Abstract
The extracellular region of the thyrotropin receptor (TSHR) can be subdivided into the leucine-rich repeat domain (LRRD) and the hinge region. Both the LRRD and the hinge region interact with thyrotropin (TSH) or autoantibodies. Structural data for the TSHR LRRD were previously determined by crystallization (amino acids Glu30–Thr257, 10 repeats), but the structure of the hinge region is still undefined. Of note, the amino acid sequence (Trp258–Tyr279) following the crystallized LRRD comprises a pattern typical for leucine-rich repeats with conserved hydrophobic side chains stabilizing the repeat fold. Moreover, functional data for amino acids between the LRRD and the transmembrane domain were fragmentary. We therefore investigated systematically these TSHR regions by mutagenesis to reveal insights into their functional contribution and potential structural features. We found that mutations of conserved hydrophobic residues between Thr257 and Tyr279 cause TSHR misfold, which supports a structural fold of this peptide, probably as an additional leucine-rich repeat. Furthermore, we identified several new mutations of hydrophilic amino acids in the entire hinge region leading to partial TSHR inactivation, indicating that these positions are important for intramolecular signal transduction. In summary, we provide new information regarding the structural features and functionalities of extracellular TSHR regions. Based on these insights and in context with previous results, we suggest an extracellular activation mechanism that supports an intramolecular agonistic unit as a central switch for activating effects at the extracellular region toward the serpentine domain.
Keywords: G-protein-coupled Receptors (GPCRs), Receptor Regulation, Receptor Structure-Function, Signal Transduction, Thyroid
Introduction
In accordance with other G-protein-coupled receptors, the general structural topology of the homologous glycoprotein hormone receptors (GPHRs)2 is characterized by an extracellular N-terminal region, seven transmembrane helices (TMHs), intracellular loops, and extracellular loops and is intracellularly terminated by a cytoplasmic tail. A common special structural feature of all GPHRs and leucine-rich repeat containing receptors 4–8 compared with other family A G-protein-coupled receptors (rhodopsin-like) is the large N-terminal extracellular region, which is responsible for hormone binding and signal transduction (reviewed in Ref. 1). This N-terminal extracellular region can be subdivided into: the extreme N-terminal tail with the signal peptide, cysteine box 1, containing the first group of four interacting cysteines, which are part of the leucine-rich repeat domain (LRRD), and the hinge region.
For GPHRs it has been demonstrated that the major binding region for the glycosylated hormones TSH, LH, CG, and FSH is located in the extracellular LRRD (2, 3) (reviewed in Ref. 4). Repeats are 20–30 amino acids long and contain a conserved 11 residue segment with the consensus sequence LXXLXLXX(N/C)XL (where X represents Leu, Val, Ile, or Phe), comprising a β-strand. Previously, the FSHR LRRD crystal structure in complex with the hormone FSH (5) (PDB entry 1XWD) and the complex between the TSHR LRRD and an activating autoantibody (6) were solved (PDB entry 3G04). The general spatial orientation of the hormone to the LRRD is described by the FSHR LRRD/FSH x-ray structure. FSH interfaces with particular β-strands of the concave inner surface of the LRRD (7–9). The LRRD crystal structures comprise nine complete repeats, constituted by parallel β-strands and loop-backbone structures, whereby the C-terminal site terminates with a 10th β-strand and, therefore, with an incomplete repeat loop around amino acid position Leu260 (TSHR). Because a large extracellular part is missing in the x-ray structures, it is not unlikely that the C terminus of the LRRD might not be captured exactly in length and structure by the crystallized isolated protein fragments. This is supported by three facts. First, the three-dimensional superimposition of the two different FSHR LRRD monomers occurring in the crystal structure shows that the C-terminal ends are different in length (FSHR: monomer 1 to Tyr250 and monomer 2 to Leu269) and folding. Second, the extracellular region following directly the crystallized part does not participate in hormone binding and might therefore be “unstructured” in the isolated FSHR fragment (9). Third, the deletion of the corresponding sequence fragment in LHCGR prior Ser277 (Ser281 in TSHR) resulted in an identical phenotype like the deletion of each single LRRD repeat in the LHCGR (10).
The hinge region, located between the LRRD and the transmembrane serpentine domain, is the most diverse amino acid sequence region between the three homologous GPHR subtypes TSHR, FSHR (follitropin receptors), and LHCGR (lutropin receptor) (11). However, common features of the hinge region consist of two N- and C-terminally located cysteine boxes (cysteine box 2 (C-b2) and cysteine box 3 (C-b3)), which are connected via the cysteine box 2/3 linker region. The hinge region of the TSHR has also been termed the cleavage domain (12), C-terminal cysteine-rich region (13), signaling and specificity domain (14), or cysteine-rich C-flanking region (15). The cysteines of C-b2 are linked to cysteines of C-b3 close to TMH1 by disulfide bonds (16). Indirect evidence permits the global assignment of disulfide bridges between the six cysteines (17–20). Especially two recent publications on TSHR (21) and LHCGR (13) suggest disulfide bridges between Cys283 (C-b2)/Cys398 (C-b3) and Cys284 (C-b2)/Cys408 (C-b3), whereas Cys301 (in C-b2) is likely to be paired with Cys390 (in C-b3). Directly related to the structure of the hinge region are important functional phenomena concerning the mechanism of initial GPHR activation, including hormone binding as well as signal transduction and amplification (13, 16, 22–25). Recently, evidence was provided that the TSH receptor transmembrane domain influences TSH binding kinetics possibly by altering the conformation of the closely associated hinge region that contributes to the TSH binding site (26). This study supports functional-structural interrelations between different regions of the TSHR. In this respect, it has been shown several times for the TSHR that the hinge region is necessary for the stabilization of a signaling-competent basal receptor conformation (17, 25, 27–31) and that interaction with the hormone leads to receptor activation, most probably by release of an intramolecular agonist (10, 32, 33).
But important questions are still only partially answered. 1) Which particular amino acids are of importance for correct TSHR function besides the already known residues? 2) Which amino acids constitute the transition between the LRRD and the hinge region, and how are they assigned structurally (Fig. 1)? Interestingly, the sequence directly following the commonly crystallized LRRD fragments comprises a typical sequence pattern of leucine-rich repeats, suggesting the possibility of a full 10th repeat and an 11th β-strand on the concave surface (24). Therefore, in this study, we investigated systematically conserved residues between positions Arg255 and Asp395 by mutagenesis. We focused especially on side chain variations of hydrophobic amino acids in the Arg255–Tyr279 region for identification of positions that are potentially mandatory for a repeat folding or other secondary structures. We also characterized conserved hydrophilic amino acids in the remaining part of the hinge region between positions Pro280 and Asp395 to identify residues of functional importance. Partially inactivating side-chain substitutions were combined with the constitutively activating mutation (CAM) S281Q to receive information concerning the spatial localization of Ser281 and the hinge region relative to the serpentine domain.
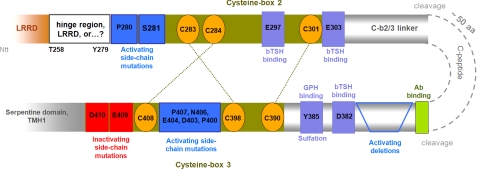
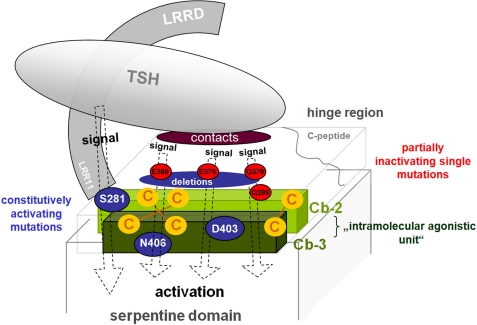
FIGURE 1.
