent B for 2 min. The mass spectrometric analyses were performed on a micrOTOF-Q (QqTOF) (Bruker) mass spectrometer equipped with an electrospray ion source operating in positive mode. The instrument parameters were as follows: capillary voltage, 4500 V; nebulizer gas, 3.0; dry gas, 6 liters/min; dry temperature, 200 °C; funnel 1RF, 400 Vpp; funnel 2RF, 400 Vpp; hexapole RF, 500 Vpp; collision energy, 10 eV; and collision RF, 1000 Vpp. Spectra were scanned from m/z 200 to m/z 3000. Nitrogen was used as the collision gas.
RedJ Assays
An LC-MS based assay was used to determine the amount of acyl-ACP hydrolysis catalyzed by RedJ. In a standard 20-μl assay, 5 pmol of RedJ was incubated in 50 mm potassium phosphate buffer, pH 7.4, with varying concentrations of an acyl-ACP for 5 min at 37 °C (reaction rate was shown to be linear under these conditions). Assays at each acyl-ACP concentration were carried out in triplicate. Reactions were quenched with 20 μl of 10% formic acid. The loss of acyl-ACP and formation of ACP were analyzed by LC-MS as described above. Standard curves for the acyl-ACP (20–0.5 μm) were generated by serial dilutions of 5–10 mg/ml stock solutions, under the same LC-MS conditions. Hystar 3.3 software was used to acquire the data. Data were integrated using the software Data Analysis version 4.0 (Bruker Daltonics).
A spectrophotometric assay was used to assess substrate specificity of RedJ with various acyl-CoAs. The assays were conducted at 37 °C in 96-well flat bottom plates (Falcon) and contained 100 mm HEPES (pH 7.4); 20 mm NaCl; 100 mm 5,5′-dithiobis(2-nitrobenzoic acid) (Sigma); various concentrations of lauroyl, decanoyl, malonyl, and acetyl-CoA (Sigma); and 100 nm RedJ. The production of 2-nitrobenzoic acid-5-thiolate resulting from disulfide exchange with 5,5′-dithiobis(2-nitrobenzoic acid) by CoASH liberated in the thioesterase reaction was measured at 412 nm (molar extinction coefficient 13,600 m−1 cm−1) using a Spectramax spectrophotometer (Molecular Devices). The amount of CoASH released corresponds to the rate of acyl-CoA hydrolysis. All data points were collected in triplicate. Nonlinear regression with GraFit 4.012 (Middlesex, UK) was used to determine kcat and Km values.
Crystallization
His6-RedJT was crystallized by vapor diffusion in 4-μl drops containing equal volumes of protein stock and well solution (500 mm NaCl, 12% polyethylene glycol 400, 100 mm HEPES, pH 7.6, 2 mm dithiothreitol). The well solution for SeMet His6-RedJT was 700 mm NaCl, 12% polyethylene glycol 2000, 100 mm HEPES, pH 7.6, 2 mm dithiothreitol, and the well solution for RedJT was 220 mm NaCl, 20% polyethylene glycol 3350, 100 mm HEPES, pH 7, 2 mm dithiothreitol. Crystals grew within 24 h at 20 °C and were harvested in loops, cryoprotected with the corresponding well solution containing 20% glycerol, and frozen in liquid nitrogen.
Data Collection and Structure Determination
Data were collected at the Advanced Photon Source, General Medicine/Cancer-Collaborative Access Team (GM/CA-CAT) beamline 23ID-D at Argonne National Laboratory (Argonne, IL). All data were processed using HKL2000 (30). SeMet His6-RedJT crystallized in space group P21, and RedJT crystallized in space group C2, both forms with four polypeptides in the asymmetric unit (Table 1). Initial phases for SeMet His6-RedJT were determined using the single-wavelength anomalous diffraction method. SOLVE (31) and RESOLVE (32) were used to locate selenium atoms, determine initial phases, perform density modification, and build a 95% complete initial model. The asymmetric unit contained 12 Met residues, but due to partial occupancy, a total of 23 selenium sites were found. Phases for RedJT were determined by molecular replacement using Phaser (33) with SeMet His6-RedJT as the search model. COOT (34) was used for model building and REFMAC5 (35) of the CCP4 (36) suite for refinement. The asymmetric unit of both crystal forms contained two RedJ dimers, which formed by domain swapping of residues 6–21. For SeMet His6-RedJT, density for additional residues from the His6 tag was observed on the N terminus of chains C and D. The final crystallographic models of RedJ are complete except for a few residues at the N termini and lid subdomains of some subunits; SeMet His6-RedJT chain A residues 6–8 and 166–175, SeMet His6-RedJT chain B residues 6–8, RedJT chain A residues 6–11, RedJT chain B residues 6–10 and 167–170, RedJT chain C residues 6–10 and 167–171, and RedJT chain D residues 165–179 were disordered. Structures were validated by MOLPROBITY (37). Sequence alignments were done with MUSCLE alignment tool (38), and molecular figures were prepared with PyMOL (The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC).
TABLE 1.
Crystallographic data
| SeMet His6-RedJT | RedJT | |
|---|---|---|
| Data collection | ||
| Space group | P21 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 74.81, 79.40, 87.44 | 155.55, 45.01, 156.40 |
| α, β, γ (degrees) | 90, 90.12, 90 | 90, 109.49, 90 |
| X-ray source | APS 23ID-D | APS 23ID-D |
| Wavelength (Å) | 0.9794 | 1.0332 |
| dmin (Å) | 2.12 (2.2-2.12)a | 2.5 (2.59-2.49) |
| Rsymm | 0.089 (.420) | 0.101 (.359) |
| Average I/σI | 22.12 (5.2) | 11.0 (2.7) |
| Completeness (%) | 100 (99.9) | 93.5 (83.7) |
| Average redundancy | 7.7 (7.5) | 2.8 (2.2) |
| Refinementb | ||
| Data range (Å) | 50–2.12 | 50–2.49 |
| No. of reflections | 57,954 | 33,414 |
| Rwork/Rfree | 0.201/0.247 | 0.215/0.284 |
| Polypeptide chains | 4 | 4 |
| No. of atoms | ||
| Protein | 8176 | 7703 |
| Water | 542 | 151 |
| Ligand | 0 | 13 |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.010 | 0.012 |
| Bond angles (degrees) | 1.18 | 1.20 |
| Average B factors (Å2) | ||
| Protein | 31.4 | 43.7 |
| Water | 34.1 | 34.5 |
| Ligand | 40.5 | |
| Ramachandran plot | ||
| Allowed (%) | 100 | 100 |
| Outliers (%) | 0 | 0 |
a Values in parentheses pertain to the outermost shell of data.
b Final structures are deposited in the Protein Data Bank with accession codes 3QMV for SeMet His6-RedJT and 3QMW for RedJT.
