Abstract
Photosystem II (PSII) is the membrane protein complex that catalyzes the photo-induced oxidation of water at a manganese-calcium active site. Light-dependent damage and repair occur in PSII under conditions of high light stress. The core reaction center complex is composed of the D1, D2, CP43, and CP47 intrinsic polypeptides. In this study, a new chromophore formed from the oxidative post-translational modification of tryptophan is identified in the CP43 subunit. Tandem mass spectrometry peptide sequencing is consistent with the oxidation of the CP43 tryptophan side chain, Trp-365, to produce N-formylkynurenine (NFK). Characterization with ultraviolet visible absorption and ultraviolet resonance Raman spectroscopy supports this assignment. An optical assay suggests that the yield of NFK increases 2-fold (2.2 ± 0.5) under high light illumination. A concomitant 2.4 ± 0.5-fold decrease is observed in the steady-state rate of oxygen evolution under the high light conditions. NFK is the product formed from reaction of tryptophan with singlet oxygen, which can be produced under high light stress in PSII. Reactive oxygen species reactions lead to oxidative damage of the reaction center, D1 protein turnover, and inhibition of electron transfer. Our results are consistent with a role for the CP43 NFK modification in photoinhibition.
Keywords: Membrane Proteins, Metalloproteins, Photosynthesis, Post-translational Modification, Protein Conformation, UV Resonance Raman, Mass Spectrometry, Photoinhibition, Tryptophan, Water Oxidation
Introduction
Oxygenic photosynthesis is the enzyme-catalyzed conversion of light energy to biochemical energy, and this process occurs in the membranes of plants, algae, and cyanobacteria. In oxygenic photosynthesis, photosystem II (PSII)4 catalyzes the light-driven oxidation of water and reduction of plastoquinone. On the acceptor side of PSII, electrons are transferred sequentially to two quinone molecules, QA and QB (1). On the donor side, a Mn4Ca active site is the binding site for water and the site of oxygen production. Each monomer is composed of 20 protein subunits, chlorophylls, carotenoids, and redox-active plastoquinones (2, 3). Calcium and chloride are required for activity under physiological conditions (4). The chloride binding site has been assigned near the active site (2, 3).
The D1, D2, CP43, and CP47 polypeptides form the intrinsic core complex of PSII. The D1 and D2 membrane spanning proteins bind the electron transfer cofactors active in water oxidation (2, 3). This central heterodimeric core is symmetrically flanked by the CP43 and CP47 proteins, which bind light-harvesting antennae chlorophyll (Chl) molecules (5). Each of these core polypeptides is composed of intrinsic membrane-spanning helices, as well as several hydrophilic loops that protrude into the interior lumen of the thylakoid membrane (2, 3). The lumenal loop regions of CP43 have been implicated as important in assembly and protection from photoinhibition (see Ref. 5 and references therein).
The active site of water oxidation, the Mn4Ca cluster, is located on the lumenal surface and is protected by three extrinsic polypeptides (6). In plants, these extrinsic proteins, the 18-kDa, 24-kDa, and psbO (or the 33-kDa, manganese stabilizing protein), are essential for maximal oxygen evolution under physiological conditions (6). Both cyanobacterial and plant PSII contain an intrinsic cytochrome b559 (7), whereas cyanobacterial PSII also contains an extrinsic cytochrome c550 (2, 8–11). The structure of cyanobacterial PSII has been solved to 1.9-Å resolution (Ref. 3, and also see Refs. 2 and 8–11). In contrast, the resolution of a plant PSII structure remains at 8-Å resolution (12).
Given its structural and functional complexity, many aspects of PSII function remain elusive. In particular, the roles of post-translational modifications (PTMs) of amino acid side chains are not thoroughly understood. The biological relevance of PTMs is evident in their wide range of functions, including roles in cellular regulation (13, 14) and catalysis (15). Modifications of the intrinsic subunits of PSII have been described previously (16, 17). For example, in the D1 subunit, the N-terminal methionine is removed, the N-terminal amino acid is acylated, and the carboxyl terminus is processed by a specific lumenal protease, CtpA (18, 19). In CP43, 14 amino acids are cleaved from the N terminus, which is then N-acetylated. In D2 and CP47, amino-terminal residues are removed, and the subunits are also N-acetylated. In addition, in a PSII reaction center preparation, in which CP47 and CP43 have been removed with chaotropes, a susceptibility to oxidation of D1/D2 has been reported (20). A proteomics-based study of Arabidopsis has shown a increased prevalence of oxidative modifications under high light stress (21). Despite improvements in PSII structure resolution, detection of PTMs based on available x-ray structures is not yet possible.
Other PTMs of PSII proteins have been identified, including oxidation of tryptophan to kynurenine (Fig. 1A) (22), reduction of aspartic acid to aspartyl aldehyde (23), acyl activation of glutamic acid to a species that binds primary amines (24), as well as numerous phosphorylations (25). Core PSII subunits contain multiple unidentified PTM residues that covalently bind amines (23, 26, 27). These reactions were attributed to reactive, carbonyl-containing amino acid side chains close to the active site, and covalent binding was proposed to occur via a Schiff base complex. The addition of chloride was observed to inhibit amine binding, suggesting that the binding sites were near the water oxidizing complex (28, 29). Importantly, amines are well known inhibitors of photosynthetic water oxidation (28, 29), this reactivity was found in plants and cyanobacteria, and experiments showed that amines were oxidized to produce aldehydes (26, 27). These experiments imply that the amine-binding residues may play a role in the structure, function, or assembly of PSII.
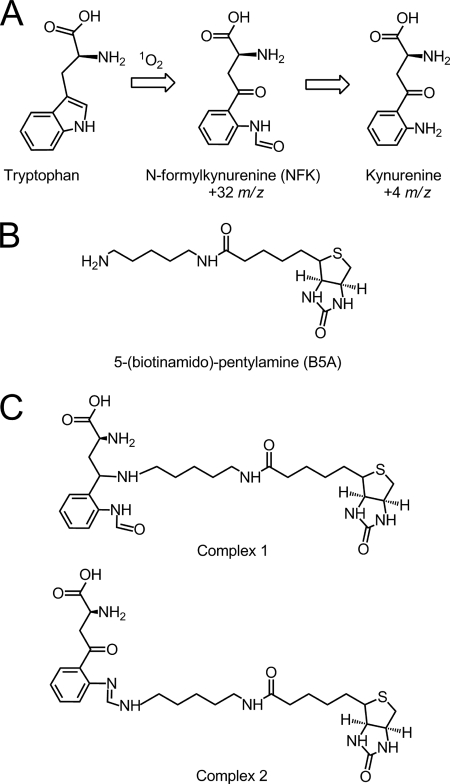
FIGURE 1.
Structures of NFK and the PSII labeling reagent. A, oxidation of tryptophan to form NFK and kynurenine. B, B5A reagent used for derivatization. C, possible covalent complexes of B5A with NFK.
In addition to kynurenine, another known PTM of tryptophan in proteins is N-formylkynurenine (NFK). NFK may bind amines and is created as a stable double oxidation intermediate in the formation of kynurenine (Fig. 1A) (21, 30–32). In this work, we use tandem mass spectrometry (MS/MS) and UV resonance Raman (UVRR) to show that PSII contains NFK. The unique ∼325 nm absorption band of NFK was employed in the purification of NFK-containing CP43 peptides. By MS/MS peptide sequencing, NFK was identified as a +32 m/z modification of Trp-365 in CP43. A vibrational band at 1044 cm−1 was observed, which is a characteristic of the oxidized indole ring in NFK. Quantitative analysis of the HPLC chromatogram was compared with the amount of inhibition under high light conditions. This comparison suggests that the CP43 NFK modification can be induced by high light stress in PSII membrane preparations.
