Abstract
Data from clinical studies, cell culture, and animal models implicate the urokinase plasminogen activator (uPA)/uPA receptor (uPAR)/plasminogen system in the development of atherosclerosis and aneurysms. However, the mechanisms through which uPA/uPAR/plasminogen stimulate these diseases are not yet defined. We used genetically modified, atherosclerosis-prone mice, including mice with macrophage-specific uPA overexpression and mice genetically deficient in uPAR to elucidate mechanisms of uPA/uPAR/plasminogen-accelerated atherosclerosis and aneurysm formation. We found that macrophage-specific uPA overexpression accelerates atherosclerosis and causes aortic root dilation in fat-fed Ldlr−/− mice (as we previously reported in Apoe−/− mice). Macrophage-expressed uPA accelerates atherosclerosis by stimulation of lesion progression rather than initiation and causes disproportionate lipid accumulation in early lesions. uPA-accelerated atherosclerosis and aortic dilation are largely, if not completely, independent of uPAR. In the absence of uPA overexpression, however, uPAR contributes modestly to both atherosclerosis and aortic dilation. Microarray studies identified S100A8 and S100A9 mRNA as the most highly up-regulated transcripts in uPA-overexpressing macrophages; up-regulation of S100A9 protein in uPA-overexpressing macrophages was confirmed by Western blotting. S100A8/A9, which are atherogenic in mice and are expressed in human atherosclerotic plaques, are also up-regulated in the aortae of mice with uPA-overexpressing macrophages, and macrophage S100A9 mRNA is up-regulated by exposure of wild-type macrophages to medium from uPA-overexpressing macrophages. Macrophage microarray data suggest significant effects of uPA overexpression on cell migration and cell-matrix interactions. Our results confirm in a second animal model that macrophage-expressed uPA stimulates atherosclerosis and aortic dilation. They also reveal uPAR independence of these actions and implicate specific pathways in uPA/Plg-accelerated atherosclerosis and aneurysmal disease.
Keywords: Atherosclerosis, Lipoprotein, Macrophage, Microarray, Plasminogen, Plasminogen Activation, Urokinase
Introduction
Atherosclerosis and aneurysm formation are common cardiovascular diseases that have a significant impact on human health. Atherosclerosis is characterized by accumulation in the inner blood vessel wall of cellular and matrix-rich plaques that cause vessel lumen narrowing and tissue ischemia. Clinical events in patients with atherosclerosis (e.g. heart attacks and strokes) are typically caused by rupture of atherosclerotic plaques with superimposed thrombosis that occludes the vessel lumen (1). In contrast, aneurysms are abnormal expansions of a blood vessel that can lead to complete disruption of the vessel wall, causing sudden death (2). The molecular mechanisms that contribute to plaque growth, plaque rupture, and aneurysm formation are incompletely understood. Elucidation of these mechanisms should enable the development of novel, targeted therapies that improve human health.
We and others (3) have hypothesized that the urokinase plasminogen activator (uPA)6/plasminogen (Plg) system contributes to plaque growth, plaque rupture, and aneurysm formation. This hypothesis is supported by observational human studies showing that uPA (expressed primarily by macrophages) and its cell-surface receptor uPAR (expressed by several vascular cell types) are present in human atherosclerotic lesions, with expression of both molecules correlated with lesion severity (4–7). Moreover, patients with evidence of increased Plg activation have an elevated risk for accelerated atherosclerosis and major cardiovascular events (8–10), and uPA expression is elevated in aneurysmal human aortae (11, 12). Important roles for uPA in atherosclerosis and aneurysm formation are supported by recent studies in our laboratory showing that 15-week-old Apoe−/− mice with macrophage-targeted uPA overexpression (SR-uPA+/0 mice, with scavenger receptor promoter-driven expression of a uPA transgene) have significantly accelerated plaque growth and aneurysm formation (13–15), older SR-uPA+/0 Apoe−/− mice have increased atherosclerotic plaque rupture (16), and recipients of bone marrow transplants from uPA null mice have less atherosclerosis than recipients of bone marrow from uPA-expressing mice (14).
These mouse experiments strongly support the hypothesis that elevated uPA expression by artery wall macrophages accelerates atherosclerosis and aneurysm formation. We also showed using Plg−/− mice that accelerated atherosclerosis and aortic dilation in SR-uPA mice were mediated by uPA activation of Plg (15). However, our experiments also raise several issues. First, the role of uPAR in production of these phenotypes, acting either to facilitate uPA-mediated cell surface Plg activation (17) or independently of Plg activation (18), remains unclear and cannot be addressed definitively in Apoe−/− mice because Plaur and Apoe are linked on mouse chromosome 7 (data not shown). Second, it is unknown whether uPA-accelerated atherosclerosis and aneurysm formation are confined to the Apoe−/− mouse model, an experimental setting that, although informative, is potentially limited by large differences in lipoprotein profiles compared with those of humans and by complete absence of apolipoprotein E (apoE), a protein with diverse biological roles (19–21). Third, because SR-uPA+/0 Apoe−/− mice die suddenly at an early age, it is difficult to use Apoe−/− mice to investigate time-dependent effects of macrophage uPA overexpression on lesion development. Fourth, the molecular and cellular events that are downstream of Plg activation in SR-uPA+/0 Apoe−/− mice and are, therefore, more directly responsible for accelerated atherosclerosis, plaque rupture, and aneurysm formation, are unknown. To address these issues and generate a more complete mechanistic picture of uPA/Plg-accelerated atherosclerosis, we bred the SR-uPA transgene into the Ldlr−/− background and performed atherosclerosis studies in transgenic and nontransgenic Ldlr−/− mice (both Plaur+/+ and Plaur−/−). In addition, we performed microarray studies on peritoneal macrophages from SR-uPA+/0 and nontransgenic Apoe−/− mice, identified gene expression patterns and transcriptional pathways associated with uPA overexpression, and confirmed certain of the array findings with in vivo and in vitro biochemical assays.
EXPERIMENTAL PROCEDURES
Mice and Atherosclerosis Studies
ApoE-null mice (22) with macrophage-specific overexpression of uPA (i.e. SR-uPA+/0 Apoe−/−mice, in which the human scavenger receptor-A promoter drives expression of a mouse uPA transgene) were generated in our laboratory (13). These SR-uPA+/0 Apoe−/− Ldlr+/+ mice (i.e. wild type for the low density lipoprotein receptor) were bred with nontransgenic mice lacking this receptor (Apoe+/+ Ldlr−/− mice; The Jackson Laboratory) (23) and offspring (all Apoe+/− Ldlr+/−) crossed to generate SR-uPA+/0 and nontransgenic Apoe+/+ Ldlr−/− mice (hereafter referred to simply as Ldlr−/−). SR-uPA+/0 Ldlr−/− mice were bred with nontransgenic mice lacking the urokinase plasminogen activator receptor (Plaur−/− Ldlr+/+ mice; The Jackson Laboratory) (24), and offspring of this mating were then intercrossed to generate SR-uPA+/0 and nontransgenic Ldlr−/− Plaur+/− mice. These mice were crossed to yield Ldlr−/− littermates with four experimental genotypes: SR-uPA+/0 Plaur−/−; SR-uPA0/0 Plaur−/−; SR-uPA+/0 Plaur+/+; SR-uPA0/0 Plaur+/+. All mice in this study were wild type at the endogenous uPA locus (Plau+/+). Mice were genotyped by PCR of tail DNA (see supplemental Table 1 for primer sequences and supplementary Methods for genotyping details). All experimental mice were progeny of C57BL/6 backcrosses for at least 11 generations. Atherosclerosis studies were carried out by initiating a “Western” diet (21% fat, 0.15% cholesterol by weight; Harlan-Teklad TD88137) at 5 weeks of age and killing the mice either at 15 or 25 weeks of age. Only female mice were used for atherosclerosis studies, plasma lipid analyses, and serum chemistries. Both genders were used for uPA expression studies and peripheral blood cell counts. We also performed new experiments on tissues and samples from mice reported previously; that is, aortic protein extracts from SR-uPA+/0 Apoe−/− mice (both plasminogen null (Plg−/−) and plasminogen wild type (Plg+/+)) (15, 25), aortic mRNA from Apoe−/− recipients of SR-uPA+/0 or nontransgenic Apoe−/− bone marrow (16), and aortic mRNA from SR-uPA+/0 or nontransgenic Apoe−/− Plg+/+ mice (15). All animal protocols were approved by the University of Washington Office of Animal Welfare.
