Abstract
How fish embryos that develop externally survive microbial attacks is poorly understood. Here, we clearly demonstrated that the embryo extract of zebrafish and its early embryo both displayed antimicrobial activity against microbes, including pathogenic Aeromonas hydrophila, and phosvitin (Pv), a nutritional protein abundant in eggs, was related to this antimicrobial activity. We also showed that recombinant Pv (rPv) acted as a pattern recognition receptor capable of recognizing the microbial signature molecules LPS, lipoteichoic acid, and peptidoglycan, as well as binding the Gram-negative and -positive microbes Escherichia coli, A. hydrophila, and Staphylococcus aureus and functioned as an antimicrobial agent capable of killing the microbes. Furthermore, we revealed that its C-terminal 55 residues (Pt5) with the functional sites Arg242 and Ala201/Ile203 were indispensable for Pv antimicrobial activity. Importantly, microinjection of rPv or Pt5 into early embryos significantly enhanced their resistance to A. hydrophila challenge, and this enhanced bacterial resistance was markedly reduced by co-injection of anti-Pv antibody plus rPv (or Pt5) but not by injection of anti-actin antibody plus rPv. Moreover, the generated mutants with in vitro antimicrobial activity, when injected into the embryos, could also promote their resistance to A. hydrophila, but those without in vitro antimicrobial activity could not. It is thus proposed that Pv participates in the protection of early embryos against pathogenic attacks via binding and disrupting potential pathogens. This work also opens a new way for the study of the immunological roles of yolk proteins in oviparous animals that rely on yolk proteins for embryonic development.
Keywords: Antimicrobial Peptides, Embryo, Mutant, Protein Purification, Zebrafish, Antimicrobial Activity, Embryogenesis, Phosvitin, Vitellogenin, Zebrafish
Introduction
Eggs of most fish are released and fertilized externally, and the resulting embryos and larvae are therefore exposed to an aquatic environment that is full of potential pathogens capable of causing various types of diseases. During the early stages of development, fish embryos have little or only limited ability to synthesize immune relevant molecules endogenously, and their lymphoid organs are not yet fully formed (1, 2). Moreover, embryogenesis of fish is one of most vulnerable stages in life history (3). How they survive the pathogenic attacks in such a hostile environment is one of the key issues for reproductive and developmental immunology, but the study as such is rather limited to date.
Fish eggs are in most cases cleidoic, i.e. a closed free-living system following fertilization; they are therefore supposed to depend upon the maternal provision of immunorelevant molecules for protection against invading pathogens before full maturation of immunological systems. Previous studies on several fish species have shown that maternal IgM is able to be transferred from mother to offspring (4–13). Likewise, maternal transfer of the innate immune factors, including the complement component C3 (14–19), lectins (20–22), protease inhibitors (23, 24), and lysozymes (25, 26), to offspring has also been reported in different teleost species. Moreover, immunization of parents results in a significant increase in IgM levels (7, 12) and anti-protease and lysozyme activities (7) in the eggs compared with control. These transferred maternal molecules have been proposed to be involved in the immune defense against pathogens in developing fish embryos and larvae. For example, Wang et al. (19, 27) have recently demonstrated that the maternal complement components operating via the alternative pathway are attributable to the protection of early embryos of zebrafish Danio rerio against microbial attacks. Besides the above substances, yolk proteins consisting of phosvitin (Pv)3 and lipovitellin are also maternally transferred molecules stored in fish eggs. Both Pv and lipovitellin are the proteolytically cleaved products of a high molecular mass lipoglycophosphoprotein, vitellogenin, and are traditionally regarded as the yolk reserves of nutrients essential for growth and development (28). Interestingly, as a major component of yolk proteins, chicken Pv has been shown to be able to inhibit the growth of the Gram-negative bacterium Escherichia coli (29) via chelating ions through its numerous phosphorylated serine residues (30, 31). In addition, vitellogenin, the precursor of Pv, has also been revealed to be an antimicrobial agent involved in immune defense in fish (32–36). Similarly, mosquito vitellogenin has recently been shown to be able to interfere with the anti-Plasmodium response in the malaria mosquito Anopheles gambiae (37). From these, it may be prudent to hypothesize that Pv, in addition to being a simple nutritional reserve, is also involved in the immune defense of developing embryos in fish. The aim of this study is thus to verify this hypothesis, using the model fish D. rerio. Clearly, such a study will definitely shed light on the mechanisms of how early embryos of fish survive the pathogenic attacks.
MATERIALS AND METHODS
Preparation of Embryo Extracts
The embryos of D. rerio were collected at about 10 h post-fertilization, washed three times with sterilized PBS (pH 7.4), and homogenized on ice. After centrifugation at 5000 × g at 4 °C for 5 min, the embryo extracts were pooled, aliquoted, and stored at −70 °C until used.
Preparation of Bacteria
The Gram-negative bacterium E. coli and the Gram-positive bacterium Staphylococcus aureus were incubated at 37 °C in LB medium for 16 h, and the Gram-negative bacterium Aeromonas hydrophila LSA 20 (pathogenic to D. rerio) was grown at 28 °C in tryptic soy broth medium for 16 h. They were harvested by centrifugation at 3000 × g at 4 °C for 15 min. The bacterial pellets were resuspended in 10 mm PBS (pH 7.4), giving a concentration of 5 × 107 cells/ml (for both E. coli and S. aureus) or a concentration of 2.5 × 109 cells/ml (for A. hydrophila), and used for the following experiments.
Assays for Antimicrobial Activity of Pv in Embryo Extracts
To inactivate the complement activity, the embryo extracts were heated at 45 °C for 30 min (27). Heating at this temperature had little effect on Pv because Pv was heat-resistant (38). To precipitate Pv in embryo extract, an aliquot of 1 ml of heated extract (with ∼3.2 μm Pv) was mixed with ∼1 μg of anti-fish Pv antibody (polyclonal antibody; E91679Fi, Uscnk) and incubated at 4 °C for 2 h, and then 20 μl of protein A/G Plus-agarose (Santa Cruz Biotechnology) was added to the mixture. After incubation overnight at 4 °C, the mixture was centrifuged at 3000 × g for 10 min, and the supernatant was pooled and used for antimicrobial activity assay. For control, 1 ml of embryo extract was mixed with ∼1 μg of anti-β-actin antibody (polyclonal antibody; AA128-1; Beyotime, Nantong, China) and processed similarly.
The antimicrobial activity of embryo extracts against E. coli, A. hydrophila, and S. aureus was assayed as described by Wang et al. (27). PBS solution instead of embryo extracts was used as control. The percent of bacterial growth inhibition was inferred from the difference between the numbers of colonies in the test and control.
Assays for Antimicrobial Activity of Pv in Developing Embryos
To test if Pv has any ability to protect developing embryos, 30 dechorionated embryos were microinjected at the 8-cell stage in the yolk sac with ∼6 nl of sterilized PBS (blank control), anti-Pv antibody solution (∼0.33 ng), anti-β-actin antibody solution (∼0.33 ng), purified recombinant Pv (rPv; see below) solution (∼0.6 ng), or BSA solution (∼0.6 ng) and challenged 1 h later by injection of ∼6 nl (∼500 cells) of live A. hydrophila suspension. The mortality was recorded, and cumulative mortality was calculated at 24 h after bacterial injection. To confirm the specificity of the antimicrobial activity of rPv in vivo, anti-Pv antibody was injected together with rPv into the embryos, which were then challenged by injection of live A. hydrophila. For control, the embryos were injected with anti-actin antibody plus rPv and treated similarly.
To verify the killing of A. hydrophila by developing embryos, 8-cell stage embryos were dechorionated and microinjected with live A. hydrophila as above. Five embryos were collected each time at 0, 6, and 12 h post-bacterial injection. The normal embryos were also collected as control. Total DNAs were isolated from each embryo according to the method of Wang et al. (27) and used for PCR. The PCR was carried out to amplify a specific region of A. hydrophila 16 S rRNA gene using the sense primer 5′-AATACCGCATACGCCCTAC-3′ and anti-sense primer 5′-AACCCAACATCTCACGACAC-3′, which were designed on the basis of A. hydrophila 16 S rRNA sequence (GenBankTM accession number DQ207728).