EXPERIMENTAL PROCEDURES
PSII Preparations, Oxygen Evolution Measurements, and Purification of PSII Peptides
PSII was isolated from spinach (33) with the modifications previously described (27). Unless otherwise noted, all procedures were performed at 4 °C and under dim green light illumination. Chlorophyll (34) and oxygen assays (35) were performed, and steady-state rates of oxygen evolution were ≥600 μmol of O2/(mg of Chl·h).
The 18- and 24-kDa extrinsic subunits were removed by treatment with 2 m NaCl for 30 min in the dark (36). In some experiments, removal of psbO and the Mn4Ca cluster (supplemental Fig. S1, step 1) was performed by incubation with 800 mm Tris-NaOH, pH 8.0, for 45 min at room temperature in the light (37). These Tris-washed PSII membranes were washed three times with a buffer of 400 mm sucrose, 50 mm HEPES-NaOH, pH 7.5, and finally resuspended in the same buffer to yield a chlorophyll concentration of 2–4 mg/ml. Samples were stored at −70 °C.
Supporting information describes the purification of PSII peptides, including derivatization with a primary amine-biotin conjugate, 5-(biotinamido)-pentylamine (B5A) (Fig. 1B), in situ trypsin digestion, high-pressure liquid chromatography (HPLC), two-dimensional electrophoresis, clear native polyacrylamide gel electrophoresis (PAGE), in-gel digestion, and avidin affinity chromatography.
Synthesis of the Model Compound NFK
NFK (Fig. 1A) was synthesized by formylation of commercially available kynurenine (95% purity, Sigma). The method has been previously described (38) and is known to produce a mixture of the single formylated NFK, and a double formylated compound, N′,Nα-formylkynurenine (39). ESI MS analysis was used to characterize the product. A Micromass Quattro LC, a triple quadrupole tandem mass spectrometer, was employed. The MH+ peaks observed were 236.8 and 265.0 m/z, consistent with the predicted MH+ masses for the singly and doubly formulated NFK at 237.2 and 265.2 m/z. The relative intensities of the two peaks were ∼1:2 (236.8:265:0), consistent with the expectation that a mixture of the singly and doubly formulated species was produced. The electronic spectra of the singly and doubly formylated compounds have been reported to be indistinguishable (39).
UV-Visible Spectrophotometry
Optical spectra in Figs. 3C and 4 were recorded at room temperature from 200 to 750 nm on a Hitachi (U3000) spectrophotometer. The quartz cuvettes contained 200 μl, the slit width was 2 nm, and the scan speed was 120 nm min−1. The optical spectra in Fig. 3, A and B, were derived from the chromatogram through the use of a Beckman System Gold® HPLC (Brea, CA), equipped with a 125 solvent module, a 168 photodiode array detector (1-cm path length, 2-nm scan interval), and 32 Karat Software, version 7.0. Peptide samples were suspended in 200 μl of 50% acetonitrile, 0.1% trifluoroacetic acid (TFA). The model compounds, 40 μm l-tryptophan (Sigma), l-kynurenine (Sigma), and NFK (synthesis described above), were suspended either in H2O or 50% acetonitrile, 0.1% TFA. Reduction of NFK with NaBH4 was performed by incubation of 40 μm NFK with 400 μm NaBH4 for 30 min at room temperature (40). Reduction of a NFK/B5A mixture with 400 μm NaBH4 was performed using 40 μm NFK and 160 μm B5A.
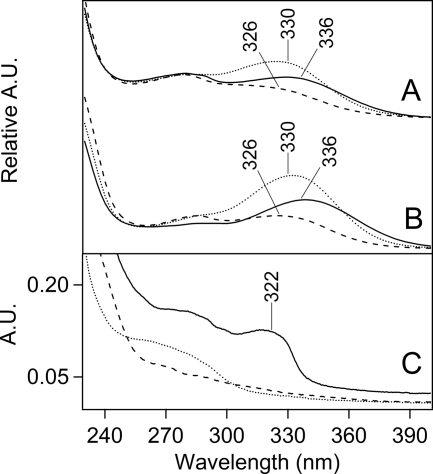
FIGURE 3.
Absorption spectra of peptides, derived from tryptic digestion of Tris-washed PSII. In A and B, the peptides were separated by HPLC. The 350-nm chromatogram exhibited three fractions with approximate retention times of 28 (fraction 1), 35 (fraction 2), and 36 (fraction 3) min (supplemental “Experimental Procedures” and Fig. S2). A, absorption spectra of fraction 1 (solid line), fraction 2 (dotted line), and fraction 3 (dashed line) from samples in which PSII was not treated with B5A. B, absorption spectra of fraction 1 (solid line), fraction 2 (dotted line), and fraction 3 (dashed line) from samples in which PSII was treated with B5A. Fraction 1 corresponds to a CP43 peptide. C, absorption spectra of two-dimensional gel-purified, B5A-labeled CP43 peptides before (dotted line) and after (solid line) affinity chromatography. The data in the dashed black line in C is the spectrum of B5A alone. Spectra shown in A and B were derived from the HPLC chromatogram and are on an arbitrary y scale. The spectra shown in C were measured on a Hitachi spectrophotometer. See “Experimental Procedures” for more information.
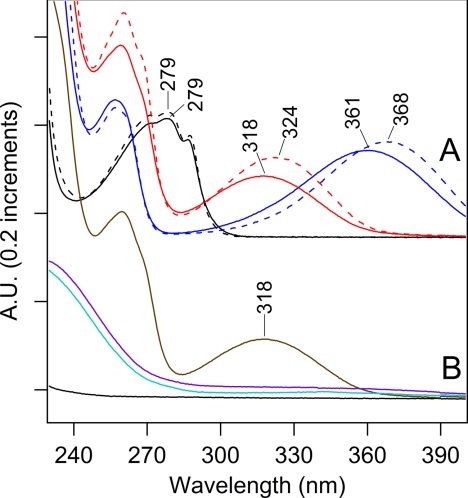
FIGURE 4.
Absorption spectra of model compounds. A, spectra of 40 μm tryptophan in water (black, solid line) and in 50% acetonitrile, 0.1% TFA (black, dashed line). Spectra of 40 μm kynurenine in water (blue, solid line) and in 50% acetonitrile, 0.1% TFA (blue, dashed line). Spectra of 40 μm NFK in water (red, solid line) and in 50% acetonitrile, 0.1% TFA (red, dashed line). B, spectra of a mixture of 40 μm NFK and 160 μm B5A (brown) and when treated with 400 μm NaBH4 in water (cyan). Spectrum of 160 μm B5A when treated with 400 μm NaBH4 in water (black) and spectrum of 40 μm NFK when treated with 400 μm NaBH4 (violet) in water. Spectra shown in A and B were displaced by an arbitrary amount on the y axis for comparison. Data were recorded on a Hitachi spectrophotometer. See “Experimental Procedures” for more information.
UVRR Spectroscopy
A Renishaw (Hoffman Estates, IL) microprobe resonance Raman spectrometer was employed, as described (41, 42). A ×15 objective was used to focus the laser beam on the sample and to collect backscattered radiation. Experiments were conducted at room temperature, and the slit width was 50 μm.
To reduce the fluorescence background, PSII peptides were both HPLC and affinity purified (supplemental Fig. S1, steps 5B and 6A). Lyophilized peptide samples were suspended in water, 0.1% TFA to increase solubility. A 3-μl peptide sample and a 360 μW 325-nm probe beam from a He-Cd laser (KIMMON, Tokyo, Japan) were used. The 325-nm probe was chosen to give resonance enhancement of the PSII chromophore. The total exposure time for each spectrum was 2 min (41), and data from three individual experiments were averaged. The spectral resolution was 6 cm−1.
A 220 μW 229-nm probe beam from a frequency-doubled argon-ion laser (Cambridge LEXEL 95, Fremont, CA) was used to record Raman spectra of the model compounds, kynurenine and NFK, which were dissolved in water. These samples were recirculated at a flow rate ∼4.5 m/s through a 120-μm diameter nozzle, which formed a jet (41). The total exposure time for each spectrum was 2 min (41). The spectral resolution was 10 cm−1.