Scheme of the TSHR hinge region with highlighted functionalities at specific structural parts. C-b2 consists of 31 amino acids. C-b2 follows the LRRD domain back-to-back. The C-terminal end of the LRRD is not definitively characterized in length by diverse terminal ends of available FSHR and TSHR LRRD crystal structures. Pathogenic constitutively activating mutations at Ser281 (blue box) underline the importance of this region for receptor function. Cys283 and Cys284 are essential for receptor protein folding, and constitutive activation by mutation at these cysteines is reported. They are disulfide-bridged to the cysteines in C-b3, probably as shown by the dotted lines. Recently, the negatively charged amino acid Glu297 (lilac box) within the C-b2 was identified as being involved in complementary charge-charge recognition to bTSH. The cysteine box 2/3 linker region (C-b2/3) comprises 88 amino acids and is N-terminally and C-terminally bordered by C-b2 and C-b3. Two cleavage sites define the so-called cleavable peptide (C-peptide, ∼50 amino acids). Antibody binding epitopes are known at the transition between the C-peptide and the C-terminal C-b2/3 linker. Sulfation at amino acid 385 is involved in the common hormone binding of GPHRs. It was found that N-terminally and C-terminally located negatively charged amino acids (Glu303 and Asp382; lilac box) participate in bTSH binding. The cysteine box 3 follows the cysteine box 2/3 linker region back-to-back and close to TMH1. The three cysteines are most likely disulfide-bridged to cysteines in cysteine box 2 and are essential for receptor fold. The N-terminal part of C-b2 and the C-terminal region of C-b3 are important for extracellular signaling processes and regulation of activity states as indicated by CAMs (blue boxes).
EXPERIMENTAL PROCEDURES
Generation of Mutants
TSHR mutants were constructed by site-directed mutagenesis using the human TSHR-pSVL (mutations at positions Gln289 to Asp395) or -pcDNA3.1(−)/hygro (mutations at positions Arg255–His282) as template as described previously (25). Mutated TSHR sequences were verified by dideoxy sequencing with dRhodamine terminator cycle Sequencing chemistry (ABI Advanced Biotechnologies, Inc., Columbia, MD).
Cell Culture and Transfection of Mutant TSHRs
COS-7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) at 37 °C in a humidified 5% CO2 incubator. Cells were transiently transfected using the GeneJammer® transfection reagent (Stratagene), whereas a transfection efficiency of 50–60% of viable cells could be achieved for each experiment. For FACS analysis and determination of inositol phosphates, 12-well plates (1 × 105 cells/well) were transfected with 1 μg of DNA/well. 24-well plates (0.5 × 105 cells/well) were transfected with 500 ng of DNA/well for measurement of intracellular cAMP accumulation and 48-well plates (0.25 × 105 cells/well) with 0.25 μg of DNA/well for TSH binding analysis. Transfection of the human TSHR wild type and the vector alone (mock transfection) was performed in each assay as control. Each binding or cAMP experiment was done simultaneously with a FACS control from the same transfection.
FACS Analyses
Determination of cell surface expression and transfection efficiency was done by FACS analysis as described previously (25), using the mouse anti-human TSHR monoclonal antibody 2C11 (MAK1281, Linaris, Wertheim-Bettingen; 0.25 μg/sample) as primary antibody and the fluorescein-conjugated F(ab′)2 rabbit anti-mouse IgG (Serotec; 0.5 μg/sample). Receptor expression was determined by the mean fluorescence intensity using the FACScan (BD Biosciences). The wild type TSHR was set at 100%, and receptor expression of the mutants was calculated according to this. The percentage of signal-positive cells corresponds to transfection efficiency.
cAMP Accumulation Assay
48 h after transfection, cells were incubated in the absence or presence of 100 milliunits/ml bTSH (Sigma) in serum-free medium supplemented with 1 mm 3-isobutyl-1-methylxanthine (Sigma) for 1 h at 37 °C in a humidified 5% CO2 incubator. Reactions were terminated by aspiration of the medium. Cells were washed once with ice-cold PBS and then lysed by incubation with 0.1 n HCl for 30 min. Supernatants were collected and dried at 54 °C. cAMP contents of the cell extracts were determined using the cAMP AlphaScreenTM assay (PerkinElmer Life Sciences) according to the manufacturer's instructions.
Activation of Inositol Phosphate (IP) Formation
Transfected COS-7 cells were incubated with 1 μCi/well myo-[3H]inositol (PerkinElmer Life Sciences) for 6 h. Thereafter, cells were incubated with serum-free DMEM containing 10 mm LiCl and 100 milliunits/ml TSH for stimulation of transfected cells. Evaluation of basal and TSH-induced increases in intracellular IP levels was performed by anion exchange chromatography as described previously (34). IP values were expressed as the percentage of radioactivity incorporated from [3H]IP-1 to -3 over the sum of radioactivity incorporated in IPs and phosphatidylinositol.
Expression Studies for the TSHR Hinge Region
Details are provided in the supplemental material.
Molecular Homology Modeling and Bioinformatics
The crystallized TSHR LRRD (Protein Data Bank code 3G04 (6)) was analyzed to find a repeat and β-strand with high similarity to the amino acid sequence 255RNTWTLKKLPLSLSFLHLTRADLSY279, which is not included in the GPHR LRRD crystal structure. A potential additional β-strand is constituted by amino acids 273TRADLSY279, whereby alanine 275 and leucine 277 are predicted to be directed inside the domain according to the repeat fold (35–37). The template sequence comprising amino acids Ser234–Asn256 of the crystal structure was found as best matching in sequence identity and we used this structural fragment as a template to complete the crystallized 10th repeat and to add an additional 11th β-strand model.
Side chains of this partial homology model were subjected to conjugate gradient minimizations (until converging at a termination gradient of 0.05 kcal/(mol·Å)) and molecular dynamics simulation (3 ns) by fixing the backbone atoms. Finally, the leucine-rich repeat model was minimized without backbone atom constraints. The Sybyl version 7.3.5 (Tripos Inc., St. Louis, MO) was used for modeling procedures. The quality and stability of the model were validated by checking the geometry using PROCHECK (38). Structure images were produced using the PyMOL Molecular Graphics System, version 1.3, Schrödinger, LLC.
RESULTS
Site-directed Mutagenesis of Amino Acids between Arg255 and Leu277 of the hTSHR Revealed Fold-sensitive Hydrophobic Residues
The crystallized TSHR LRRD (6) is composed of amino acids Glu30–Thr257. Little is known concerning the functional and structural contribution of amino acids between Thr257 and cysteine box 2 (defined by Cys283, Cys284, and Cys301) (39–41). Participation in hormone binding is most probably excluded based on the crystal structure of the FSHR LRRD-FSH complex (4, 5). We therefore mutated conserved hydrophilic and hydrophobic amino acids in this region to evaluate their potential involvement in receptor functions. It is noteworthy that specific hydrophobic residues of this particular region, Leu260, Leu263, Leu272, Ala275, and Leu277, are predicted to point inside the LRRD core and to stabilize the repeat-like fold by hydrophobic interactions. Especially Ala275 and Leu277 are suggested to be constituents of a β-strand conformation. Therefore, it was speculated that these residues might have an impact on receptor fold.