Titration of Pv Content in Eggs/Embryos
A total of 60 fertilized eggs and 120 embryos collected at 12 and 24 h post-fertilization were washed three times with sterilized 0.9% saline, homogenized, and centrifuged at 5000 × g at 4 °C for 5 min. The supernatants were pooled and used to measure the contents of Pv in the extracts using fish Pv ELISA kit (E91679Fi, Uscnk) according to manufacturer's protocol. The mean diameter of D. rerio eggs measured was ∼600 μm, and therefore the egg/embryo volume was approximately ∼1.1 × 10−5 cm3. Accordingly, the content of Pv in each egg/embryo was inferred as described by Liang et al. (16).
Expression and Purification of rPv
The cDNA region encoding zebrafish Pv was amplified by PCR with the upstream primer 5′-GCCATATGGTCAGAAACATTGAAG-3′ (NdeI site is underlined) and the downstream primer 5′-CGGAATTCTTATGGAATATCATTTC-3′ (EcoRI site is underlined). The PCR product was digested with NdeI and EcoRI and subcloned into the plasmid expression vector pET28a (Novagen) previously cut with the same restriction enzymes. The identity of the insert was verified by sequencing, and the plasmid was designated pET28a/Pv.
The cells of E. coli BL21 were transformed with the plasmid pET28a/Pv and cultured overnight in LB broth containing 50 μg/ml kanamycin. The culture was diluted 1:1000 with LB broth and subjected to further incubation at 37 °C for 2 h, and the expression of rPv was induced by adding isopropyl β-d-thiogalactoside to the culture at a final concentration of 0.1 mm. The rPv was purified and electrophoresed on 12% SDS-PAGE. MALDI/TOF MS analysis was performed on Bruker Ultraflex MALDI/TOF MS mass spectrometer (Bremen, Germany). Protein concentrations were determined by the method of Bradford using bovine serum albumin as a standard.
Antimicrobial Activity Assay of rPv
The antimicrobial activity of rPv against the Gram-negative bacteria E. coli and A. hydrophila and the Gram-positive bacterium S. aureus was measured by the colony-forming unit assay as described by Fan et al. (39). The percent of bacterial growth inhibition by rPv was calculated as follows: (number of colonies (control − test)/number of colonies (control)) × 100 (n = 3).
Scanning Electron Microscopy
Aliquots of 150 μl of E. coli (a representative Gram-negative bacterium) and S. aureus (a representative Gram-positive bacterium) suspensions containing 5 × 107 cells/ml were mixed with 150 μl of 3.2 μm rPv in 50 mm Tris-HCl buffer (TB; pH 8.0) or with 150 μl of TB alone as control. The mixtures were incubated at 25 °C for 1 h, fixed in 2.5% glutaraldehyde in 100 mm PBS (pH 7.4), and dropped onto 24 × 24-mm cover glasses. The samples were post-fixed in 1% osmium tetroxide, dehydrated with graded ethanol, dried by the critical point method, and coated with gold. Observation was made under a JEOL JSM-840 scanning electron microscope.
Iron Binding Activity Assay
Chrome azurols (CAS) assay buffer was prepared as described by Schwyn and Neilands (40). The iron binding activity of rPv was assayed spectrophotometrically as described by Liu et al. (41). The iron binding activity of rPv was expressed as A630 ratio, which indicates the relative absorbance of the samples in comparison with control.
Labeling of rPv with Fluorescein Isothiocyanate (FITC)
Purified rPv (∼32 μm) and BSA were labeled as described by Li et al. (33). The purity of the conjugate, FITC-labeled rPv, was confirmed by SDS-PAGE and Coomassie Blue staining, and its F/P ratio (F/P ratio is defined as the ratio of moles of FITC to moles of protein in the conjugate) was calculated by the equation F/P = (2.77 × A495)/(A280 −(0.35 × A495)) from the absorbance readings of the conjugated samples (42). To assess if rPv activity is affected by labeling with FITC, colony-forming unit assay was conducted to compare the growth inhibition rates of E. coli by FITC-labeled rPv and nonlabeled rPv.
Assay for Binding of FITC-labeled rPv to Microbial Cells
Binding of rPv to the microbes E. coli, A. hydrophila, and S. aureus was assayed by the method of Li et al. (33). The binding of FITC-labeled rPv to microbial cells was observed under an Olympus BX51 fluorescence microscope. The microbes treated with FITC-labeled BSA under the same conditions were used as control. To test if nonlabeled rPv could competitively inhibit the binding of FITC-labeled rPv to the microbes, nonlabeled rPv was mixed with labeled rPv at a ratio of 1:1. The mixture was then incubated with the microbes and the binding observed as above.
Assay for Binding of rPv to Various Ligands
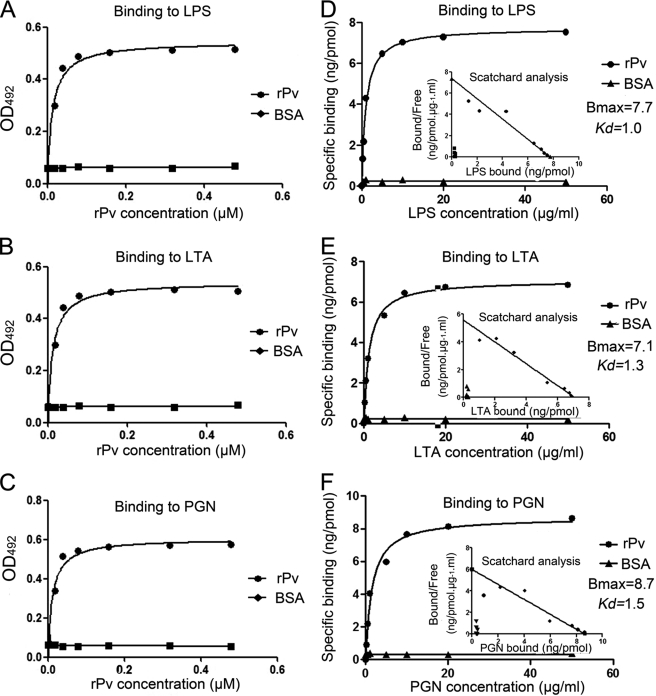
LPS from E. coli and lipoteichoic acid (LTA) and peptidoglycan (PGN) from S. aureus (all from Sigma) were individually dissolved in re-distilled water, all giving a concentration of 40 μg/ml. A volume of 50 μl (2 μg) of each solution was applied to each well of a 96-well microplate and air-dried at room temperature overnight. The plates were incubated at 60 °C for 30 min to fix the ligands, and the wells were each blocked with 200 μl of 1 mg/ml BSA in 10 mm PBS (pH 7.4) at 37 °C for 2 h. After washing four times with 200 μl of 10 mm PBS supplemented with 1% Tween 20, 50 μl of digoxigenin-labeled rPv solutions with different concentrations of digoxigenin-labeled rPv (0, 0.02, 0.04, 0.08, 0.16, 0.32, and 0.5 μm) was added to the wells. After incubation at room temperature for 3 h, the wells were each rinsed four times with 200 μl of 10 mm PBS supplemented with 1% Tween 20, and 100 μl of anti-digoxigenin Fabs (Roche Applied Science) diluted to 1:1000 with 10 mm PBS (pH 7.4) containing 1% dry milk powder were added to the wells. The wells were incubated at 37 °C for 2 h, washed, and reacted with 75 μl of 0.4 mg/ml O-phenylenediamine (Amresco) in 51.4 mm Na2HPO4, 24.3 mm citric acid, and 0.045% H2O2 (pH 5.0) at 37 °C for 20 min. Subsequently, 25 μl of 2 mm H2SO4 was added to each well to terminate the reactions, and absorbance at 492 nm was monitored by a microplate reader (GENios Plus, Tecan). For control, BSA at the same concentrations was treated similarly.
For Scatchard analysis, LPS, LTA, and PGN were labeled with biotin hydrazide (Sigma) by the method of Gotoh et al. (43). rPv was dissolved in re-distilled water, giving a concentration of 40 μg/ml. A volume of 50 μl (2 μg) of rPv solution was applied to each well of a 96-well microplate and air-dried at room temperature overnight. The plates were incubated at 60 °C for 30 min to fix rPv, and the wells were each blocked with 200 μl of 20 mg/ml BSA in 10 mm PBS (pH 7.4) at 37 °C for 2 h. After washing four times with 200 μl of 10 mm PBS supplemented with 1% Tween 20, a total of 50 μl of solution with different concentrations (0, 0.02, 0.04, 0.08, 0.16, 0.32, and 0.5 μg/ml) of biotin-labeled LPS, LTA, or PGN was added each well and then processed as above. For control, BSA instead of rPv at the same concentrations was treated similarly. The equilibrium association constant (Kd) and apparent maximum number of bindings (Bmax) were determined according to the Scatchard plot using the software GraphPad Prism 5.01.