Peptide Sequencing with MS/MS
Lyophilized peptide samples were reconstituted in 50 μl of buffer A (99.9% water, 0.1% TFA) and analyzed on a Waters nano-HPLC C18 column (75 μm × 150 mm, 130 Å, 1.7 μm). For reverse phase chromatography, a gradient of buffer A (99.9% H2O, 0.1% TFA) and buffer B (99.9% acetonitrile, 0.1% TFA) was used. For MS analysis, a Thermo LTQ Orbitrap mass spectrometer was operated in a duty cycle consisting of one 400–2000 m/z Fourier-transform-MS and four MS/MS LTQ scans.
MS/MS Data Analysis
For analysis of the LC-MS/MS data, the Sequest algorithm (43), implemented in the Bioworks software (Thermo Scientific, Waltham, MA), was applied for peptide identification versus a data base. The data base consisted of all spinach protein sequences present in National Center for Biotechnology Information (NCBI) database. For detection of modified peptides, a tryptophan modification of 31.98928 m/z was used as a parameter during the search.
Photoinhibition Experiments
Photoinhibition experiments were conducted with intact PSII (22, 44, 45). Samples were illuminated with white light from a Dolan-Jenner (Boxborough, MA) Fiber-Lite illuminator. The applied light intensity was ∼9,000 μmol of photons/(m2·s) when measured with a Li-Cor (Lincoln, NE) Light Meter (model LI-189, with a ∼8 cm diameter sensor) before the sample. The light intensity was ∼7,000 μmol of photons/(m2·s) when measured after an empty sample tube. During illumination, PSII samples were maintained at 25 °C by immersion in a water bath. The same 2-h illumination experiment was also conducted without the water bath. During this time, the temperature was observed to increase to 37 °C. As dark controls, PSII samples were incubated for 2 h either at room temperature (∼25 °C) or at 37 °C.
These conditions are similar to those described in the literature. For example, in spinach PSII membranes, at 25 °C, and a light intensity of 4,000 μmol of photons/(m2·s), the half-time for oxygen evolution was reported as ∼30 min (46). In spinach thylakoid membranes, at 20 °C, and a light intensity of 7,000 μmol of photons/(m2·s), the half-time was ∼25 min (47). A light intensity of 5,000 μmol of photons/(m2·s) at 25 °C was used for studies of photoinhibition and degradation of the spinach CP43 subunit in spinach PSII membranes (48).
For quantitation of the amount of NFK induced by photoinhibition, the intact PSII samples were digested with trypsin, and an HPLC assay was performed (see supplemental “Experimental Procedures”). Briefly, tryptic peptides were injected onto a C18 column, and the elution was monitored with a diode array detector, as described above. To quantitate the yield of the NFK-containing peptide, the area of the 350-nm peak was calculated using instrument software. This value was normalized to the total integrated area in the 220-nm chromatogram (0–50 min). This normalization provides an internal standard and corrects for any changes in the yield of tryptic peptides. Experiments were performed 3–7 times, and the values were averaged. Oxygen evolution experiments (35) were performed under the same conditions, i.e. after a 2-h dark incubation or a 2-h illumination (with water bath at 25 °C). Measurements were performed six times, and the values were averaged.
RESULTS
Isolation of the Amine-binding Chromophore
Following purification from spinach (33), PSII membranes were depleted of the 18-kDa, 24-kDa, and psbO extrinsic polypeptides, as well as the Mn4Ca cluster (36, 37). These modifications allow access to the sterically hindered core complex where covalent amine-binding occurs (supplemental Fig. S1, step 1) (26, 27). Tris-washed PSII membranes were reacted with a primary amine-biotin conjugate, B5A (Fig. 1B), by incubation in the light (supplemental Fig. S1, step 2). Previous work demonstrated that PSII core subunits form stable covalent adducts with amines under these conditions, and binding was attributed to reactive carbonyl groups in PTMs (23, 26, 27). In our experiments, the biotin-linked amine allowed the selective purification of peptides by avidin affinity chromatography (supplemental Fig. S1, steps 6A and B). Binding of B5A was confirmed by Western blot of a SDS-PAGE of B5A-derivatized PSII and by detection with an avidin-alkaline phosphatase conjugate (data not shown) (49).
Following derivatization, in situ digestion was employed to release modified, surface-exposed peptides. This method was used previously to identify surface-exposed phosphorylation sites in Arapidopsis thylakoid membranes (50). B5A-derivatized Tris-washed PSII was trypsin digested overnight (supplemental Fig. S1, step 4B), cleaved peptides were separated from undigested PSII by centrifugation, and the peptides were subjected to HPLC (supplemental Fig. S1, step 5B). When a 10–60% acetonitrile, 0.1% TFA gradient was used and the peptide elution was monitored at 350 nm, three fractions with unique red-shifted absorption peaks were observed (supplemental Fig. S2). The fractions had retention times of ∼28 (fraction 1), ∼35 (fraction 2), and ∼36 min (fraction 3). Unlabeled peptides gave a similar 350-nm chromatogram (supplemental Fig. S2). These peaks were also present in intact PSII membranes (Fig. 2).
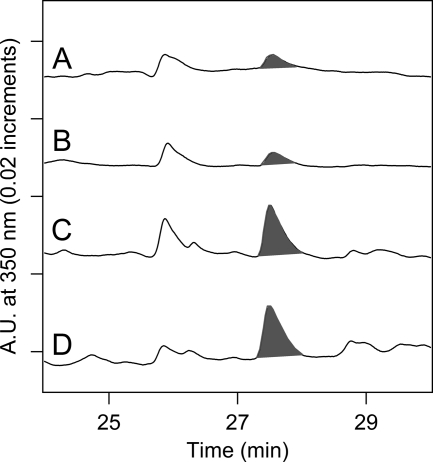
FIGURE 2.
HPLC chromatograms of tryptic peptides from intact PSII membranes. The shaded peak at ∼27 min corresponds to fraction 1 and contains a CP43 peptide with Trp-365 modified to NFK. Elution was monitored at 350 nm. PSII was maintained in the dark at room temperature (∼25 °C) (A) or 37 °C (B) for 2 h. PSII was illuminated with white light (C and D) at a light intensity of ∼7,000 μmol of photons/(m2·s) for 2 h. In C, the temperature under illumination was maintained at 25 °C. In D, the temperature under illumination was allowed to increase to 37 °C (see “Experimental Procedures”). Chromatograms were displaced in the y direction for comparison, and the y axis marks correspond to 20 milliabsorbance units. As an internal standard, the chromatograms were normalized to the total 220 nm absorption, which was integrated from 0 to 50 min. The 350-nm peak at 26 min is not observed in tryptic digests of Tris-washed PSII (supplemental “Experimental Procedures”).
The absorption spectra of these unlabeled and B5A-labeled fractions are shown in Fig. 3, A and B, respectively. As derived from the HPLC detector, the spectra all exhibited maxima between 326 and 336 nm. Small shifts may be due to overlap with a 280-nm shoulder, which is indicative of tyrosine absorption (51). The spectra of the fractions also showed strong 220 nm absorption from the peptide bond (52), but no visible absorption, which is characteristic of photosynthetic pigments. The observation of these 220- and 280-nm absorption bands supports the conclusion that the fractions contain peptides, which have been post-translationally modified to produce a chromophore with a ∼325-nm absorption maximum.
To identify the 325-nm chromophore in the peptide samples, comparison was made to model compounds. Some oxidative tryptophan products, such as hydroxytryptophan, dioxyindolylalanine, and oxyindolylalanine, absorb near 295 nm, only slightly red-shifted from the tryptophan absorption band (53, 54). However, other PTMs of tryptophan, including NFK and kynurenine (Fig. 1A), show more red-shifted absorption (40, 55). To compare with the peptide spectra, NFK was synthesized by formylation of commercially available kynurenine. As shown in Fig. 4A, NFK had an absorption peak at ∼320 nm (Fig. 4A, red), whereas kynurenine had a longer wavelength absorption maximum at ∼365 nm (Fig. 4A, blue). Both spectra were red-shifted compared with the tryptophan absorption maximum at ∼280 nm (Fig. 4A, black). The absorption maximum of NFK was slightly solvent dependent, showing a shift from 318 to 324 nm when water (Fig. 4A, solid red) was compared with 50% acetonitrile, 0.1% TFA (Fig. 4A, dashed red). The peptide spectra (Fig. 3, A and B) exhibited a clear similarity with the NFK spectrum (Fig. 4A, red), making NFK a candidate for the PTM.