All positions were substituted by alanine and/or valine (Table 1). Wild type Ala275 was mutated to leucine to modify length and bulkiness of side chains. Mutations showing decreased cell surface expression like L260A or L277A have further been varied in side chains to investigate the sensitivity and tolerance for substitutions at these specific positions (Table 1). Moreover, Ala275 was substituted with glutamic acid to evaluate the influence of hydrophilic properties.
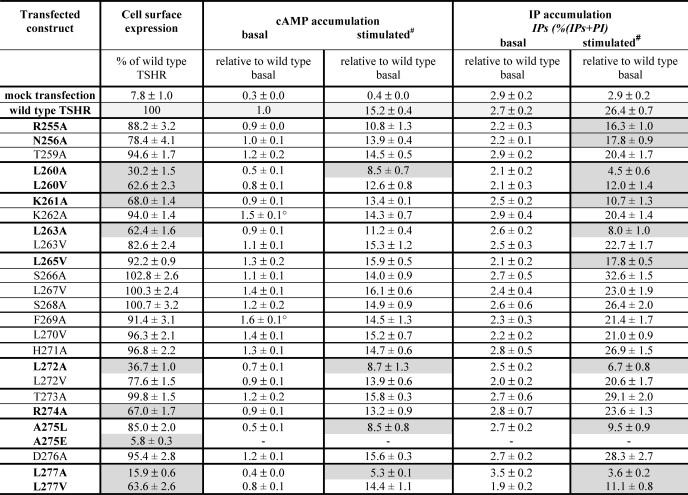
TABLE 1.
Functional characterization of TSHR mutants in the region between the crystallized leucine-rich repeat domain and cysteine box 2
The mutated TSH receptors were transiently expressed in COS-7 cells. Characterization of mutants was performed by determination of cell surface expression, basal and maximal cAMP, and IP accumulation (see “Experimental Procedures“). The discussed functional data are highlighted in gray, and particular amino acids are in boldface type. The empty vector was used as a mock-transfected control. Data are given as mean ± S.E. of at least three independent experiments, each carried out in duplicate.
# Maximal cAMP; 100 milliunits of bTSH.
o Basal cAMP accumulation slightly increased compared with wild type.
Cell Surface Expression
The majority of the mutants revealed a cell surface expression in the range of 78–103% of wild type (Table 1). Mutants K261A (68%) and R274A (67%) showed a decreased cell surface expression compared with wild type. Alanine mutations and even slight side chain alterations like the introduction of valine at the four hydrophobic residues (L260A, L260V (30%, 63%); L263A (62%); L272A (37%); and L277A, L277V (16%, 64%)) showed a decreased cell surface expression. A275E is expressed at the cell surface with only 6%.
bTSH Binding
Mutated TSHRs showed a bTSH maximum binding similar to wild type or in a range comparable with the cell surface expression level measured by FACS (data not shown). This is in accordance with previous experiments in which few of the positions we mutated and tested here were also investigated regarding ligand binding (e.g. Arg274 and Asp276 (40, 41)).
cAMP Accumulation
The basal activity for cAMP accumulation was similar to wild type for all constructs with the exception of mutants K262A and F269A with a slightly increased ligand-independent activity of 1.5- and 1.6-fold over wild type, respectively (Table 1). The maximum cAMP accumulation after stimulation with bTSH revealed a decrease for the mutants with an impaired cell surface expression: L260A (56%), L272A (57%), and L277A (35%) (Table 1).
IP Accumulation
Mutations L260A, L272A, and L277A with a cell surface expression level between 15 and 37% showed an impairment of the TSH-induced maximal IP signal. Mutations with a decreased cell surface expression level of around 60% of wild type (L260V, K261A, L263A, and L277V) are characterized also by a decreased maximum of IP accumulation of around 40% of wild type TSHR (Table 1). R255A, N256A, L265V, and A275L showed a decreased IP accumulation also, whereas the cell surface expression was only slightly decreased or comparable with wild type.
Taken together, most of the mutants characterized by a cell surface expression comparable with wild type are also similar to the wild type in their signaling properties (Table 1). Interestingly, a decrease in TSH-induced IP signaling but only a mild effect on cAMP production was observed for the well expressed mutants R255A and N256A. Substitutions to a small and hydrophobic side chain of alanine might interrupt hydrophilic interactions between Arg255 and Asn256 to so far unknown interaction partners. Their selective involvement in the activation of the Gq/11-IP pathway remains unclear, but these data together with the partial inactivation of IP accumulation by the L265V mutant reflect the principle involvement of this region in signaling regulation at the TSHR. For the signaling-sensitive A275L mutant that diminishes maximum production of cAMP and IP, it can be speculated that the substitution from a small hydrophobic to a large, branched, and bulky hydrophobic side chain might cause spatial effects on interior interactions with neighboring and interacting hydrophobic amino acids.
Structure of the TSHR Leucine-rich Repeat Domain Combined with a Structural Model of a Completed Repeat 10th and 11th β-Strand
The TSHR LRRD crystal structure is composed of nine complete repeats, constituted by parallel β-strands and loop-backbone structures (6). The N-terminal site of the LRRD starts with an antiparallel β-strand and terminates with a 10th β-strand, therefore with an incomplete leucine-rich repeat (Fig. 2A). An appended homologous loop-backbone model of a potential 10th LRR and an 11th β-strand reflects a possible interpretation of the fact that the amino acid sequence following directly the crystallized LRRD is characterized by high conservation among the GPHRs (Fig. 3) and shows a typical LRR sequence pattern profile. Our experimental data showed for mutants of the conserved hydrophobic amino acids Leu260, Leu263, Leu272, Ala275, and Leu277 cell surface expression deficiency. This is in agreement with the hypothesized additional repeat (Fig. 2B) because exactly these specific hydrophobic residues stabilize the fold by tight van der Waals side chain interaction with neighboring hydrophobic residues in the interior core of this domain.
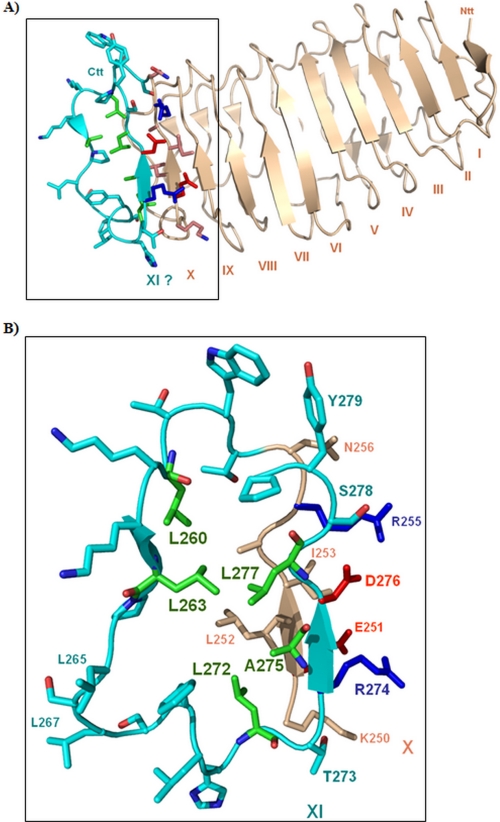
FIGURE 2.