Tissue Harvest, Processing, and Analysis of Atherosclerosis
Mice were deeply anesthetized and perfused with formalin-based fixative (13). Aortic roots were processed into optimal cutting temperature medium and aortae pinned and stained with Sudan IV (13, 14). Optimal cutting temperature medium blocks were sectioned (10-μm thickness), and sections were stained with hematoxylin and eosin, oil red O, and the macrophage-specific antibodies MOMA-2 (BIOSOURCE) or Mac-2 (Cedarlane Labs) (13, 14). For each stain, a total of 5–7 sections per mouse (at 50-μm steps) were analyzed. Observers blinded to genotype used image analysis software (ImagePro) to analyze digital images of histologic sections and pinned aortae. Aortic root atherosclerosis was quantified by measuring intimal area. Oil red O-, MOMA-2, and Mac-2 areas were measured using color thresholding and planimetry, and the percentage of stained intimal area was calculated by dividing the stained area by the total intimal area measured on an adjacent section. Total and percentage of Sudan IV-positive aortic surface area were measured similarly. We also counted the number of individual lesions on the surface of pinned aortae of 15-week-old mice. Lesions were counted by two independent observers using a dissecting microscope. The observer counts were highly correlated (r2 = 0.8), and the mean of their counts was used as the final determination. Aortic root circumference was measured at the level of the internal elastic lamina. The maximal percent luminal stenoses of proximal coronary artery segments were calculated using measurements made on oil red O-stained sections (13).
Measurement of Plasma Lipids, Serum Chemistries, Peripheral Blood Counts, and uPA Antigen
Plasma obtained by retro-orbital bleed after a 4-h fast was used for measurement of total cholesterol and triglycerides (13). Plasma lipoproteins were also analyzed by fast protein liquid chromatography (FPLC) (13). Plasma LDL and HDL were determined by measuring cholesterol in the respective pooled FPLC fractions. Retro-orbital blood drawn at 8–10 weeks of age was used for complete blood counts (Phoenix Central Laboratory). Plasma obtained by retro-orbital bleed and centrifugation of blood at 2000 × g for 5 min was frozen at −80 °C and thawed, and uPA antigen was measured by ELISA (Innovative Research).
Macrophage and Aortic Plasminogen Activator Activity
Bone marrow-derived macrophages were obtained as described (15). On day 10 after plating, serum-free M199 medium was added and collected 20 h later. Cells were counted with a hemocytometer, and cell lysate protein was measured with the BCA method (Bio-Rad). Thoracic aortae were rinsed, then incubated in M199 for 4 h, and conditioned medium was collected. Medium was frozen at −80 °C and thawed, and Plg activator (PA) activity was measured with a chromogenic assay using human Plg (American Diagnostica) and the plasmin substrate S-2251 (Diapharma) (14). PA activity measured in tissues from SR-uPA+/0 mice is Plg-dependent (14).
RNA Extraction, Purification, and Quantification
Six SR-uPA+/0 and 6 nontransgenic mice (all Apoe−/−) were fed the Western diet from 5 to 15 weeks of age, then injected intraperitoneally with thioglycollate (13). Peritoneal macrophages were harvested in ice-cold PBS 5 days later, centrifuged (300 × g for 5 min), resuspended in DMEM without phenol red (Invitrogen), and plated onto tissue culture dishes. After 2 h, nonadherent cells were removed, and attached cells were harvested using TRIzol reagent (Invitrogen). Total RNA was extracted and purified using the Total RNA PureLink Micro-to-Midi Total RNA Purification System (Invitrogen) and quantified using a Nanodrop spectrophotometer (Thermo Scientific). RNA integrity was analyzed using the Agilent 2010 Bioanalyzer; the RNA integrity number for all samples was 8.9- 9.7 with the majority >9.5.
Microarray Analysis of Gene Expression
Gene expression was analyzed using Sentrix Mouse Ref-8 Expression BeadChips (Illumina) at the Center for Array Technologies at University of Washington. cRNA was amplified and labeled using Illumina® TotalPrep RNA Amplification kit (Illumina). Briefly, single-stranded cDNA was synthesized from total RNA using T7 Oligo(dT) Primer and ArrayScript. The single-stranded cDNA was then converted to double-stranded cDNA using DNA polymerase and RNase H. The double-stranded cDNA was purified with a cDNA filter cartridge and transcribed to biotin-labeled cRNA by in vitro transcription using T7 Enzyme Mix and biotin-NTP. cRNA was purified with a cRNA filter cartridge, quantified with a Nanodrop spectrophotomoter, quality-checked by Agilent Bioanalyzer, and hybridized to Sentrix Mouse Ref-8 Expression BeadChips (Illumina). After hybridization, the chips were washed, stained with streptavidin-Cy3, and scanned for fluorescence intensity.
Microarray Data Analysis
A total of 16 RNA samples were analyzed (all from Apoe−/− mice); 6 from individual SR-uPA+/0 mice, 6 from individual nontransgenic mice, 2 samples of pooled RNA from the 6 SR-uPA+/0 mice, and 2 samples of pooled RNA from the 6 nontransgenic mice. Trimmed mean intensities for each bead type were reported by the Illumina BeadArray software. For our initial analysis, the intensities were imported into R (Version 2.5.0) and processed using several BioConductor packages. Signals were normalized using the quantile normalization function of the AFFY package (26). Hybridization control probes showed appropriate intensities and indicated no problems with RNA quality or hybridization in any of the arrays. Differential expression between groups was determined using the linear model and empirical Bayes methods in the limma package (27). All p values were corrected for multiple hypothesis testing using the method of Benjamini and Hochberg and a false discovery rate of 0.05 (28). Gene annotations were taken from the illuminaMousev1 annotation package, which assigns gene symbols to probes that have a RefSeq identifier.
Gene ontology (GO) term overrepresentation analysis was performed with GOstat (29) using RefSeq identifiers and the Mouse Genome Informatics data base. For each GO term, a p value was calculated (using Fisher's exact test) to represent the probability that random distribution could account for the number of times this term appears in the tested group relative to the reference group (all genes on the array). Correction for multiple testing was carried out using the Benjamini and Hochberg algorithm (28). Gene set enrichment analysis (30) was also used to discover sets of genes that were regulated in association with uPA overexpression. The algorithm was implemented using the Babelomics website (31) and KEGG mouse pathway gene sets. An enrichment score was calculated for each pathway and normalized for the size of the gene set. Permutation analysis within gene sets was used to determine p values. A false discovery rate q value was generated by comparing the enrichment scores for individual gene sets against the permuted values and against the enrichment scores of all the gene sets.
For added confidence, we reanalyzed the gene array data independently. Probe-level intensities were quantile normalized using the preprocessCore package in BioConductor. Of the 24,611 probes on the BeadArray chip, 21,279 probes were successfully mapped to one or more mouse UniGene clusters using the illuminaMousev1p1 software package in BioConductor. For each mouse gene probe, a two-tailed t test was used to generate a p value for differential log2 intensity between the SR-uPA+/0 and nontransgenic samples. The log2 of the ratio of the average intensity of the SR-uPA+/0 samples to the average intensity in the nontransgenic samples was also computed for each probe. Probes were selected that had an absolute log2 ratio greater than 0.6 (∼1.5-fold) and a p value less than 0.05. This method was used to preferentially identify genes for which differential expression between the sample groups was large enough that differential protein expression between the groups is probable.
Pathway-level Analysis Using MetaCore
To identify pathways that may be activated or inhibited in macrophages by urokinase overexpression, the transcriptional profiling data were analyzed using the MetaCore software program from GeneGo. Proteins encoded by genes differentially expressed in SR-uPA+/0 macrophages were extended into protein networks of up to 50 molecules based on protein-protein, protein-metabolite, and protein-DNA interactions from a large interaction data base curated from the molecular biology literature. Networks were constructed using a shortest paths algorithm based on two factors, (i) relative enrichment in the SR-uPA+/0 versus nontransgenic samples and (ii) degree of overlap of networks with canonical pathways. The resulting ranked networks were analyzed for enrichment of biological process and molecular function annotations. For this enrichment test, gene annotations were obtained from the built-in data base in MetaCore, and p values were obtained using the hypergeometric distribution.
Gene Function/Pathway Enrichment Analysis
To identify groups of Gene Ontology functional annotations and molecular pathway annotations that are enriched within the up-regulated and down-regulated gene sets, the gene lists were analyzed using the Web-based gene annotation software program DAVID (32, 33). This analysis identifies pathway and gene functional annotations that are enriched within either of the two sample groups relative to their genome-wide average frequencies. Enriched annotations were then clustered by similarity, and the resulting clusters were ordered based on the median of the enrichment p value (Fisher's exact test) for all of the annotations within a cluster.
Transcription Factor Enrichment Analysis of Differentially Expressed Genes
Promoters of the differentially expressed genes were analyzed to identify transcription factors that were enriched within the set of differentially expressed genes (analyzed by MetaCore) or that have known binding site motifs that are over-represented within the gene promoters compared with the expected frequencies of such motifs within promoters of randomly selected genes (analyzed using the web tool oPOSSUM) (34) using Fisher's exact test for significance testing.
Western Blotting
ApoA-I was detected by Western analysis of extracts of aortae of 3–4-month-old SR-uPA+/0 Apoe−/− mice (either Plg+/+ or Plg−/−; n = 5 each). Western analysis of uPA in these same extracts was reported earlier (15). S100A9 was detected by Western analysis of extracts of peritoneal macrophages of 10-week-old SR-uPA+/0 and nontransgenic mice (both Apoe−/−). These macrophages were obtained exactly as described above for the microarray study. Either 30 μg (for the apoA-I blots) or 15 μg (for the S100A9 blots) of protein were separated by SDS/PAGE and transferred to PVDF membranes (15). ApoA-I was detected with a rabbit antibody to mouse apoE that also reacts with apoA-I (1:1000; reference (35)), and bound antibody was detected with peroxidase-conjugated anti-rabbit IgG (1:10,000, Santa Cruz). S100A9 was detected using a goat antibody to mouse S100A9 (1:1000, R&D Systems) and peroxidase-conjugated anti-goat IgG (1:10,000, Rockland). To control for protein loading, membranes were stripped and re-probed with a mouse monoclonal antibody to GAPDH (1:7,000; Ambion).