Expression of Truncated Pv and Bioactivity Assay
To determine the structure-activity relationship, the N-terminal 55, 102, 134, 174, and 194 residues of Pv were deleted, respectively. The upstream primers used were as follows: 55d, 5′-GCCATATGGTCCCATACATTGAA-3′; 102d, 5′-GCCATATGAGACTGGAGTTTGAAGT-3′; 134d, 5′-GCCATATGTTCTTGTTGAAACTGAG-3′; 174d, 5′-GCCATATGAGCTCAAGCTTAAGG-3′; 194d, 5′-GCCATATGCTCTCGTATGTCTAAGA-3′ (NdeI site is underlined); and the downstream primer used was all the same, 5′-CGGAATTCTTATGGAATATCATTTC-3′ (EcoRI site is underlined). The C-terminal 55 residues of Pv were also deleted using the primer pair 5′-GCCATATGGTCAGAAACATTGAAG-3′ (NdeI site is underlined) and 5′-CGGAATTCAGCCTGTCCTTCTGGAG-3′ (EcoRI site is underlined).
Construction of the expression vector plasmid, transforming into E. coli BL21, as well as expression and purification of recombinant proteins were all performed as above. The plasmids constructed were verified by sequencing and designated Pt1 (N-terminal 55 residues deleted), Pt2 (N-terminal 102 residues deleted), Pt3 (N-terminal 134 residues deleted), Pt4 (N-terminal 174 residues deleted), Pt5 (N-terminal 194 residues deleted), and Pt6 (C-terminal 55 residues deleted).
Antimicrobial activity and ligand binding assays were performed as above. Pt5 and Pt6 were also microinjected into the early embryos of D. rerio, respectively, to test their antimicrobial activity in vivo. To verify the specificity of the antimicrobial activity of Pt5 in vivo, the embryos were co-injected with Pt5 plus anti-Pv antibody and then challenged by injection of live A. hydrophila.
Site-directed Mutation of Pv and Bioactivity Assay
The plasmid Pt5 was used for mutational analyses. Mutants were generated using Takara MutanBEST kit (Takara, Dalian, China) according to the kit's protocol. The specific primers used are listed in Table 1. The mutations were confirmed by DNA sequencing. Expression and purification of the mutated recombinant proteins, antimicrobial activity, and ligand binding assays were carried out as above. Pt5 and Pt6 were used as positive and negative controls, respectively. To test their antimicrobial activity in vivo, the mutants m1, m5, and m9 with in vitro antimicrobial activity and the mutants m12 and m13 with no in vitro activity (see below) were selected and microinjected into the early embryos of D. rerio, respectively, followed by the challenge with A. hydrophila.
TABLE 1.
Primers used for site-directed mutagenesis
The mutations are shown in boldface and underlined.
| Mutation | Sequence (5′ to 3′) |
|---|---|
| I203M | Sense, GACTGCCACCATGATTGAGCCTT |
| Antisense, TTAGACATACGAGAGCTGGACATAG | |
| K309E | Sense, CAGGAAATTCCACAAGGATCGGT |
| Antisense, AAAGGCTCAATGATGGTGGCAGTC | |
| H211L | Sense, CTCAAAGATCGGTACTTGGCACAC |
| Antisense, GAATTTCCTGAAAGGCTCAATGATGG | |
| K212E | Sense, CACGAAGATCGGTACTTGGCACAC |
| Antisense, GAATTTCCTGAAAGGCTCAATGATGG | |
| R214G | Sense, CACAAAGATGGGTACTTGGCACAC |
| Antisense, GAATTTCCTGAAAGGCTCAATGATGG | |
| Y225F | Sense, GATCGGTACTTGGCACACCATAG |
| Antisense, TTTGTGGAATTTCCTGAAAGGCT | |
| F234S | Sense, GCAGCTAGCTCTGAACAAATGCA |
| Antisense, AGCACTTCCACTGCTAGTATCCT | |
| F234L | Sense, GCAGCTAGCTTGGAACAAATGCA |
| Antisense, AGCACTTCCACTGCTAGTATCCT | |
| E235K | Sense, GCAGCTAGCTTTAAACAAATGCA |
| Antisense, AGCACTTCCACTGCTAGTATCCT | |
| K239E | Sense, GAACAAATGCAGGAACAGAATAGA |
| Antisense, AAAGCTAGCTGCAGCACTTCCACT | |
| Q240H | Sense, AACATAATAGATTCCTTGGAAATG |
| Antisense, TCTGCATTTGTTCAAAGCTAGCTG | |
| R242G | Sense, AACAGAATGGATTCCTTGGAAATG |
| Antisense, TCTGCATTTGTTCAAAGCTAGCTG | |
| A201G/I203M | Sense, GACTGCCACCATGATTGAGCCTT |
| Antisense, TTAGACATACGAGAGCTGGACATAG |
Statistical Analysis
All the experiments were conducted at least three times. Statistical analyses were performed using the computer program SPSS 13.0. The statistical significance of difference between mean values was calculated by analysis of variance, and difference at p < 0.05 was considered significant. All the data were expressed as mean ± S.D.
RESULTS
Antimicrobial Activity of Pv in Embryo Extracts
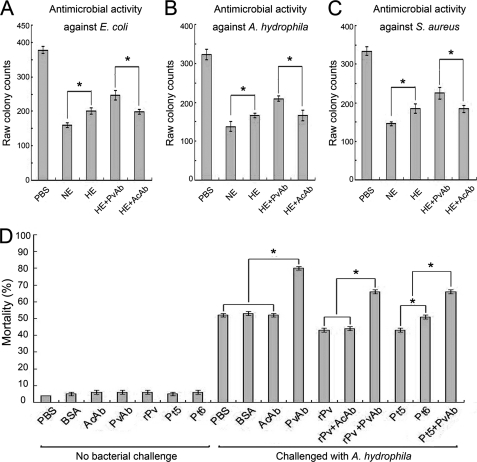
We first sought to examine if Pv in the embryo extract of D. rerio has any antimicrobial activity. The protein concentration of the extract prepared from the embryos collected at ∼10 h post-fertilization was ∼20 mg/ml. As shown in Fig. 1, the embryo extract exhibited conspicuous antimicrobial activities against the Gram-negative bacteria E. coli and A. hydrophila as well as the Gram-positive bacterium S. aureus. When the complement activities, which had been demonstrated to be present in the egg cytosol and heat-sensitive (19), were inactivated by heating, the antimicrobial activities of the embryo extracts against both E. coli and A. hydrophila as well as S. aureus were remarkably reduced. However, some antimicrobial activities remained in the heated extracts. As Pv was heat-resistant (38), we therefore thought that it might be related to the remaining antimicrobial activities. As expected, the remaining antimicrobial activities of the heated extracts against E. coli, A. hydrophila, and S. aureus were significantly reduced by the preincubation with anti-fish Pv antibody but not by the preincubation with anti-actin antibody (Fig. 1, A–C). These data suggested that in addition to complement, Pv was also a factor responsible for the antimicrobial activity in the embryo extract.
FIGURE 1.
Antimicrobial activities of Pv in vitro and in vivo. A, antimicrobial activity of the embryo extract against E. coli; B, antimicrobial activity of the embryo extract against A. hydrophila; C, antimicrobial activity of the embryo extract against S. aureus; D, effects of microinjected recombinant Pv, Pt5, and Pt6 as well as anti-fish Pv antibody on the antimicrobial activity of the embryos. The early (8-cell stage) embryos were first microinjected and then challenged by injection of live A. hydrophila 1 h later. The mortality was recorded, and the cumulative mortality was calculated at 24 h following the bacterial injection. About 95% of the nonbacterial challenge control embryos developed normally. The challenge with live A. hydrophila resulted in a significant increase in the mortality of the embryos microinjected with anti-Pv antibody but not the embryos injected with anti-actin antibody or BSA or PBS. Microinjection of rPv or Pt5 into the embryos was able to increase their resistance to A. hydrophila, whereas the injection of Pt6 into the embryos was not. Co-injection of anti-Pv antibody together with rPv or Pt5 was able to counteract their activity to enhance the embryonic resistance to A. hydrophila. All data were expressed as mean values ± S.E. (n = 3). The bars represent the mean ± S.E. The asterisk means a significant difference (p < 0.05). NE, embryo extract; HE, heated embryo extract; PvAb, anti-fish Pv antibody; AcAb, anti-actin antibody; BSA, bovine serum albumin; Pt1, Pt2, Pt3, Pt4, Pt5, and Pt6 are the six truncated products of Pv (see Fig. 6).
