Abstract
Chlorella virus DNA ligase (ChVLig) is a minimized eukaryal ATP-dependent DNA sealing enzyme with an intrinsic nick-sensing function. ChVLig consists of three structural domains, nucleotidyltransferase (NTase), OB-fold, and latch, that envelop the nicked DNA as a C-shaped protein clamp. The OB domain engages the DNA minor groove on the face of the duplex behind the nick, and it makes contacts to amino acids in the NTase domain surrounding the ligase active site. The latch module occupies the DNA major groove flanking the nick. Residues at the tip of the latch contact the NTase domain to close the ligase clamp. Here we performed a structure-guided mutational analysis of the OB and latch domains. Alanine scanning defined seven individual amino acids as essential in vivo (Lys-274, Arg-285, Phe-286, and Val-288 in the OB domain; Asn-214, Phe-215, and Tyr-217 in the latch), after which structure-activity relations were clarified by conservative substitutions. Biochemical tests of the composite nick sealing reaction and of each of the three chemical steps of the ligation pathway highlighted the importance of Arg-285 and Phe-286 in the catalysis of the DNA adenylylation and phosphodiester synthesis reactions. Phe-286 interacts with the nick 5′-phosphate nucleotide and the 3′-OH base pair and distorts the DNA helical conformation at the nick. Arg-285 is a key component of the OB-NTase interface, where it forms a salt bridge to the essential Asp-29 side chain, which is imputed to coordinate divalent metal catalysts during the nick sealing steps.
Keywords: ATP, DNA-Protein Interaction, DNA Repair, Enzyme Catalysis, Enzyme Structure
Introduction
DNA ligases are ubiquitous enzymes essential for DNA replication, recombination, and repair. They seal 3′-OH and 5′-PO4 ends via three sequential nucleotidyl transfer reactions (1, 2). First, nucleophilic attack on the α-phosphorus of ATP or NAD+ by ligase results in release of PPi or NMN and formation of a covalent ligase-adenylate intermediate in which AMP is linked via a P–N bond to Nζ of a lysine. Second, the AMP is transferred to the 5′ end of the 5′-phosphate-terminated DNA strand to form DNA-adenylate, AppDNA. Finally, ligase catalyzes attack by the 3′-OH of the nick on DNA-adenylate to join the polynucleotides and liberate AMP.
The ATP-dependent Chlorella virus DNA ligase (ChVLig)2 is the smallest eukaryal ligase known (298 amino acids) (3). ChVLig has an intrinsic nick-sensing function that depends on the nick 5′-PO4 group and covalent adenylylation of the ligase (3–5). ChVLig consists of three structural modules that envelop the nicked DNA as a C-shaped protein clamp, including: a nucleotidyltransferase (NTase) domain and an OB domain (these two are common to all DNA ligases) and a distinctive latch module that inserts into the major groove flanking the nick (6–8). The NTase domain (amino acids 1–189; see Fig. 1, cyan) performs the chemical steps of nick sealing (9). The NTase domain binds to the broken and intact DNA strands in the major groove flanking the nick and also in the minor groove on the 3′-OH side of the nick (see Fig. 1A). The NTase active site is composed of a cage of β strands and interstrand loops that includes six peptide motifs (I, Ia, III, IIIa, IV, and V) that define a superfamily of polynucleotide 5′-end-modifying enzymes that act through lysyl-Nζ-NMP intermediates (10). NTase motif I (25TPKIDGIR32 in ChVLig) contains the lysine to which AMP becomes covalently linked in the first step of the ligation pathway (6). The motif I Lys-27 nucleophile (see Fig. 1, B and C, K27-AMP) is located in a loop between the two antiparallel β sheets that form the adenylate-binding pocket. Other amino acids in NTase motifs I (Asp-29 and Arg-32), Ia (Ser-41 and Arg-42), III (Asp-65 and Glu-67), IIIa (Phe-98), IV (Glu-161 and Arg-166), and V (Lys-186 and Lys-188) contact AMP, the DNA nick, or neighboring catalytic residues (see Fig. 1C) and thereby play essential roles in one or more steps of the ligation pathway, as surmised from mutational effects on ChVLig activity in vitro or in vivo (4–9, 11–13). Additional essential NTase domain side chains comprise part of the DNA-binding surface (Phe-75, Met-83, Thr-84, Lys-173, and Arg-176) or aid in forming the ligase clamp around the nicked DNA (Phe-44; see Fig. 1B) (13).
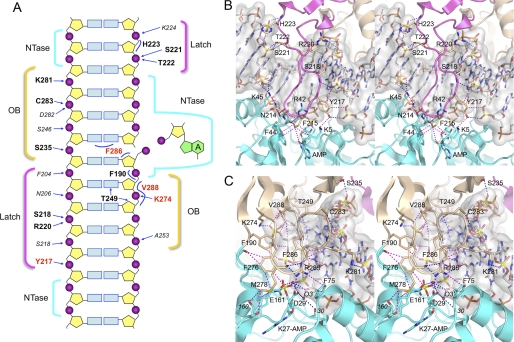
FIGURE 1.
Interface of the ChVLig OB and latch domains with nicked DNA. A, schematic summary of the DNA interface and OB and latch domain atomic contacts to the nicked DNA substrate. The nicked duplex DNA is depicted as a two-dimensional graphic, with the continuous template strand on the left and the nicked strands on the right. The lysyl-adenylate is shown at the right. The regional DNA contacts of the NTase, latch, and OB domains are denoted by cyan, yellow, and magenta brackets, respectively. Electrostatic and hydrogen-bond contacts of NTase domain side chains (residue identity in plain text) and main-chain amides or carbonyls (residue identity in italics) with DNA are indicated by arrows. Selected van der Waals interactions are indicated by curved lines. Residues defined herein as essential are highlighted in bold red font, whereas those defined presently as nonessential are in bold black font. B, stereo view of the ChVLig latch and NTase domains (shown as magenta and cyan ribbon traces with selected amino acids as stick models) bound to the nicked DNA, which is rendered as a gray transparent surface over a stick model. The image highlights how the latch occupies the major groove flanking the nick and makes contacts to the phosphates. The tip of the latch makes kissing contacts with the NTase domain that close the ligase clamp around the nick. Atomic contacts are depicted as blue dashed lines (hydrogen bonds) or magenta dashed lines (van der Waals contacts). C, a stereo view of the OB (beige) and NTase (cyan) domains highlighting the DNA contacts of the OB domain and the atomic contacts at the OB-NTase interface surrounding the nick and the adenylate-binding pocket.
The OB domain of ChVLig (see Fig. 1, beige) is composed of a five-stranded antiparallel β barrel and an α-helix. The OB domain plays a dual role in ligase catalysis. Its C-terminal peptide motif VI (293RHEEDR298) is essential for ligase adenylylation but dispensable for phosphodiester synthesis at a pre-adenylylated nick (9). Motif VI is thought to bind and orient the PPi leaving group during the attack of Lys-27 on the ATP α-phosphorus that results in ligase adenylylation. Motif VI is disordered in the crystal structures of free ChVLig-AMP and the ChVLig-AMP·DNA complex (6, 8). In the DNA-bound ligase structure, the OB domain is a major component of the DNA-binding surface. It interacts with the DNA across the minor groove on the face of the duplex behind the nick (see Fig. 1, A and C), making extensive hydrophilic and van der Waals contacts to the DNA nucleosides and phosphates (see Fig. 1C). The OB domain also contributes interdomain contacts to amino acids in the NTase domain surrounding the active site (see Fig. 1C).