Structure of the TSHR LRRD combined with a structural model of a completed repeat 10 and an 11th β-strand. A, the TSHR LRRD crystal structure (light orange) is composed of nine complete repeats, constituted by parallel β-strands and loop-backbone structures (6). The N-terminal site of the LRRD starts with an antiparallel β-strand (I) and terminates with a 10th β-strand (X), therefore with an incomplete leucine-rich repeat. B, an appended homologous loop-backbone structure model of a potential 10th LRR (cyan) and an 11th β-strand (XI), which is not observable in the crystal structure reflects a possible interpretation of the fact that the amino acid sequence following directly the crystallized LRRD is characterized by high conservation among the GPHRs. It is also conserved in length (no gaps) and shows a typical LRR sequence pattern profile, which is in accordance with a potential 11th β-strand. Our experimental data showed for the conserved hydrophobic amino acids Leu260, Leu263, Leu272, Ala275, and Leu277 cell surface expression-deficient mutants (green). The hydrophobic residues (green) identified in this study by cell surface-increased mutants stabilize the fold of a repeat by tight van der Waals side chain interaction to neighboring hydrophobic residues in the interior core of this domain.
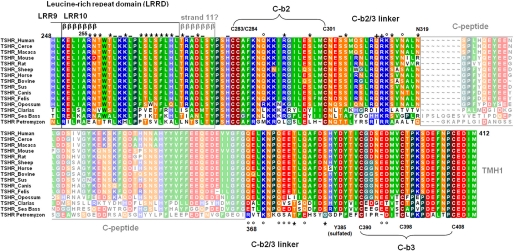
FIGURE 3.
Partial alignment of the extracellular TSHR region with amino acid conservation, annotated structural implications, and assigned functional insights from the current study. This alignment shows parts of the extracellular region of several TSHR subspecies from the C-terminal end of the LRRD to the beginning of TMH1 (amino acid Met412). Colors reflect similarities (Blossum 62 matrix) and specific biophysical properties: green, hydrophobic; brown, cysteines; red, negatively charged; blue, positively charged; orange, hydrophilic uncharged; black, prolines; light blue, histidines. Subregions of the hinge region like the cysteine boxes, the potential C-peptide (cleavable peptide), or β-strand regions of the LRRD (βββββββ) are annotated. The residues investigated in this study are marked with symbols according to their functional properties: *, comparable with wild type; °, decreased maximum cAMP production; +, hyperstimulation induced by bTSH; −, decreased cell surface expression.
Systematic Single Side-chain Variations at Conserved Amino Acids between His282 and Asp395 Revealed Hydrophilic Signaling-sensitive Residues in the Hinge Region of the hTSHR
It has been shown previously on chimeric GPHRs (13, 16, 42, 43), deletion constructs (13, 28, 32, 44, 45), single side-chain substitutions (22, 24, 25, 32, 33, 46–49), or double mutations (50, 51) that the hinge region of the GPHRs is of enormous importance for hormone binding or antibody interaction and signal transmission (reviewed in Refs. 1, 52, and 53). But specific determinants or mechanisms involved in these processes are only partially known. Therefore, we investigated systematically single-side chain substitutions of conserved hydrophilic amino acids in the TSHR hinge region (Table 2) with the exception of residues located in the C-peptide.
TABLE 2.
Functional characterization of mutations at hydrophilic residues in the TSHR hinge region
After transient transfection, mutated TSH receptors were characterized regarding cell surface expression and cAMP signaling as described under “Experimental Procedures.” The discussed functional data are highlighted in boldface type. The empty vector was used as mock-transfected control. Data are given as mean ± S.E. of at least three independent experiments, each carried out in duplicate.
| Transfected construct | Cell surface expression (percentage of wild type TSHR) | cAMP accumulation, relative to wild type basal |
|
|---|---|---|---|
| Basal | Stimulated | ||
| % | |||
| Mock transfection | 9.3 ± 0.1 | 0.6 ± 0.2 | 0.4 ± 0.2 |
| Wild type TSHR | 100 | 1 | 13.7 ± 0.7 |
| H282A | 38.6 ± 1.7 | 1.1 ± 0.1 | 9.3 ± 0.7 |
| H282F | 79.8 ± 1.6 | 0.6 ± 0.1 | 7.5 ± 0.3 |
| Q289A | 78.3 ± 0.4 | 1.0 ± 0.1 | 8.6 ± 0.5 |
| S298A | 94.3 ± 1.9 | 1.3 ± 0.1 | 10.8 ± 0.9 |
| S304A | 67.5 ± 0.9 | 1.3 ± 0.1 | 13.4 ± 0.4 |
| S305A | 104.8 ± 3.1 | 1.2 ± 0.1 | 10.8 ± 0.6 |
| Q311A | 91.2 ± 0.9 | 1.4 ± 0.1 | 19.8 ± 1.2 |
| R312A | 94.5 ± 3.2 | 1.6 ± 0.1 | 18.6 ± 2.3 |
| K313A | 80.8 ± 2.0 | 0.9 ± 0.1 | 8.4 ± 1.1 |
| S314A | 93.2 ± 3.0 | 1.0 ± 0.1 | 10.3 ± 0.7 |
| N316A | 86.5 ± 2.6 | 0.9 ± 0.1 | 8.2 ± 2.0 |
| N319A | 79.1 ± 0.3 | 1.0 ± 0.1 | 10.7 ± 1.8 |
| Q368A | 72.1 ± 2.9 | 0.8 ± 0.2 | 8.3 ± 1.9 |
| E369A | 64.8 ± 1.8 | 0.8 ± 0.3 | 8.7 ± 1.0 |
| K371A | 94.9 ± 2.7 | 1.1 ± 0.1 | 8.9 ± 1.0 |
| Q374A | 79.5 ± 3.1 | 1.0 ± 0.0 | 7.7 ± 0.6 |
| E375A | 83.6 ± 2.8 | 0.7 ± 0.1 | 6.5 ± 0.4 |
| E376A | 85.7 ± 2.7 | 0.9 ± 0.0 | 7.2 ± 0.3 |
| T377A | 94.3 ± 2.1 | 1.3 ± 0.2 | 12.0 ± 0.6 |
| Q379A | 86.3 ± 2.5 | 0.9 ± 0.1 | 7.4 ± 0.8 |
| S383A | 95.8 ± 3.5 | 1.1 ± 0.1 | 11.5 ± 0.8 |
| E394A | 92.8 ± 2.8 | 1.0 ± 0.1 | 6.6 ± 0.5 |
| D395A | 85.4 ± 2.6 | 1.0 ± 0.1 | 5.7 ± 1.1 |
Cell Surface Expression
Most of the mutations revealed a cell surface expression comparable with wild type in the range of 72–100% (Table 2), except substitutions H282A (39%), S304A (68%), and E369A (65%).
bTSH Binding
All mutated TSHRs were similar to wild type in their capability for bTSH binding, or they were in a range comparable with the cell surface expression level (data not shown). For several of the mutants (R312A, K313A, E369A, K371A, E375A, E376A, E394A, and D395A), hormone binding data were previously published (25).
cAMP Accumulation
The mutations showed a basal cAMP accumulation comparable with wild type TSHR. Measurement of maximal cAMP accumulation after stimulation with bTSH revealed for 14 of 23 mutations a decrease of maximum signaling compared with wild type TSHR in a range between 42 and 67% of the wild type (Table 2). Interestingly, Q311A and R312A showed a slightly increased capability for TSH-induced maximum of cAMP stimulation with 135 and 145% of wild type, respectively.