Involvement of Pv in Antimicrobial Activity in Early Embryos
To test if Pv plays any protective role in early development, 8-cell stage embryos were each microinjected with the antibody against fish Pv to block Pv action, followed by injection with live A. hydrophila (pathogenic to D. rerio). The majority of the nonbacterial challenge control embryos (∼95%) developed normally (Fig. 1D). Interestingly, the challenge with live A. hydrophila resulted in a significant increase in the mortality of the embryos microinjected with anti-Pv antibody, with the 24-h cumulative mortality of ∼80%, whereas the same challenge caused only ∼52% cumulative mortality at 24 h in the embryos injected with either anti-actin antibody or BSA or PBS alone (Fig. 1D). Moreover, anti-Pv antibody-induced embryonic death rate was remarkably reduced by the co-injection of the antibody plus purified rPv (see below), with the 24-h cumulative mortality of ∼65%, but not by the injection of anti-actin antibody plus rPv (Fig. 1D). All these data suggested that Pv was involved in the antimicrobial activity of developing embryos.
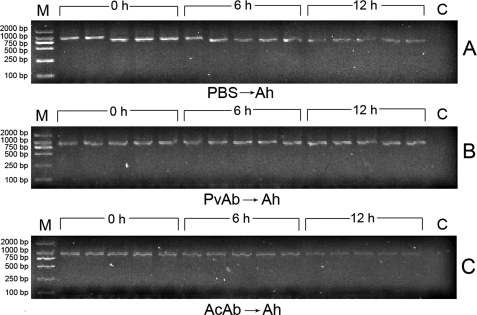
To prove the killing of live A. hydrophila by the embryos, PCR analysis was performed to amplify a specific region of A. hydrophila 16 S rRNA gene using the total DNAs isolated from each embryo as template (27). As shown in Fig. 2A, no band was observed in the control sample (from an embryo without injected A. hydrophila), but intense bands were found in the embryos collected soon after bacterial injection (0 h), and the band intensities apparently decreased with time (at 6 and 12 h), suggesting the lysis of the bacterium by the embryos. Moreover, the microinjection of anti-Pv antibody into the embryos blocked the decrease in the band intensity during the initial 12 h (Fig. 2B), that is little A. hydrophila lysis took place in the embryos, whereas the injection of anti-actin antibody into the embryos failed to block reduction of the band intensity at 6 and 12 h, that is A. hydrophila lysis continued in the embryos (Fig. 2C). These data implicated the presence of a link between Pv and lysis of the bacterium in the early embryos.
FIGURE 2.
PCR analysis of A. hydrophila 16 S rRNA gene. The 8-cell stage embryos were microinjected with live A. hydrophila, and five embryos were collected each time at 0, 6, and 12 h after the bacterial injection. Total DNAs were isolated from each embryo and used as template to amplify the specific region of A. hydrophila (Ah) 16 S rRNA gene. All the PCR products were electrophoresed in 1% agarose, and the bands were recorded using the gel imaging system. A, embryos injected with A. hydrophila only. B, embryos injected with anti-fish Pv antibody, followed by injection with A. hydrophila. C, embryos injected with anti-actin antibody, followed by injection with A. hydrophila. M, marker; C, control.
Microbicidal Activity of rPv
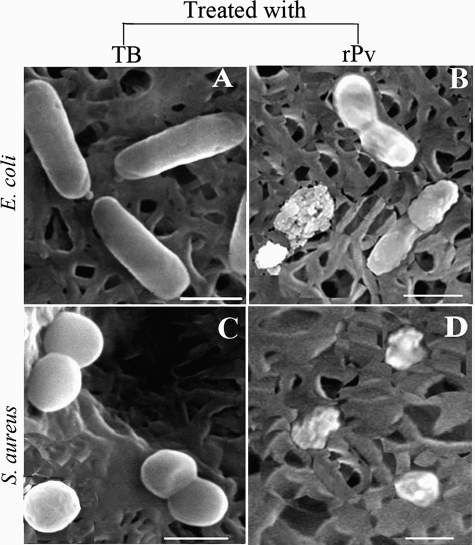
To verify that Pv is indeed involved in antimicrobial activity in developing embryos, we expressed zebrafish Pv in E. coli and examined its microbicidal activity directly in vitro. An expression vector, including the cDNA encoding Pv of D. rerio and 5′ additional tag of pET28a, was constructed and transformed into E. coli cells. The recombinant peptide was induced by isopropyl β-d-thiogalactoside and was purified by affinity chromatography on a nickel-nitrilotriacetic acid resin column. The purified rPv with the thioredoxin-His tag yielded a single band of about 29 kDa on SDS-polyacrylamide gel after Coomassie Blue staining, corresponding to the expected size. MALDI/TOF MS analysis showed that the protein purified was zebrafish Pv (supplemental Fig. S1). The purified rPv was then used to detect its antimicrobial activity against the Gram-negative bacteria E. coli and A. hydrophila and the Gram-positive bacterium S. aureus by colony-forming unit assay. As expected, the growth of both E. coli and A. hydrophila as well as S. aureus was all inhibited by rPv in a dose-dependent manner (Table 2). rPv was able to significantly inhibit the growth of E. coli, A. hydrophila, and S. aureus, with the half-inhibitory concentrations (IC50) of ∼3.1, ∼3.0, and ∼3.0 μm, respectively. In contrast, the peptide thioredoxin-His tag expressed did not suppress the bacterial growth (data not shown; also see An et al. (44)), implicating that the thioredoxin-His tag extra part in rPv had little effect on its antimicrobial activity. Moreover, scanning electron microscopy examination revealed that rPv caused damage to the cells of both E. coli and S. aureus. The cells of E. coli and S. aureus incubated with rPv were severely damaged, with the appearance of collapsed architecture (Fig. 3, B and D), but the bacterial cells incubated with TB alone remained intact, and their cell surfaces were smooth (Fig. 3, A and C). All these suggested that consistent with its involvement in antimicrobial activity in vivo, Pv was also a microbicidal factor in vitro, being capable of killing the Gram-negative and -positive bacteria such as E. coli and S. aureus.
TABLE 2.
Antimicrobial activity of recombinant Pv against E. coli, S. aureus, and A. hydrophila
| Bacteria | rPv | Colony counts | Inhibition | IC50 |
|---|---|---|---|---|
| μm | Colony-forming units | % | μm | |
| E. coli | 0 (control) | 307.3 ± 21.1 | 3.1 | |
| 1.6 | 244.0 ± 13.3 | 20 ± 3 | ||
| 3.2 | 125.3 ± 31.3 | 59 ± 6 | ||
| 8.0 | 6.3 ± 1.8 | 98 ± 2 | ||
| A. hydrophila | 0 (control) | 298.7 ± 16.6 | 3.0 | |
| 1.6 | 236.7 ± 9.7 | 21 ± 2 | ||
| 3.2 | 119.3 ± 13.3 | 60 ± 3 | ||
| 8.0 | 12.0 ± 1.6 | 96 ± 3 | ||
| S. aureus | 0 (control) | 313.3 ± 35.3 | 3.0 | |
| 1.6 | 260.0 ± 26.6 | 18 ± 5 | ||
| 3.2 | 132.7 ± 20.6 | 57 ± 4 | ||
| 8.0 | 9.0 ± 1.0 | 97 ± 3 |
FIGURE 3.
Micrographs showing E. coli and S. aureus incubated with TB (A and C) or with recombinant Pv (B and D) at 25 °C for 1 h. Scale bars represent 1 μm.