The latch module (see Fig. 1, magenta) consists of a β-hairpin loop (amino acids 203–231) that emanates from the OB domain (8). The latch occupies the major groove flanking the nick and completes the circumferential clamp via contacts between the tip of the loop and the surface of the NTase domain (see Fig. 1, A and B). The latch is an important determinant of nick sensing (8). The latch module is disordered in the absence of nicked DNA (5, 6). Comparing the structures of free ligase-AMP and the ligase-AMP·DNA complex revealed how DNA binding triggers a massive conformational switch in the OB and latch modules, entailing a nearly 180° rotation of the OB domain around a swivel at residue Gln-189 so that the concave surface of the OB β barrel fits into the DNA minor groove (8). This transition results in a 63 Å movement of the OB domain and places the now structured latch module in direct contact with the DNA backbone in the major groove. The OB and latch constituents essential for these transitions and for attaining a productive binding mode with nicked DNA are entirely uncharted.
Here we used the ChVLig-AMP·DNA crystal structure to guide a mutational analysis of the OB and latch domains. We performed an alanine scan of OB domain side chains that make atomic contacts to the nicked DNA duplex and to the NTase active site. We similarly interrogated the roles of latch residues that bind the major groove or comprise the kissing interface with the NTase domain. We thereby identified seven new amino acids essential for nick sealing in vivo. Structure-activity relations were then clarified by introducing conservative substitutions. New mechanistic insights were gleaned from an analysis of mutational effects on individual steps of the ligation pathway.
EXPERIMENTAL PROCEDURES
Recombinant ChVLig
Missense mutations were introduced by PCR into the pET-ChVLig expression plasmid as described previously (4). The entire ChVLig insert was sequenced in every case to confirm the desired mutation and exclude the acquisition of unwanted changes during PCR amplification and cloning. The expression plasmids were transformed into Escherichia coli BL21(DE3). Mutant and wild-type ligases were purified from the soluble lysates of isopropyl β-d-thiogalactopyranoside-induced BL21(DE3) cells by nickel-agarose chromatography and phosphocellulose chromatography as described (4). The purity and concentration of the ChVLig preparations were determined as described previously (13). The detailed methods and purity data are documented in the supplemental Experimental Procedures and supplemental Fig. S1.
Assays of Nick Ligation
Reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 7.5), 5 mm DTT, 10 mm MgCl2, 1 mm ATP, 1 pmol of singly nicked 36-bp duplex DNA substrate (5′ 32P-labeled at the nick; prepared as described (14)), and wild-type or mutant ChVLig as specified were incubated for 10 min at 22 °C. Single-turnover nick ligation mixtures lacked exogenous ATP. The reactions were initiated by the addition of ChVLig and quenched by the addition of 10 μl of 90% formamide, 40 mm EDTA. The samples were heated at 95 °C for 5 min. The products were resolved by electrophoresis through a 15-cm 18% polyacrylamide gel containing 7 m urea in 45 mm Tris borate, 1.25 mm EDTA. The extents of ligation were determined by scanning the gel with a Fujix BAS2500 imager. Ligation was plotted as a function of input ChVLig, with each datum being the average of three separate enzyme titration experiments. The specific activities of the wild-type and mutant ligases were determined from the slopes of the titration curves in the linear range of enzyme dependence. The activities of the mutant ligases were normalized to the specific activity of wild-type ChVLig protein that was purified in parallel with that mutant and assayed in parallel using the same preparation of radiolabeled DNA substrate (see Table 1).
TABLE 1.
Mutational effects on in ChVLig activity in vivo and in vitro
+++, colony size indistinguishable from a cdc9Δ strain expressing wild-type ChVLig; ++, smaller colony size; −, no growth.
| Mutant | Nick sealing (% of WT) | cdc9Δ complementation | ||
|---|---|---|---|---|
| 18 °C | 30 °C | 37 °C | ||
| F190A | 31 | +++ | +++ | ++ |
| S235A | 112 | +++ | +++ | +++ |
| T249A | 92 | +++ | +++ | +++ |
| K274A | 34 | − | − | − |
| K274Q | 72 | − | − | − |
| K274R | 67 | − | − | − |
| F276A/M278A | 42 | ++ | ++ | ++ |
| K281A/C283A | 73 | +++ | +++ | +++ |
| S235A/K281A/C283A | 49 | +++ | +++ | ++ |
| R285A | 7 | − | − | − |
| R285K | 6 | − | − | − |
| R285Q | 8 | − | − | − |
| F286A | 4 | − | − | − |
| F286L | 61 | − | − | − |
| V288A | 58 | − | − | − |
| V288I | 91 | − | − | − |
| V288T | 74 | − | − | − |
| N214A | 51 | − | − | − |
| N214D | 94 | − | − | − |
| N214L | 113 | +++ | ++ | ++ |
| N214Q | 98 | − | − | − |
| F215A | 70 | − | − | − |
| F215L | 75 | − | − | − |
| Y217A | 44 | − | − | − |
| Y217F | 112 | − | − | − |
| Y217L | 84 | − | − | − |
| Y217S | 85 | − | − | − |
| S218A/R220A | 49 | +++ | +++ | +++ |
| S221A/T222A/H223A | 42 | +++ | +++ | ++ |
Assay of ChVLig Activity in Vivo by Complementation of Yeast cdc9Δ
NdeI-BamHI restriction fragments containing the WT and mutated ChVLig ORFs were excised from their respective pET-ChVLig plasmids and inserted into the yeast expression plasmid pYX1 (CEN TRP1), wherein the ChVLIG gene is driven by the yeast TPI1 promoter. The pYX1-ChVLig plasmids were transformed into a yeast cdc9Δ strain bearing a CEN URA3 CDC9 plasmid (15). Trp+ isolates were streaked on agar plates containing 0.75 mg/ml 5-fluoroorotic acid (FOA). Lethal mutations were those that formed no FOA-resistant colonies at 18, 30, or 37 °C (scored as − at all temperatures in Table 1). The viable cdc9Δ ChVLIG yeast strains were tested for growth on YPD agar at 18, 30, and 37 °C. +++ indicates colony size indistinguishable from a cdc9Δ strain expressing wild-type ChVLig; ++ indicates smaller colony size; − indicates no growth (Table 1).
RESULTS
Mutagenesis Strategy
The OB domain occupies the minor groove flanking the nick and interacts extensively with a 5-nucleotide segment of the 5′-PO4 strand (Fig. 1A). Thr-249 makes a bifurcated hydrogen bond to the deoxyribose O4′ atom of the third nucleoside and to the adenine N3 atom of the second nucleoside. Val-288 makes van der Waals contacts with the second and third deoxynucleoside sugars and the intervening phosphate. Lys-274 coordinates the third phosphate of the 5′-PO4 strand. Phe-190 makes van der Waals contacts with the phosphate and the deoxyribose C5′ atoms of the second nucleotide of the 5′-PO4 strand. At the nick, Phe-286 packs against 5′-terminal deoxynucleoside sugar and forces the terminal base pair to adopt an RNA-like A-helical conformation (Fig. 1C). OB domain side chains also engage several phosphate groups of the template DNA strand on the 3′-OH side of the nick (Fig. 1A), via hydrogen bonds from Lys-281 Nζ, Cys-283 Sγ, and Ser-235 Oγ (Fig. 1C). The NTase and OB domains interface with each other through contacts emanating from the loop connecting the fourth and fifth β strands of the OB domain. These include: a salt bridge from Arg-285 to Asp-29; hydrogen bonding from Arg-285 to the Met-278 carbonyl; a cation-π interaction between Arg-285 and Phe-75; and van der Waals interactions between Glu-161 (NTase motif III) and both Phe-276 and Met-278 (Fig. 1C). Here we mutated OB domain constituents Ser-235, Thr-249, Lys-274, Arg-285, Phe-286, and Val-288 individually to alanine. We also made compound mutations as follows: F276A/M278A; K281A/C283A and S235A/K281A/C283A.