Construction of S. coelicolor W35 (redJ::oriT-apr Mutant of S. coelicolor M511)
Disruption of the redJ gene was carried out using a PCR targeting methodology (39). PCR amplification of the oriT-apr gene replacement cassette from pIJ773 was carried out with the forward primer 5′-GCGCCCATGTCGCCCGCTGACCTGCTCTCCCAGCGTTCCATTCCGGGGATCCGTCGACC-3′ (P1) and the reverse primer 5′-CCAGGCTCAGAATGTCCATGTTGCTTCCCTAGTTGCCTTTGTAGGCTGGAGCTGCTTC-3′ (P2) using Expand High Fidelity polymerase (Roche Applied Science). The resulting PCR product was used to replace redJ in cosmid SC3F7. Forward primer 5′-TGCTGGGCAAGCAGATGGTG-3′ (P3) and reverse primer 5′-CTTGGCCAGGCTCAGAAT-GTCC-3′ (P4), designed to prime ∼200 bp upstream/downstream, respectively, of redJ, were used in a PCR to confirm correct replacement of the gene with the oriT-apr cassette in the cosmid. The modified Sc3F7/redJ::oriT-apr cosmid was transferred by intergenic conjugation from E. coli ET12567/pUZ8002 to S. coelicolor M511. Genomic DNA was isolated from kanS and aprR colonies grown on SFM agar using the Fast DNA SPIN kit for soil (MB Biolabs). Correct replacement of the redJ gene on the chromosome of S. coelicolor with the oriT-apr cassette in these mutants was verified by PCR with the P3 and P4 primers described above and by Southern blot hybridization using labeled Sc3F7 as a probe. A spore stock of one verified mutant was prepared according to standard procedures (40) and stored at −20 °C.
Genetic Complementation of S. coelicolor W35
PCR amplification of a DNA fragment containing redJ was carried out with forward 5′-AAAGGAAGCTTAGGAGGGCGCCCATGTCGCCCG-3′ (P5) and reverse 5′-CCCTTTCTCGAGGCTCGACGAAGCCCTTGG-3′ (P6) primers. The forward primer contains a 5′-HindIII restriction site (boldface type) and was designed to anneal 12 bp upstream of the start codon of redJ to include the natural ribosome binding site. The reverse primer contains a 5′-XhoI restriction site (boldface type) and was designed to anneal around 100 bp downstream of the stop codon of redJ. The PCR used cosmid SC3F7 as a template and conditions described previously (39). The HindIII- and XhoI-digested amplimer was cloned into HindIII- and XhoI-digested pOSV556t (kindly provided by Dr. Jean-Luc Pernodet, Orsay) using the Rapid DNA Ligation Kit (Roche Applied Science) following the manufacturer's instructions. 2 μl of the ligation mixture was used to transform E. coli DH5α electrocompetent cells following standard procedures (41). Plasmids were purified from ampicillin-resistant transformants, and the presence of the desired insert was determined by restriction digestion and agarose gel electrophoresis analysis. One correct clone was used to transform E. coli ET12567 containing pUZ8002 by electroporation. An ampicillin-resistant colony was picked and used to transfer the plasmid from E. coli to S. coelicolor W35 by intergenic conjugation following a standard procedure (39). A spore stock of one hygromycin-resistant transconjugant was prepared using standard procedures (40) and stored at −20 °C.
Growth and Harvesting of Mycelia and Extraction and Analysis of Prodiginines
R5 agar plates were overlaid with sterile permeable membranes (12,000–14,000 molecular weight cut off, size 20). 10 μl of spore suspensions of S. coelicolor M511, W35, and the genetically complemented W35 strains were spread on the membranes. After 5–7 days of incubation at 30 °C, three separate mycelia samples were scraped off each plate and placed in separate microcentrifuge tubes. Prodiginines were extracted from the harvested mycelia by shaking for 2 h with 1 ml of methanol acidified with 10 μl of 2 n HCl. Samples were centrifuged, and the absorbance at 533 nm of the supernatant was measured. The mycelia were dried overnight at 70 °C, and the dry cell weight was measured. To calculate prodiginine concentrations, the absorbance value was converted into μg of prodiginine/mg of dry cell weight using the known extinction coefficient of 100,500 m−1 cm−1 for prodiginine absorbance at 533 nm (40).
Prodiginines in the extracts after 5 days of growth were analyzed using LC-MS/MS monitoring absorbance at 533 nm. 20 μl of each extract was injected onto an Eclipse XDB-C18 column (150 × 4.6 mm, 5 μm, column temperature 25 °C; Agilent) connected to an Agilent 1100 HPLC instrument equipped with a binary pump and a diode array detector and eluted using the method described previously (5). The HPLC outflow was connected via a splitter (10% flow to MS, 90% flow to waste) to a Bruker HCTultra mass spectrometer equipped with an electrospray source operated in positive ion mode with parameters as follows: nebulizer flow, 40 p.s.i.; dry gas flow, 10.0 liter/min; dry temperature, 300 °C; capillary, −4 kV; skimmer, 40 V; capillary exit, 106 V; ion charge control target, 100,000; spectral averages, 3.