Contents of Pv in Eggs/Embryos
To evaluate if early embryos possess sufficient Pv in vivo to fulfill antimicrobial activity, we measured the endogenous content of Pv. An ELISA analysis revealed that the concentrations of Pv in each of the newly fertilized eggs as well as 12- and 24-h embryos were ∼36.0, ∼9.0, and ∼1.3 μm, respectively. This indicated that at least at the initial 12 h post-fertilization, the concentration of Pv in each embryo was significantly higher than IC50 and was therefore sufficient enough to kill invading pathogens in vivo.
Binding of rPv to Bacteria
Next we explored the mechanisms by which Pv inhibits bacterial growth. As Pv isolated from chicken eggs was able to chelate ions through its phosphorylated serine residues (30, 31), we therefore determined whether rPv has any capacity to bind iron, which is necessary for bacterial growth and proliferation (45). When chrome azurols (CAS) assay solution was mixed with rPv, no color change was found in the mixture media, and the A630 values did not change with rPv concentrations (supplemental Fig. S2A). Moreover, the growth of both E. coli and S. aureus in the presence or absence of excess iron showed little difference (supplemental Fig. S2, B and C). This suggested that rPv did not bind Fe3+. As the phosphorylated serine residues of chicken Pv were shown to contribute to its chelating effect, including binding to Fe3+ (46), the inability of zebrafish rPv to bind to Fe3+ might be due to the lack of phosphorylation of the serine residues in the rPv expressed in E. coli.
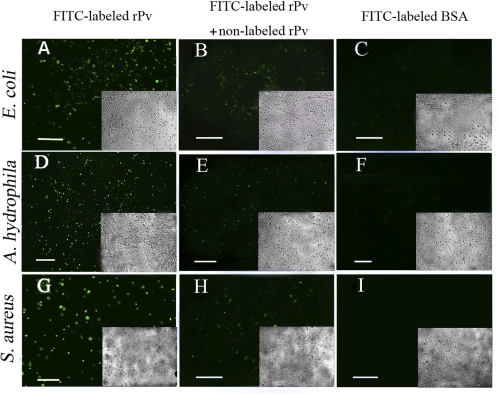
We then tried to test if rPv is able to bind to microbes. To do this, we labeled rPv and BSA with FITC. The F/P ratios of FITC-labeled rPv and BSA were 4.9 and 5.4, respectively, showing that they had similar F/P ratios. Antimicrobial activity examination by colony-forming unit assay revealed that the growth of E. coli preincubated with 1.6 μm of FITC-labeled rPv and nonlabeled rPv was both significantly inhibited, with the similar inhibitory rates of ∼21 and ∼22%, respectively. In addition, FITC-labeled rPv appeared as a single band on SDS-PAGE, suggesting that FITC-labeled rPv remained as monomer (supplemental Fig. S3). The data indicated that rPv activity was not affected by the labeling with FITC. Preincubation of FITC-labeled rPv with the Gram-negative bacteria E. coli and A. hydrophila and the Gram-positive bacterium S. aureus showed that rPv was able to bind to E. coli, A. hydrophila, and S. aureus (Fig. 4, A, D, and G). Similarly, FITC-labeled chicken Pv also bound to the microbes (data not shown), suggesting that the phosphorylation of serine residues in Pv was not necessary for its binding to the microbes. Notably, the binding of FITC-labeled rPv to the microbes was found to be competitively inhibited by nonlabeled rPv (Fig. 4, B, E, and H). In contrast, FITC-labeled BSA employed as a control did not bind to the microbial cells tested (Fig. 4, C, F, and I). All this suggested that both zebrafish rPv and chicken Pv had the capacity to bind to microbes.
FIGURE 4.
Binding of FITC-labeled recombinant Pv to microbial cells. A, D, and G, binding of FITC-labeled recombinant Pv to E. coli, A. hydrophila, and S. aureus. B, E, and H, binding of FITC-labeled rPv to E. coli, A. hydrophila, and S. aureus was weakened in the presence of nonlabeled rPv. C, F and I, no binding of FITC-labeled BSA to E. coli, A. hydrophila and S. aureus. Insets show the corresponding bright field images. Scale bars represent 20 μm.
Binding of rPv to Various Ligands
To better understand the mechanisms by which rPv binds microbes, an ELISA was carried out to investigate what molecules on the microbial surfaces are recognized by rPv. It was found that rPv was able to bind to all the immobilized ligands tested, including LPS, LTA, and PGN, in a dose-dependent manner, although BSA did not (Fig. 5, A–C). To directly characterize the affinities of rPv to the ligands, we further performed the Scatchard analysis. The Bmax (ng/pmol protein) and the Kd values (μg/ml) of the affinity of rPv to LPS, LTA, and PGN were ∼7.7 and 1.0, 7.1, and 1.3 as well as 8.7 and 1.5 (Fig. 5, D–F). It was clear that the binding of rPv to LPS, LTA, and PGN was specific and saturable. No specific binding of BSA to LPS, LTA, and PGN was detected. This indicated that rPv was capable of binding to the microbial signature molecules LPS, LTA, and PGN, thereby establishing it as a multivalent pattern recognition molecule.
FIGURE 5.
Interaction of recombinant Pv with various ligands. A–C, binding of recombinant Pv to various ligands. LPS, LTA, and PGN dissolved in re-distilled water was applied to wells of a 96-well microplate and air-dried overnight at room temperature, followed by ELISA. BSA instead of recombinant Pv was used as control. D–F, saturation curve of the binding of rPv to LPS, LTA, and PGN. rPv dissolved in re-distilled water was applied to wells of a 96-well microplate and air-dried overnight at room temperature, followed by Scatchard analysis. BSA was treated at the same concentrations as control. Data were expressed as mean values ± S.E. (n = 3).
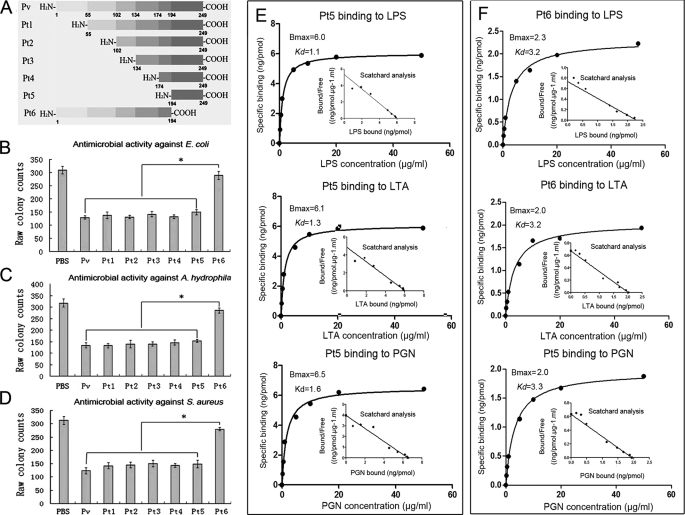
Effects of N- and C-terminal Deletion on Antimicrobial Activity of rPv
To determine the structure-activity relationship, various truncated phosvitins (Pt1–Pt6), with the N or C terminus deleted, were expressed in E. coli (supplemental Fig. S4) and subjected to functional analyses. As shown in Fig. 6, deletion of the N-terminal 55, 102, 134, 174, and 194 residues had little effect on antimicrobial activity against E. coli, A. hydrophila, and S. aureus, but deletion of the C-terminal 55 residues resulted in a remarkable decrease in antimicrobial activity (Fig. 6, B–D). To detect if the reduction of antimicrobial activity of truncated peptides was related to any alteration in ligand binding, their affinity to LPS, LTA, and PGN was assayed. Consistent with the fact that Pt6 displayed little antimicrobial activity, its affinity to LPS, LTA, and PGN was significantly reduced (Fig. 6F). In contrast, Pt5 retaining the large antimicrobial activity still possessed the affinity to the ligands similar to that of full rPv (Fig. 6E). These suggested that the antimicrobial activity of Pv and its truncated products were correlated with their capacity to bind the ligands. Scatchard analysis revealed that the Bmax and Kd values of the binding of Pt5 to LPS, LTA, and PGN were about 6.0 and 1.1, 6.1, and 1.3 as well as 6.5 and 1.6 (Fig. 6E), whereas the Bmax and Kd values of the binding of Pt6 to LPS, LTA, and PGN were 2.3 and 3.2, 2.0 and 3.2 as well as 2.0 and 3.3 (Fig. 6F). Compared with Pt5, both the Bmax and Kd values of Pt6 to bind LPS, LTA, and PGN were markedly decreased. All of this indicated that the C-terminal 55 residues of rPv were indispensable for both antimicrobial activity and binding to the ligands.