The latch contacts a 5-nucleotide segment of the template strand in the major groove on the 5′-PO4 side of the nick (Fig. 1A). The Tyr-217, Ser-218, and Arg-220 side chains and Phe-204, Asn-206, and Ser-218 main-chain amides donate hydrogen bonds to the five backbone phosphates. Ser-218 and Arg-220 coordinate the same phosphodiester, via the nonbridging O2P and O1P atoms, respectively (Fig. 1B). Tyr-217 makes additional van der Waals contacts to the DNA backbone (Fig. 1B). The latch also engages a 2-nucleotide segment of the broken 3′-OH strand (Fig. 1A) via hydrogen bonds to the vicinal phosphates from Ser-221 Oγ, Thr-222 Oγ, and the Lys-224 main-chain amide and via van der Waals contacts of His-223 to the bridging nucleoside sugar (Fig. 1B). The clamp-closing interface between the tip of the latch loop and the NTase domain is depicted in Fig. 1B. It entails a network of van der Waals contacts from Phe-215 and Tyr-217 of the latch and Phe-44 and Lys-5 of the NTase domain, van der Waals contacts of Phe-215 to the Arg-42 main-chain carbonyl and Thr-43 Cα, and van der Waals contacts from Asn-214 to Lys-45 Nζ and the Thr-43 main-chain carbonyl. Here we introduced single alanine mutations at Asn-214, Phe-215, and Tyr-217, a double-alanine change at S218A/R220A, and a triple-alanine substitution at S221A/T222A/H223A.
Effects of Alanine Mutations on ChVLig Function in Vivo in Yeast
A deletion of the gene encoding the essential Saccharomyces cerevisiae ATP-dependent DNA ligase Cdc9 can be complemented by expression of ChVLig (15). Viability of the yeast cdc9Δ strain is contingent on maintenance of an extrachromosomal CDC9 gene on a CEN URA3 plasmid. Hence, cdc9Δ cells cannot grow on medium containing FOA (a drug that selects against the URA3 CDC9 plasmid), but they can grow on FOA if the cells have been transformed with a CEN TRP1 plasmid expressing wild-type ChVLIG under the control of the constitutive yeast TPI1 promoter. Here we tested by the plasmid shuffle assay (15) whether the 15 ChVLig-Ala mutants were functional in yeast. We found that seven single-alanine mutations were lethal in vivo, i.e. the ChVLIG-Ala alleles were unable to support growth of cdc9Δ on FOA at 18, 30, or 37 °C (scored as − in Table 1). The lethal changes were: K274A, R285A, F286A, and V288A in the OB domain and N214A, F215A, and Y217A in the latch (Table 1).
Eight of the ChVLIG-Ala alleles supported growth of cdc9Δ cells on FOA at one or more of the selection temperatures. Four of the viable cdc9Δ ChVLIG-Ala strains grew as well as the wild-type cdc9Δ ChVLIG strain on YPD agar medium at 18, 30, and 37 °C, as gauged by colony size (scored as +++ in Table 1). The fully functional mutants were S235A, T249A, and K281A/C283A in the OB domain and S218A/R220A in the latch. Four of the viable mutants displayed mild to modest hypomorphic growth phenotypes. F190A, S235A/K281A/C283A, and S221A/T222A/H223A cells grew as well as wild-type ChVLIG yeast at 18° and 30 °C but formed smaller colonies (++) at 37 °C. ChVLIG-(F276A/M278A) cells formed smaller colonies than wild-type ChVLIG yeast at all temperatures (Table 1). These results signify that none of the contacts of the OB domain and latch domain side chains with the template and broken DNA strands on the 3′-OH side of the nick are essential for ligase activity in vivo.
Structure-Activity Relationships in Vivo at Essential Residues of the OB and Latch Domains
We tested the effects of conservative substitutions for the seven essential amino acids identified in the alanine scan. Lys-274 was replaced with arginine and glutamine; Arg-285 was replaced with lysine and glutamine; Phe-215 and Phe-286 were replaced with leucine; Val-288 was replaced with isoleucine and threonine; Asn-214 was replaced with aspartate, glutamine, and leucine; and Tyr-217 was replaced with phenylalanine, leucine, and serine. The conservative mutants were tested for activity in yeast cdc9Δ complementation (Table 1).
Lys-274 was strictly essential (i.e. conservative changes did not restore function), implying that its ionic interaction with the third phosphodiester of the 5′-PO4 strand seen in the crystal structure is the pertinent property, which cannot be fulfilled by the larger arginine side (presumably because of steric issues). Arg-285 was also strictly essential, thereby attesting to the importance of its multivalent ionic and hydrogen binding interactions to surrounding amino acids in the ligase active site, which could not be sustained by lysine (Fig. 1C). Phe-215 and Phe-286 could not be functionally substituted with leucine (an aliphatic γ-branched partial isostere of Phe), signifying that the van der Waals contacts of the Cϵ and Cζ atoms of the phenyl rings of Phe-215 (Fig. 1B) and Phe-286 (Fig. 1C) are important for ChVLig activity in vivo. Val-288 could not be substituted by the β-branched hydrophobe isoleucine (presumably because of hindrance by the extra methyl group) or by the polar isostere threonine (Table 1). Replacing the Val-288 Cγ2 atom with a threonine hydroxyl might perturb its van der Waals contact to the deoxyribose C4 atom of the penultimate nucleoside of the 5′-PO4 strand (Fig. 1C). It is noteworthy that Lys-274 and Val-288, located in neighboring β strands of the OB domain, are both essential in vivo and contact the same DNA phosphate (Fig. 1C). Replacing Asn-214 with the apolar γ-branched leucine restored ChVLig activity in vivo, whereas aspartate and glutamine substitutions did not (Table 1); this result underscores the pertinence of the van der Waals interactions of Asn-214 at the latch-NTase interface (Fig. 1B). Finally, replacing Tyr-217 with phenylalanine failed to restore yeast growth, implying that the hydrogen bond from the phenolic hydroxyl to the DNA backbone is essential for activity in vivo.
Mutational Effects on Nick Sealing Activity in Vitro
The wild-type and mutated ChVLig proteins were produced as N-terminal His10 fusions and purified from soluble bacterial extracts by nickel-agarose and phosphocellulose chromatography (supplemental Fig. S1). The extent of ligation of singly nicked 36-bp DNA substrate (labeled with 32P at the 5′-PO4 of a centrally placed 3′-OH/5′PO4 nick) by wild-type ChVLig and each mutant was gauged as a function of input enzyme. To determine specific activities, the nick sealing data from three independent enzyme titration experiments were averaged, and the mean values for ligation (fmol of nicks sealed) were plotted versus fmol of input ChVLig. The specific activities were calculated (in Prism) from the slopes of the titration curves in the linear range of enzyme dependence and then normalized to the wild-type specific activity of 82 fmol of nicks sealed per fmol of ChVLig (defined as 100%). The results are compiled in Table 1. Our operational definition (13) of a functionally important residue is one at which alanine substitution reduced specific activity in nick joining to ≤25% of wild-type ChVLig. An essential residue is one at which alanine mutation reduced activity to ≤10%. By these criteria, 2 of the 19 targeted residues were deemed essential: Arg-285 and Phe-286, with 7 and 4% of wild-type activity, respectively (Table 1). None of the targeted residues was deemed important. Although the two alanine changes that defined essentiality for nick sealing under standard conditions in vitro were associated with lethality in vivo in yeast (Table 1), there were several instances in which alanine mutations with relatively little impact on nick sealing in vitro resulted in loss of function in vivo (e.g. V288A, N214A, and F215A).