MS/MS Identifies a NFK Modification in the CP43 Subunit
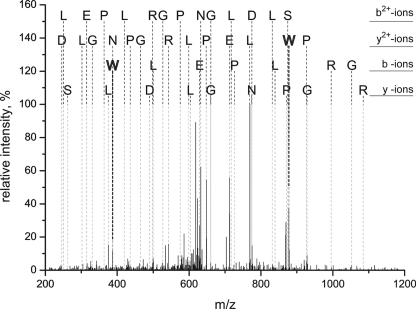
HPLC fractions 1–3 were purified by a second round of chromatography, avidin affinity (supplemental Fig. S1, step 6A). Applying LC-MS/MS analysis to the purified peptide samples resulted in unambiguous identification of a CP43 peptide, 363AP(W*)LEPLRGPNGLDLSR379, in fraction 1 with a p value of 10−7 and displaying a mass shift of +32 m/z on Trp-365 (Fig. 5). These results are indicative of an NFK modification. In fraction 2, a peptide of CP24, 169PDSQSVE(W*)ATPWSR184, with a NFK modification was observed (data not shown). There were no spectra detected consistent with the B5A-labeled peptides, however, suggesting that the adduct of NFK and B5A was not stable under the conditions employed for mass spectrometry.
FIGURE 5.
MS/MS spectrum assigned to the triply charged CP43 peptide, 363AP(W*)LEPLRGPNGLDLSR379. The labels in the figure indicate the N-terminal amino acids for the b-fragments and the C-terminal amino acids for the y-fragments. W is boldface because this residue carries a +32 m/z modification. The mass shift of +32 m/z can be unambiguously assigned to Trp-365 due to the y2+-ion and b-ion series. All relevant signals in the MS/MS spectra, which are complex due to the presence of singly, doubly, and triply charged fragment ions, can be explained by the peptide sequence.
Two-dimensional Gel Electrophoresis
To confirm that the chromophore arises from a PTM in a PSII peptide, PSII core peptides were purified by two-dimensional gel electrophoresis. For these experiments, B5A-labeled PSII was solubilized and electrophoresized in the first dimension by non-denaturing clear native-PAGE (56), which separates the PSII membranes into dimer complexes with varying amounts of light-harvesting proteins (24). All gels were run without Coomassie Blue or bromphenol blue to eliminate possible spectral artifacts from the dyes. The PSII dimer complex, which is deficient in light-harvesting proteins (24), was excised and resolved into individual polypeptides in the second dimension (supplemental Fig. S1, step 3) (57). Due to their similarity in electrophoretic mobility, the CP43 and CP47 protein bands were not fully resolved. MS/MS analysis, following in-gel digestion and peptide extraction (supplemental Fig. S1, step 4C), validated the band identities (data not shown). The identities were also substantiated by previous work (24, 58). Although in situ these polypeptides bind many pigment molecules, the non-covalently bound pigments were separated from the proteins under denaturing gel conditions in the second dimension. Therefore, this experiment eliminated chlorophyll or carotenoid (or their degradation products) as possible sources of the optical absorption.
Before affinity chromatography (supplemental Fig. S1, step 6B), the absorption spectrum of the gel-extracted sample exhibited only absorption characteristic of tyrosine containing peptides, with a 280-nm absorption band (51) (Fig. 3C, dotted line). However, upon affinity purification of the gel-extracted peptide mixture, a ∼322 nm peak was observed (Fig. 3C, solid line). This band resembled the chromophore absorption observed in the HPLC-purified peptides (Fig. 3, A and B). The λmax was similar to the spectra recorded from the HPLC fractions, given the increased scattering background in the gel-extracted samples. Fig. 3C also shows that the B5A compound itself does not contribute to the optical absorption (Fig. 3C, dashed line), although affinity chromatography was essential for the selection of the modified peptides.
Proposed Structure of the B5A Adduct
NFK is expected to react with hydrazines, hydrazides, and amines (59–61). Fig. 1C presents two possible structures for the B5A-NFK adduct formed in our experiments. The small 2 m/z mass difference between the two structures may not be distinguishable by low resolution peptide mass spectrometry. Fig. 1C, complex 1, is a covalent adduct, formed by nucleophilic addition of the B5A label at the C4 position of NFK and a subsequent reduction reaction. Fig. 1C, complex 2, is another possibility for the formation of a B5A-derived amidine, in which the nucleophilic addition occurs at the N-formyl carbon. Although complex 1 should be quite stable during MS and should be observed in MS/MS spectra, complex 2 is expected to break preferentially at the NFK-B5A interface and escape MS/MS detection.
To distinguish between these two possible structures, we considered the effect of reduction on the optical spectrum. As shown in Fig. 4B, addition of sodium borohydride (NaBH4) and reduction of the C4 carbonyl group eliminated the 318 nm absorption of NFK (compare Fig. 4, A, red and B, violet). Addition of sodium borohydride to a mixture of NFK and B5A had a similar effect (Fig. 4B, brown and cyan). This result suggests that reduction of the carbonyl group of NFK will eliminate the 322 nm absorption. Because the 322-nm band was observed in the labeled peptides, consideration of these optical properties supports an assignment of complex 2 as the covalent adduct.
UVRR Spectroscopy of the Chromophore
To obtain more information concerning the structure of the B5A peptide complex, UVRR was employed (62). Fig. 6, A and B, are the Raman spectra of kynurenine and NFK model compounds, respectively. These data were obtained with 229 nm excitation. The Raman spectrum of kynurenine could not be acquired at 325 nm, due to a large fluorescence background. The Raman spectrum of NFK, obtained either with 229- or 325-nm probe beams, displayed a band at 1242 cm−1. There was no observable signal from the B5A label alone, either at 229 or 325 nm, due to lack of resonance enhancement (data not shown).
FIGURE 6.
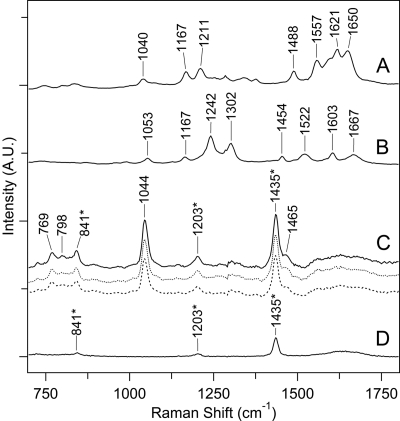
UVRR spectra of chromophore-containing PSII peptides and model compounds. Spectra of kynurenine (A) and NFK (B) in water, recorded with 220 μW 229-nm laser excitation. C, spectra of the chromophore-containing peptides from HPLC fractions 1 (solid line), 2 (dotted line), and 3 (bold dotted line) (see supplemental Fig. S2B), recorded with 360 μW 325-nm laser excitation. The peptides were B5A-derivatized, purified both by HPLC and affinity chromatography, and suspended in H2O, 0.1% TFA. D shows the UVRR spectrum of H2O, 2% TFA only, and the bands assigned to TFA are indicated with a asterisk. The spectra were displaced by an arbitrary amount on the y axis for comparison. Each y axis mark corresponds to 6,500 arbitrary intensity units.