FIGURE 6.
Antimicrobial activity of truncated Pv peptides and their affinities to various ligands. A, diagram showing Pv truncation. B, antimicrobial activity against E. coli. C, antimicrobial activity against A. hydrophila. D, antimicrobial activity against S. aureus. E, saturation curve of the binding of Pt5 to LPS, LTA, and PGN. F, saturation curve of the binding of Pt6 to LPS, LTA, and PGN. BSA instead of recombinant Pv was used as control. The numbers in A indicated the positions at which Pv was truncated. Pt1(55–249), Pt2(102–249), Pt3(134–249), Pt4(174–249), Pt5(194–249),and Pt6(1–194) represented the truncated peptides. Data are expressed as mean values ± S.E. (n = 3). The bars represent the mean ± S.E. The asterisk means a significant difference (p < 0.05).
To investigate if Pt5 and Pt6 function similarly in vivo as in vitro, the embryos were each microinjected with Pt5 or Pt6, followed by challenge with the injection of live A. hydrophila. About 95% of the embryos microinjected with Pt5 and Pt6 developed normally. Notably, the challenge with live A. hydrophila caused a significant increase in the mortality of the embryos injected with Pt6 or PBS, with the 24-h cumulative mortality of ∼52%, but the same challenge induced only ∼42% cumulative mortality at 24 h in the embryos injected with Pt5 (Fig. 1D). Moreover, the embryo-protecting role of Pt5 was counteracted by the co-injection of Pt5 plus anti-Pv antibody (Fig. 1D). These showed that like rPv, Pt5 with in vitro antimicrobial activity was also involved in the antimicrobial activity of developing embryos.
Effects of Amino Acid Replacement on Antimicrobial Activity
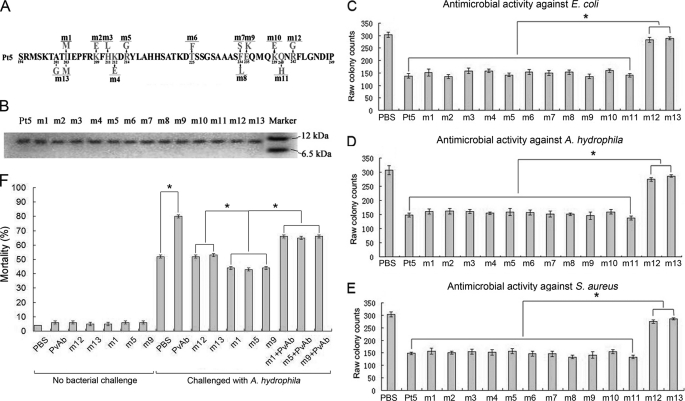
Antimicrobial peptides are generally accepted to exert antimicrobial activities via their positively charged surface and amphipathicity (47, 48). Therefore, inverse PCR was used to generate mutants using the plasmid Pt5 harboring the C-terminal 55 residues as a template to assay the effects of charged and hydrophobic residue replacement on antimicrobial activity (Fig. 7A). All the mutants had a closely similar molecular mass (Fig. 7B), and we thus compared the antimicrobial activity of each mutant with that of control under a same concentration. Compared with the control, the single mutation R242G (m12; positively charged Arg to neutral Gly) and the double mutation A201G/I203M (m13; hydrophobic Ala and Ile to hydrophilic Gly and Met) resulted in a significant decrease in their antimicrobial activity against E. coli, A. hydrophila, and S. aureus (Fig. 7, C–E) as well as their affinity to LPS, LTA, and PGN (supplemental Fig. S5). For all the other mutants, including mutations I203M (m1), K209E (m2), H211L (m3), K212E (m4), R214G (m5), T225F (m6), F234S (m7), F234L (m8), E235K (m9), K239E (m10), and Q240H (m11), no marked decrease in antimicrobial activity against E. coli, A. hydrophila, and S. aureus was observed (Fig. 7, C–E). Consistently, the Scatchard analysis showed that the binding affinities of m12 and m13 to LPS, LTA, and PGN were significantly reduced, whereas the affinities of the mutants such as m1, m3, m5, m7, and m9 to LPS, LTA, and PGN remained comparable with that of Pt5 (Table 3). These indicated that the positively charged residue Arg242 and the hydrophobic residues Ala201/Ile203 were the functional determinants contributing to the antimicrobial activity of rPv.
FIGURE 7.
Antimicrobial activity of Pt5 mutants. A, diagram showing the site-directed mutagenesis. B, SDS-PAGE of the mutants, showing that they all have a similar molecular mass. C, antimicrobial activity against E. coli. D, antimicrobial activity against A. hydrophila. E, antimicrobial activity against S. aureus. BSA was used as control. F, effects of microinjected m1, m5, m9, m12, or m13 as well as anti-fish Pv antibody on the antimicrobial activity of the embryos. The early (8-cell stage) embryos were first microinjected and then challenged by injection of live A. hydrophila 1 h later. The mortality was recorded, and the cumulative mortality was calculated at 24 h following the bacterial injection. About 95% of the nonbacterial challenge control embryos developed normally. The challenge with live A. hydrophila resulted in a significant increase in the mortality of the embryos microinjected with anti-Pv antibody but not the embryos injected with PBS. Microinjection of m1, m5, or m9 into the embryos was able to increase their resistance to A. hydrophila, whereas the injection of m12 or m13 into the embryos was not. Co-injection of anti-Pv antibody with m1, m5, or m9 was able to counteract their activity to enhance the embryonic resistance to A. hydrophila. Data were expressed as mean values ± S.E. (n = 3). The bars represent the mean ± S.E. The asterisk means a significant difference (p < 0.05). PvAb, anti-fish Pv antibody; AcAb, anti-actin antibody.
TABLE 3.
Bmax and Kd values of the binding of m1, m3, m5, m7, m9, m12, and m13 to LPS, LTA and PGN, respectively
| Mutants | Binding to LPS |
Binding to LTA |
Binding to PGN |
|||
|---|---|---|---|---|---|---|
| Kd | Bmax | Kd | Bmax | Kd | Bmax | |
| μg/ml | ng/pmol | μg/ml | ng/pmol | μg/ml | ng/pmol | |
| m1 | 1.1 | 5.9 | 1.3 | 6.0 | 1.5 | 6.3 |
| m3 | 1.1 | 6.0 | 1.3 | 6.1 | 1.6 | 6.3 |
| m5 | 1.1 | 5.8 | 1.3 | 5.9 | 1.6 | 6.1 |
| m7 | 1.2 | 5.8 | 1.2 | 5.8 | 1.6 | 6.1 |
| m9 | 1.2 | 5.7 | 1.3 | 5.9 | 1.7 | 6.1 |
| m12 | 2.3 | 2.4 | 2.2 | 2.5 | 2.7 | 2.4 |
| m13 | 2.8 | 2.1 | 2.6 | 2.1 | 3.0 | 2.2 |
To test if the mutants play similar roles in vivo, the embryos were each microinjected with m12 and m13, both of which had little antimicrobial activity in vitro, as well as with m1, m5, and m9 that retained antimicrobial activity comparable with Pt5 in vitro, followed by the challenge with injection of live A. hydrophila. The majority of the embryos (∼95%) microinjected with all the mutants above developed normally. Interestingly, the challenge with live A. hydrophila induced a significant increase in the mortality of the embryos injected with m12 or m13, with the 24-h cumulative mortality of more than 50%, whereas the same challenge caused ∼42% cumulative mortality at 24 h in the embryos injected with m1 or m5 or m9 (Fig. 7F). It was obvious that the mutants with in vitro antimicrobial activity were able to protect the developing embryos, whereas the mutants without in vitro antimicrobial activity could not.