Feeding experiments with dodecanoic acid and analogues (decanoic acid, pentadecanoic acid, and 10-undecynoic acid) were performed on agar plates and in liquid culture. Agar plates were inoculated with W35 strain, and after 2 or 3 days of incubation, 50 ml of 0.5 m fatty acid in methanol was dripped on the plate (∼20 drops). Incubation was carried out for a further 3–5 days, mycelia from whole plate were scraped and analyzed for prodiginine production as described above. Feeding experiments in liquid medium followed a published protocol (3).
RESULTS
Kinetic Characterization of the RedJ Thioesterase
The redJ gene encodes a 280-amino acid protein homologous to type II thioesterases. To test alternative possibilities for RedJ function (Fig. 1B), redJ was expressed in E. coli, recombinant RedJ was purified, and its catalytic activity was investigated. The purified His6-RedJ (RedJ) has the expected molecular mass of ∼35 kDa as judged by SDS-PAGE and migrates as a monomer during gel filtration chromatography (supplemental Fig. S1). Assays with purified RedJ demonstrated that it has thioesterase activity with both acyl-CoA and acyl-ACP substrates (Table 2).
TABLE 2.
Kinetic properties of RedJ with acyl substrates
| Substrate | Km | kcat | kcat/Km |
|---|---|---|---|
| mm | min−1 | mm−1min−1 | |
| Decanoyl-RedQ | 5.2 ± 2.8 | 138 ± 12 | 27.6 |
| Decanoyl-AcpP | 5.0 ± 3.5 | 198 ± 18 | 39.6 |
| Dodecanoyl-AcpP | 2.5 ± 1.2 | 498 ± 10 | 199 |
| Acetyl-AcpP | 60 ± 4.5 | 200 ± 22 | 3.3 |
| Malonyl-AcpP | NDa | ND | ND |
| Malonyl-RedQ | ND | ND | ND |
| Decanoyl-CoA | 70.0 ± 2.7 | 3.0 ± 0.16 | 0.04 |
| Dodecanoyl-CoA | 16.7 ± 1.2 | 2.0 ± 0.19 | 0.125 |
| Acetyl-CoA | 1050 ± 310 | 1.2 ± 0.13 | 0.0012 |
| Malonyl-CoA | ND | ND | ND |
a ND, not determined.
The activity of RedJ with acetyl-, malonyl-, decanoyl-, and dodecanoyl-CoA was determined using a 5,5′-dithiobis(2-nitrobenzoic acid)-based continuous spectrophotometric assay. The enzyme has 10–30-fold greater catalytic efficiency with the long-chain acyl substrates than with acetyl-CoA and no detectable activity with malonyl-CoA (Table 2). Nearly all of the differences in catalytic efficiency are due to Km values, which are 60-fold greater for acetyl-CoA relative to dodecanoyl-CoA (Table 2). Thus, RedJ has a significant preference for longer-chain acyl-CoA substrates.
The hydrolysis of acyl-ACPs by RedJ was carried out using a low volume (20-μl) LC-MS assay. The acyl-ACP substrates were generated from either apo-RedQ or apo-AcpP (the E. coli fatty acid synthase ACP) and the appropriate acyl-CoA, using Sfp (a permissive phosphopantetheinyl transferase from B. subtilis (28)). In this way, acetyl-, malonyl-, decanoyl-, and dodecanoyl-AcpP were generated. Sfp was less effective with RedQ, and attempts to generate acetyl- and dodecanoyl-RedQ, the proposed native substrate of RedJ, were complicated by the instability of RedQ during the several-h Sfp reaction. This problem precluded generation of dodecanoyl-RedQ and permitted only small quantities of acetyl-RedQ to be obtained. However, decanoyl-RedQ was readily generated (supplemental Fig. S2).
Like the spectrophotometric assay, the LC-MS assay demonstrated strong substrate preference of RedJ toward long acyl chains. The kcat/Km is 60-fold greater for dodecanoyl-AcpP than for acetyl-AcpP (Table 2). In addition, no thioesterase activity was detected for RedJ when malonyl-RedQ or malonyl-AcpP was utilized as a substrate, indicating strong substrate selectivity of RedJ for non-carboxylated acyl-thioester substrates. Moreover, RedJ has a marked preference for ACP thioesters over CoA thioesters; the overall kinetic efficiency is almost 3 orders of magnitude greater for decanoyl-RedQ than for decanoyl-CoA, due predominantly to increases in kcat (Table 2).
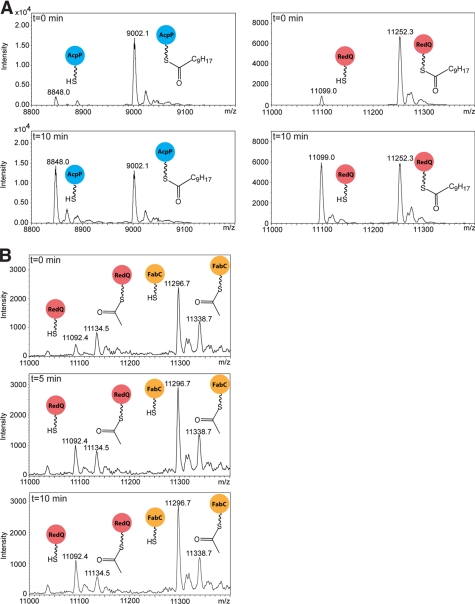
Interestingly, there is no significant difference in the catalytic efficiency of RedJ with decanoyl-AcpP versus decanoyl-RedQ (Table 2). We tested this further in a competition experiment in which RedJ was incubated with an equimolar mixture of decanoyl-RedQ and decanoyl-AcpP (Fig. 2A) and found that the two acyl-ACPs were hydrolyzed at comparable rates. This observation posed the question of whether acyl groups tethered to the native streptomycete fatty acid synthase ACP (FabC) are substrates for RedJ. An alternative method was needed to generate acetyl-FabC for this assay; the recombinant FabC is expressed in E. coli exclusively in the holo form, and thus the apo-FabC required for the Sfp-catalyzed reaction with an acyl-CoA was not readily available. Instead, the malonyl-ACP decarboxylase activity of FabH (29) was employed. This method provided a mixture of acetyl-FabC and holo-FabC (data not shown). In a competition experiment, RedJ was presented with an equimolar mixture of acetyl-FabC and acetyl-RedQ, and after 10 min, a selective hydrolysis of acetyl-RedQ was observed with no detectable hydrolysis of acetyl-FabC (Fig. 2B). Similarly, in a competition experiment assay with acetyl-AcpP and acetyl-FabC, hydrolysis of only the acetyl-AcpP was observed (data not shown). These analyses clearly demonstrate that RedJ selectively hydrolyzes the acyl-RedQ intermediate from the prodiginine biosynthetic pathway over acyl-FabC intermediates from fatty acid biosynthesis in streptomycetes. In contrast, RedJ does not discriminate against AcpP, the ACP from E. coli fatty acid biosynthesis.