Structure-activity relations for nick sealing in vitro were gauged from the specific activities of conservative mutants (Table 1). In some cases, the in vitro data correlated nicely with the in vivo effects on ChVLig function. For example, the lethal R285K and R285Q substitutions afforded no improvement in their nick sealing activities in vitro (which were 6 and 8% of wild type, respectively) when compared with R285A (Table 1). However, it was more often the case that in vitro and in vivo activities did not track together, e.g. F286L elicited a significant gain of function in vitro (to 61% of wild type versus 4% for F286A) but did not restore activity in yeast. The K274Q and K274R mutations resulted in 2-fold increases in nick sealing (to 72 and 67% of wild type) versus K274A (34%), but this did not suffice to confer cdc9Δ complementation (Table 1), notwithstanding that other ChVLig mutants with lower specific activity in vitro were functional in yeast (e.g. S218A/R220A, F190A). Similarly, the Y217F change restored nick sealing to wild-type levels (112% versus 44% for Y217A) but did not rescue ChVLig function in yeast. The only case in which gains of function tracked together in vitro and in vivo was N214L (113% of wild-type specific activity versus 51% for N214A), although similar increases in the N214D and N214Q activities (to 94 and 98%, respectively) did not revive function in yeast. Possible reasons for some of these discrepancies are considered below.
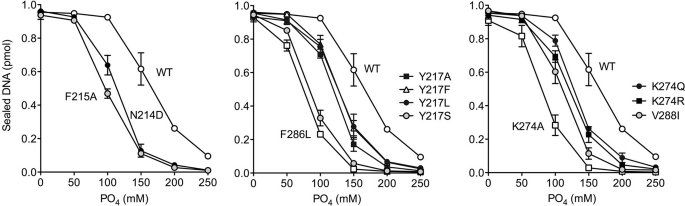
Assay of Nick Sealing under More Stringent Reaction Conditions Exacerbates Functional Deficits of ChVLig Mutants
Our standard nick sealing reactions contained 50 mm Tris-HCl buffer, 10 mm MgCl2, 1 mm ATP, and no added salt above the 1–2 mm NaCl contributed by the enzyme solution. We showed previously (13) that such relatively nonstringent hypotonic conditions can mask the effects of mutations at the ligase-DNA interface on sealing in vitro, i.e. the loss of one of many protein-DNA contacts might have little impact at low ionic strength in vitro but still exert significant effects in vivo in yeast, where the intracellular monovalent cation concentration during exponential phase growth on YPD medium is reported to be 330 mm and the concentration of inorganic phosphate is ∼25 mm (16, 17). To explore this issue here, we gauged the effects of increasing phosphate concentration on the extent of nick sealing by wild-type ChVLig and selected ChVLig mutants that were inactive in yeast, despite retaining substantial ligase activity in vitro under standard conditions. The amounts of each enzyme added were varied to attain similar extents of sealing in the control reactions containing no added phosphate. The results are shown in Fig. 2. Wild-type ChVLig activity was reduced by 4, 35, 73, and 91% at 100, 150, 200, and 250 mm phosphate, respectively. Mutants F215A, N214D, F286L, Y217S, Y217L, Y217F, K274A, K274Q, K274R, and V288I were hypersensitive to inhibition by 100–250 mm phosphate concentrations (Fig. 2). Thus, increasing the stringency of the in vitro assay conditions does exacerbate the functional impact of many of the ChVLig OB and latch mutations and helps account for their lethality in yeast. (Other lethal ChVLig mutants did not display phosphate hypersensitivity, e.g. N214A, N214Q, V288A, V288T, and F215L; not shown.)
FIGURE 2.
Sensitivity of nick sealing to inhibition by phosphate. Reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 7.5), 5 mm DTT, 10 mm MgCl2, 1 mm ATP, 1 pmol of 5′ 32P-nick-labeled DNA substrate, Na3PO4 as specified, and wild-type or mutant ChVLig were incubated at 22 °C for 10 min. The amounts of each ChVLig preparation in the reaction mixtures (1.7 ng of WT; 3.3 ng of K274A; 4 ng of K274Q; 3.3 ng of K274R; 3.3 ng of F286L; 1.7 ng of V288I; 2.8 ng of N214D; 2.7 ng of F215A; 3.5 ng of Y217A; 3.5 ng of Y217F; 3.3 ng of Y217L; and 3.33 ng of Y217S) were adjusted to attain comparable levels of nick sealing in the control reactions lacking added phosphate. The extents of nick sealing by each ChVLig preparation are plotted as a function of added Na3PO4 concentration. Each datum is the average of three separate phosphate titration experiments ± S.E.
Mutational Effects on Ligase Adenylylation
Recombinant ChVLig purified from bacteria comprises a mixture of ligase apoenzyme and preformed enzyme-AMP intermediate. Only the apoenzyme is available to react in vitro with [α-32P]ATP (in the absence of DNA) to form a covalent ChVLig-[32P]adenylate adduct. The autoadenylylation activities of the wild-type and mutant ChVLig preparations were assayed in parallel in the presence of 100 μm [α-32P]ATP; the results are depicted in supplemental Fig. S2 and expressed in Table 2 as the percentage of total input enzyme that was labeled in vitro with [32P]AMP. In the case of wild-type ChVLig, 37% of the available enzyme was labeled.
TABLE 2.
Mutational effects on ChVLig adenylylation
| Mutant | Preformed Lig–AMP in vivo (% of total enzyme) | Lig–[32P] AMP in vitro (% of total enzyme) |
|---|---|---|
| WT | 60 | 37 |
| F190A | 48 | 12 |
| S235A | 48 | 20 |
| T249A | 53 | 44 |
| K274A | 46 | 28 |
| K274Q | 27 | 45 |
| K274R | 35 | 46 |
| F276A/M278A | 60 | 31 |
| K281A/C283A | 36 | 41 |
| S235A/K281A/C283A | 75 | 22 |
| R285A | 45 | 34 |
| R285K | 18 | 32 |
| R285Q | 51 | 45 |
| F286A | 14 | 7 |
| F286L | 23 | 34 |
| V288A | 38 | 54 |
| V288I | 47 | 41 |
| V288T | 49 | 44 |
| N214A | 56 | 39 |
| N214D | 40 | 41 |
| N214L | 56 | 33 |
| N214Q | 53 | 36 |
| F215A | 53 | 39 |
| F215L | 60 | 40 |
| Y217A | 57 | 41 |
| Y217F | 57 | 41 |
| Y217L | 51 | 42 |
| Y217S | 56 | 42 |
| S218A/R220A | 37 | 44 |
| S221A/T222A/H223A | 58 | 39 |
The level of preformed Lig-AMP can be determined by assaying nick sealing in the absence of exogenous ATP, such that there is a 1:1 correspondence between the molar yield of ligated DNA and the molar amount of catalytically active Lig-AMP in the reaction. Here we gauged the extent of ligation of singly nicked 36-bp DNA substrate by wild-type ChVLig and each mutant as a function of input enzyme in the absence of ATP. The nick sealing data from three independent enzyme titration experiments were averaged, and the mean values for ligation (fmol of nicks sealed) were plotted versus fmol of input ChVLig. The concentration of active Lig-AMP was calculated (in Prism) from the slopes of the titration curves in the linear range of enzyme dependence and then expressed in Table 2 as the percentage of the total enzyme comprising preformed Lig-AMP. In the case of wild-type ChVLig, 60% of the available enzyme was pre-adenylylated and active in single-turnover nick sealing. Summing 60% preformed Lig-AMP and 37% reactive apoenzyme, we conclude that ∼97% of the wild-type preparation was catalytically active in one or more steps of the ligation pathway.