DISCUSSION
Pv, a major component of yolk proteins, is a maternally transferred molecule abundant in fish eggs. The physiological role known for Pv so far is to provide necessary nutrients for early developing embryos and larvae. Here, we clearly demonstrated that in D. rerio, both the embryo extract and the developing embryo displayed antimicrobial activity against microbes, including A. hydrophila (pathogenic to D. rerio), and all the findings point to Pv being one of the most important factors involved in the antimicrobial activity. First, the antimicrobial activity of the embryo extract was significantly decreased by the preincubation with anti-fish Pv antibody, a process that causes the precipitation of Pv. Second, the microinjection of anti-Pv antibody into the early developing embryos (which were then challenged by injection of live A. hydrophila) resulted in a significantly increased mortality, whereas the antibody-induced embryonic death rate was markedly reduced by the co-injection of purified rPv plus anti-Pv antibody. Third, rPv was capable of directly inhibiting the growth of E. coli, A. hydrophila, and S. aureus. Fourth, injection of exogenous rPv or its truncated product Pt5 into the embryos markedly promoted their resistance to A. hydrophila, and this promoted bacterial resistance was significantly reduced by the co-injection of anti-Pv antibody plus rPv (or Pt5) but not by the injection of anti-actin antibody plus rPv. Fifth, like rPv and Pt5, the generated mutants with in vitro antimicrobial activity, when injected into the embryos, were able to protect the developing embryos against A. hydrophila challenge, but those without in vitro antimicrobial activity could not. Finally, the early embryos contained sufficient Pv, which is high enough to defend against potential pathogens in vivo. Taken together, these indicate that Pv, in addition to being a simple nutritional reserve, also has an antimicrobial activity, a novel function assigned to Pv. This nature of Pv possibly has an important physiological significance. Fish embryos develop externally in most cases and are exposed to an aquatic environment full of potential pathogens, but they have little or only limited ability to mount an efficient and protective response. How they survive the pathogenic attacks in such a hostile environment is poorly understood. Our results suggest it is highly likely that Pv may be physiologically involved in the antimicrobial defense of early embryos from zebrafish.
Nonself-recognition is a pivotal process in the innate immune response. Recognition of nonself-recognition in innate immunity is mediated by a set of host's molecules known as pattern recognition receptors (49) that recognize the microbial cell wall constituents called pathogen-associated molecular patterns present on the surface of microbes but absent in the host, such as LPS, LTA, and PGN. We showed that zebrafish rPv had a strong affinity for LPS from Gram-negative bacteria, LTA from Gram-positive bacteria, and PGN from both Gram-positive and negative bacteria. In agreement, rPv also bound to Gram-negative bacteria E. coli and A. hydrophila as well as Gram-positive bacterium S. aureus. Recently, significant advances have been made in identifying nonself-recognition molecules, and a few of them have been tested for the existence of multiple specificities recognizing pathogen-associated molecular patterns (50). It is clear that Pv is a pattern recognition receptor with a wide spectrum of specificity capable of identifying the microbial signature molecules LPS, LTA, and PGN.
An important part of innate immunity is that a group of proteins have microbicidal activity in addition to their immune recognition function. These proteins all play essential roles in the host nonspecific defenses by preventing or limiting infections via their ability to recognize potential pathogens; most of these proteins also exert their antimicrobial effects by interacting with and destabilizing either the microbial plasma membrane or cell wall, eventually leading to cell death (51). In this study, we found that rPv was able to cause direct damage to E. coli and S. aureus, as evidenced by scanning electron microscopy, suggesting that rPv can kill both the Gram-negative bacterium E. coli and the Gram-positive bacterium S. aureus. It is obvious that in addition to being a pattern recognition receptor, Pv is also an effector (microbicidal) molecule capable of killing microbes. Because microinjection of exogenous rPv into early embryos promoted their resistance to pathogenic A. hydrophila challenge, and this promoted bacterial resistant activity was markedly reduced by the co-injection of anti-Pv antibody plus rPv, we thus propose that Pv may also defend early developing embryos in vivo against pathogenic attacks by the same mechanisms by binding to and disrupting invading pathogens. However, this demands further study at the host-pathogen interaction level.
Most antimicrobial proteins are cationic and amphipathic molecules with a net positive charge ranging from +2 to +9 and a high percentage of hydrophobic residues (47). Zebrafish Pv had a net positive charge +2.2 (pH 7.0) and 78 hydrophobic residues (∼32%), conforming to the characteristics of antimicrobial proteins. To gain insight into the structure-activity relationship, N- and C-terminal deletion was carried out. Deletion of the N-terminal 194 residues did not impair the antimicrobial activity of the resultant peptide (Pt5), but truncation of the C-terminal 55 residues resulted in almost complete loss of the antimicrobial activity of the resultant peptide (Pt6). In addition, the microinjection of Pt5 into the embryos can enhance their resistance to pathogenic A. hydrophila, but the injection of Pt6 cannot. The data suggest that the C-terminal 55 residues are the “core” structure contributing to antimicrobial activity of rPv. This is also corroborated by the fact that Pt5 with antimicrobial activity had a net positive charge +4.4 (pH 7.0), whereas Pt6 without antimicrobial activity possessed a net negative charge −2.3 (pH 7.0).
Site-directed mutagenesis of Pt5 showed that the single mutation R242G (m12) and the double mutation A201G/I203M (m13) resulted in a marked decrease in its antimicrobial activity against E. coli, A. hydrophila, and S. aureus as well as its affinity to LPS, LTA, and PGN, but the replacement of the other positively charged and hydrophobic residues such as I203M (m1; hydrophobic Ile to hydrophilic Met), R214G (m5; positively charged Arg to negatively charged Gly), and E235K (m9; negatively charged Glu to positively charged Lys) caused no changes in their antimicrobial activity. These were also supported by in vivo studies showing that a markedly greater number of the embryos injected with m1, m5, or m9 survived the challenge with live A. hydrophila than the embryos injected with m12 or m13. It is clear that the positively charged residue Arg242 and the hydrophobic residues Ala201 and Ile203 are the functional determinants critical for Pt5, but the other positively charged and hydrophobic residues mutated are not. This suggests that the distribution of positively charged and hydrophobic residues rather than net positive charges and hydrophobicity could be the major factors of Pt5 (also rPv) to mediate specific electrostatic surface and amphipathicity, consistent with the mechanism for specific high affinity binding between proteins proposed by Sinha and Smith-Gill (52). Of note, the antimicrobial activity of Pt5 (also rPv) against E. coli, A. hydrophila and S. aureus was correlated with its affinity to LPS, LTA, and PGN, and consistent with the loss of antimicrobial activity of the mutants m12 and m13, their affinity to LPS, LTA, and PGN was also lost. The correlation between antimicrobial activity and ligand binding suggests that the functional sites Arg248 and Ala201/Ile203 may be simultaneously involved in multiple activities, including binding to microbial signature molecule LPS, LTA, and PGN and destabilizing/disrupting microbial cell membranes.
In summary, this study first demonstrates that Pv is associated with the immune defense of developing embryos in zebrafish and then elucidates that Pv is a multivalent pattern recognition receptor capable of identifying LPS, LTA, and PGN as well as an antimicrobial effector capable of damaging Gram-negative and -positive microbes. It also reveals that the C-terminal 55 residues with the functional sites Arg242 and Ala201/Ile203 are critical for Pv antimicrobial activity. As the deposit of Pv in eggs is widespread in oviparous animals, the Pv-mediated immune defense may be widely present in the early embryos of different species. This work also opens a new avenue for the study of the immunological roles of yolk proteins in animals that rely on yolk proteins for embryonic development.
Supplementary Material
This work was supported by Grants 30972274 and 30730072 from the Natural Science Foundation of China (to S. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- Pv
- phosvitin
- rPv
- recombinant Pv
- LTA
- lipoteichoic acid
- PGN
- peptidoglycan.