FIGURE 2.
Competition assays of RedJ with acyl-RedQ versus acyl-ACPs from fatty acid biosynthesis. A, LC/MS analysis of a mixture of decanoyl-RedQ (right spectra, red circles) and decanoyl-AcpP (left spectra, blue circles) incubated with RedJ at 37 °C. Deconvoluted ESI mass spectra are shown at t = 0 and 10 min. RedJ hydrolyzed the two decanoyl-ACP substrates with equal efficiency. B, deconvoluted ESI mass spectra of a mixture of acetyl-RedQ (red circles) and acetyl-FabC (orange circles) incubated with RedJ at 37 °C and resolved with LC. Incubation times of t = 0, 5, and 10 min are shown. The peak corresponding to acetyl-RedQ decreased significantly after a 10-min incubation of the ACP mixture with RedJ, whereas the acetyl-FabC was undiminished.
Structure of RedJ
The full-length RedJ (RedJFL) used for the activity assays crystallized readily both with and without the His6 tag, but crystals diffracted to only 8 Å. Full-length RedJ has an additional 4 residues at the N terminus and 19 at the C terminus compared with RifR, the characterized thioesterase with the highest sequence identity (38%) to RedJ (supplemental Fig. S3) (13). On this basis, we made a truncated RedJ variant, RedJT, with 5-residue N-terminal and 19-residue C-terminal truncations. Interestingly, RedJT is dimeric in solution, whereas RedJFL is monomeric (supplemental Fig. S1). Crystal structures were determined for selenomethionyl RedJT at 2.12 Å with an N-terminal His6 fusion (SeMet His6-RedJT) and at 2.49 Å without the His6 tag (RedJT) (Table 1). Both forms of RedJT crystallized as dimers with domain-swapped N termini (supplemental Fig. S4, A and B). In the RedJT dimer, residues 6–21 of each monomer insert into the N-terminal position of the partner monomer. The domain swap is probably an artifact of the 19-residue C-terminal truncation because the truncated C terminus is at the interface of the domain-swapped dimer, and RedJFL is monomeric. Nevertheless, His6-RedJT and RedJT have levels of activity similar to that of RedJFL, indicating that neither the truncations nor the domain swapping affect activity (supplemental Fig. S5). The dimer contact and domain-swapped residues are far from the active site.
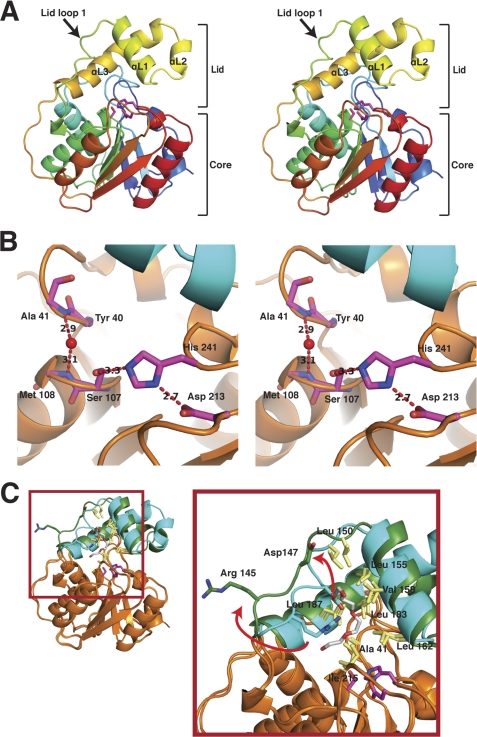
RedJ adopts the expected α/β-hydrolase fold with core and lid domains (Fig. 3A). The core is composed of a six-stranded parallel β-sheet surrounded by five α-helices, and the lid (residues 144–192) is composed of three α-helices inserted between strands β4 and β5 of the core. The active site of RedJ is composed of a conserved catalytic triad of Ser107, Asp213, and His241 (Fig. 3B). Ser107, the catalytic nucleophile, is located between strand β3 and helix α3 in the conserved Gly-His-Ser-Xaa-Gly motif. The backbone amides of Met108 and Ala41 form an oxyanion hole, which is occupied by a water molecule in the crystal structures.
FIGURE 3.
Structure of RedJ. A, trace of the polypeptide chain from the N terminus (blue) to the C terminus (red). The three helices and mobile loop of the lid domain are labeled in the stereo image. Side chains of the catalytic triad are shown as sticks with carbon (magenta), nitrogen (blue), and oxygen (red). B, RedJ active site. Side chains of the catalytic triad (Ser107, Asp213, and His241) and backbone atoms of the oxyanion hole residues (Ala41 and Met108) are shown as sticks, colored as in A in this stereo view. The ribbon diagram of the polypeptide has an orange core and cyan lid. C, RedJ hydrophobic pocket. RedJ in the open entrance conformation (cyan lid, orange core) is superimposed on the closed entrance form (green lid, orange core) (root mean square deviation = 0.457 Å). Lid loop 1 shifts 4.5 Å in the transition between forms, exposing more hydrophobic surface to the polyethylene glycol ligand (sticks with carbon (white) and oxygen (red)). In contrast to lid loop 1, the hydrophobic side chains in the pocket (yellow sticks, labeled in the zoom view) do not move in the structural transition.