In Fig. 6B, unique bands assignable to the NFK N-formyl group were observed at 1667 and 1242 cm−1. A band between 1040 and 1053 cm−1 was observed both in the kynurenine and the NFK Raman spectra (Fig. 6, A and B). These bands are characteristic of the oxidized indole group (31, 63). These spectral features were not observed in the UV Raman spectra of the aromatic amino acids, histidine, tryptophan, tyrosine, and phenylalanine (supplemental Fig. S3). For example, unmodified tryptophan exhibited a benzene breathing mode at 1009 cm−1 (supplemental Fig. S3). In addition, these bands were not observed in spectra derived from Chl a or a carotenoid, after correction for solvent scattering (supplemental Fig. S4).
Previous Fourier-transform-Raman measurements on NFK assigned a band at 1050 cm−1 to the ring system, a band at 1604 cm−1 to a ring stretching mode, and bands at 1239 and 1685 cm−1 to the N-formyl group (63). In addition, bands at 1052 and 1050 cm−1 were observed for NFK modifications in lysozyme (63) and egg white ovalbumin (31), respectively. Formamide gave rise to Raman bands at 1670, 1599, 1391, 1313, 1098, 1048, and 983 cm−1 (64). In anilides, the carbonyl band was observed at higher frequency (1704 cm−1 in formanilide), due to delocalization of the unpaired electrons on the formamide nitrogen into the phenyl ring (65).
Because the UV Raman spectrum is specific for the contribution of NFK, the Raman spectrum of derivatized peptide samples was obtained with a 325-nm probe (Fig. 6C), which resonantly enhances the chromophore. In peptides purified by HPLC only (supplemental Fig. S1, step 5B), a strong fluorescence background obscured the Raman signal. Therefore, samples were subjected to purification by affinity chromatography (supplemental Fig. S1, step 6A), which reduced the background fluorescent signal. The resulting Raman spectra of all three HPLC 350-nm absorbing fractions were similar (Fig. 6C). These results imply that all three fractions contain the same PTM. Comparison with the TFA buffer spectrum (Fig. 6D) showed that bands at 841, 1203, and 1435 cm−1 arise from TFA. The Raman spectra of all three peptide fractions displayed a band at 1044 cm−1, which is consistent with the presence of NFK in all three fractions. The shift from the 1053 cm−1 frequency observed in NFK alone (Fig. 6B) may be due to reaction with the B5A label. No vibrational bands from the N-formyl group (1242 and 1667 cm−1) were observed, supporting the interpretation that complex 2 (Fig. 1C) is the stable structure.
Yield of NFK in Tris-washed PSII
Using an extinction coefficient of 3750 m−1 cm−1 at 321 nm (55), the yield of NFK in Tris-washed PSII can be estimated. For the HPLC experiment, fraction 1 was collected, the sample was concentrated to 200 μl, and the absorption spectrum was measured on a Hitachi spectrophotometer (see “Experimental Procedures”). Starting with 6 mg of Chl or 24 nmol of PSII reaction center (66), the NFK yield (on a reaction center basis) was estimated as ∼7% in the HPLC method. For the two-dimensional gel experiment, peptides were extruded from the gel, concentrated to 200 μl, and the absorption spectrum was measured as described above (Fig. 3C). Starting with 13 mg of Chl or 52 nmol of PSII reaction center (66), the NFK yield was estimated as ∼6% in the two-dimensional gel method.
Photoinhibition Increases the Yield of NFK in Intact PSII
Our MS/MS data support the interpretation that NFK is formed by PTM of Trp-365 in the CP43 subunit. NFK can be generated from tryptophan by ROS (67, 68). These species, including singlet oxygen (1O2,), hydrogen peroxide (H2O2), superoxide anion (O2˙̄), and hydroxyl radical (•OH) (69), have been proposed to be involved with photoinhibition in PSII. However, the mechanism of their involvement remains controversial (reviewed in Refs. 70 and 71). High light conditions have been linked to oxidative modification of Arabidopsis PSII in proteomic studies (21). We have previously reported that substitutions at Trp-365 increase the rate of photoinhibition (22). The magnitude of the change depends on light intensity (data not shown) and will be described in a future publication.
To probe for a possible connection between the yield of NFK and photoinhibition, we compared 350-nm chromatograms of tryptic peptides obtained from intact PSII (Fig. 2). The integrated area of the 350-nm peak, derived from fraction 1, was corrected for the total integrated absorption at 220 nm. This normalization is an internal standard, which corrects for any change in the total yield of tryptic peptides. In this experiment, intact PSII samples were maintained in the dark at room temperature (∼25 °C) (Fig. 2A) or in the dark at 37 °C (Fig. 2B). These dark-maintained, intact PSII samples gave a 350-nm fraction, with a similar retention time to Tris-washed PSII fraction 1 (Fig. 2, shaded peaks, and supplemental Fig. S2). The fraction 1 yield in the dark-maintained PSII was estimated as 0.3% on a reaction center basis. The NFK yield was not significantly altered by an increase in temperature in the dark. Comparison of the integrated areas (Fig. 2, shaded peaks, fraction 1), derived from dark-maintained PSII at 25 and 37 °C, gave a ratio of 1.1 ± 0.1.
Intact PSII samples were also illuminated with white light for 2 h under temperature-controlled conditions at 25 °C (Fig. 2C), or under conditions (Fig. 2D) in which the temperature of the sample increased to 37 °C. An increase in peak height was observed after 2 h of illumination. This increase was observed when the temperature was controlled at 25 °C (ratio 2.4 ± 0.8, Fig. 2C, shaded peak) or when the temperature was allowed to increase to 37 °C (ratio 2.2 ± 0.5, Fig. 2D, shaded peak).
The steady-state rate of oxygen evolution was also measured under the same conditions (35). Before illumination, the average rate was 740 ± 50 μmol/mg of Chl·h. After 2 h in the dark, the rate was 630 ± 30 μmol/mg of Chl·h. However, with a 2-h illumination, the rate declined to 270 ± 60 μmol/mg of Chl·h. The 2.4 ± 0.5-fold decrease in activity is similar to the increase observed in NFK yield. Therefore, these results suggest that the NFK modification at Trp-365 is induced by illumination and high light stress in intact PSII preparations.
DISCUSSION
Summary
In this paper, we provide evidence that PSII contains a modified form of tryptophan, NFK. Mass spectrometry on purified peptides shows a mass shift of +32 m/z for the 363AP(W*)LEPLRGPNGLDLSR379 peptide from CP43. This mass shift and peptide sequencing by MS/MS are consistent with a double oxidation of Trp-365. Optical absorption and UV resonance Raman data support the conclusion that CP43 peptides contain NFK. In these experiments, NFK was observed following in situ and in-gel tryptic digestion. The yield of NFK in Tris-washed PSII was significant, and the yield increased when intact PSII was subjected to photoinhibitory conditions.
Generation of NFK and NFK in Other Proteins
NFK has been identified in other proteins by mass spectrometry, including bovine heart mitochondrial proteins (72), rat skeletal muscle proteins (73), bovine α-crystalline (74), and spinach LHCII (32). NFK is formed by the reaction of ROS with tryptophan side chains in proteins (67). One potential reactive species is singlet oxygen, 1O2 (68). Initial reaction of tryptophan with 1O2 has been proposed to form one of two unstable intermediates. A dioxetane derivative intermediate can form across the C2-C3 indole ring bond; subsequent ring cleavage gives NFK. On the other hand, an intermediate hydroperoxide, formed at the C3 position on the indole ring, can also decompose to form NFK (75). Therefore, we propose that NFK is formed by a reaction between Trp-365 and 1O2.
Other Modifications at Trp-365
Other modifications of Trp-365 in the lumenal loop of CP43 (Trp-352 in Synechocystis sp. PCC 6803) have been reported previously (22). The data were obtained by tandem mass spectrometry (22) and were consistent with modification of the side chain to kynurenine (+4 m/z), oxindolalanine (+16 m/z), and a hydroxy-indole derivative (+18 m/z). The oxindolalanine and hydroxy-indole derivatives were proposed to be intermediates produced during oxidative cleavage to give kynurenine (22). None of these species are expected to show an absorption maximum at 325 nm (53, 54), as observed here for the NFK-containing peptides. NFK can be formed as a stable intermediate during the production of kynurenine (Fig. 1A) (38). In previous work, kynurenine was observed to be present in PSII, which had not been Tris washed and had not been subjected to gel electrophoresis (22). Kynurenine was also observed in PSII, which had been maintained in the dark. Taken together with the results described here, these data support the conclusion that oxidative modification of Trp-365 is relevant in vivo.