Quantitative Reverse Transcriptase(qRT)-mediated PCR
The same macrophage RNA samples used for the array studies were used for qRT-PCR to measure mRNA encoding S100A8, S100A9, and CX3CR1 (for primer and probe sequences, see supplemental Table 1). Reaction and thermal cycling conditions were as described (14). RNA levels were normalized to GAPDH expression in the same sample, also determined by qRT-PCR (14). S100A8 and S100A9 mRNA levels were also measured by qRT-PCR of RNA extracted from aortae of SR-uPA+/0 and nontransgenic mice (both Apoe−/− and Ldlr−/−) and from aortae of nontransgenic Apoe−/− recipients of SR-uPA+/0 Apoe−/− or nontransgenic Apoe−/− bone marrow (14).
Regulation of Macrophage S100A9 Expression in Vitro
Peritoneal macrophages were isolated from SR-uPA+/0 and nontransgenic Apoe−/− mice as described above, centrifuged (300 × g for 5 min), resuspended in DMEM (Invitrogen), and plated onto 100-mm tissue culture dishes at a density of 107 cells/plate. After 2 h, nonadherent cells were removed, and 10 ml of fresh DMEM was added. Conditioned medium was collected 48 h later and used immediately to treat nontransgenic macrophages that had been plated in 6-well plates 2 h earlier (1 × 106 cells/well; 1 ml conditioned medium/well). Some macrophages were treated with DMEM only as an additional control. To test whether Plg activation was sufficient to up-regulate S100A9, macrophages were also treated with DMEM plus plasmin (Hematologic Technologies) or with nontransgenic conditioned medium plus plasmin (100 nm, a concentration at which plasmin activates signaling in both endothelial and dendritic cells) (36, 37). After 6 h, the medium was aspirated, and the treated macrophages were washed twice with PBS to remove any cells or debris that might have been carried over with the conditioned medium. Macrophage RNA was extracted with Trizol (Invitrogen), quantified as described above, and S100A9 and GAPDH mRNA were measured by qRT-PCR.
Statistical Methods
Data are the mean ± S.D. except as indicated. Groups were compared with the unpaired t test or with the Mann-Whitney rank-sum test for non-normally distributed data or when group variances were unequal. Two-way ANOVA on ranks was used to discern separate effects of the SR-uPA transgene and uPAR and to test for possible interactions between these two factors in determining the extent of atherosclerosis, aortic root dilation, and proximal coronary artery stenosis in Ldlr−/− mice (38). The raw data were rank-transformed before analysis by 2-way ANOVA to satisfy the requirement of the test for normally distributed data. These tests were all carried out with the SigmaStat program (Systat Software). The microarray data were analyzed separately using the “R” statistical environment and limma analysis (27) and as described above.
RESULTS
Expression of the SR-uPA Transgene in 15-week-old Ldlr−/− Mice
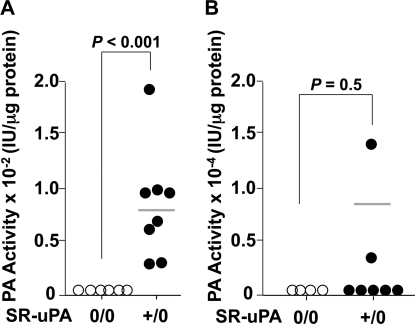
We bred the SR-uPA transgene into the Ldlr−/− background, fed the mice a western diet from 5 until 15 weeks of age, and measured PA activity in medium conditioned by either bone marrow-derived macrophages or thoracic aortae of nontransgenic and SR-uPA+/0 Ldlr−/− mice. PA activity was increased in medium conditioned by SR-uPA+/0 macrophages (Fig. 1A; p < 0.001). However, PA activity was not significantly increased in medium conditioned by SR-uPA+/0 aortae (Fig. 1B; p = 0.5). Expression of the SR-uPA+/0 transgene did not significantly affect plasma cholesterol, triglycerides, or FPLC profiles of Ldlr−/− mice (Table 1 and supplemental Fig. 1). Similarly, peripheral blood monocyte and percent monocyte counts were not affected by the SR-uPA transgene (Table 1).
FIGURE 1.
PA activity of medium conditioned by bone marrow-derived macrophages and explanted aortae. Medium conditioned by macrophages (A) or aortae (B) of Ldlr−/− mice (either nontransgenic (0/0) or hemizygous for the SR-uPA transgene (+/0)) was assayed for PA activity by the addition of plasminogen and the plasmin substrate S-2251. Data points are values from individual mice; bars are group means. p values are from the Mann-Whitney test.
TABLE 1.
Peripheral blood and vascular measurements in 15-wk old Ldlr−/− mice
Measurements are from transgenic mice with macrophage-specific uPA overexpression (SR-uPA+/0) and nontransgenic (SR-uPA0/0) littermates, all fed a high fat diet from 5–15 weeks of age. n = number of mice in each group. Data are the mean ± S.D. p values are from t tests except for monocyte data (Mann-Whitney test). IEL = internal elastic lamina.
| SR-uPA0/0 | n | SR-uPA+/0 | n | P | |
|---|---|---|---|---|---|
| Plasma cholesterol (mg/dl) | 1267 ± 277 | 13 | 1246 ± 158 | 13 | 0.8 |
| Plasma triglycerides (mg/dl) | 150 ± 77 | 13 | 139 ± 40 | 13 | 0.6 |
| Peripheral blood monocytes (cells/mm3) | 98 ± 40 | 8 | 169 ± 182 | 10 | 1.0 |
| Monocytes (% of total leukocytes) | 2.4 ± 1.3 | 8 | 2.6 ± 2.9 | 10 | 0.6 |
| Aortic root ORO area (mm2) | 0.15 ± 0.08 | 9 | 0.3 ± 0.14 | 10 | 0.007 |
| ORO area (% intimal area) | 51 ± 10 | 9 | 62 ± 7 | 10 | 0.02 |
| Aortic root macrophage area (mm2) | 0.08 ± 0.03 | 7 | 0.07 ± 0.03 | 10 | 0.7 |
| Macrophage area (% intimal area) | 32 ± 8 | 7 | 21 ± 6 | 10 | 0.006 |
| Pinned aorta lesion (%) | 4.1 ± 1.4 | 13 | 4.7 ± 1.5 | 13 | 0.3 |
| Aortic surface lesions (n) | 20.0 ± 7 | 5 | 17.7 ± 5 | 6 | 0.5 |
| IEL circumference (mm) | 3.9 ± 0.3 | 7 | 4.5 ± 0.7 | 10 | 0.1 |
Macrophage-expressed uPA Accelerates Lesion Growth and Lipid Accumulation in Aortic Roots of 15-Week-old Ldlr−/− Mice
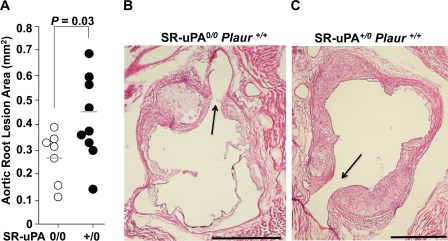
15-Week-old SR-uPA+/0 Ldlr−/− mice had significantly more aortic root atherosclerosis than nontransgenic mice (Fig. 2; 0.44 ± 0.17 versus 0.27 ± 0.1 mm2; a 60% increase; p = 0.03). Intimal lesions of SR-uPA+/0 mice also had more ORO-positive area than lesions of nontransgenic mice (Table 1; p < 0.007) and had a higher percentage of ORO-positive intimal area (Table 1; p = 0.02). The total lesion area occupied by macrophages did not differ significantly in SR-uPA+/0 and nontransgenic Ldlr−/− mice, resulting in a lower percentage of macrophage-stained area in the (larger) intimal lesions of the SR-uPA+/0 mice (Table 1; p = 0.006). In contrast, neither the percentage of aortic surface area that stained with Sudan IV nor the number of individual Sudan IV-positive lesions per aorta differed between SR-uPA+/0 and nontransgenic mice (Table 1). There was also no significant difference in the aortic root internal elastic lamina circumferences of SR-uPA+/0 and nontransgenic mice (Table 1; p = 0.01).
FIGURE 2.
Aortic root atherosclerosis in 15-week-old mice. Aortic roots from Ldlr−/− Plaur+/+ mice (either nontransgenic (0/0) or hemizygous for the SR-uPA transgene (+/0)) were sectioned and stained, and lesions were measured. A, shown is the intimal lesion area. Data points are values from individual mice; bars are group means. B and C, shown are representative sections from each of the genotypes with a hematoxylin and eosin stain; arrows = coronary ostia; size bars = 500 μm. p value is from a t test.