Structural Basis for Substrate Specificity
The structure of RedJ provides an explanation of the observed selectivity for substrates with long acyl chains. A large hydrophobic pocket lined with the side chains of Ala41, Leu150, Leu155, Val158, Leu162, Leu183, Leu187, and Ile215 is positioned in the active site cavity 8 Å above the catalytic serine (Fig. 3C). Substrates with long acyl chains (e.g. dodecanoyl thioesters) could react with the catalytic serine while contacting the hydrophobic pocket, but substrates with shorter acyl chains (e.g. acetyl thioesters) would be unable to simultaneously contact the hydrophobic pocket and the catalytic serine. Thus, the hydrophobic pocket may confer selectivity on RedJ toward substrates with long acyl chains, consistent with the kinetic data (Table 2).
The RedJ hydrophobic pocket can bind long-chain ligands. Density for a long-chain ligand was present in one of the eight RedJ molecules in the two crystal structures (supplemental Fig. S6). This density was interpreted as a portion of a polyethylene glycol molecule, based on the components of the protein and crystallization solutions. Ligand binding in the hydrophobic pocket is associated with movement of lid loop 1 (residues 144–151) (Fig. 3C). Without ligand, lid loop 1 partially covers the hydrophobic pocket and is stabilized by a hydrogen bond between the Arg145 and Asp147 side chains. In the presence of a long-chain ligand, the loop moves to uncover the hydrophobic pocket, and the hydrogen bond is broken (Fig. 3C). The movement of lid loop 1 increases the surface area of the hydrophobic pocket by 70% (from 185 Å2 without ligand to 312 Å2 with ligand).
To test the importance of the hydrophobic pocket for hydrolysis of long-chain acyl substrates, polar Asn substitutions were introduced at hydrophobic sites in RedJFL, including Leu150, Leu162, Leu187, and Ile215. In addition, a more conservative substitution of Thr was made at Val158. The activity of these RedJ variants was determined with both acetyl- and decanoyl-RedQ substrates and compared with the activity of wild type RedJFL with the same substrates. All of the Asn substitutions resulted in no detectable activity with either substrate, whereas the Thr substitution at Val158 resulted in 2-fold decreased activity with acetyl-RedQ and nearly 3-fold decreased activity with decanoyl-RedQ (supplemental Table S1). The significant decrease in activity of the hydrophobic pocket variants suggests that the integrity of the hydrophobic pocket is critical to catalysis.
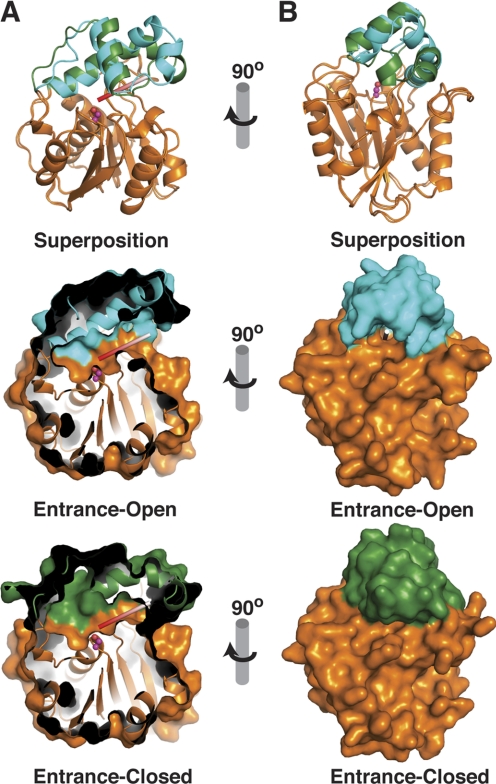
Active Site Entrance Channel
A flexible entrance channel for phosphopantetheine-linked substrates has been identified between the core and lid of other monomeric TEs (13, 21–23, 25). The analogous region of RedJ also forms a channel into the active site (Fig. 4). The RedJ channel has a highly flexible “entrance flap” (residues 163–179), consisting of lid helix αL2 and residues 173–179 of αL3. The flexibility of this flap is evident in its high temperature factors (52.2 Å2) compared with the core domain (28.9 Å2) and its eight different positions captured in the two RedJ crystal structures (supplemental Fig. S7). At one extreme, the flap is fully open (open entrance conformation), and the active site is accessible through a narrow channel (Fig. 4, cyan lid). At the other extreme, the flap fully closed (closed entrance conformation), and the active site is inaccessible (Fig. 4, green lid). The other six views of RedJ captured the entrance flap in a variety of intermediate positions, some with a few disordered residues.
FIGURE 4.
Conformational changes of RedJ. A, front view of the Ppant channel and active site cavity (view similar to Fig. 3C). In the ribbon diagram at the top left, RedJ in the open entrance conformation (cyan lid, orange core) is superimposed on the closed entrance form (green lid, orange core). Open entrance (left center) and closed entrance (left bottom) forms are shown separately with the molecular surfaces. The images are cut away to illustrate the Ppant cleft. A white and red rod mimics the Ppant arm of the substrate, and the catalytic serine is in magenta. The Ppant cleft leading to the catalytic serine is closed only at its entrance in the bottom image. B, side view into the Ppant entrance. Images are rendered as in A with full molecular surfaces in the center and bottom images.
The phosphopantetheine entrance flap is at the opposite side of the lid from the lid loop 1 (Fig. 3C). Despite the motion of both ends of the lid, the hydrophobic pocket has the same structure and is in the same position relative to the catalytic triad in all eight views of RedJ in the two crystal structures. Interestingly, the closed entrance flap occurs only in the RedJ molecule with a long-chain ligand in the hydrophobic pocket (Fig. 3C). However, the ligand does not contact the entrance flap, and we thus view the flap and loop motions as independent.