In summary, several mutants of conserved hydrophilic amino acids in the hinge region decrease the signaling capability of the TSHR (Table 2). It is of note that most of them are located upstream of the C-peptide region between Gln368 and Asp395. Interestingly, none of the single side-chain substitutions constitutively activates the receptor.
Multiple Substitutions Combining Partially Inactivating Mutations in the Hinge Region Do Not Amplify the Inactivating Effect of Single Mutations
We were interested in effects on cAMP production induced by combinations of partially inactivating mutations and hypothesized that double or triple mutations in the hinge region might potentially amplify TSHR inactivation and might lead to complete signaling inhibition. The double mutant Q289A/K313A (N-terminal part of the hinge region) and the triple mutant Q374A/E375A/E376A (C-terminal part of the hinge region) are comparable with the single mutations in their cAMP signaling properties and cell surface expression levels (Table 3). In summary, these double and triple mutations that combine partially inactive single mutants do not amplify inhibition of cAMP signaling.
TABLE 3.
Cell surface expression and cAMP signaling of partially inactivating single TSHR mutations combined to multiple mutants
The mutated TSH receptors were transiently expressed in COS-7 cells and functionally characterized as described under “Experimental Procedures.” The empty vector was used as mock-transfected control. Data are given as mean ± S.E. of at least three independent experiments, each carried out in duplicate.
| Transfected construct | Cell surface expression (percentage of wild type TSHR) | cAMP accumulation, relative to wild type basal |
|
|---|---|---|---|
| Basal | Stimulated | ||
| % | |||
| Wild type TSHR | 100 | 1 | 13.4 ± 0.7 |
| Mock transfection | 9.3 ± 0.1 | 0.6 ± 0.2 | 0.4 ± 0.2 |
| Q289A | 78.3 ± 0.4 | 1.0 ± 0.1 | 8.6 ± 0.5 |
| K313A | 80.8 ± 2.0 | 0.9 ± 0.2 | 8.4 ± 1.1 |
| Q289A/K313A | 80. 0 ± 3.2 | 0.8 ± 0.2 | 10.3 ± 0.6 |
| Q374A | 79.5 ± 3.1 | 1.0 ± 0.2 | 7.7 ± 0.6 |
| E375A | 83.6 ± 2.8 | 0.7 ± 0.1 | 6.5 ± 0.4 |
| E376A | 85.7 ± 2.7 | 0.9 ± 0.0 | 7.2 ± 0.3 |
| Q374A/E375A/E376A | 86.4 ± 5.2 | 1.4 ± 0.1 | 10.9 ± 0.2 |
A Partially Inactivating Mutation at Cysteine Box 3 of the Hinge Region Suppresses Constitutive Activation Induced by S281Q at Cysteine Box 2
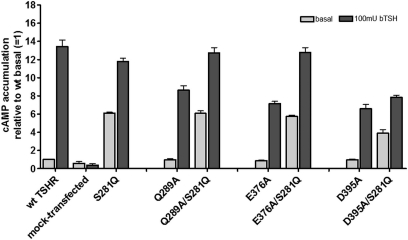
We combined partially inactivating mutations located in different parts of the hinge region with the CAM S281Q in double mutants to evaluate dominant effects on signaling induced either by the CAM or by the inactivating mutations (Fig. 4). In the double mutants Q289A/S281Q (N-terminal hinge region) and E376A/S281Q (central hinge region), the constitutive activation of S281Q was not suppressed, and the maximum cAMP signal induced by bTSH was comparable with wild type. This led us to propose that the signaling properties of mutant S281Q are dominant in these two mutant combinations. However, in the combination of S281Q with D395A (C-terminal hinge region, C-b3), the constitutive activity caused by S281Q as well as the bTSH-induced cAMP response were suppressed.
FIGURE 4.
cAMP signaling of the constitutively activating mutation S281Q in combination with partially inactivating mutations as double mutants. Determination of basal and stimulated (100 milliunits of bTSH) cAMP values after transient transfection of COS-7 cells was performed as described under “Experimental Procedures.” The figure shows the summary of three independent experiments, each carried out in duplicate. The mutation S281Q is known as a naturally occurring CAM. In this mutant, the basal TSHR activity is increased about 500% compared with wild type, whereby the capability for TSH-induced maximum of signaling is not modified. In contrast, we identified several partially inactivating mutations for TSH-induced signaling in the hinge region and combined a few of them with CAM S281Q in double mutants. Q289A is located at the N terminus, and Glu376 is located more C-terminally at the hinge region. Mutant D395A is located in proximity to cysteine box 3 close to TMH1. Double mutations of S281Q with Q289A and E375A as well show signaling properties comparable with the single S281Q mutant, whereas D395A blocks constitutive and TSH-induced signaling in combination with S281Q. Error bars, S.E.
Expression Studies for TSHR Hinge Region Constructs That Are Different in Length
We also approached the question of whether the isolated hinge region is a self-folding (stable) domain or an unfolded region. We performed extensive expression studies of extracellular TSHR fragments. By this we also aimed to obtain hints concerning the potential definition in length of the hinge region. The expression studies were performed in different expression systems, and the constructs varied in length or constitution. None of these hinge region constructs could be observed as a self-folding domain. Details and results of these approaches are provided in the supplemental material.
DISCUSSION
Hydrophobic Amino Acids at the Junction between the Leucine-rich Repeat Domain and the Hinge Region Are Important for the Structure of the TSHR
Pathogenic mutations in the hinge region that constitutively activate the human TSHR were reported for amino acid Ser281 (46, 54). The conserved residue Ser281 in the TSHR (LHCGR Ser277, FSHR Ser273) is located at the N terminus of C-b2 (Fig. 1). Mutants at Ser281 have indicated an involvement of this region in regulation of signaling activity; therefore, they were a starting point for more detailed investigation of this extracellular region. Previous in vitro studies of the TSHR and the LHCGR have shown that several mutations of this serine lead to constitutive activation (e.g. S281N and S281T), but few mutations also cause receptor inactivation (e.g. S281D and S281R) (55, 47, 56). Substitution of Pro276 in the LHCGR (in TSHR, Pro280) adjacent to Ser277 (in TSHR, Ser281) with a glycine leads also to constitutive receptor activation (33). The cysteines of C-b2 (Cys283 and Cys284) close to Ser281 in TSHR are linked to cysteines of C-b3 close to TMH1 by disulfide bonds (reviewed in Ref. 16) (Fig. 1). Mutation of residues Cys283 and Cys284 to serine leads to a 2-fold increase of basal TSHR signaling activity (55). Finally, it was hypothesized that residue Ser281 is spatially close to the extracellular loop 1, where also activating and inactivating mutations are found by mutagenesis or as naturally occurring mutants (47). Furthermore, it has been suggested that this region around Ser281 is attached directly to the LRRD and therefore should function as an interface between the LRRD and the transmembrane serpentine domain (24). This scenario would imply that amino acids between the LRRD and C-b2 (peptide 260–278) are structurally defined and assigned either to the LRRD or the hinge region. This sequence is conserved in amino acid similarity and length between the GPHRs (also no gaps) and shows a sequence pattern of a typical LRRD profile, which would be in accordance with a potential 11th β-strand (Figs. 2 and 3). There are only fragmental hints available concerning the function of amino acids between positions 261 and 278 (40, 41).