As one might expect, given that the NTase domain embraces the active site, almost all of the OB and latch domain mutants displayed wild-type or near wild-type summed levels of reactive preformed Lig-AMP plus apoenzyme, as follows: F190A (60%); S325A (68%); T249A (97%); K274A (74%); K274Q (72%); K274R (81%); F276A/M278A (91%); K281A/C283A (77%); S235A/K281A/C283A (97%); R285A (79%); R285K (50%); R285Q (96%); F286L (57%); V288A (92%); V288I (88%); V288T (93%); N214A (95%); N214D (81%); N214L (89%); N214Q (89%); F215A (92%); F215L (100%); Y217A (98%); Y217F (98%); Y217L (93%); Y217S (98%); S218A/R220A (81%); S221A/T222A/H223A (97%). We can surmise from these data that the defects of the R285A, R285K, and R285K mutant in overall nick sealing (Table 1) are attributable to feeble execution of one or more steps subsequent to ligase adenylylation.
The lone exception was F286A, which was defective in adenylylation in vitro (7% AMP labeling) and for which active preformed Lig-AMP comprised only 14% of the enzyme preparation (Table 2). These results suggest a role for Phe-286 during step 1 of the ligation reaction, which can be fulfilled in good measure when Phe-286 is substituted by leucine, but not alanine. Although we do not have a crystal structure of ChVLig bound to ATP (prior to step 1 chemistry), we speculate that Phe-286 (which is located close to motif VI) might comprise part of the interdomain interface generated when the ligase adopts the “closed” conformation proposed for step 1 catalysis (6, 10, 18). This transition is thought to entail a large motion of the OB domain that places it directly over the adenylate site, where it contacts the PPi leaving group of ATP and orients it apically to the attacking motif I lysine.
Kinetics of Single-turnover Nick Sealing by Lig-AMP and Mutational Effects on Step 2 Catalysis
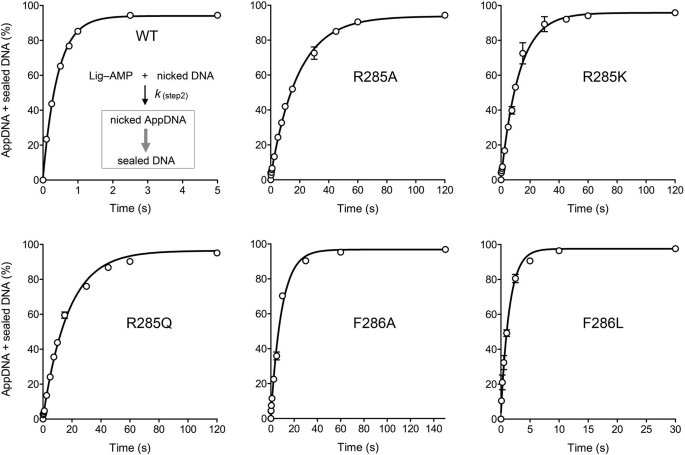
We subjected wild-type ChVLig and the mutants that were nonfunctional in yeast to transient-state kinetic analysis of single-turnover nick sealing by preformed Lig-AMP in the absence of added ATP (13). A rapid chemical quench apparatus was used to assay the reaction of nicked DNA with a 10-fold molar excess of ChVLig-AMP in the range of 0.1–5-s reaction times. (Longer time points were assayed manually, where warranted.) The products were analyzed by denaturing PAGE, and the distributions of 32P-labeled DNAs (as sealed 36-mer DNA product, 18-mer AppDNA intermediate, and residual 18-mer pDNA substrate) were quantified. The rate of step 2 catalysis (DNA adenylylation) was gauged by plotting the sum of AppDNA plus sealed DNA as function of reaction time for the wild-type and mutant ChVLig preparations (Fig. 3 and supplemental Fig. S3). Virtually all the pDNA substrate was converted to ligated product by wild-type ChVLig; the kinetic profile conformed to a single exponential function by nonlinear regression curve fitting (Fig. 3). The apparent rate constant for DNA adenylylation by wild-type ChVLig-AMP was 2.37 s−1 (Table 3). Because halving the concentration of ChVLig-AMP did not significantly affect this value (not shown), we surmise that Lig-AMP binding to the nick is not rate-limiting under our reaction conditions and that kobs plausibly reflects the rate of the step 2 chemistry.
FIGURE 3.
Kinetics of single-turnover DNA adenylylation and nick sealing by pre-adenylylated ChVLig-AMP. A Kintek RQF3 rapid chemical quench apparatus was used to assay the reaction of nicked DNA with a 10-fold molar excess of preformed ChVLig-AMP at 22 °C in the absence of added ATP, in the range of 0.1–5-s reaction times. Where applicable, longer reaction times were assayed manually. The reaction mixtures contained 50 mm Tris-HCl (pH 7.5), 5 mm DTT, 10 mm MgCl2, 500 nm wild-type or mutant ChVLig-AMP, and 50 nm 5′ 32P-nick-labeled DNA substrate. The rapid kinetic measurements were initiated by mixing two buffer solutions (20 μl each of 50 mm Tris-HCl, pH 7.5, 5 mm DTT, 10 mm MgCl2) containing 100 nm nicked DNA substrate and 1 μm ChVLig-AMP, respectively. The reactions were quenched by rapid mixing with 110 μl of 90% formamide, 50 mm EDTA. The products were analyzed by denaturing PAGE, and the distributions of 32P-labeled DNAs (as sealed 36-mer DNA product, 18-mer AppDNA intermediate, and residual 18-mer pDNA substrate) were quantified by scanning the gels. The rate of step 2 catalysis (DNA adenylylation) was gauged by plotting the sum of AppDNA plus sealed DNA (see the reaction scheme in the top left panel) as a function of reaction time for the wild-type and mutant ChVLig preparations. Each datum in the graphs is the average of three separate kinetic experiments ±S.E. Nonlinear regression curve fitting of the data to a single exponential was performed in Prism; the calculated step 2 rate constants are compiled in Table 3.
TABLE 3.
Mutational effects on single-turnover DNA adenylylation and phosphodiester synthesis
| ChVLig | Step 2 rate constanta | Step 3 rate constantb |
|---|---|---|
| s−1 | s−1 | |
| WT | 2.37 ± 0.16 | 24.1 ± 1.69 |
| K274A | 2.44 ± 0.16 | 11.5 ± 0.38 |
| K274Q | 2.84 ± 0.24 | 11 ± 0.61 |
| K274R | 2.29 ± 0.12 | 14 ± 0.8 |
| R285A | 0.05 ± 0.003 | 0.2 ± 0.01 |
| R285K | 0.08 ± 0.006 | 0.46 ± 0.01 |
| R285Q | 0.06 ± 0.006 | 0.24 ± 0.01 |
| F286A | 0.11 ± 0.01 | 2.01 ± 0.35 |
| F286L | 0.69 ± 0.08 | 17.5 ± 2 |
| V288A | 1.16 ± 0.06 | 17.7 ± 0.58 |
| V288I | 1.61 ± 0.16 | 31.5 ± 2 |
| V288T | 2.33 ± 0.14 | 30.8 ± 2.18 |
| N214A | 1.88 ± 0.19 | 23.5 ± 1.45 |
| N214D | 1.79 ± 0.13 | 21.8 ± 0.83 |
| N214Q | 2.48 ± 0.31 | 22.7 ± 1.38 |
| F215A | 1.46 ± 0.1 | 21.1 ± 1.5 |
| F215L | 2.34 ± 0.15 | 27.4 ± 1.26 |
| Y217A | 1.72 ± 0.09 | 22.5 ± 1.87 |
| Y217F | 1.79 ± 0.1 | 23.3 ± 2.27 |
| Y217L | 1.9 ± 0.17 | 21.5 ± 2.32 |
| Y217S | 1.64 ± 0.16 | 28.3 ± 1.65 |
a The step 2 rate constants were derived by fitting the kinetic data in Fig. 3 and supplemental Fig. S3 to a single exponential in Prism.
b The step 3 rate constants were calculated in MATLAB from the kinetic profiles in Fig. 4 and supplemental Fig. S4 according to the reaction scheme depicted in Fig. 4 (see supplemental Table S1).