Prodiginine Production in an S. coelicolor redJ Deletion Mutant
To investigate the role of RedJ in prodiginine biosynthesis in vivo, the redJ gene was replaced on the S. coelicolor M511 chromosome with an oriT-apr cassette using PCR targeting (39). Production in the resulting W35 mutant (M511/redJ::oriT-apr) was significantly reduced to about 25% of the level produced by the wild type M511 strain (supplemental Fig. S8). In addition, the W35 mutant produces no unnatural analogues of undecylprodiginine or streptorubin B. Wild type levels of prodiginine production were restored by genetic complementation of the W35 mutant by integration of a plasmid containing redJ under the control of the constitutive ermE* promoter into its chromosome, showing that the drop in prodiginine production in the W35 strain does not result from a polar effect on the expression of genes downstream of redJ. Zeylas et al. (14) independently constructed an in-frame deletion of redJ in S. coelicolor M511 and reported that prodiginine production was reduced to ∼10% of the wild type level, broadly consistent with our results. However, they did not genetically complement their mutant. Thus, it is not possible to discern whether the drop in prodiginine production they observed is due solely to the loss of redJ or due also to the unintended introduction of a second mutation.
The experiments with purified recombinant RedJ suggest that it plays a role in production of the dodecanoic acid starter unit for RedL in 2-undecylpyrrole biosynthesis. To investigate this role in vivo, we fed dodecanoic acid to the redJ mutant on agar plates and in liquid culture. Surprisingly, this did not boost the level of prodiginine production in the mutant. We also fed several dodecanoic acid analogues (decanoic acid, pentadecanoic acid, and 10-undecanoic acid) to the redJ mutant, which did not result in the production of any prodiginine analogues.
DISCUSSION
The redJ gene within the prodiginine biosynthetic (red) gene cluster of S. coelicolor encodes a protein homologous to TE IIs of PKSs and NRPSs. RedJ has strong selectivity for substrates with long acyl chains, demonstrated by a 70-fold increase in catalytic efficiency with dodecanoyl-ACP compared with acetyl-ACP and no activity with malonyl-ACP or malonyl-CoA (Table 2). In addition, the catalytic efficiency of RedJ is ∼1000-fold greater with acyl-ACPs than with the corresponding acyl-CoAs, indicating strong selectivity for ACP-thioesters over CoA-thioesters. Furthermore, RedJ is specific for particular ACPs, exhibiting activity with acetyl-RedQ and E. coli acetyl-AcpP but no activity with acetyl-FabC (Streptomyces fatty acid biosynthesis ACP). Thus, ACP selectivity allows prodiginine biosynthesis to occur without impacting fatty acid biosynthesis. RedJ has significantly stronger substrate selectivity than does RifR, an editing TE II with 38% sequence identity to RedJ (13). For example, RifR hydrolyzes both long-chain and carboxylated acyl-CoAs and has at most 30-fold selectivity for ACP- over CoA-thioesters. The low activity and low substrate selectivity of RifR are consistent with its proposed editing function, which requires the hydrolysis of a range of aberrant intermediates that stall natural product biosynthesis (13, 16, 17). The stronger substrate selectivity of RedJ suggests that editing is not its primary function.
The crystal structures of RedJ provide a structural explanation for the kinetic data. The large hydrophobic pocket above the serine-histidine-aspartate catalytic triad explains the selectivity of RedJ toward long-chain acyl thioesters, and site-directed mutagenesis results confirm the importance of the hydrophobic pocket. By comparison, the editing thioesterase, RifR, lacks a hydrophobic pocket. Instead, the RifR pocket is lined with side chains of Arg141, Glu145, Tyr174, and Arg202 in similar positions relative to the catalytic triad to Val158, Leu162, Leu187, and Ile215, respectively, in the hydrophobic pocket of RedJ. Such a contrast in the hydrophobicity of the RedJ and RifR pockets is interesting considering the significant sequence identity of the lid domains of RedJ and RifR (32%). Consistent with this level of sequence identity, the small lid domains of RedJ and RifR have identical folds (for the lids alone, root mean square deviation of backbone atoms is 0.383 Å). However, there are small differences, particularly in αL1 and in the orientation of the lid with respect to the core. These differences result in the hydrophobic pocket of RedJ and a more polar pocket in RifR. Therefore, the substrate selectivity of TE IIs and thus their function may be dictated by subtle variations in the lid domains that are not detectable by sequence analysis.
A polyethylene glycol molecule in the hydrophobic pocket of one of the eight RedJ molecules in the two crystal structures demonstrates that the pocket can accommodate long-chain ligands (supplemental Fig. S6). In the absence of a long-chain ligand, lid loop 1 covers about 40% of the surface of the hydrophobic pocket. In the presence of a long-chain ligand, lid loop 1 shifts toward the outside of the protein, fully uncovering the hydrophobic pocket (Fig. 3C). We predict a similar motion upon binding of long-chain substrates. The position of the RedJ hydrophobic pocket with respect to the catalytic triad remains fixed as other parts of the lid move, implying fixed subsites for thioester and acyl-chain binding.
Crystal and NMR structures of TE IIs and NRPS TE Is demonstrate that the helical lid domain in these monomeric TEs is mobile and may control access of substrates to the active site (13, 21–23, 25). The crystal structures of RedJ also demonstrate lid mobility. In the eight RedJ molecules from the two crystal structures, a substrate channel entrance flap (αL2 and residues 173–179 of αL3 of the lid domain) occurred in eight different conformations (supplemental Fig. S7), including fully open and fully closed positions (Fig. 4).