Atherosclerosis Studies in 25-Week-old SR-uPA+/0 Ldlr−/− Mice; Investigating the Role of Time and uPAR
Because sudden death associated with the SR-uPA transgene occurs later in Ldlr−/− mice than in Apoe−/− mice (Ref. 13 and data not shown), we were able to set up an atherosclerosis study of longer duration in Ldlr−/− mice. This study was designed to simultaneously test three hypotheses; 1) the effects of uPA on atherosclerosis will be accentuated in older versus younger Ldlr−/− mice, 2) accelerated atherosclerosis in SR-uPA+/0 mice is dependent on uPAR, and 3) uPAR is atherogenic in nontransgenic Ldlr−/− mice. For this experiment we bred SR-uPA+/0 and nontransgenic Plaur+/− Ldlr−/− mice to generate four experimental genotypes: SR-uPA+/0 and nontransgenic Plaur+/+ mice as well as SR-uPA+/0 and nontransgenic Plaur−/− mice (all littermates, all Ldlr−/−). The mice were fed a high fat diet beginning at 5 weeks of age, and atherosclerosis was assessed 20 weeks later.
Neither Plaur genotype nor the SR-uPA transgene affected either peripheral blood monocyte counts or percent peripheral blood monocytes in these 25-week-old mice (Table 2). Total plasma cholesterol was unaffected by the SR-uPA transgene in Plaur+/+ mice; however, in Plaur−/− mice plasma cholesterol was modestly increased in SR-uPA+/0 versus nontransgenic mice (Table 2; p = 0.02). Nevertheless, FPLC profiles of plasma lipids were similar in all four groups (supplemental Fig. 2) as were the plasma concentrations of LDL, HDL, and triglycerides (Table 2 and supplemental Fig. 2).
TABLE 2.
Peripheral blood and vascular measurements in 25-week-old Ldlr−/− mice
Measurements are from nontransgenic mice (SR-uPA0/0) or littermate transgenic mice with macrophage-specific uPA overexpression (SR-uPA+/0) and were either wild type or null for the uPA receptor (Plaur+/+ or −/−). All mice were fed a high fat diet from 5–25 weeks of age. n = number of mice in each group. Data are the mean ± S.D. except for % coronary stenosis, which is ±S.E. p values are for comparison of nontransgenic and transgenic mice within each Plaur genotype and are from t tests except for peripheral blood monocytes, triglycerides, ORO-positive area, and coronary stenosis (Mann-Whitney tests).; IEL = internal elastic lamina.
|
Plaur+/+ |
Plaur−/− |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SR-uPA0/0 | n | SR-uPA+/0 | n | P | SR-uPA0/0 | n | SR-uPA+/0 | n | P | |
| Peripheral blood monocytes (cells/mm3) | 83 ± 91 | 6 | 180 ± 158 | 5 | 0.2 | 81 ± 111 | 8 | 237 ± 201 | 6 | 0.08 |
| Monocytes (% of total leukocytes) | 2.0 ± 1.7 | 6 | 3.0 ± 1.4 | 5 | 0.3 | 2.1 ± 2.3 | 8 | 4.7 ± 3.8 | 6 | 0.3 |
| Plasma total cholesterol (mg/dl) | 1182 ± 154 | 16 | 1252 ± 328 | 8 | 0.5 | 1027 ± 203 | 16 | 1218 ± 220 | 13 | 0.02 |
| Plasma LDL cholesterol (mg/dl) | 189 ± 46 | 5 | 216 ± 36 | 3 | 0.4 | 195 ± 52 | 6 | 217 ± 48 | 5 | 0.5 |
| Plasma HDL cholesterol (mg/dl) | 89 ± 20 | 5 | 100 ± 8 | 3 | 0.4 | 103 ± 13 | 6 | 122 ± 24 | 5 | 0.1 |
| Plasma triglycerides (mg/dl) | 155 ± 86 | 16 | 144 ± 79 | 8 | 0.8 | 134 ± 89 | 16 | 127 ± 70 | 13 | 0.9 |
| Aortic root ORO-positive area (mm2) | 0.4 ± 0.1 | 11 | 0.8 ± 0.2 | 7 | <0.001 | 0.3 ± 0.1 | 12 | 0.7 ± 0.1 | 11 | <0.001 |
| ORO area (% intimal area) | 47 ± 5 | 11 | 46 ± 8 | 7 | 0.8 | 47 ± 7 | 12 | 46 ± 5 | 11 | 0.9 |
| Aortic root macrophage area (mm2) | 0.2 ± 0.06 | 9 | 0.5 ± 0.2 | 7 | <0.004 | 0.2 ± 0.06 | 10 | 0.4 ± 0.1 | 10 | <0.001 |
| Macrophage area (% intimal area) | 30 ± 8 | 9 | 32 ± 13 | 7 | 0.7 | 32 ± 7 | 10 | 29 ± 7 | 10 | 0.3 |
| Aortic total surface area (mm2) | 0.8 ± 0.07 | 15 | 0.78 ± 0.05 | 8 | 0.6 | 0.79 ± 0.07 | 11 | 0.76 ± 0.06 | 11 | 0.6 |
| IEL circumference (mm) | 5.6 ± 0.3 | 12 | 7.4 ± 0.5 | 8 | <0.001 | 5.3 ± 0.4 | 12 | 7 ± 0.3 | 6 | <0.001 |
| Maximal proximal coronary stenosis (%) | 47.3 ± 8.0 | 17 | 75.0 ± 7.4 | 20 | 0.05 | 17.5 ± 7.8 | 18 | 58.8 ± 8.0 | 17 | 0.002 |
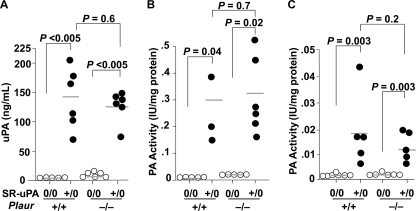
uPA Antigen and Activity Are Increased in 25-Week-old SR-uPA+/0 Ldlr−/− Mice
The SR-uPA transgene elevated plasma uPA antigen substantially in both Plaur+/+ and Plaur−/− mice (Fig. 3A). Elevated PA activity was present in medium conditioned by bone marrow-derived macrophages of both SR-uPA+/0 Plaur+/+ and SR-uPA+/0 Plaur−/− mice (all 25 weeks old; Fig. 3B). In contrast to results obtained with the aortae of 15-week-old mice (Fig. 1B), PA activity was also significantly elevated in medium conditioned by aortae of 25-week-old SR-uPA+/0 mice (Fig. 3C). Plaur genotype did not significantly affect plasma uPA antigen or secreted PA activity (Fig. 3, A–C).
FIGURE 3.
Plasma uPA antigen and PA activity in medium conditioned by bone marrow-derived macrophages and explanted aortae. A, plasma uPA was measured by ELISA. Medium conditioned by macrophages (B) or aortae (C) from Ldlr−/− mice (either nontransgenic (0/0) or hemizygous for the SR-uPA transgene (+/0) and either Plaur+/+ or Plaur−/−) was assayed for PA activity by the addition of plasminogen and the plasmin substrate S-2251. Data points are values from individual mice; bars are group means. p values comparing SR-uPA+/0 versus SR-uPA0/0 groups are from Mann-Whitney tests; p values comparing Plaur+/+ and Plaur−/− groups are from t tests.
uPA-accelerated Atherosclerosis Is Time-dependent and Does Not Require uPAR; uPAR Is Atherogenic
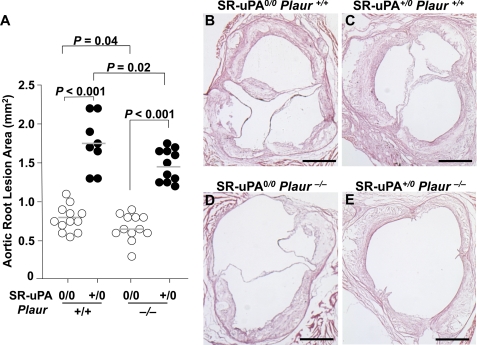
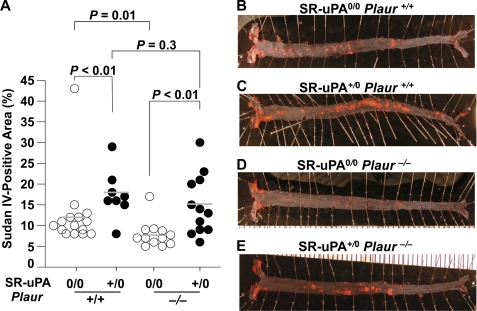
In 25-week-old Plaur+/+ mice, the SR-uPA transgene increased the aortic root intimal lesion area by 2.3-fold (Fig. 4; 1.8 ± 0.34 versus 0.79 ± 0.16 mm2; p < 0.001). Total oil red O- and macrophage-positive areas were both significantly increased in lesions of SR-uPA+/0 versus nontransgenic Plaur+/+ mice; however, the percentage of lesion area occupied by lipid and macrophages was not affected by the SR-uPA transgene (Table 2). In 25-week-old Plaur−/− mice, the SR-uPA transgene increased the aortic root intimal area by 2.1-fold (Fig. 4; 1.4 ± 0.21 versus 0.66 ± 0.15 mm2; p < 0.001). Intimas of SR-uPA+/0 Plaur−/− mice had significantly more total lesion lipid and macrophage area than intimas of nontransgenic Plaur−/− mice; however, as with the Plaur+/+ mice, the percentage of lesion occupied by lipid and macrophages was not affected by the SR-uPA transgene (Table 2). In contrast to the 15-week results (Table 1), the SR-uPA transgene increased the percentage of aortic surface coverage by Sudan IV-stained lesions in both Plaur+/+ mice and Plaur−/− mice (18 ± 6.8 versus 12 ± 8.6 and 16 ± 7.2 versus 8.1 ± 3.3%, respectively, Fig. 5; p < 0.01 for both).