Analysis of Three Different HPLC Fractions
In this work, three 350-nm absorbing fractions, with reproducible retention times, were observed with HPLC purification of Tris-washed PSII peptides. UV resonance Raman studies suggest that all three fractions contained the same, B5A-derivatized NFK chromophore. The NFK modification in CP43 was confirmed by MS/MS of fraction 1. An NFK modification in a light-harvesting protein (CP24) was observed in fraction 2. However, the modified peptide detected in fraction 1 was the result of incomplete tryptic digestion. Therefore, it is possible that other fractions contain a different cleavage product of the same CP43 peptide. In addition to 325 nm absorption, all three fractions exhibited a 280-nm peak, which is indicative of tyrosine absorption. It should be noted that the NFK containing CP43 sequence does not contain tyrosine (363AP(W*)LEPLRGPNGLDLSR379). Thus, other tyrosine-containing peptides must be present in all three eluting fractions. Our MS analysis of the fractions provided evidence for ∼40 peptides, even after HPLC and affinity purification (data not shown). Such complexity can be attributed to the challenges of MS/MS as applied to membrane-associated peptides.
Proposed Structure of the B5A-labeled NFK Complex
In some of our experiments, a biotinylated amine was used to label the NFK-containing peptide. Amines and hydrazines are expected to label activated carbonyl groups (see Refs. 26 and references therein). In other proteins, it has been suggested that kynurenine and NFK react with hydrazine. These proteins include low-density lipoprotein (LDL) (76), cucumber microsomal membrane proteins (60), and ribulose-1,5-bisphosphate carboxylase oxygenase (60). In bovine serum albumin (59) and α-crystallin (59), NFK was proposed to cross-link with lysine residues. On the other hand, in PSII, a complex between kynurenine and a hydrazide labeling reagent was not observed by MS/MS (22).
NFK may react with amines to form an adduct at the C4 position (Fig. 1C, complex 1). Reaction of the amine and electrophilic carbon produces a Schiff base, which can be reduced to give the stable product shown in complex 1. Previous work has indicated that reducing equivalents are produced during PSII light reactions. These reducing equivalents were observed to stabilize amine-PTM complexes (27). On the other hand, reaction of the amine with the NFK formamide group would give the structure shown in Fig. 1C, complex 2. Although amide groups do not usually react with amines, a similar product complex was observed between N-acetyl-formylkynurenine and dimethyl-p-benzoquinonediimine in solution (77). Delocalization of the unpaired electrons on the nitrogen into the aromatic ring may help to activate the formamide carbon (65). It should be noted that we have not observed the labeled NFK adduct by MS/MS. However, our optical and resonance Raman data support binding of the amine label, B5A, at the formamide group, as shown in complex 2. An addition at the N-formyl carbon would provide a breaking point during collision-induced dissociation, which could lead to the neutral loss of the B5A moiety during MS analysis.
Photoinhibition and High Light Stress
Examination of CP43 protein sequences in other organisms, both prokaryotes and eukaryotes, indicates strict sequence conservation of Trp-365. This conservation suggests a functional role for this residue. We hypothesize that Trp-365 may play a role in protection from photoinhibition (22). In PSII, photoinhibition is the light-induced inactivation of photosynthetic activity induced by excess light energy (reviewed in Ref. 78). The results are a decreased efficiency in electron transfer, damage, and degradation of the D1 and other PSII subunits, and finally repair by de novo protein synthesis. When the rate of repair is slower than the rate of degradation, a loss of PSII activity is observed (78). This cycle of damage and repair must be coordinated, and the mechanisms of these reactions have not yet been elucidated.
Photoinhibition may occur by two different mechanisms (reviewed in Refs. 71 and 78–80). In the acceptor side photoinhibition model, damage is initiated by charge separation in reaction centers that contain a reduced quinone, QA−. Recombination leads to the product of triplet chlorophyll, 3Chl, species (81). Changes in the midpoint potential of QA− may alter the susceptibility of PSII to photoinhibition (reviewed in Ref. 79). In donor side photoinhibition, damage is initiated by inactivation of the OEC and water oxidation. Under these conditions, P680+, which has a high potential and a long lifetime, can act as an oxidant for prosthetic groups and amino acid residues (reviewed in Ref. 79).
ROS and High Light Stress in PSII
We attribute the formation of NFK to the reaction of Trp-365 with 1O2. In PSII, 1O2 can be formed by photoexcitation of chlorophyll molecules, which results in formation of 3Chl (82). ROS and 3Chl are generated by charge recombination in acceptor side inhibition (81, 83). 1O2 has been detected by spin trapping in photoinhibited PSII (70, 84) and by a fluorescence sensor in Arabidopsis leaves (85). Chemical trapping in PSII reaction center preparations, which lack the quinone acceptors, has also detected 1O2 (86). Other ROS species may be formed in donor side photoinhibition (70, 84).
Although a correlation between ROS production and photodamage is largely accepted, there is no consensus on the specific role of 1O2 (see Ref. 84 and references therein). One view suggests direct involvement of ROS in damage and increased turnover of the reaction center D1 protein (87). This can occur directly, because ROS damage can lead to peptide bond cleavage (67). Alternatively, ROS-induced modifications could cause a protein conformational change, which allows access of specific proteases to the D1 subunit (22, 88, 89). Finally, 1O2 may inhibit the D1 repair cycle (for example, see Ref. 90). These roles of ROS are not mutually exclusive.
Proposed Role for NFK-365 in Photoinhibition
In this paper, we provide evidence that NFK is present in dark-maintained PSII, and that the yield of NFK increases by a factor of 2 under high light intensity in intact PSII preparations. This change was accompanied by a 2-fold decrease in steady-state oxygen evolution rate. Previous preliminary characterization of site-directed mutations at Trp-365 reported that Trp provides photoprotection (22). The 1.9-Å resolution crystal structure (3) of cyanobacterial PSII shows that Trp-365 is ∼17 Å from the water oxidizing complex (Fig. 7). Trp-365 is found in a CP43 loop region, which is in close proximity to the D1 subunit.
FIGURE 7.
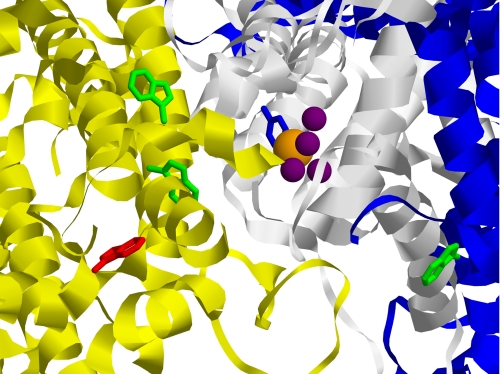
Location of CP43 Trp-365, a site of NFK modification, in the PSII structure from the cyanobacterium, Thermosynechococcus vulcanus (3) (Protein Data Bank code 3ARC). The CP43 protein backbone is shown in yellow, and the D1 and D2 proteins are shown in white and blue, respectively. The Mn4Ca cluster is displayed in purple and orange; the Tyrz side chain is shown in blue. Trp residues located within 20 Å of the Mn4Ca cluster are shown. The side chain of CP43 Trp-365, which is modified to NFK, is shown in red. Trp-359 (CP43), Trp-291 (CP43), and Trp-328 (D2) are shown in green. The measured distances to the Mn4Ca are 17 Å for Trp-365 (CP43), 16 Å for Trp-359 (CP43), 11 Å for Trp-291 (CP43), and 15 Å for Trp-328 (D2).