In addition to the entrance flap, RedJ controls access of substrates to the active site by selecting for certain ACPs. As discussed above, S. coelicolor prevents wasteful hydrolysis of dodecanoyl fatty acid intermediates by strong selection of RedJ against the streptomycete ACP of fatty acid biosynthesis, FabC. In contrast, RedJ has no selection against the foreign ACP of E. coli fatty acid biosynthesis, AcpP. Structural and mutagenesis studies of the enterobactin biosynthetic assembly line (Ent) have provided a coherent picture of the productive interaction of a monomeric TE with its cognate carrier domains in which the C terminus of helix 3 in the carrier domain contacts the surface of the TE near the entrance channel (22, 42, 43). Additional TE contacts occur with the C terminus of helix 1 in the carrier domains (22). In light of the results from the Ent system, the structures of FabC (44) and AcpP (45) are informative about the basis of the ACP selectivity of RedJ. The surface of the helix 3 C terminus is positively charged in FabC and negatively charged in RedQ and AcpP (supplemental Fig. S9). The surface of the RedJ TE that would engage the ACP helix 3, based on the EntF structure, is positively charged. Therefore, charge-charge interactions between the RedJ surface and the helix 3 C terminus of ACPs may confer selectivity of RedJ toward RedQ and discrimination against FabC. Furthermore, the C terminus of helix 1 in FabC is one turn longer than that of RedQ and AcpP. The extension of helix 1 may create a steric clash that prevents interaction between RedJ and FabC.
The kinetic and structural data indicate that RedJ is selective for long-chain substrates tethered to RedQ. This substrate selectivity indicates that the primary function of RedJ is the hydrolysis of dodecanoyl-RedQ to provide dodecanoic acid in UP synthesis. Consistent with this primary function, deletion of the redJ gene in S. coelicolor resulted in a 75% decrease in prodiginine production. This effect on prodiginine biosynthesis is similar to S. coelicolor deletion mutants of redP and redR (3). Functional complementation by fatty acid biosynthetic enzymes or slow direct transfer of the dodecanoyl intermediate from RedQ to RedL may prevent complete loss of prodiginine production in the mutant. Genetic complementation restored prodiginine production to WT levels in the redJ deletion mutant. However, feeding dodecanoic to the redJ deletion mutant did not increase prodiginine production. This result was unexpected because if RedJ hydrolyzes dodecanoyl-RedQ to form dodecanoic acid as the data suggest, then feeding of dodecanoic acid was expected to restore prodiginine production to wild type levels in the mutant. However, a similar inability to restore prodiginine production by feeding dodecanoic acid was observed for redP and redQ mutants of S. coelicolor.6 Of the red genes involved in the early steps of UP production, only a redR deletion mutant was complemented by dodecanoic acid feeding (3). Like RedJ, both RedP and RedQ are proposed to function in the biosynthesis of dodecanoic acid for the prodiginine pathway (Fig. 1B) (3, 10). Therefore, the interplay of fatty acid and prodiginine biosynthesis in S. coelicolor may complicate interpretation of the dodecanoic acid feeding experiments in redJ, redP, and redQ deletion mutants. Nevertheless, the results from the feeding experiment are consistent with a secondary editing function for RedJ to remove non-cognate intermediates from the Red ACP/PCP domains to maintain maximal efficiency of the prodiginine assembly line. The low activity of RedJ toward acetyl-ACP substrates (Table 2) supports this additional editing function.
The kinetic and structural analyses presented here demonstrate that RedJ is a unique thioesterase with selectivity toward substrates with long acyl chains tethered to RedQ. These data support a primary role for RedJ in facilitating transfer of a dodecanoyl group from one pathway protein (RedQ) to another (RedL), via hydrolysis of dodecanoyl-RedQ to form the free intermediate, dodecanoic acid. Further experiments will be required to determine whether RedJ plays additional roles in prodiginine biosynthesis (e.g. as an editing enzyme that removes aberrant acyl groups from carrier proteins).
Supplementary Material
Acknowledgments
We thank the staff at the General Medicine/Cancer (GM/CA) beamlines (supported by NIGMS, National Institutes of Health (NIH), Grant GM Y1-GM-1104 and NCI, NIH, Grant CA Y1-CO-1020), the Advanced Photon Source (supported by the United States Department of Energy), and Argonne National Laboratory. We thank Bruker Daltonics for providing the Maximum Deconvolution algorithm to prepare Fig. 2.
This work was supported, in whole or in part, by National Institutes of Health Grants GM77147 (to K. A. R. and G. L. C.) and DK42303 (to J. L. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S9.
The atomic coordinates and structure factors (codes 3QMV and 3QMW) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
K. A. Reynolds, unpublished results.
- PKS
- polyketide synthase
- NRPS
- non-ribosomal peptide synthetase
- Ppant
- phosphopantetheine
- ACP
- acyl carrier protein
- PCP
- peptidyl carrier protein
- TE
- TE I, and TE II, thioesterase, thioesterase type I, and thioesterase type II, respectively
- OAS
- α-oxoamine synthase
- UP
- 2-undecylpyrrole
- A domain
- adenylation domain
- KS
- ketosynthase
- IPTG
- isopropyl β-d-thiogalactopyranoside
- SeMet
- selenomethionyl
- TCEP
- tris(2-carboxyethyl)phosphine
- ESI
- electrospray ionization
- Vpp
- voltage peak to peak.