We observed a hierarchy of mutational effects on step 2 catalysis, ranging from: (i) no significant impact (kobs within a factor of two of wild type), which was the case for K274A, K274Q, K274R, V288I, V288T, N214A, N214D, N214Q, F215L, Y217A, Y217F, Y217L, and Y217S; to (ii) modest (2–3-fold) rate reductions in the cases of V288A and F286L, with kobs values of 1.16 and 0.69 s−1, respectively; to (iii) severe (20–50-fold) rate decrements, in the cases of F286A (0.11 s−1), R285A (0.05 s−1), R285K (0.08 s−1), and R285Q (0.06 s−1). The step 2 defect of F286A attested to the imputed role of this residue in distorting the local DNA conformation at the nick into an RNA-like A helix (8) by packing against the terminal deoxyribose sugar of the 5′-PO4 strand and the terminal nucleobase pair on the 3′-OH side of the nick (Fig. 1C) (14). The step 2 defect was only partially alleviated by introducing leucine at position 286, a maneuver that would be expected, in light of the crystal structure (Fig. 1C), to elicit less of the requisite distortion of the 5′-PO4 nucleoside. The profound step 2 defects of the Arg-285 mutants highlight the essentiality of this residue in organizing the active site for adenylate transfer to the nick.
Requirements for Arg-285 and Phe-286 during DNA Adenylylation (Step 2) and Phosphodiester Synthesis (Step 3)
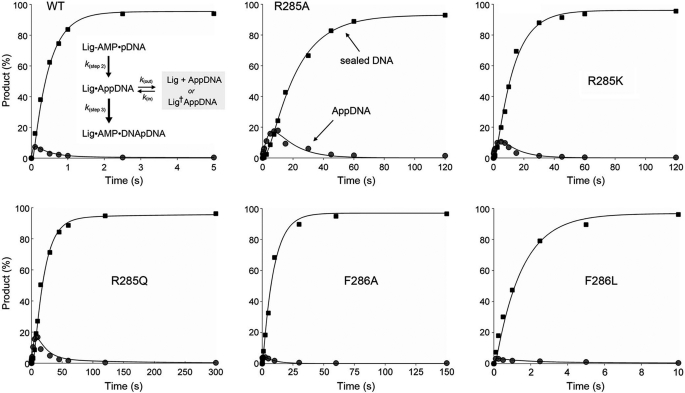
With the use of a rapid mix-quench protocol (13), we were able to visualize flux through AppDNA en route to the ligated end-product (Fig. 4). For wild-type ChVLig, the AppDNA intermediate peaked at 0.1 s, at which time AppDNA comprised 7% of the labeled DNA and sealed 36-mer DNA comprised 16%. AppDNA decayed steadily thereafter as the sealed product accumulated. The wild-type ligation reaction proceeded to completion by 2.5 s. As reported previously (13), the kinetic profiles for AppDNA and ligated product in the single-turnover reactions catalyzed by wild-type ChVLig-AMP (and various mutated versions thereof) fit well to the kinetic scheme shown in Fig. 4, which incorporates a reversible transition of the step 2 product (AppDNA) from an active “in pathway” state to an inactive “out of pathway” state (described by rate terms kout and kin). The scheme posits that ligation steps 2 and 3 are effectively unidirectional under our assay conditions; these assumptions are consistent with available data (5, 9, 19). The first-order differential equations for the kinetic scheme (13) were solved in MATLAB (version R2010b, The MathWorks, Inc.). Simulations and curve fitting to the experimental data were also performed in MATLAB by importing the experimentally derived step 2 rate constants (Table 3) and stipulating only that k(step 3), kout, and kin must be greater than zero. The modeled curves for the wild-type ChVLig and the OB and latch mutants are plotted in Fig. 4 and supplemental Fig. S4 and fit well to the experimental data. The sets of rate constants calculated in MATLAB are compiled in supplemental Table S1. The step 3 rate constants are included in Table 3.
FIGURE 4.
Flux through DNA-adenylate to ligated product during single-turnover nick sealing. The distributions of 18-mer AppDNA intermediate (gray circles) and sealed 36-mer DNA (black squares) during single-turnover nick sealing by the indicated ChVLig-AMP proteins are plotted as a function of reaction time. Each datum in the graphs is the average of three separate kinetic experiments. The data were modeled in MATLAB to the kinetic scheme depicted in the top left panel, and the resulting curve fits are shown.
The apparent step 3 rate constant for wild-type ChVLig was 24.1 s−1; thus catalysis of the attack of the nick 3′-OH on AppDNA was 10-fold faster than the formation of AppDNA. Most of the mutant ligases displayed step 3 rates similar to wild type (Table 3 and supplemental Table S1). The K274A, K274Q, and K274R mutations elicited modest (2-fold) step 3 rate decrements (11.5, 11, and 14 s−1, respectively), but no effect on step 2 rate, which resulted in a higher transient accumulation of the nicked DNA-adenylate intermediate (to 12–18% of the DNA at 0.1–0.25 s) than in the wild-type reaction. Mutant F286A caused a 12-fold rate decrement in the step 3 rate constant (2 s−1), thereby underscoring the key role of the Phe-286-mediated distortion of the nick in attaining an optimal catalytic conformation for both DNA adenylylation and phosphodiester synthesis. The F286A change exerted a relatively greater impact on step 2 than step 3, such that its k(step 3)/k(step 2) ratio was 18 (versus 10 for wild-type ChVLig). Installing leucine at position 286 restored the step 3 rate to 17.5 s−1 (Table 3).
Mutating Arg-285 to alanine, glutamine, and lysine slowed step 3 catalysis by factors of 120, 100, and 50, respectively. The k(step 3) values were 0.2, 0.24, and 0.46 s−1 for R285A, R285Q, and R285K (Table 3). Although the network of Arg-285 atomic contacts is clearly critical for both step 2 and step 3 reactions at the nick, the alanine and glutamine mutations exerted a relatively greater impact on the rate of phosphodiester synthesis than on DNA adenylylation, i.e. the k(step 3)/k(step 2) ratios of R285A (0.2/0.05 = 4) and R285Q (0.24/0.06 = 4) were lower than that of wild-type ChVLig. This was reflected in a higher transient accumulation of the nicked DNA-adenylate intermediate (to 17–18% of the DNA) during single-turnover ligation (Fig. 4).
DISCUSSION
Here we conducted a structure-guided mutational analysis of the ChVLig OB and latch domains, surveying the effects of 29 mutations in 19 residues on DNA ligase activity in vivo and in vitro. Alanine scanning defined seven individual amino acids as essential in vivo (Lys-274, Arg-285, Phe-286, Val-288, Asn-214, Phe-215, and Tyr-217), after which structure-activity relations were clarified by conservative substitutions. The contributions of Arg-285 and Phe-286 to catalysis of nick 5′ adenylylation and phosphodiester synthesis were illuminated by transient state kinetic analyses. The other residues mutated, even those essential in vivo, contributed little to the chemical steps of the nick ligation pathway under routine in vitro conditions, consistent with the remoteness of these residues from the ligase active site. Our results highlight the distinct roles of the latch and OB domains, as discussed below.