Here, we investigated this region in more detail by mutagenesis, functional characterization of mutants, and homology modeling (Table 1 and Fig. 2). Most of the mutants characterized by a cell surface expression comparable with wild type are also similar to the wild type in their signaling properties. However, the introduction of the hydrophilic and negatively charged glutamate instead of Ala275 abolishes receptor cell surface expression, probably by a strong effect on the receptor fold. Moreover, side-chain substitutions at highly conserved hydrophobic amino acids Leu260, Leu263, Leu272, and Leu277 impair receptor cell surface expression and subsequently receptor signaling (Table 1). The high conservation of these amino acids in contrast to other hydrophobic residues in this region, like Leu265, Leu267, or Leu270, indicates their evolutionary importance for the TSHR. Screening of available mutagenesis data for the LRRDs of the GPHRs (see the SSFA-GPHR database on the World Wide Web) (57, 58) revealed that mutations at specific hydrophobic key positions of the typical LRR profile always lead to an impaired receptor expression level. The hydrophobic positions that were identified here as fold-sensitive match exactly this repeat pattern of hydrophobic residues by pointing into the interior of the LRR domain model (Fig. 2B) and should stabilize the fold by interaction with neighboring hydrophobic residues (hydrophobic LRRD core) of the prior repeat. In contrast, mutants of non-conserved hydrophobic residues in this region, such as Leu265 and Leu267, did not affect cell surface expression. Therefore, a repeat fold including an additional β-strand might be an option for this particular sequence region between Arg255 and Tyr279 (Fig. 2). The investigated amino acids are not involved in hormone interactions, which is not necessarily a prerequisite of repeats because it has been observed for many LRRDs of other proteins (35). However, although the functional data match the model, a definitive answer for the existing structure of the sequence between the crystallized LRRDs and C-b2 does not yet exist. Based on our data, we conclude that this particular sequence region is folded rather than that it constitutes an unstructured region.
The TSHR Hinge Region Is Not an Autonomously Self-folding Domain
The demonstrated functional importance of the hinge region (reviewed in Ref. 1) and the occurrence of disulfide-bridged cysteines (13, 16, 21) has suggested that a defined fold might be necessary for hormone binding and signal transduction. Crystallization studies with isolated entire extracellular regions of the FSHR and TSHR have not resulted in the elucidation of the structure of the hinge region (5, 6). Therefore, we asked the question of whether the hinge region is an autonomously self-folding domain. We performed extensive expression studies in different systems with constructs that varied in length or in constitution to reveal insights in the folding properties of different extracellular TSHR fragments (for details, see the supplemental material).
We were not able to produce stable hinge region constructs in a soluble form, neither by significant construct extensions at the N-terminal (LRRD fragments) or C-terminal (TMH1) end nor by expression in Escherichia coli or in Pichia pastoris. At least in our hands, the hinge region in its isolated form is not correctly folded, and therefore, it is most likely not a structural domain by itself. On one hand, the majority of the hinge region might be flexible because the cleavage process indicates that a specific part of the hinge region must be accessible for proteases. On the other hand, it is likely that the hinge region bordered in the TSHR by the LRR domain and the serpentine domain needs additional parameters like tight intra- or intermolecular contact partners to achieve the correct fold. This hypothesis would be in agreement with the idea of a sandwich-like arrangement of the three main TSHR components: LRR domain, hinge region, and serpentine domain. Alternatively, an oligomeric packing of monomers might be important regarding the fold of the hinge region.
Signaling-sensitive Amino Acids in the Hinge Region Constitute Determinants of an Intramolecular Signal Transmitter
The cysteine boxes 2 and 3 are connected by a linker region that is most diverse between the homologous GPHR subtypes (11). Between the Ser281–Cys284 region at the N terminus of the hinge region and the C terminus, C-b3-specific amino acids and peptides are known to be sensitive for hormone binding. For Tyr385 (27) in the TSHR and tyrosines in the LHCGR and the FSHR in corresponding sequence motifs, posttranslational sulfation (increased negative charge) was shown to be required for hormone binding and signal induction (22, 23). Other recently identified negatively charged residues (Glu297, Glu303, and Asp382) located in the hinge region of the TSHR refined an extended hormone binding site in the hinge region of the TSHR (Fig. 1), especially for bTSH or superagonistic derivatives compared with human TSH (25, 48). The importance of the C-b2/3 linker in the maintenance of the signaling competence of the receptor was revealed by several studies (10, 31, 32, 45, 51, 59, 60). Stepwise deletions of fragments in the hinge region have shown that particularly the absence of sequence region Lys371–His384 causes the highest constitutive activation of the TSHR (28).
Here, we investigated conserved hydrophilic amino acids in the hinge region by single-side chain substitutions that have not been investigated so far to obtain detailed information regarding participation of particular determinants for signal transduction. By this and in contrast to our findings for the region Arg255–Ser278 (see above), several conserved hydrophilic amino acids decreased the signaling capability of the TSHR by mutation (Table 2 and Fig. 3). It is of note that most of them were located upstream of the C-peptide region between Gln368 and Asp395. This observation is in general agreement with studies from Mizoturi et al. (28), where the region between Lys371 and His384 was identified as most important for TSHR function. Deletions at this region lead to the strongest constitutive TSHR activation. Here, we show that single side-chain substitutions in this region are not able to constitutively activate the receptor; in contrast, they prevent partially TSH-induced signal transduction. This finding is in accordance with a recent study by the group of Rapoport and co-workers (61), which suggests that especially downstream hinge residues 377–384 contribute to the interdependence between affinity of ligand binding and cAMP signal transduction.
A Refined Scenario of Thyrotropin Receptor Activation at the Extracellular Region
Taking our data and previously published findings into account, we suggest the extended mechanism for extracellular TSHR activation and regulation shown in Fig. 5).
FIGURE 5.
Scheme of the “intramolecular signal transmitter” at the thyrotropin receptor. Cysteine boxes 2 (light green) and 3 (dark green) are tightly linked by cysteine bridges and are located close to the serpentine domain. They function together as an intramolecular agonistic unit (green). Single-side chain substitutions (e.g. S281Q (47) and D403A (24), blue full circles) are inducing the activation mechanism between this unit and the serpentine domain. The partially inactivating mutations (significant examples only, red full circles) suggest that parts of the TSHR hinge region function as a signal transmitter for TSH-induced activation. Deletions of fragments in the TSHR hinge region like del371–384 (28) lead to constitutive receptor activation (blue large full ellipse), but none of the here identified single mutations in the deletion-sensitive hinge regions (blue ellipse) causes constitutive TSHR activation. The potential LRR11 studied here attaches the LRRD directly to the hinge region which enables on one hand a direct signaling (dotted arrows) via modifications of the axis LRRD-agonistic hinge unit-serpentine domain by TSH binding and, on the other hand, via binding and signaling-sensitive residues (e.g. Glu297 and Asp382 (25) or Tyr385 (22, 23)) in the hinge region. The received signal from the hormone at the hinge region is also conducted through the agonistic unit of C-b2/C-b3 as a central extracellular switch toward the serpentine domain.
As suggested by our new data, the TSHR region between Leu260 and Pro280 might have a specific fold, probably as an additional LRRD repeat. This region is tightly attached to the subsequent amino acid Ser281 and therewith to cysteine box 2, which was shown to be sensitive for activation of the TSHR and other GPHRs (1).
We suggest that this N-terminal part of C-b2 (around Ser281) together with the C terminus of C-b3 is part of an intramolecular agonistic unit, because particular amino acids like Pro280-Ser281 and peptide Asp403–Pro407 are tightly linked due to cysteine bridges (Fig. 1), and mutations at these residues (e.g. S281Q and D403A) induce activation from the extracellular region toward the serpentine domain (24, 62). This can be described as a signaling-sensitive interface between the extracellular and transmembrane TSHR components (24). The existence of an activation-sensitive part close to TMH1 was also proposed by Zhang et al. in 2000 (45).