REFERENCES
- 1. Newman D. J., Cragg G. M. (2007) J. Nat. Prod. 70, 461–477 [DOI] [PubMed] [Google Scholar]
- 2. Tsao S. W., Rudd B. A., He X. G., Chang C. J., Floss H. G. (1985) J. Antibiot. 38, 128–131 [DOI] [PubMed] [Google Scholar]
- 3. Mo S., Sydor P. K., Corre C., Alhamadsheh M. M., Stanley A. E., Haynes S. W., Song L., Reynolds K. A., Challis G. L. (2008) Chem. Biol. 15, 137–148 [DOI] [PubMed] [Google Scholar]
- 4. Thomas M. G., Burkart M. D., Walsh C. T. (2002) Chem. Biol. 9, 171–184 [DOI] [PubMed] [Google Scholar]
- 5. Haynes S. W., Sydor P. K., Stanley A. E., Song L., Challis G. L. (2008) Chem. Commun. 16, 1865–1867 [DOI] [PubMed] [Google Scholar]
- 6. Mortellaro A., Songia S., Gnocchi P., Ferrari M., Fornasiero C., D'Alessio R., Isetta A., Colotta F., Golay J. (1999) J. Immunol. 162, 7102–7109 [PubMed] [Google Scholar]
- 7. Trudel S., Li Z. H., Rauw J., Tiedemann R. E., Wen X. Y., Stewart A. K. (2007) Blood 109, 5430–5438 [DOI] [PubMed] [Google Scholar]
- 8. Williamson N. R., Simonsen H. T., Ahmed R. A., Goldet G., Slater H., Woodley L., Leeper F. J., Salmond G. P. (2005) Mol. Microbiol. 56, 971–989 [DOI] [PubMed] [Google Scholar]
- 9. Stanley A. E., Walton L. J., Kourdi Zerikly M., Corre C., Challis G. L. (2006) Chem. Commun. 38, 3981–3983 [DOI] [PubMed] [Google Scholar]
- 10. Mo S., Kim B. S., Reynolds K. A. (2005) Chem. Biol. 12, 191–200 [DOI] [PubMed] [Google Scholar]
- 11. Cerdeño A. M., Bibb M. J., Challis G. L. (2001) Chem. Biol. 8, 817–829 [DOI] [PubMed] [Google Scholar]
- 12. Trivedi O. A., Arora P., Sridharan V., Tickoo R., Mohanty D., Gokhale R. S. (2004) Nature 428, 441–445 [DOI] [PubMed] [Google Scholar]
- 13. Claxton H. B., Akey D. L., Silver M. K., Admiraal S. J., Smith J. L. (2009) J. Biol. Chem. 284, 5021–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zelyas N., Tahlan K., Jensen S. E. (2009) Gene 443, 48–54 [DOI] [PubMed] [Google Scholar]
- 15. Yeh E., Kohli R. M., Bruner S. D., Walsh C. T. (2004) Chembiochem 5, 1290–1293 [DOI] [PubMed] [Google Scholar]
- 16. Kim B. S., Cropp T. A., Beck B. J., Sherman D. H., Reynolds K. A. (2002) J. Biol. Chem. 277, 48028–48034 [DOI] [PubMed] [Google Scholar]
- 17. Heathcote M. L., Staunton J., Leadlay P. F. (2001) Chem. Biol. 8, 207–220 [DOI] [PubMed] [Google Scholar]
- 18. Schwarzer D., Mootz H. D., Linne U., Marahiel M. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14083–14088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tsai S. C., Miercke L. J., Krucinski J., Gokhale R., Chen J. C., Foster P. G., Cane D. E., Khosla C., Stroud R. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14808–14813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsai S. C., Lu H., Cane D. E., Khosla C., Stroud R. M. (2002) Biochemistry 41, 12598–12606 [DOI] [PubMed] [Google Scholar]
- 21. Samel S. A., Wagner B., Marahiel M. A., Essen L. O. (2006) J. Mol. Biol. 359, 876–889 [DOI] [PubMed] [Google Scholar]
- 22. Frueh D. P., Arthanari H., Koglin A., Vosburg D. A., Bennett A. E., Walsh C. T., Wagner G. (2008) Nature 454, 903–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bruner S. D., Weber T., Kohli R. M., Schwarzer D., Marahiel M. A., Walsh C. T., Stubbs M. T. (2002) Structure 10, 301–310 [DOI] [PubMed] [Google Scholar]
- 24. Scaglione J. B., Akey D. L., Sullivan R., Kittendorf J. D., Rath C. M., Kim E. S., Smith J. L., Sherman D. H. (2010) Angew Chem. Int. Ed. Engl. 49, 5726–5730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koglin A., Löhr F., Bernhard F., Rogov V. V., Frueh D. P., Strieter E. R., Mofid M. R., Güntert P., Wagner G., Walsh C. T., Marahiel M. A., Dötsch V. (2008) Nature 454, 907–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Donnelly M. I., Zhou M., Millard C. S., Clancy S., Stols L., Eschenfeldt W. H., Collart F. R., Joachimiak A. (2006) Protein Expr. Purif. 47, 446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guerrero S. A., Hecht H. J., Hofmann B., Biebl H., Singh M. (2001) Appl. Microbiol. Biotechnol. 56, 718–723 [DOI] [PubMed] [Google Scholar]
- 28. Quadri L. E., Weinreb P. H., Lei M., Nakano M. M., Zuber P., Walsh C. T. (1998) Biochemistry 37, 1585–1595 [DOI] [PubMed] [Google Scholar]
- 29. Smirnova N., Reynolds K. A. (2001) J. Bacteriol. 183, 2335–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Otwinowski Z., Minor V. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 31. Terwilliger T. C. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang J. W., Chen J. R., Gu Y. X., Zheng C. D., Jiang F., Fan H. F., Terwilliger T. C., Hao Q. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1244–1253 [DOI] [PubMed] [Google Scholar]
- 33. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 36. Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 37. Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) Nucleic Acids Res. 32, W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edgar R. C. (2004) Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gust B., Kieser T., Chater K. F. (2002) REDIRECT Technology: PCR-targeting System in Streptomyces coelicolor, John Innes Centre Foundation, Norwich, UK [Google Scholar]
- 40. Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. (2000) Practical Streptomyces Genetics, 2nd Ed., pp. 43–60, The John Innes Centre Foundation, Norwich, UK [Google Scholar]
- 41. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., pp. 1.119–1.123, Cold Spring Harbor Laboratory Press, New York, NY [Google Scholar]
- 42. Lai J. R., Fischbach M. A., Liu D. R., Walsh C. T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5314–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhou Z., Lai J. R., Walsh C. T. (2006) Chem. Biol. 13, 869–879 [DOI] [PubMed] [Google Scholar]
- 44. Płoskoń E., Arthur C. J., Kanari A. L., Wattana-amorn P., Williams C., Crosby J., Simpson T. J., Willis C. L., Crump M. P. (2010) Chem. Biol. 17, 776–785 [DOI] [PubMed] [Google Scholar]
- 45. Qiu X., Janson C. A. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1545–1554 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.