Clamp Closure Is a Key Function of the Latch Module via Contacts to the NTase Domain
It was surprising to us that subtraction of most of the amino acid side-chain contacts between the latch and the DNA sugar phosphate backbone had little effect on ChVLig activity in vivo or in vitro. On the 3′-OH side, simultaneous removal of all three DNA-binding latch side chains (Ser-221, Thr-222, and His-223) was benign. On the 5′-PO4 side, the DNA-binding Ser-218 and Arg-220 side chains were collectively inessential. We had assumed, based on the crystal structure, that the latch side-chain DNA contacts were instrumental in templating the disordered-to-ordered transition of the latch upon the encounter of ChVLig with nicked DNA, which was posited to be a key step in clamp closure (8). The present functional analysis makes it clear that most of the side-chain DNA contacts are not required for effective clamp closure. The default hypothesis is that the hydrogen bonds between main-chain amides of the latch module and the DNA phosphates (via Phe-204, Asn-206, and Ser-218 to the template strand and Lys-224 to the 3′-OH strand; Fig. 1A) are sufficient to tack down the latch over the DNA major groove.
The salient positive findings here are that the Asn-214, Phe-215, and Tyr-217 side chains at the tip of the latch hairpin that make the kissing contacts to the NTase domain are each essential for ChVLig activity in vivo. van der Waals contacts between the latch residues and both main-chain and side-chain atoms of the NTase domain predominate at the latch-NTase interface (Fig. 1B). Although Phe-215 and Tyr-217 both contact the Lys-5 side chain, we surmise that this interaction is not contributory to ChVLig activity in yeast, insofar as a K5A mutation did not affect cdc9Δ complementation or nick sealing in vitro (13). Rather, we suspect that the interactions of Phe-215 with the Phe-44 phenyl ring and Arg-42 and Thr-43 main-chain atoms, plus the contacts of Asn-214 with the Lys-45 side chain and Thr-43 main chain, are the critical clamp-closing contacts that make latch residues Phe-215 and Asn-214 essential in vivo. The “latch receptor” segment in the NTase domain corresponds to the motif Ia loop (41SRTFKPIR48) that is itself critical for ligase activity in vivo and in vitro. The motif Ia residues Thr-43 and Lys-45 that contact the second and third phosphates from the 3′-OH end, respectively, are essential pairwise in vivo, although functionally redundant. The Thr-43-Lys-45 dyad contributes to DNA binding (13). Motif I residue Ser-41, which is essential per se (13), contacts the same DNA phosphate as nonessential Thr-43, but they donate hydrogen bonds to different nonbridging phosphate oxygens. Arg-42 in motif Ia is essential in vivo and in vitro; Arg-42 is located adjacent to the nick 5′-phosphate and 3′-terminal phosphodiester and accelerates catalysis of steps 2 and 3 of the nick sealing pathway by an order of magnitude (13). Finally, motif Ia side-chain Phe-44 (which is contacted by Phe-215 of the latch) is essential for ChVLig function in vivo, i.e. the F44A mutant did not complement cdc9Δ (13). Viability of F44L underscored the contributions of the van der Waals contacts of Phe-44 to the latch Phe-215 side chain seen in the crystal structure. The F44A mutation sensitized ChVLig to inhibition by increasing ionic strength (13); we saw a similar sensitization in the present study by F215A (Fig. 2). The key contribution of latch residue Tyr-217 appears to be at the level of the DNA interface, via its hydrogen bond to a DNA phosphate and its surface complementarity to a dinucleotide segment of the template strand, to which it makes several van der Waals contacts (Fig. 1B). Loss of the Tyr-217 atomic contacts sensitized nick sealing to inhibition by phosphate anion. In sum, the available structural and mutational data highlight the tip of the latch as its business end, which, via its contacts to DNA and the NTase domain, provides topological stability to the ligase clamp and promotes an active conformation of the motif Ia loop at the nick.
Essential Functions of the OB Domain in Binding and Distorting the DNA Nick
Binding of ChVLig-AMP to nicked duplex DNA induces a 12° bend in the DNA centered at the nick (8). The DNA helix has typical B-form secondary structure throughout, except at the two base pairs on either side of the nick, which adopt an A-like conformation. The OB domain occupies the minor groove and is thought to promote these structural transitions. Here we found that loosening of specific OB domain contacts (via Lys-274 and Val-288) to the third phosphate of the 5′-PO4 strand compromised ChVLig function in vivo and nick sealing in vitro under conditions of increased ionic strength. By contrast, simultaneously mutating several other OB domain residues (Lys-281, Cys-283, and Ser-235) that contact the template strand phosphodiester backbone did not affect cdc9Δ complementation. It is conceivable that the several hydrogen bonds to consecutive template strand phosphates from main-chain amide nitrogens of the OB domain (via Asp-282 and Ser-246; Fig. 1A) suffice to establish the minor groove interface of the OB domain with the template strand.
Closer to the nick, the OB domain makes essential DNA contacts via Phe-286, which packs against and distorts the 5′-phosphate terminal nucleotide and the 3′-OH base pair. It also makes essential NTase domain contacts in the active site via Arg-285, which forms a salt bridge to the motif I Asp-29 side chain and also intercalates between the Phe-286 and Phe-75 aromatic rings (Fig. 5, top panel). Phe-75 is an essential residue that packs against the nick 3′-OH nucleotide and perturbs the helical conformation of the 3′-OH strand. Collectively, the conformational changes at the nick elicited by Phe-75, Arg-285, and Phe-286 prepare the DNA substrate for nick sensing by ChVLig and for the formation of catalytically proficient Lig-AMP·pDNA and Lig·AppDNA binary complexes. As discussed previously (13), a mechanism entailing induced fit of the nicked DNA into a bent/distorted conformation when bound within the ligase clamp can account for the selective binding of ChVLig-AMP to nicked DNA versus an otherwise identical duplex. The nick allows the DNA conformational freedom to adapt to the ligase interface, whereas the closed duplex is less free to do so.
FIGURE 5.
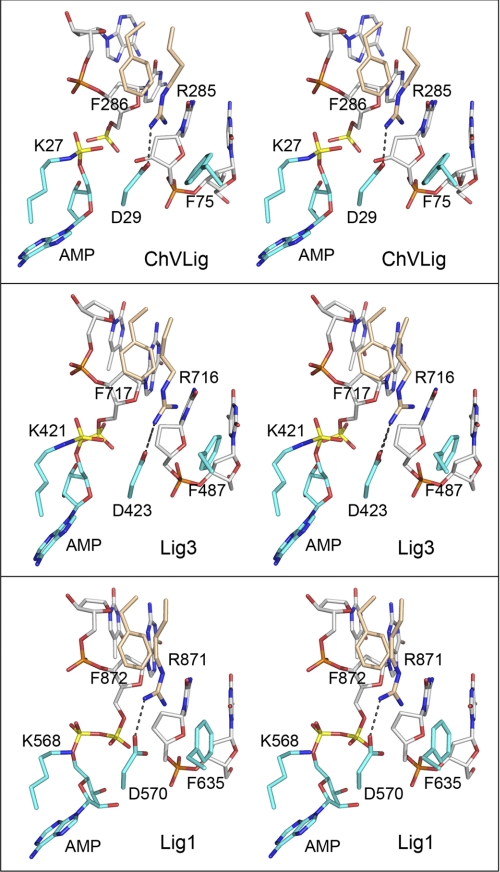
The ChVLig Arg-285-Phe-286, Phe-75, and Asp-29 interaction network at the nick is conserved in human DNA ligases. Aligned stereo images of DNA-bound ChVLig (from 2Q2T; top panel), human Lig3 (from 3L2P; middle panel), and human Lig1 (from 1X9N; bottom panel) highlighting the terminal dinucleotides of the nicked strands (a 5′-PO4/3′-OH in ChVLig, a 5′-PO4/3′-H in Lig3, or an AppDNA/3′-H in Lig1), the motif I lysine (lysyl-AMP in ChVLig and Lig3; lysine in Lig1), the motif I aspartate, the OB domain Arg-Phe dipeptide, and a phenylalanine of the NTase domain are shown. NTase residues are rendered with cyan carbons; OB residues are rendered with beige carbons, and DNA is rendered with gray carbons. The adenylate and nick 5′-phosphorus atoms are colored yellow. The Arg-Asp salt bridges are denoted by dashed lines.