Taken together, these observations suggest a role for modification of Trp-365 in the high light-induced, damage/repair cycle. This could be accomplished by two different mechanisms. In the first, Trp-365 may act as a ROS scavenger, and in the second, Trp-365 may serve as signal, which facilitates reaction center repair. It should be noted that Trp-359 and -291 in CP43, as well as Trp-328 in D2, are also within 17 Å of the oxygen-evolving complex (Fig. 7). In our MS/MS experiments, we have seen no evidence for NFK modifications of these side chains, although Trp-359 was observed in the +16 m/z form (data not shown).
Conclusions
We have identified an oxidative modification of tryptophan in the CP43 subunit of PSII. This NFK is a UV absorbing chromophore, which is formed by oxidation of a Trp side chain by ROS. We propose that NFK plays a role in protection and repair during photoinhibition. The evolutionarily conserved residue may act as a 1O2 scavenger. Alternatively, oxidation of the tryptophan may promote repair by signaling for degradation or enabling efficient removal of the damaged D1.
Supplementary Material
Acknowledgments
We thank Prof. Cindy Putnam-Evans for helpful discussions concerning the photoinhibition experiments. We are also grateful to Dr. Adam Offenbacher for the UV resonance Raman spectra of the aromatic amino acid residues.
This work was supported, in whole or in part, by Grant MCB 08-42246 from the National Science Foundation (to B. A. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and “Experimental Procedures.”
- PSII
- photosystem II
- B5A
- 5-(biotinamido)-pentylamine
- Chl
- chlorophyll
- NFK
- N-formylkynurenine
- PTM
- post-translational modification
- ROS
- reactive oxygen species
- TFA
- trifluoroacetic acid
- UVRR
- ultraviolet resonance Raman.
REFERENCES
- 1. Nelson N., Yocum C. F. (2006) Annu. Rev. Plant Biol. 57, 521–565 [DOI] [PubMed] [Google Scholar]
- 2. Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009) Nat. Struct. Mol. Biol. 16, 334–342 [DOI] [PubMed] [Google Scholar]
- 3. Umena Y., Kawakami K., Shen J. R., Kamiya N. (2011) Nature 473, 55–60 [DOI] [PubMed] [Google Scholar]
- 4. Yocum C. F. (2008) Coord. Chem. Rev. 252, 296–305 [Google Scholar]
- 5. Bricker T. M., Frankel L. K. (2002) Photosynth. Res. 72, 131–146 [DOI] [PubMed] [Google Scholar]
- 6. Miyao M., Murata N. (1989) Biochim. Biophys. Acta 977, 315–321 [DOI] [PubMed] [Google Scholar]
- 7. MacDonald G. M., Boerner R. J., Everly R. M., Cramer W. A., Debus R. J., Barry B. A. (1994) Biochemistry 33, 4393–4400 [DOI] [PubMed] [Google Scholar]
- 8. Zouni A., Witt H. T., Kern J., Fromme P., Krauss N., Saenger W., Orth P. (2001) Nature 409, 739–743 [DOI] [PubMed] [Google Scholar]
- 9. Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., Iwata S. (2004) Science 303, 1831–1838 [DOI] [PubMed] [Google Scholar]
- 10. Kamiya N., Shen J. R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 98–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. (2005) Nature 438, 1040–1044 [DOI] [PubMed] [Google Scholar]
- 12. Rhee K. H., Morris E. P., Barber J., Kühlbrandt W. (1998) Nature 396, 283–286 [DOI] [PubMed] [Google Scholar]
- 13. Allen J. F. (1992) Biochim. Biophys. Acta 1098, 275–335 [DOI] [PubMed] [Google Scholar]
- 14. Stadtman E. R. (1990) Biochemistry 29, 6323–6331 [DOI] [PubMed] [Google Scholar]
- 15. Janes S. M., Mu D., Wemmer D., Smith A. J., Kaur S., Maltby D., Burlingame A. L., Klinman J. P. (1990) Science 248, 981–987 [DOI] [PubMed] [Google Scholar]
- 16. Whitelegge J. P., Faull K. F., Gundersen C. B., Gómez S. M. (1999) in Photosynthesis: Mechanisms and Effects (Garab G. ed) pp. 4381–4384, Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 17. Whitelegge J. P., Gundersen C. B., Faull K. F. (1998) Protein Sci. 7, 1423–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bowyer J. R., Packer J. C., McCormack B. A., Whitelegge J. P., Robinson C., Taylor M. A. (1992) J. Biol. Chem. 267, 5424–5433 [PubMed] [Google Scholar]
- 19. Liao D. I., Qian J., Chisholm D. A., Jordan D. B., Diner B. A. (2000) Nat. Struct. Biol. 7, 749–753 [DOI] [PubMed] [Google Scholar]
- 20. Sharma J., Panico M., Shipton C. A., Nilsson F., Morris H. R., Barber J. (1997) J. Biol. Chem. 272, 33158–33166 [DOI] [PubMed] [Google Scholar]
- 21. Galetskiy D., Lohscheider J. N., Kononikhin A. S., Popov I. A., Nikolaev E. N., Adamska I. (2011) Rapid Commun. Mass Spectrom. 25, 184–190 [DOI] [PubMed] [Google Scholar]
- 22. Anderson L. B., Maderia M., Ouellette A. J., Putnam-Evans C., Higgins L., Krick T., MacCoss M. J., Lim H., Yates J. R., 3rd, Barry B. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14676–14681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Anderson L. B., Ouellette A. J., Eaton-Rye J., Maderia M., MacCoss M. J., Yates J. R., 3rd, Barry B. A. (2004) J. Am. Chem. Soc. 126, 8399–8405 [DOI] [PubMed] [Google Scholar]
- 24. Rexroth S., Wong C. C., Park J. H., Yates J. R., 3rd, Barry B. A. (2007) J. Biol. Chem. 282, 27802–27809 [DOI] [PubMed] [Google Scholar]
- 25. Mamedov F., Rintamäki E., Aro E. M., Andersson B., Styring S. (2002) Photosynth. Res. 74, 61–72 [DOI] [PubMed] [Google Scholar]
- 26. Ouellette A. J., Anderson L. B., Barry B. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2204–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Anderson L. B., Ouellette A. J., Barry B. A. (2000) J. Biol. Chem. 275, 4920–4927 [DOI] [PubMed] [Google Scholar]
- 28. Sandusky P. O., Yocum C. F. (1984) Biochim. Biophys. Acta 766, 603–611 [Google Scholar]
- 29. Sandusky P. O., Yocum C. F. (1986) Biochim. Biophys. Acta 849, 85–93 [Google Scholar]
- 30. Previero A., Coletti-Previero M. A., Jollès P. (1967) J. Mol. Biol. 24, 261–268 [DOI] [PubMed] [Google Scholar]
- 31. Rokos H., Wood J. M., Hasse S., Schallreuter K. U. (2008) J. Raman Spectrosc. 39, 1214–1218 [Google Scholar]
- 32. Rinalducci S., Campostrini N., Antonioli P., Righetti P. G., Roepstorff P., Zolla L. (2005) J. Proteome Res. 4, 2327–2337 [DOI] [PubMed] [Google Scholar]
- 33. Berthold D. A., Babcock G. T., Yocum C. F. (1981) FEBS Lett. 134, 231–234 [Google Scholar]
- 34. Lichtenthaler H. K. (1987) Methods Enzymol. 148, 350–382 [Google Scholar]
- 35. Barry B. A. (1995) Methods Enzymol. 258, 303–319 [DOI] [PubMed] [Google Scholar]
- 36. Ghanotakis D. F., Topper J. N., Babcock G. T., Yocum C. F. (1984) FEBS Lett. 170, 169–173 [Google Scholar]