REFERENCES
- 1. Ellis A. E. (1988) in Fish Vaccination (Ellis A. E. ed) pp. 20–31, Academic Press, London [Google Scholar]
- 2. Zapata A., Diez B., Cejalvo T., Gutiérrez-de Frías C., Cortés A. (2006) Fish Shellfish Immunol 20, 126–136 [DOI] [PubMed] [Google Scholar]
- 3. Zhang S. C., Haimanti B., Li H. Y. (2009) in Reproductive Biology and Phylogeny of Fishes (Agnathans and Bony Fishes) (Jamieson B. G. ed) pp. 485–517, Science Publishers, Enfield, NH [Google Scholar]
- 4. Bly J. E., Grimm A. S., Morris I. G. (1986) Comp. Biochem. Physiol. A 84, 309–313 [DOI] [PubMed] [Google Scholar]
- 5. Breuil G., Vassiloglou B., Pepin J. F., Romestand B. (1997) Fish Shellfish Immunol. 7, 29–43 [Google Scholar]
- 6. Castillo A., Sánchez C., Dominguez J., Kaattari S. L., Villena A. J. (1993) Dev. Comp. Immunol. 17, 419–424 [DOI] [PubMed] [Google Scholar]
- 7. Hanif A., Bakopoulos V., Dimitriadis G. J. (2004) Fish Shellfish Immunol. 17, 411–435 [DOI] [PubMed] [Google Scholar]
- 8. Mor A., Avtalion R. R. (1990) J. Fish Biol. 37, 249–255 [Google Scholar]
- 9. Olsen Y. A., Press C. M. (1997) Fish Shellfish Immunol. 7, 81–91 [Google Scholar]
- 10. Picchietti S., Taddei A. R., Scapigliati G., Buonocore F., Fausto A. M., Romano N., Mazzini M., Mastrolia L., Abelli L. (2004) Cell Tissue Res. 315, 259–270 [DOI] [PubMed] [Google Scholar]
- 11. Picchietti S., Abelli L., Buonocore F., Randelli E., Fausto A. M., Scapigliati G., Mazzini M. (2006) Fish Shellfish Immunol. 20, 398–404 [DOI] [PubMed] [Google Scholar]
- 12. Swain P., Dash S., Bal J., Routray P., Sahoo P. K., Sahoo S. K., Saurabh S., Gupta S. D., Meher P. K. (2006) Fish Shellfish Immunol. 20, 519–527 [DOI] [PubMed] [Google Scholar]
- 13. Van Loon J. J. A., van Oosterom R., van Muiswinkel W. B. (1981) Comp. Dev. Immunol. 1, 469–470 [Google Scholar]
- 14. Ellingsen T., Strand C., Monsen E., Bøgwald J., Dalmo R. A. (2005) Fish Shellfish Immunol. 18, 351–358 [DOI] [PubMed] [Google Scholar]
- 15. Huttenhuis H. B., Grou C. P., Taverne-Thiele A. J., Taverne N., Rombout J. H. (2006) Fish Shellfish Immunol. 20, 586–596 [DOI] [PubMed] [Google Scholar]
- 16. Liang Y., Zhang S., Wang Z. (2009) PLoS ONE 4, e4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Løvoll M., Kilvik T., Boshra H., Bøgwald J., Sunyer J. O., Dalmo R. A. (2006) Immunogenetics 58, 168–179 [DOI] [PubMed] [Google Scholar]
- 18. Løvoll M., Johnsen H., Boshra H., Bøgwald J., Sunyer J. O., Dalmo R. A. (2007) Fish Shellfish Immunol. 23, 542–552 [DOI] [PubMed] [Google Scholar]
- 19. Wang Z., Zhang S., Wang G., An Y. (2008) PLoS ONE 3, e1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bildfell R. J., Markham R. J. F., Johnson G. R. (1992) J. Aquat. Anim. Health 4, 97–105 [Google Scholar]
- 21. Jung W. K., Park P. J., Kim S. K. (2003) Int. J. Biochem. Cell Biol. 35, 255–265 [DOI] [PubMed] [Google Scholar]
- 22. Tateno H., Yamaguchi T., Ogawa T., Muramoto K., Watanabe T., Kamiya H., Saneyoshi M. (2002) Dev. Comp. Immunol. 26, 543–550 [DOI] [PubMed] [Google Scholar]
- 23. Choi J. H., Park P. J., Kim S. K. (2002) Fish Sci. 68, 1367–1373 [Google Scholar]
- 24. Yamashita M., Konagaya S. (1996) J. Biol. Chem. 271, 1282–1284 [DOI] [PubMed] [Google Scholar]
- 25. Yousif A. N., Albright L. J., Evelyn T. P. (1991) Dis. Aquat. Org. 10, 45–49 [Google Scholar]
- 26. Yousif A. N., Albright L. J., Evelyn T. P. (1994) Dis. Aquat. Org. 19, 15–19 [Google Scholar]
- 27. Wang Z., Zhang S., Tong Z., Li L., Wang G. (2009) PLoS ONE 4, e4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Finn R. N., Fyhn H. J. (2010) Aquac. Res. 41, 684–716 [Google Scholar]
- 29. Sattar Khan M. A., Nakamura S., Ogawa M., Akita E., Azakami H., Kato A. (2000) J. Agric. Food Chem. 48, 1503–1506 [DOI] [PubMed] [Google Scholar]
- 30. Losso J. N., Nakai S. A. (1994) in Egg Uses and Processing Technologies: New Developments (Sim J. S., Nakai S. eds) pp. 150–157, CAB International, Wallingford, United Kingdom [Google Scholar]
- 31. Taborsky G. (1983) Adv. Inorg. Biochem. 5, 235–279 [PubMed] [Google Scholar]
- 32. Garcia J., Munro E. S., Monte M. M., Fourrier M. C., Whitelaw J., Smail D. A., Ellis A. E. (2010) Fish Shellfish Immunol. 29, 293–297 [DOI] [PubMed] [Google Scholar]
- 33. Li Z., Zhang S., Liu Q. (2008) PLoS ONE 3, e1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li Z., Zhang S., Zhang J., Liu M., Liu Z. (2009) Mol. Immunol. 46, 3232–3239 [DOI] [PubMed] [Google Scholar]
- 35. Tong Z., Li L., Pawar R., Zhang S. C. (2010) Immunobiology 215, 898–902 [DOI] [PubMed] [Google Scholar]
- 36. Zhang S., Wang S., Li H., Li L. (2011) Int. J. Biochem. Cell Biol. 43, 303–305 [DOI] [PubMed] [Google Scholar]
- 37. Rono M. K., Whitten M. M., Oulad-Abdelghani M., Levashina E. A., Marois E. (2010) PLoS Biol. 8, e1000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Itoh T., Abe Y., Adachi S. (1983) J. Food Sci. 48, 1755–1757 [Google Scholar]
- 39. Fan C., Zhang S., Li L., Chao Y. (2008) Mol. Immunol. 45, 3338–3346 [DOI] [PubMed] [Google Scholar]
- 40. Schwyn B., Neilands J. B. (1987) Anal. Biochem. 160, 47–56 [DOI] [PubMed] [Google Scholar]
- 41. Liu J., Zhang S., Li L. (2009) Mol. Immunol. 46, 3117–3124 [DOI] [PubMed] [Google Scholar]
- 42. Roque A. C., Taipa M. A., Lowe C. R. (2004) J. Mol. Recognit. 17, 262–267 [DOI] [PubMed] [Google Scholar]
- 43. Gotoh M., Takamoto Y., Kurosaka K., Masuda J., Ida M., Satoh A., Takayama E., Kojima-Aikawa K., Kobayashi Y., Matsumoto I. (2005) Immunol. Lett. 98, 297–302 [DOI] [PubMed] [Google Scholar]
- 44. An Y., Fan N., Zhang S. C. (2009) Mol. Immunol. 46, 2666–2670 [DOI] [PubMed] [Google Scholar]
- 45. Ong S. T., Ho J. Z., Ho B., Ding J. L. (2006) Immunobiology 211, 295–314 [DOI] [PubMed] [Google Scholar]
- 46. Grogan J., Taborsky G. (1987) J. Inorg. Biochem. 29, 33–47 [DOI] [PubMed] [Google Scholar]
- 47. Wiesner J., Vilcinskas A. (2010) Virulence 1, 440–464 [DOI] [PubMed] [Google Scholar]
- 48. Yeaman M. R., Yount N. Y. (2003) Pharmacol. Rev. 55, 27–55 [DOI] [PubMed] [Google Scholar]
- 49. Janeway C. A., Jr., Medzhitov R. (2002) Annu. Rev. Immunol. 20, 197–216 [DOI] [PubMed] [Google Scholar]
- 50. Akira S., Uematsu S., Takeuchi O. (2006) Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 51. Cash H. L., Whitham C. V., Behrendt C. L., Hooper L. V. (2006) Science 313, 1126–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sinha N., Smith-Gill S. J. (2002) Curr. Protein Pept. Sci. 3, 601–614 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.