The unifying principal of the interaction network depicted in Fig. 5 (top panel) is that each of the four component side chains is important for step 2 and step 3 chemistry. As shown here and previously (13), Phe-286 and Phe-75 enhance the rate of step 2 DNA adenylylation by 62- and 22-fold, respectively; they accelerate step 3 phosphodiester synthesis by 54- and 12-fold, respectively. Arg-285 accelerates step 2 and 3 by factors of 47 and 120, respectively. We think it likely that the essentiality of Arg-285 for step 2 + step 3 chemistry is due, in no small part, to its interaction with Asp-29, an essential constituent of motif I that is conserved in virtually all DNA ligases. Changing Asp-29 to alanine has no apparent effect on ligase adenylylation or nick sensing, but it slows the rate of single-turnover nick sealing to 3.5 × 10−5 s−1 (4, 5), which is a 67,000-fold decrement when compared with the rate of 2.37 s−1 measured presently.
The ChVLig OB domain Arg-285-Phe-286 dipeptide and NTase domain residues Asp-29 and Phe-75 are conserved in the OB and NTase domains of human Lig3 (as Arg-716-Phe-717, Asp-423, and Phe-487), where they engage in similar interactions with each other and the nucleotides flanking the nick in the crystal structure of Lig3-AMP bound at a 5′-PO4/3′-H nick in duplex DNA (Fig. 5, middle panel) (20). The Tomkinson laboratory (21) reported that Lig3 OB domain mutations R716G and F717L eliminated the capacity of Lig3 to complement growth of an E. coli ligA-ts mutant and inhibited overall nick sealing in vitro without affecting ligase adenylylation in vitro. Moreover, they showed that the Lig3 R716G and F717L mutants were impaired in their ability to transfer AMP from the Lig-AMP adduct to a nicked DNA substrate (21). Their early results anticipated what we report here for the impact of the analogous ChVLig OB domain mutations at Arg-285 and Phe-286 on ligase activity in vivo and step 1, step 2, and step 3 catalysis in vitro. The structure of human Lig1 bound at an adenylylated 5′-PO4/3′-H nick further underscores the conservation of the interaction network of the OB Arg-Phe dipeptide, motif I Asp, and NTase phenylalanine side chains with each other and the nick terminal nucleotides (Fig. 5, bottom panel) (22).
How does this conserved interaction network promote catalysis of nick 5′ adenylylation and phosphodiester synthesis? The ChVLig-AMP·DNA crystal structure provides clues, in the forms of two water molecules in the active site (Fig. 1C, red spheres) that plausibly mimic the positions of divalent cation cofactors. Note that the ChVLig-AMP·DNA crystals were grown in the absence of metals to preclude adenylate transfer from ChVLig-AMP to the nick and that exposing the preformed crystals to 5 mm manganese immediately before cryoprotection triggered catalysis of nick sealing in situ (8). The chemistry of phosphodiester synthesis by DNA ligase is similar to that of DNA polymerase, which exploits a two-metal mechanism of nucleotidyl transfer. One of the waters in the ChVLig structure engages the O3′ atom of nick 3′-OH strand; a metal ion in this position would assist step 3 catalysis by lowering the pKa of the O3′ nucleophile, thereby activating it for attack on the 5′-phosphate of AppDNA. The other water engages the phosphate of Lig-AMP; a metal in this position could stabilize the transition state during step 2 adenylate transfer to the nick 5′-PO4 and aid in expulsion of the AMP leaving group during step 3 phosphodiester synthesis. Both waters (putative metals) are coordinated by Asp-29, the residue that, among the side chains surveyed by mutagenesis, makes the single biggest kinetic contribution to catalysis of steps 2 and 3. We infer that the conserved Arg-285-Asp-29 salt bridge maintains the proper orientation of the metal-binding aspartate.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM63611 (to S. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Figs. S1–S4, and Table S1.
- ChVLig
- Chlorella virus DNA ligase
- NTase
- nucleotidyltransferase
- FOA
- 5-fluoroorotic acid.
REFERENCES
- 1. Ellenberger T., Tomkinson A. E. (2008) Annu. Rev. Biochem. 77, 313–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shuman S. (2009) J. Biol. Chem. 284, 17365–17369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ho C. K., Van Etten J. L., Shuman S. (1997) J. Virol. 71, 1931–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sriskanda V., Shuman S. (1998) Nucleic Acids Res. 26, 525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Odell M., Shuman S. (1999) J. Biol. Chem. 274, 14032–14039 [DOI] [PubMed] [Google Scholar]
- 6. Odell M., Sriskanda V., Shuman S., Nikolov D. B. (2000) Mol. Cell. 6, 1183–1193 [DOI] [PubMed] [Google Scholar]
- 7. Odell M., Malinina L., Sriskanda V., Teplova M., Shuman S. (2003) Nucleic Acids Res. 31, 5090–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nair P. A., Nandakumar J., Smith P., Odell M., Lima C. D., Shuman S. (2007) Nat. Struct. Mol. Biol. 14, 770–778 [DOI] [PubMed] [Google Scholar]
- 9. Sriskanda V., Shuman S. (1998) Nucleic Acids Res. 26, 4618–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shuman S., Lima C. D. (2004) Curr. Opin. Struct. Biol. 14, 757–764 [DOI] [PubMed] [Google Scholar]
- 11. Sriskanda V., Shuman S. (2002) J. Biol. Chem. 277, 9661–9667 [DOI] [PubMed] [Google Scholar]
- 12. Sriskanda V., Shuman S. (2002) Nucleic Acids Res. 30, 903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Samai P., Shuman S. (2011) J. Biol. Chem. 286, 13314–13326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shuman S. (1995) Biochemistry 34, 16138–16147 [DOI] [PubMed] [Google Scholar]
- 15. Sriskanda V., Schwer B., Ho C. K., Shuman S. (1999) Nucleic Acids Res. 27, 3953–3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mulet J. M., Leube M. P., Kron S. J., Rios G., Fink G. R., Serrano R. (1999) Mol. Cell Biol. 19, 3328–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Auesukaree C., Homma T., Tochio H., Shirakawa M., Kaneko Y., Harashima S. (2004) J. Biol. Chem. 279, 17289–17294 [DOI] [PubMed] [Google Scholar]
- 18. Håkansson K., Doherty A. J., Shuman S., Wigley D. B. (1997) Cell 89, 545–553 [DOI] [PubMed] [Google Scholar]
- 19. Crut A., Nair P. A., Koster D. A., Shuman S., Dekker N. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6894–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cotner-Gohara E., Kim I. K., Hammel M., Tainer J. A., Tomkinson A. E., Ellenberger T. (2010) Biochemistry 49, 6165–6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mackey Z. B., Niedergang C., Ménissier-de Murcia J., Leppard J., Au K., Chen J., de Murcia G., Tomkinson A. E. (1999) J. Biol. Chem. 274, 21679–21687 [DOI] [PubMed] [Google Scholar]
- 22. Pascal J. M., O'Brien P. J., Tomkinson A. E., Ellenberger T. (2004) Nature 432, 473–478 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.