FIGURE 4.
Aortic root atherosclerosis in 25-week-old mice. Aortic roots from Ldlr−/− Plaur+/+ or Ldlr−/− Plaur−/− mice (either nontransgenic (0/0) or hemizygous for the SR-uPA transgene (+/0)) were sectioned and stained, and lesions were measured. A, the intimal lesion area is shown. Data points are values from individual mice; bars are group means. B–E, representative sections are from each of the genotypes and hematoxylin- and eosin-stained; size bars = 500 μm. p values are from t tests except for comparison of SR-uPA+/0 Plaur+/+ versus SR-uPA0/0 Plaur+/+ groups (Mann-Whitney test).
FIGURE 5.
Aortic surface atherosclerosis in 25-week-old mice. Aortae from Ldlr−/− Plaur+/+ or Ldlr−/− Plaur−/− mice (either nontransgenic (0/0) or hemizygous for the SR-uPA transgene (+/0)) were pinned and stained with Sudan IV, and lesions were measured. A, the percentage of surface of each aorta staining positive with Sudan IV is shown. Data points are values from individual mice; bars are group means. B–E, shown are representative aortae from each of the genotypes. p values are from t tests.
This experiment also revealed a role for uPAR in atherogenesis. This role was most apparent in the nontransgenic mice, in which the absence of uPAR reduced both the aortic root intimal lesion area and Sudan IV-positive aortic surface coverage (Figs. 4 and 5; p ≦ 0.04 for both). In SR-uPA+/0 mice, the absence of uPAR reduced the aortic root intimal lesion area (Fig. 4; p = 0.02) but did not significantly change aortic surface coverage by Sudan IV-positive lesions (Fig. 5; p = 0.3). 2-Way ANOVA (which analyzes data from all four groups; see “Statistical Methods” under “Experimental Procedures”) revealed 1) overall significant effects of the SR-uPA transgene (independently of uPAR) on both the aortic root intimal area and aortic surface atherosclerosis (p < 0.001) and 2) overall significant effects of uPAR genotype (independently of the SR-uPA transgene) on both aortic root intimal area (p = 0.003) and aortic surface atherosclerosis (p = 0.01).
Both Macrophage-expressed uPA and uPAR Independently Contribute to Aortic Root Dilation and Accelerate Proximal Coronary Artery Disease in Ldlr−/− Mice
Similar to results obtained with younger Apoe−/− mice (13), the SR-uPA transgene also increased aortic root circumference in 25-week-old Ldlr−/− mice (both Plaur+/+ and Plaur−/−; Table 2; p < 0.001 for an overall contribution of the SR-uPA transgene by two-way ANOVA). uPAR genotype made a small independent contribution to aortic root circumference (p = 0.03 by two-way ANOVA). Proximal coronary artery stenoses were also more severe in SR-uPA+/0 mice, with a higher mean percentage coronary stenosis in SR-uPA+/0 Plaur+/+ and SR-uPA+/0 Plaur−/− mice (75.0 ± 7.4 and 58.8 ± 8.0%, respectively) compared with nontransgenic Plaur+/+ and Plaur−/− mice (47.3 ± 8.0 and 17.5 ± 7.8%, respectively; Table 2; p ≤ 0.05 for both comparisons of SR-uPA to nontransgenic mice). uPAR also contributed to the development of proximal coronary artery stenoses. The contribution of uPAR is seen by comparing mean maximal coronary stenosis in SR-uPA0/0 Plaur+/+ versus SR-uPA0/0 Plaur−/− mice (47.3 ± 8.0 versus 17.5 ± 7.8%; Table 2) and in SR-uPA+/0 Plaur+/+ versus SR-uPA+/0 Plaur−/− mice (75.0 ± 7.4 versus 58.8 ± 8.0%; Table 2 and supplemental Fig. 3). Two-way ANOVA confirmed significant, independent effects of both the SR-uPA transgene and uPAR on coronary stenosis severity (p ≤ 0.005 for both).
ApoA-I Protein Is Not Degraded in the Aortae of SR-uPA Mice
We next focused on identifying the molecular mechanisms through which uPA, acting via Plg (15) and with at most only minor involvement of uPAR (Figs. 4 and 5 and Table 2), accelerates atherosclerosis in both Apoe−/− and Ldlr−/− mice. Plasmin can cleave apoA-I in vitro (39, 40), and plasmin-mediated loss of apoA-I in aortae of SR-uPA+/0 mice would likely impair reverse cholesterol transport, increase lipid accumulation, and accelerate atherosclerosis. To test the “candidate gene” hypothesis that apoA-I degradation by plasmin in aortae of SR-uPA+/0 mice contributes to accelerated atherosclerosis, we performed Western analysis of aortic extracts of SR-uPA+/0 Plg+/+ and SR-uPA+/0 Plg−/− mice (all Apoe−/−), available from a previous experiment (15). There was no difference in apoA-I protein abundance between Plg+/+ and Plg−/− aortae and no evidence of plasmin-mediated apoA-I cleavage (supplemental Fig. 4).
Analysis of Differential Gene Expression in SR-uPA+/0 Versus Nontransgenic Macrophages
Rather than continue a candidate gene approach, we used microarray-based global analysis of gene expression as a means of identifying both individual molecular mediators of uPA-accelerated atherosclerosis and atherogenic pathways activated specifically in SR-uPA+/0 mice. Accelerated atherosclerosis in SR-uPA+/0 mice is macrophage-dependent (14); therefore, we designed our microarray studies to detect transcriptional changes in this cell type. Our initial analysis revealed differential expression of 78 genes in SR-uPA+/0 versus nontransgenic macrophages, including 14 up-regulated genes and 64 down-regulated genes (supplemental Table 2). S100A9 (calgranulin B) and S100A8 (calgranulin A) were the two most highly up-regulated transcripts. CX3CR1, which plays a significant role in atherosclerosis (41), was also up-regulated in SR-uPA+/0 macrophages. Several collagens were among the most highly down-regulated transcripts. GOstat analysis of the up-regulated genes revealed 11 overrepresented GO terms (p = 0.04–0.05); however, these were represented by only 1–2 transcripts each (supplemental Table 3 and data not shown). GOstat analysis of down-regulated transcripts revealed 19 over-represented GO terms (p = 0.03- 4 × 10−11) with 2–26 transcripts per term (supplemental Table 3 and data not shown). Overrepresented GO terms for which at least 5 down-regulated transcripts were observed included several terms related to the extracellular matrix, cell adhesion, or proteinase inhibition. Gene set enrichment analysis revealed several sets of genes that appeared to be regulated in association with macrophage uPA overexpression (p < 0.05 with most false discovery rate q values <0.25; supplemental Table 4), including sets related to tight junctions, cell communication, extracellular matrix-receptor interaction, and regulation of actin cytoskeleton.
To help focus subsequent validation studies on the most likely candidate genes, we performed a second, independent analysis of the array data using different normalization and statistical testing. This analysis revealed 75 probes with an absolute log2 ratio >0.6 (∼1.5-fold) and p < 0.05, including 17 with higher intensities in the SR-uPA+/0 group and 54 with lower intensities in the SR-uPA+/0 group (supplemental Table 5). As before, S100A9 and S100A8 were the most highly up-regulated transcripts in SR-uPA+/0 macrophages. A significant (p = 0.02) up-regulation of the atherogenic cytokine IL-1β (42) was detected in this analysis (supplemental Table 2). Nine up-regulated genes and 34 down-regulated genes were common to both lists (supplemental Tables 2 and 5).
Pathway-level Analysis Using MetaCore
We used the MetaCore network analysis software to identify protein interaction networks that are enriched for molecules differentially expressed in SR-uPA+/0 versus nontransgenic macrophages. One network (supplemental Fig. 5) was identified that connected S100A8 and S100A9 to pathogen-associated and damage-associated molecular pattern receptors Toll-like receptor 4 (TLR4) and the receptor for advanced glycation end products (RAGE) and connected these receptors to two transcription factors associated with macrophage activation, NF-κB and c-Jun, a component of AP-1.
Gene Function/Pathway Enrichment Analysis
Analysis of the microarray data with DAVID (supplemental Table 6) revealed significant up-regulation of genes related to inflammation (including taxis, chemotaxis, response to external stimulus, and locomotory behavior) in SR-uPA+/0 macrophages. In agreement with the GOstat analysis, down-regulated transcripts in SR-uPA+/0 macrophages again included groups related to the extracellular space, extracellular matrix, cell adhesion, and protease inhibition.