The partially inactivating TSHR hinge mutations for conserved hydrophilic residues that we describe here suggest that parts of the hinge region may function as a signal transmitter for TSH-induced activation, which is in accordance to previous and recent findings of other groups (22, 26, 28). The GPH binding-sensitive amino acid Tyr385 (22, 23) is located in the C-terminal hinge region where we found most of our inactivating mutations.
Interestingly, constitutive activation of mutant S281Q cannot be decreased by combination with inactivating mutations in the N-terminal or middle part of the hinge region but by a mutant which is located at C-b3 (Asp395), a spatially close peptide at the interface between the hinge region and the serpentine domain. This implies that the wild type amino acids of inactivating mutants, especially those at positions between residues Glu369 and Glu394, are spatially arranged between the LRRD and the intramolecular agonistic unit close to the serpentine domain.
In this sandwich-like arrangement of TSHR components (Fig. 5), the bTSH interaction with the LRRD and hinge region leads to different activation paths: first a direct signaling via modifications of the axis (LRRD (+LRR11)-agonistic unit-serpentine domain) and, second, via binding at sensitive residues (e.g. Glu297, Asp382, or Tyr385) in the hinge region transducing the signal also to the agonistic unit as a central switch.
Of note, deletions in the TSHR hinge region like del371–384 (28) lead to constitutive receptor activation in contrast to deletions in the LRRD (32). We refer to our findings that none of our single mutations in the deletion-sensitive hinge regions causes constitutive TSHR activation as shown for CAMs at Ser281. In consequence, our single-side chain substitutions could not confirm a “silencing effect” (45) of the extracellular region on the serpentine domain. Therefore, an alternative scenario might be that also deletions in the hinge region cause an activation of the above described intramolecular agonistic unit as the central extracellular switch toward the serpentine domain via structural rearrangements (release).
Taking this information into consideration, we postulate an intramolecular signal transmitter constituted by an intramolecular agonistic unit (positions of CAMs) and determinants of signal propagation in the hinge region (positions of inactivating mutations). An important, but still unanswered question in this regard is the difference between constitutive activation by deletions at the hinge regions of TSHR (28, 32) or FSHR (44), which cannot be observed by similar approaches at corresponding regions of the LHCGR (28, 10). In fact, the available structural and functional information are not yet sufficient to reasonably answer this question, but we would like to reduce the potential difference between the TSHR/FSHR versus the LHCGR down to the release mechanism(s) or arrangement of the intramolecular agonistic unit.
Supplementary Material
This work was supported, in whole or in part, by the National Institutes of Health, NIDDK, Intramural Research Program. This work was also supported by Medical Faculty, University of Leipzig, Grant NBL Formel.1-108 and by Deutsche Forschungsgemeinschaft Grants KL2334/2-1, KR1273/1-2,3, and Pa 423/15-1,2.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. 1 and 2.
- GPHR
- glycoprotein hormone receptor
- CAM
- constitutively activating mutation
- LHCGR
- lutropin/choriogonadotropin receptor
- FSHR
- follicle-stimulating hormone receptor
- TSH
- thyroid-stimulating hormone
- TSHR
- thyroid-stimulating hormone receptor
- bTSH
- bovine thyroid-stimulating hormone
- LH
- luteinizing hormone
- CG
- choriogonadotropin
- FSH
- follicle-stimulating hormone
- TMH
- transmembrane helix
- LRRD
- leucine-rich repeat domain
- C-b2
- cysteine box 2
- C-b3
- cysteine box 3
- IP
- inositol phosphate.
REFERENCES
- 1. Kleinau G., Krause G. (2009) Endocr. Rev. 30, 133–151 [DOI] [PubMed] [Google Scholar]
- 2. Braun T., Schofield P. R., Sprengel R. (1991) EMBO J. 10, 1885–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nagayama Y., Russo D., Chazenbalk G. D., Wadsworth H. L., Rapoport B. (1990) Biochem. Biophys. Res. Commun. 173, 1150–1156 [DOI] [PubMed] [Google Scholar]
- 4. Caltabiano G., Campillo M., De Leener A., Smits G., Vassart G., Costagliola S., Pardo L. (2008) Cell Mol. Life Sci. 65, 2484–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fan Q. R., Hendrickson W. A. (2005) Nature 433, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sanders J., Chirgadze D. Y., Sanders P., Baker S., Sullivan A., Bhardwaja A., Bolton J., Reeve M., Nakatake N., Evans M., Richards T., Powell M., Miguel R. N., Blundell T. L., Furmaniak J., Smith B. R. (2007) Thyroid 17, 395–410 [DOI] [PubMed] [Google Scholar]
- 7. Dias J. A. (2005) Nature 433, 203–204 [DOI] [PubMed] [Google Scholar]
- 8. Fan Q. R., Hendrickson W. A. (2007) Mol. Cell. Endocrinol. 260, 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fan Q. R., Hendrickson W. A. (2008) Proteins 72, 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sangkuhl K., Schulz A., Schultz G., Schöneberg T. (2002) J. Biol. Chem. 277, 47748–47755 [DOI] [PubMed] [Google Scholar]
- 11. Vassart G., Pardo L., Costagliola S. (2004) Trends Biochem. Sci. 29, 119–126 [DOI] [PubMed] [Google Scholar]
- 12. Núñez Miguel R., Sanders J., Jeffreys J., Depraetere H., Evans M., Richards T., Blundell T. L., Rees Smith B., Furmaniak J. (2004) Thyroid 14, 991–1011 [DOI] [PubMed] [Google Scholar]
- 13. Bruysters M., Verhoef-Post M., Themmen A. P. (2008) J. Biol. Chem. 283, 25821–25828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moyle W. R., Xing Y., Lin W., Cao D., Myers R. V., Kerrigan J. E., Bernard M. P. (2004) J. Biol. Chem. 279, 44442–44459 [DOI] [PubMed] [Google Scholar]
- 15. Kaczur V., Puskas L. G., Nagy Z. U., Miled N., Rebai A., Juhasz F., Kupihar Z., Zvara A., Hackler L., Jr., Farid N. R. (2007) J. Mol. Recognit. 20, 392–404 [DOI] [PubMed] [Google Scholar]
- 16. Rapoport B., Chazenbalk G. D., Jaume J. C., McLachlan S. M. (1998) Endocr. Rev. 19, 673–716 [DOI] [PubMed] [Google Scholar]