- 37. Yamamoto Y., Doi M., Tamura N., Nishimura M. (1981) FEBS Lett. 133, 265–268 [Google Scholar]
- 38. Simat T., Meyer K., Steinhart H. (1994) J. Chromatogr. A 661, 93–99 [Google Scholar]
- 39. Jacobson K. B. (1978) Arch. Biochem. Biophys. 186, 84–88 [DOI] [PubMed] [Google Scholar]
- 40. Pirie A. (1971) Biochem. J. 125, 203–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen J., Barry B. A. (2008) Photochem. Photobiol. 84, 815–818 [DOI] [PubMed] [Google Scholar]
- 42. Chen J., Bender S. L., Keough J. M., Barry B. A. (2009) J. Phys. Chem. B 113, 11367–11370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eng J. K., McCormack A. L., Yates J. R., 3rd (1994) J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 44. Rosenberg C., Christian J., Bricker T. M., Putnam-Evans C. (1999) Biochemistry 38, 15994–16000 [DOI] [PubMed] [Google Scholar]
- 45. Knoepfle N., Bricker T. M., Putnam-Evans C. (1999) Biochemistry 38, 1582–1588 [DOI] [PubMed] [Google Scholar]
- 46. Henmi T., Miyao M., Yamamoto Y. (2004) Plant Cell Physiol. 45, 243–250 [DOI] [PubMed] [Google Scholar]
- 47. Virgin I., Styring S., Andersson B. (1988) FEBS Lett. 233, 408–412 [Google Scholar]
- 48. Yamamoto Y., Akasaka T. (1995) Biochemistry 34, 9038–9045 [DOI] [PubMed] [Google Scholar]
- 49. Towbin H., Staehelin T., Gordon J. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vener A. V., Harms A., Sussman M. R., Vierstra R. D. (2001) J. Biol. Chem. 276, 6959–6966 [DOI] [PubMed] [Google Scholar]
- 51. Edelhoch H. (1967) Biochemistry 6, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 52. Ham J. S., Platt J. R. (1952) J. Chem. Phys. 20, 335–336 [Google Scholar]
- 53. Zhao H., Sagert J., Hwang D. S., Waite J. H. (2009) J. Biol. Chem. 284, 23344–23352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang H. V., Bond M. W., Hunkapiller M. W., Hood L. E. (1983) Methods Enzymol. 91, 318–324 [DOI] [PubMed] [Google Scholar]
- 55. Mehler A. H., Knox W. E. (1950) J. Biol. Chem. 187, 431–438 [PubMed] [Google Scholar]
- 56. Schägger H., von Jagow G. (1991) Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 57. Schägger H., Cramer W. A., von Jagow G. (1994) Anal. Biochem. 217, 220–230 [DOI] [PubMed] [Google Scholar]
- 58. Kügler M., Jänsch L., Kruft V., Schmitz U. K., Braun H.-P. (1997) Photosynth. Res. 53, 35–44 [Google Scholar]
- 59. Fujimori E. (1981) FEBS Lett. 135, 257–260 [Google Scholar]
- 60. Caldwell C. R. (1993) Plant Physiol. 101, 947–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang C. Y., Gu Z. W., Yang H. X., Yang M., Gotto A. M., Jr., Smith C. V. (1997) Free Radic. Biol. Med. 23, 82–89 [DOI] [PubMed] [Google Scholar]
- 62. Asher S. A. (1993) Anal. Chem. 65, A59–66 [DOI] [PubMed] [Google Scholar]
- 63. Bieker L., Schmidt H. (1979) FEBS Lett. 106, 268–270 [DOI] [PubMed] [Google Scholar]
- 64. Puranik P. G., Ramiah K. V. (1959) J. Mol. Spectrosc. 3, 486–495 [Google Scholar]
- 65. Chalapathi V. V., Ramiah K. V. (1968) J. Mol. Spectrosc. 26, 444–453 [Google Scholar]
- 66. Patzlaff J. S., Barry B. A. (1996) Biochemistry 35, 7802–7811 [DOI] [PubMed] [Google Scholar]
- 67. Berlett B. S., Stadtman E. R. (1997) J. Biol. Chem. 272, 20313–20316 [DOI] [PubMed] [Google Scholar]
- 68. Gracanin M., Hawkins C. L., Pattison D. I., Davies M. J. (2009) Free Radic. Biol. Med. 47, 92–102 [DOI] [PubMed] [Google Scholar]
- 69. Asada K. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 601–639 [DOI] [PubMed] [Google Scholar]
- 70. Krieger A., Rutherford A. W., Vass I., Hideg E. (1998) Biochemistry 37, 16262–16269 [DOI] [PubMed] [Google Scholar]
- 71. Nishiyama Y., Allakhverdiev S. I., Murata N. (2006) Biochim. Biophys. Acta 1757, 742–749 [DOI] [PubMed] [Google Scholar]
- 72. Hunzinger C., Wozny W., Schwall G. P., Poznanović S., Stegmann W., Zengerling H., Schoepf R., Groebe K., Cahill M. A., Osiewacz H. D., Jägemann N., Bloch M., Dencher N. A., Krause F., Schrattenholz A. (2006) J. Proteome Res. 5, 625–633 [DOI] [PubMed] [Google Scholar]
- 73. Fedorova M., Todorovsky T., Kuleva N., Hoffmann R. (2010) Proteomics 10, 2692–2700 [DOI] [PubMed] [Google Scholar]
- 74. Finley E. L., Dillon J., Crouch R. K., Schey K. L. (1998) Protein Sci. 7, 2391–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ronsein G. E., Oliveira M. C., Miyamoto S., Medeiros M. H., Di Mascio P. (2008) Chem. Res. Toxicol. 21, 1271–1283 [DOI] [PubMed] [Google Scholar]
- 76. Yang C., Gu Z. W., Yang M., Lin S. N., Siuzdak G., Smith C. V. (1999) Biochemistry 38, 15903–15908 [DOI] [PubMed] [Google Scholar]
- 77. Eilstein J., Giménez-Arnau E., Duché D., Rousset F., Lepoittevin J. P. (2006) Chem. Res. Toxicol. 19, 1248–1256 [DOI] [PubMed] [Google Scholar]
- 78. Adir N., Zer H., Shochat S., Ohad I. (2003) Photosynth. Res. 76, 343–370 [DOI] [PubMed] [Google Scholar]
- 79. Krieger-Liszkay A., Fufezan C., Trebst A. (2008) Photosynth. Res. 98, 551–564 [DOI] [PubMed] [Google Scholar]
- 80. Nixon P. J., Michoux F., Yu J., Boehm M., Komenda J. (2010) Ann. Bot. 106, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Vass I., Styring S., Hundal T., Koivuniemi A., Aro E., Andersson B. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 1408–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Knox J. P., Dodge A. D. (1985) Phytochemistry 24, 889–896 [Google Scholar]
- 83. Keren N., Berg A., van Kan P. J., Levanon H., Ohad I. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1579–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hideg É., Spetea C., Vass I. (1994) Biochim. Biophys. Acta 1186, 143–152 [Google Scholar]
- 85. Flors C., Fryer M. J., Waring J., Reeder B., Bechtold U., Mullineaux P. M., Nonell S., Wilson M. T., Baker N. R. (2006) J. Exp. Bot. 57, 1725–1734 [DOI] [PubMed] [Google Scholar]
- 86. Telfer A., Bishop S. M., Phillips D., Barber J. (1994) J. Biol. Chem. 269, 13244–13253 [PubMed] [Google Scholar]
- 87. Mishra N. P., Francke C., van Gorkom H. J., Ghanotakis D. F. (1994) Biochim. Biophys. Acta 1186, 81–90 [Google Scholar]
- 88. Aro E. M., Virgin I., Andersson B. (1993) Biochim. Biophys. Acta 1143, 113–134 [DOI] [PubMed] [Google Scholar]
- 89. Prasil O., Adir N., Ohad I. (1992) in The Photosystems: Structure, Function and Molecular Biology (Barber J. ed) pp. 295–348, Elsevier, Amsterdam [Google Scholar]
- 90. Nishiyama Y., Allakhverdiev S. I., Yamamoto H., Hayashi H., Murata N. (2004) Biochemistry 43, 11321–11330 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.