Transcription Factor Enrichment Analysis of Differentially Expressed Genes
We used the list of differentially expressed genes generated by the second analysis of the microarray data to help identify transcription factors that may mediate transcriptional differences in SR-uPA+/0 versus nontransgenic macrophages. This analysis identified several transcription factors (supplemental Table 7) for which binding site motif matches are enriched within the differentially regulated genes listed in supplemental Table 5 or for which published promoter binding or gene regulation data suggest a role in mediating the differential gene expression (supplemental Table 8). These factors include: the AP-1 components c-Fos and c-Jun; the NF-κB components c-Rel, RelA, and NF-κB1; C/EBPβ; Sp1; NFYA. RelA, Sp1, and NF-κB1 were identified by both approaches.
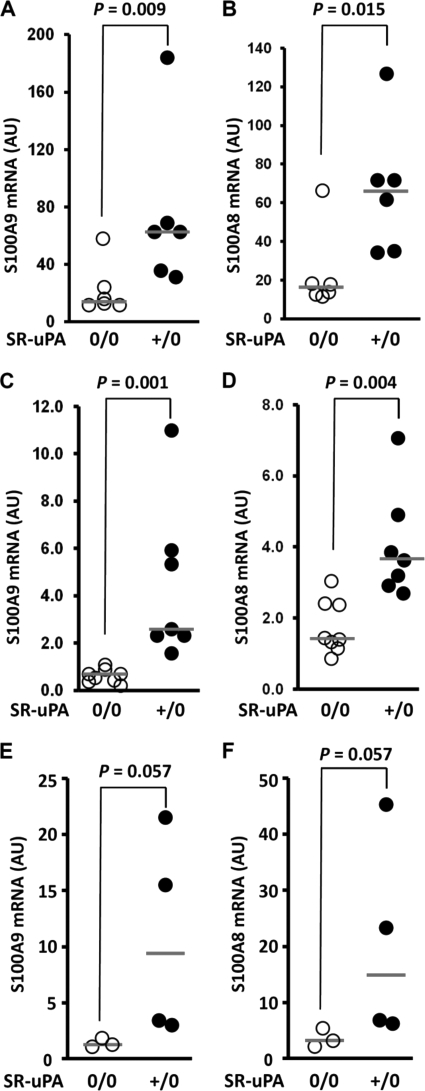
qRT-PCR Reveals Up-regulation of S100A9 and S100A8 mRNA in SR-uPA+/0 Macrophages and Aortae
The significant up-regulation of S100A9 and S100A8 mRNA was perhaps the most interesting quantitative result from the microarray experiments. We further tested this finding by qRT-PCR performed on the macrophage RNA samples used for the array study. This analysis confirmed significantly increased S100A9 and S100A8 mRNA in SR-uPA+/0 versus nontransgenic peritoneal macrophages (p ≤ 0.015 for both; Fig. 6, A and B). In contrast, qRT-PCR of CX3CR1 mRNA showed only a trend toward increased mRNA in SR-uPA+/0 macrophages (p = 0.1; data not shown). To investigate whether S100A9/A8 mRNA were also up-regulated in atherosclerotic aortae of SR-uPA+/0 mice (and, therefore, would be more likely to contribute to atherogenesis), we repeated the qRT-PCR of S100A9 and S100A8 using aortic RNA from 45-week-old nontransgenic Apoe−/− recipients of SR-uPA+/0 or nontransgenic bone marrow (available from a previous study) (16). Both transcripts were significantly more abundant in the aortae of SR-uPA+/0 bone marrow recipients (p ≤ 0.004 for both; Fig. 6, C and D). S100A9 and S100A8 mRNA were also elevated in the aortae of germ line transgenic SR-uPA+/0 Apoe−/− versus nontransgenic Apoe−/− mice (p = 0.057 for both; Fig. 6, E and F). The magnitude of increased S100A8 and A9 mRNA (i.e. the relative increases in the medians of the SR-uPA+/0 groups compared with the medians of the corresponding SR-uPA0/0 groups in Fig. 6) range from 2.2- to 6.4-fold. This degree of increased S100A8 and S100A9 expression is numerically larger than the magnitude of increased macrophage accumulation in these aortae (1.7- 2.0-fold; see the primary data in Refs. 15 and 16), which supports the hypothesis that increased S100A8 and A9 expression in SR-uPA+/0 aortae is not due simply to an increase in aortic wall macrophages.
FIGURE 6.
S100A9 and S100A8 mRNA in macrophages and aortae of Apoe−/− mice. Total RNA was extracted from peritoneal macrophages or aortae; S100A9 and S100A8 mRNA were measured by qRT-PCR, with normalization to GAPDH mRNA. A and B, S100A9 and S100A8 mRNA in peritoneal macrophages from SR-uPA0/0 or SR-uPA+/0 mice are shown. C and D, S100A9 and S100A8 mRNA in aortae of SR-uPA0/0 recipients of either SR-uPA0/0 or SR-uPA+/0 bone marrow are shown. E and F, S100A9 and S100A8 mRNA in aortae of SR-uPA0/0 or SR-uPA+/0 mice are shown. Data points are individual mice; bars are group medians. p values are from Mann-Whitney tests. AU, arbitrary units.
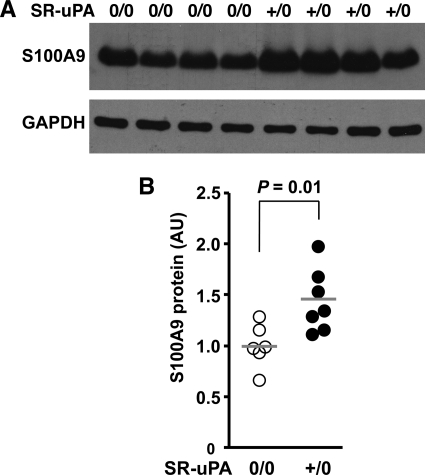
Up-regulation of Macrophage S100A9 Protein in Association with uPA Overexpression in Vivo and in Vitro
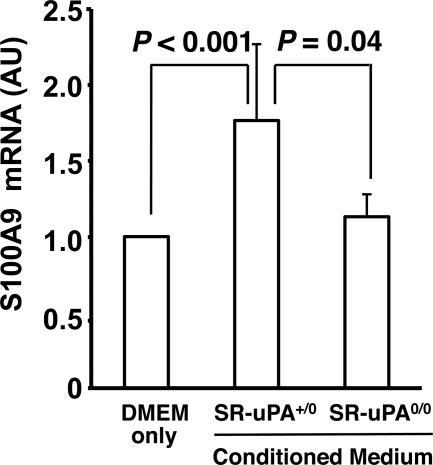
To test whether uPA overexpression also increases S100A9 protein, we performed Western analysis of extracts of peritoneal macrophages of SR-uPA+/0 and nontransgenic Apoe−/− mice. We found 40% more S100A9 protein in SR-uPA+/0 than in nontransgenic macrophages (1.4 ± 0.29 versus 1.0 ± 0.21 absorbance units; p = 0.01; Fig. 7, A and B). To further investigate whether macrophage overexpression of uPA is sufficient to up-regulate expression of S100A9, we harvested media conditioned by either SR-uPA+/0 or nontransgenic peritoneal macrophages and added these conditioned media (or DMEM alone) to nontransgenic macrophages. We used macrophage-conditioned medium for these experiments rather than purified uPA because uPA overexpression in macrophages has biologically important downstream effects (including activation of both plasminogen and matrix metalloproteinases) (16) that would not be accurately modeled simply by the addition of uPA. Medium conditioned by SR-uPA+/0 macrophages up-regulated S100A9 mRNA expression compared with both of the control groups (p ≤ 0.04 for both; Fig. 8). Increased S100A9 mRNA was not due solely to plasminogen activation because plasmin (100 nm) in either DMEM alone or in nontransgenic conditioned medium did not up-regulate S100A9 expression in nontransgenic macrophages (S100A9 expression = 93–98% of DMEM alone values; n = 4 for both; data not shown).
FIGURE 7.
S100A9 protein in extracts of peritoneal macrophages of nontransgenic versus SR-uPA+/0 mice (all Apoe−/−). Mice were fed a high fat diet from 5 until 10 weeks of age, peritoneal macrophages were harvested, and protein was extracted and analyzed. A, shown is a Western blot of macrophage extracts from individual nontransgenic and SR-uPA+/0 mice (upper panel). The blot was stripped and reprobed to detect GAPDH (lower panel). B, densities of the S100A9 bands in A and from three additional mice in each group, normalized to the corresponding GAPDH densities are shown. Data points are from individual mice from two independent experiments; bars are group means, with the mean of the nontransgenic group in each experiment set to 1.0. The p value is from t tests. AU, arbitrary units.
FIGURE 8.