- 17. Bernard M. P., Myers R. V., Moyle W. R. (1998) Biochem. J. 335, 611–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bozon V., Couture L., Pajot-Augy E., Richard F., Remy J. J., Salesse R. (2002) Protein Expr. Purif. 25, 114–123 [DOI] [PubMed] [Google Scholar]
- 19. Couet J., Sar S., Jolivet A., Hai M. T., Milgrom E., Misrahi M. (1996) J. Biol. Chem. 271, 4545–4552 [DOI] [PubMed] [Google Scholar]
- 20. Zhang R., Buczko E., Dufau M. L. (1996) J. Biol. Chem. 271, 5755–5760 [DOI] [PubMed] [Google Scholar]
- 21. Ho S. C., Goh S. S., Li S., Khoo D. H., Paterson M. (2008) Thyroid 18, 1313–1319 [DOI] [PubMed] [Google Scholar]
- 22. Bonomi M., Busnelli M., Persani L., Vassart G., Costagliola S. (2006) Mol. Endocrinol. 20, 3351–3363 [DOI] [PubMed] [Google Scholar]
- 23. Costagliola S., Panneels V., Bonomi M., Koch J., Many M. C., Smits G., Vassart G. (2002) EMBO J. 21, 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kleinau G., Jäschke H., Neumann S., Lättig J., Paschke R., Krause G. (2004) J. Biol. Chem. 279, 51590–51600 [DOI] [PubMed] [Google Scholar]
- 25. Mueller S., Kleinau G., Jaeschke H., Paschke R., Krause G. (2008) J. Biol. Chem. 283, 18048–18055 [DOI] [PubMed] [Google Scholar]
- 26. Chen C. R., McLachlan S. M., Rapoport B. (2011) J. Biol. Chem. 286, 6219–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kosugi S., Ban T., Akamizu T., Kohn L. D. (1991) J. Biol. Chem. 266, 19413–19418 [PubMed] [Google Scholar]
- 28. Mizutori Y., Chen C. R., McLachlan S. M., Rapoport B. (2008) Mol. Endocrinol. 22, 1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nagayama Y., Wadsworth H. L., Chazenbalk G. D., Russo D., Seto P., Rapoport B. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 902–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nagayama Y., Rapoport B. (1992) Endocrinology 131, 548–552 [DOI] [PubMed] [Google Scholar]
- 31. Nurwakagari P., Breit A., Hess C., Salman-Livny H., Ben-Menahem D., Gudermann T. (2007) J. Mol. Endocrinol. 38, 259–275 [DOI] [PubMed] [Google Scholar]
- 32. Vlaeminck-Guillem V., Ho S. C., Rodien P., Vassart G., Costagliola S. (2002) Mol. Endocrinol. 16, 736–746 [DOI] [PubMed] [Google Scholar]
- 33. Zeng H., Phang T., Song Y. S., Ji I., Ji T. H. (2001) J. Biol. Chem. 276, 3451–3458 [DOI] [PubMed] [Google Scholar]
- 34. Berridge M. J. (1983) Biochem. J. 212, 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Enkhbayar P., Kamiya M., Osaki M., Matsumoto T., Matsushima N. (2004) Proteins 54, 394–403 [DOI] [PubMed] [Google Scholar]
- 36. Kajava A. V. (1998) J. Mol. Biol. 277, 519–527 [DOI] [PubMed] [Google Scholar]
- 37. Kajava A. V. (2001) J. Struct. Biol. 134, 132–144 [DOI] [PubMed] [Google Scholar]
- 38. Laskowski R. A., Moss D. S., Thornton J. M. (1993) J. Mol. Biol. 231, 1049–1067 [DOI] [PubMed] [Google Scholar]
- 39. Chen C. R., McLachlan S. M., Rapoport B. (2009) Endocrinology 150, 3401–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen C. R., McLachlan S. M., Rapoport B. (2010) Endocrinology 151, 1940–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sanders J., Bolton J., Sanders P., Jeffreys J., Nakatake N., Richards T., Evans M., Kiddie A., Summerhayes S., Roberts E., Miguel R. N., Furmaniak J., Smith B. R. (2006) Thyroid 16, 1195–1206 [DOI] [PubMed] [Google Scholar]
- 42. Chazenbalk G. D., Tanaka K., Nagayama Y., Kakinuma A., Jaume J. C., McLachlan S. M., Rapoport B. (1997) Endocrinology 138, 2893–2899 [DOI] [PubMed] [Google Scholar]
- 43. Ho S. C., Goh S. S., Su Q., Khoo D. H. (2005) Mol. Cell. Endocrinol. 245, 158–168 [DOI] [PubMed] [Google Scholar]
- 44. Agrawal G., Dighe R. R. (2009) J. Biol. Chem. 284, 2636–2647 [DOI] [PubMed] [Google Scholar]
- 45. Zhang M., Tong K. P., Fremont V., Chen J., Narayan P., Puett D., Weintraub B. D., Szkudlinski M. W. (2000) Endocrinology 141, 3514–3517 [DOI] [PubMed] [Google Scholar]
- 46. Duprez L., Parma J., Costagliola S., Hermans J., Van Sande J., Dumont J. E., Vassart G. (1997) FEBS Lett. 409, 469–474 [DOI] [PubMed] [Google Scholar]
- 47. Jaeschke H., Neumann S., Kleinau G., Mueller S., Claus M., Krause G., Paschke R. (2006) Endocrinology 147, 1753–1760 [DOI] [PubMed] [Google Scholar]
- 48. Mueller S., Kleinau G., Szkudlinski M. W., Jaeschke H., Krause G., Paschke R. (2009) J. Biol. Chem. 284, 16317–16324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nakabayashi K., Kudo M., Hsueh A. J., Maruo T. (2003) Mol. Cell. Endocrinol. 202, 139–144 [DOI] [PubMed] [Google Scholar]
- 50. Kleinau G., Jaeschke H., Mueller S., Raaka B. M., Neumann S., Paschke R., Krause G. (2008) FASEB J. 22, 2798–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Neumann S., Claus M., Paschke R. (2005) Eur. J. Endocrinol. 152, 625–634 [DOI] [PubMed] [Google Scholar]
- 52. Latif R., Morshed S. A., Zaidi M., Davies T. F. (2009) Endocrinol. Metab. Clin. North Am. 38, 319–341, viii [DOI] [PubMed] [Google Scholar]
- 53. Rapoport B., McLachlan S. M. (2007) Thyroid 17, 911–922 [DOI] [PubMed] [Google Scholar]
- 54. Kopp P., Muirhead S., Jourdain N., Gu W. X., Jameson J. L., Rodd C. (1997) J. Clin. Invest. 100, 1634–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ho S. C., Van Sande J., Lefort A., Vassart G., Costagliola S. (2001) Endocrinology 142, 2760–2767 [DOI] [PubMed] [Google Scholar]
- 56. Nakabayashi K., Kudo M., Kobilka B., Hsueh A. J. (2000) J. Biol. Chem. 275, 30264–30271 [DOI] [PubMed] [Google Scholar]
- 57. Kleinau G., Brehm M., Wiedemann U., Labudde D., Leser U., Krause G. (2007) Mol. Endocrinol. 21, 574–580 [DOI] [PubMed] [Google Scholar]
- 58. Kleinau G., Kreuchwig A., Worth C. L., Krause G. (2010) Hum. Mutat. 31, E1519–E1525 [DOI] [PubMed] [Google Scholar]
- 59. Karges B., Gidenne S., Aumas C., Haddad F., Kelly P. A., Milgrom E., de Roux N. (2005) Mol. Endocrinol. 19, 2086–2098 [DOI] [PubMed] [Google Scholar]
- 60. Zhang M. L., Sugawa H., Kosugi S., Mori T. (1995) Biochem. Biophys. Res. Commun. 211, 205–210 [DOI] [PubMed] [Google Scholar]
- 61. Hamidi S., Chen C. R., Mizutori-Sasai Y., McLachlan S. M., Rapoport B. (2011) Mol. Endocrinol. 25, 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jäschke H., Neumann S., Moore S., Thomas C. J., Colson A. O., Costanzi S., Kleinau G., Jiang J. K., Paschke R., Raaka B. M., Krause G., Gershengorn M. C. (2006) J. Biol. Chem. 281, 9841–9844 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.