S100A9 mRNA, measured in nontransgenic macrophages. Peritoneal macrophages harvested from nontransgenic Apoe−/− mice were treated for 6 h with DMEM alone or with DMEM previously conditioned by SR-uPA+/0 or SR-uPA0/0 Apoe−/− macrophages. RNA was extracted, and S100A9 and GAPDH mRNA were measured by qRT-PCR. Data are the means ± S.E. of three independent experiments performed with a total of 4–8 wells of cultured macrophages for each group. The mean of the DMEM-only group in each experiment was set to 1.0. p values are from t tests. AU, arbitrary units.
DISCUSSION
To further elucidate the mechanisms of uPA/Plg-accelerated atherosclerosis, we bred the SR-uPA transgene into the Ldlr−/− and Plaur−/− backgrounds and measured atherosclerosis and aortic dimensions in SR-uPA+/0 mice and nontransgenic littermate controls. We also performed microarray studies on macrophages of SR-uPA+/0 and nontransgenic Apoe−/−mice. Our major findings are the following. (i) Macrophage-expressed uPA accelerates atherosclerosis and causes aortic root dilation in the Ldlr−/− background. (ii) Macrophage-expressed uPA does not increase lesion initiation but instead accelerates growth of established lesions. (iii) Vascular pathologies that result from macrophage uPA overexpression are largely, if not completely, independent of uPAR. (iv) uPAR is atherogenic and also contributes to aortic dilation. (v) uPA-overexpressing macrophages express increased S100A8 and S100A9 and release a soluble factor that up-regulates S100A9 expression in nontransgenic macrophages. (vi) S100A8 and S100A9 expression are increased in the aortae of mice with macrophage-specific uPA overexpression. (vii) Global gene expression analysis suggests that altered cell migration and cell-matrix interactions may contribute to uPA/Plg-accelerated atherosclerosis.
We found that SR-uPA+/0 Ldlr−/− mice developed the same vascular disease phenotypes we described previously in SR-uPA+/0 Apoe−/− mice (13), including accelerated aortic atherosclerosis, severe proximal coronary stenoses, aortic root dilation, and premature sudden death (Figs. 2, 4, and 5, Tables 1 and 2, and data not shown). Reproduction of these phenotypes in a second mouse model strengthens the conclusion that macrophage-expressed uPA is atherogenic and causes aortic dilation. Notably, however, both aortic surface atherosclerosis and aortic root dilation developed more slowly in SR-uPA+/0 Ldlr−/− mice, requiring 25 weeks versus 15 weeks in the Apoe−/− background (13). Possible explanations for the more aggressive disease in SR-uPA+/0 Apoe−/− mice despite plasma cholesterol levels similar to those of SR-uPA+/0 Ldlr−/− mice include the lack of apoE-mediated reverse cholesterol transport, absence of the anti-oxidant and anti-inflammatory activities of apoE, (43–45) and earlier onset hyperlipidemia in Apoe−/− mice.
The increased longevity of SR-uPA+/0 Ldlr−/− mice versus SR-uPA+/0 Apoe−/− mice allowed us to quantify uPA-accelerated atherosclerosis over a more extended time course, providing clues to the mechanisms of uPA/Plg-accelerated atherosclerosis. For example, our finding of equivalent numbers of aortic lesions in 15-week-old SR-uPA+/0 and nontransgenic Ldlr−/− mice (Table 1) suggests that macrophage overexpression of uPA does not increase lesion initiation but instead affects lesion progression. A preferential effect of uPA on lesion progression is also supported by our finding of more substantially accelerated atherosclerosis in the aortic roots of 25-week-old versus 15-week-old SR-uPA+/0 Ldlr−/− mice (supplemental Fig. 6A) and by the equivalent Sudan IV stained area of aortae of 15-week-old SR-uPA+/0 and nontransgenic mice compared with the significantly increased Sudan IV stained area of aortae of 25-week-old SR-uPA+/0 versus nontransgenic mice (supplemental Fig. 6B). An effect of uPA/Plg on atherosclerosis progression versus initiation is consistent with the expression pattern of the SR-uPA transgene, which is transcribed in macrophages (within the lesions) but not in peripheral blood monocytes (13, 14). Our finding of accelerated lipid accumulation in early lesions of SR-uPA+/0 versus nontransgenic Ldlr−/− mice (Table 1) provides another clue to the mechanisms of uPA/Plg-accelerated atherosclerosis, suggesting that increased artery wall Plg activation enhances lipid entry, retards lipid efflux, or both. The enhanced lipid accumulation does not appear to be due to uPA/Plg-mediated proteolysis of apoA-I, which would interfere with reverse cholesterol transport (39, 40) (supplemental Fig. 4). Future work will be aimed at identifying the mechanisms of uPA/Plg-mediated lipid accumulation. These mechanisms could include increased artery wall permeability (due to uPA/Plg-stimulated proteolysis of either endothelial basement membrane or tight junction-forming proteins) or via S100A8/A9-mediated increases in artery wall permeability (see below). Alternatively, proteolysis of vascular matrix proteins could enhance lipid retention by exposing vessel wall neoepitopes that have lipoprotein binding activity (46).
We also investigated a role for uPAR both in uPA/Plg-accelerated atherosclerosis and independently of uPA expression. Considerable published data suggest that uPAR is atherogenic. In humans, uPAR is expressed in atherosclerotic plaques in direct relation to lesion severity (7), and patients with acute myocardial infarction have increased expression of uPAR on their peripheral blood monocytes (47). In vitro studies reveal roles for uPAR in cell adhesion, migration, lipid accumulation, and oxidative stress (18, 48, 49), and in vivo animal studies demonstrate roles for uPAR in platelet adhesion to activated endothelium, foam cell formation, and macrophage accumulation in the artery wall (50–52). uPAR also dramatically increases the efficiency of Plg activation by uPA (17). All of these activities would promote atherogenesis; however, a role for uPAR in modulating atherosclerotic lesion growth has not previously been tested experimentally.
Here we found that genetic deletion of uPAR in nontransgenic Ldlr−/− mice decreased aortic root and aortic surface atherosclerosis and inhibited both aortic root dilation and the development of coronary stenoses. Deletion of uPAR in SR-uPA+/0 Ldlr−/− mice decreased aortic root atherosclerosis, coronary artery stenosis severity, and aortic dilation similarly as in nontransgenic mice. These data suggest that uPAR itself makes modest contributions to atherogenesis and aneurysm formation but that, unlike Plg (15), it plays at most a minor role in uPA-accelerated atherosclerosis. The pathways through which uPAR contributes to atherogenesis and aneurysm formation, including whether this contribution depends on uPA-uPAR binding (48), are not addressed here.
Microarray analysis revealed significant up-regulation of S100A8 and S100A9 in macrophages; qRT-PCR confirmed this finding in macrophages as well as aortae of SR-uPA mice and Western blotting confirmed up-regulation of S100A9 protein in SR-uPA macrophages (Figs. 6 and 7). A causal connection between macrophage uPA overexpression and S100A9 up-regulation is also supported by ex vivo experiments (Fig. 8), although the molecular pathways that lead from uPA expression to S100A8/A9 up-regulation remain to be defined. S100A8/A9 are excellent candidate mediators of accelerated atherosclerosis in SR-uPA mice and of uPA/Plg-mediated atherogenesis in humans because they are present in human and mouse lesions and are atherogenic in mice (53–55). S100A8/A9 potentially accelerate atherosclerosis via chemoattractant and proinflammatory activities that include binding to the RAGE and TLR4, activation of NF-κB, and up-regulation of the adhesion molecule CD11b in circulating monocytes (56–58). A recent study, however, suggested that atherogenic effects of S100A8/A9 may result from expression in non-hematopoietic cells, including endothelial cells (59). S100A8/A9 can impair endothelial integrity by disrupting cell-cell contacts and promoting apoptosis (60), actions that would likely increase lipid entry to the artery wall and could facilitate monocyte entry as well. Therefore S100A8/A9 could mediate enhanced accumulation of both lipids and macrophages in lesions of SR-uPA+/0 mice. S100A8/A9 are also associated with plaque rupture in both humans and diabetic mice (53, 61), and older SR-uPA+/0 mice have significantly increased plaque rupture (16). It will be interesting to generate SR-uPA+/0 mice that are deficient in S100A8/A9 and test whether accelerated atherosclerosis and plaque rupture in SR-uPA+/0 mice are dependent on S100A8/A9.
In addition to identifying up-regulation of S100A8/A9, the microarray studies provide insights into other pathways through which uPA overexpression may yield a more atherogenic macrophage. These pathways include increased cell adhesion and cell motility (suggested by GO, Gene set enrichment analysis, and DAVID terms such as taxis, chemotaxis, extracellular matrix-receptor interaction, regulation of actin cytoskeleton, response to external stimuli, and locomotory behavior) and altered extracellular proteolysis (suggested by terms such as proteinase inhibition, extracellular matrix, and extracellular space). Notably, an unbiased network constructed from the array data (supplemental Fig. 5) included S100A8/A9, the pattern recognition receptors TLR4 and RAGE, and transcription factors (AP-1 and NF-κB) that regulate several genes that are differentially regulated in SR-uPA+/0 macrophages. These molecules are all associated with inflammatory diseases, including atherosclerosis. Taken together, our data suggest a multifaceted model of pathways through which macrophage uPA expression could accelerate atherosclerosis (Fig. 9). Experiments that test this model and pinpoint the pathways through which macrophage-expressed uPA and Plg act to accelerate atherosclerosis could reveal new targets for atheroprotective therapies.
FIGURE 9.
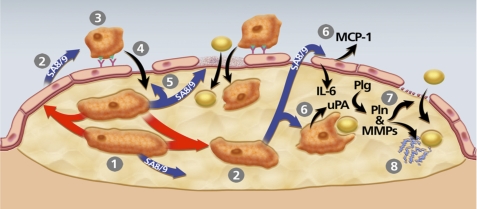
Model of pathways through which macrophage uPA expression could accelerate atherosclerosis. The model is constructed based on data from the present study and others showing that macrophage overexpression of uPA promotes early lesion lipid accumulation as well as later lesion macrophage accumulation, increases macrophage migration, enhances vascular matrix metalloproteinase (MMP) activity, accelerates lesion progression, and stimulates macrophage S100A8/A9 expression (13–16). 1, uPA-expressing macrophages begin to accumulate within an early lesion. 2, These macrophages secrete S100A8/A9 (SA8/9) themselves and can also up-regulate S100A8/A9 expression in other plaque cells (red arrows; see also Fig. 8) including macrophages and potentially in endothelial cells and smooth muscle cells, both of which can express S100A8/A9 (62, 63). 3, S100A8/A9 secreted by lesion cells increases monocyte CD11b expression and adhesion to endothelial ICAM-1 (57, 58). 4, as monocytes differentiate to macrophages, they up-regulate uPA expression, which enhances their migration into the lesion (14). 5, S100A8/A9 stimulation of endothelium also loosens tight junctions and promotes endothelial apoptosis (60, 64), effects that facilitate entry of both monocyte/macrophages and lipids to the artery wall. 6, lesion-derived S100A8/A9 binds to RAGE on endothelial cells and TLR4 on macrophages and smooth muscle cells, up-regulating expression of atherogenic cytokines MCP-1 and IL-6 in endothelium, and increasing lipid uptake by cells within the lesion (63, 65). 7, increased endothelial basement membrane proteolysis by uPA/Plg-activated matrix metalloproteinases (MMPs) (16) also increases artery wall permeability. 8, proteolysis of matrix protein within lesions exposes neoepitopes with apolipoprotein-binding activity, increasing lesion lipid retention.
Supplementary Material
Acknowledgments
We thank Alyssa Wu-Zhang, Katherine Slezicki, Shay Hai-Kun, Mia Jaffe, and Dr. Kun Qian for excellent technical assistance, Dr. Lynn Amon for assistance with bioinformatics, and Margo Weiss for administrative assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R01HL080597. This work was also supported by the John L. Locke, Jr. Charitable Trust.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–8 and Figs. 1–6.
The array hybridization data are available online at http://www.ncbi.nlm.nih.gov/geo/ with accession number GSE29028.
- uPA
- urokinase-type plasminogen activator
- Plg
- plasminogen
- uPAR
- urokinase-type plasminogen activator receptor
- PA
- plasminogen activator
- GO
- gene ontology
- RAGE
- receptor for advanced glycation end products
- TLR4
- toll-like receptor 4
- Apoe
- Ldlr, Plau, Plaur: mouse genes for apoE, low-density lipoprotein receptor, urokinase-type plasminogen activator, and urokinase-type plasminogen activator receptor
- ORO
- oil red O
- qRT
- quantitative real-time
- ANOVA
- analysis of variance.
REFERENCES
- 1. Virmani R., Burke A. P., Farb A., Kolodgie F. D. (2002) Prog. Cardiovasc. Dis. 44, 349–356 [DOI] [PubMed] [Google Scholar]
- 2. Guo D. C., Papke C. L., He R., Milewicz D. M. (2006) Ann. N.Y. Acad. Sci. 1085, 339–352 [DOI] [PubMed] [Google Scholar]
- 3. Lijnen H. R. (2002) Biochem. Soc. Trans. 30, 163–167 [DOI] [PubMed] [Google Scholar]
- 4. Padró T., Emeis J. J., Steins M., Schmid K. W., Kienast J. (1995) Arterioscler. Thromb. Vasc. Biol. 15, 893–902 [DOI] [PubMed] [Google Scholar]
- 5. Kienast J., Padró T., Steins M., Li C. X., Schmid K. W., Hammel D., Scheld H. H., van de Loo J. C. (1998) Thromb. Haemost. 79, 579–586 [PubMed] [Google Scholar]
- 6. Falkenberg M., Björnheden T., Lindnér P., Risberg B. (1998) Cardiovasc. Pathol. 7, 223–231 [DOI] [PubMed] [Google Scholar]
- 7. Steins M. B., Padró T., Schwaenen C., Ruiz S., Mesters R. M., Berdel W. E., Kienast J. (2004) Blood Coagul. Fibrinolysis. 15, 383–391 [DOI] [PubMed] [Google Scholar]
- 8. Folsom A. R., Aleksic N., Park E., Salomaa V., Juneja H., Wu K. K. (2001) Arterioscler. Thromb. Vasc. Biol. 21, 611–617 [DOI] [PubMed] [Google Scholar]
- 9. Cushman M., Lemaitre R. N., Kuller L. H., Psaty B. M., Macy E. M., Sharrett A. R., Tracy R. P. (1999) Arterioscler. Thromb. Vasc. Biol. 19, 493–498 [DOI] [PubMed] [Google Scholar]
- 10. Sakkinen P. A., Cushman M., Psaty B. M., Rodriguez B., Boineau R., Kuller L. H., Tracy R. P. (1999) Arterioscler. Thromb. Vasc. Biol. 19, 499–504 [DOI] [PubMed] [Google Scholar]
- 11. Schneiderman J., Bordin G. M., Engelberg I., Adar R., Seiffert D., Thinnes T., Bernstein E. F., Dilley R. B., Loskutoff D. J. (1995) J. Clin. Invest. 96, 639–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lijnen H. R. (2001) Thromb. Haemost. 86, 324–333 [PubMed] [Google Scholar]
- 13. Cozen A. E., Moriwaki H., Kremen M., DeYoung M. B., Dichek H. L., Slezicki K. I., Young S. G., Véniant M., Dichek D. A. (2004) Circulation 109, 2129–2135 [DOI] [PubMed] [Google Scholar]
- 14. Krishnan R., Kremen M., Hu J. H., Emery I., Farris S. D., Slezicki K. I., Chu T., Du L., Dichek H. L., Dichek D. A. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 1737–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kremen M., Krishnan R., Emery I., Hu J. H., Slezicki K. I., Wu A., Qian K., Du L., Plawman A., Stempien-Otero A., Dichek D. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17109–17114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu J. H., Du L., Chu T., Otsuka G., Dronadula N., Jaffe M., Gill S. E., Parks W. C., Dichek D. A. (2010) Circulation 121, 1637–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ellis V., Behrendt N., Danø K. (1991) J. Biol. Chem. 266, 12752–12758 [PubMed] [Google Scholar]
- 18. Blasi F., Carmeliet P. (2002) Nat. Rev. Mol. Cell Biol. 3, 932–943 [DOI] [PubMed] [Google Scholar]
- 19. Wouters K., Shiri-Sverdlov R., van Gorp P. J., van Bilsen M., Hofker M. H. (2005) Clin. Chem. Lab. Med. 43, 470–479 [DOI] [PubMed] [Google Scholar]
- 20. Grainger D. J., Reckless J., McKilligin E. (2004) J. Immunol. 173, 6366–6375 [DOI] [PubMed] [Google Scholar]
- 21. Shamshiev A. T., Ampenberger F., Ernst B., Rohrer L., Marsland B. J., Kopf M. (2007) J. Exp. Med. 204, 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Piedrahita J. A., Zhang S. H., Hagaman J. R., Oliver P. M., Maeda N. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4471–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ishibashi S., Brown M. S., Goldstein J. L., Gerard R. D., Hammer R. E., Herz J. (1993) J. Clin. Invest. 92, 883–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bugge T. H., Suh T. T., Flick M. J., Daugherty C. C., Rømer J., Solberg H., Ellis V., Danø K., Degen J. L. (1995) J. Biol. Chem. 270, 16886–16894 [DOI] [PubMed] [Google Scholar]
- 25. Bugge T. H., Flick M. J., Daugherty C. C., Degen J. L. (1995) Genes Dev. 9, 794–807 [DOI] [PubMed] [Google Scholar]
- 26. Irizarry R. A., Gautier L., Cope L. M. (2003) in The Analysis of Gene Expression Data: Methods and Software (Parmigiani G., Garrett E. S., Irizarry R. A., Zeger S. L. eds) pp. 102–119, Springer-Verlag New York Inc., New York [Google Scholar]
- 27. Smyth G. K. (2004) Stat. Appl. Genet. Mol. Biol. 3, Article 3, 1–25 [DOI] [PubMed] [Google Scholar]
- 28. Benjamini Y., Hochberg Y. (1995) J. Royal Stat. Soc. Series B (Methodological) 57, 289–300 [Google Scholar]
- 29. Beissbarth T., Speed T. P. (2004) Bioinformatics 20, 1464–1465 [DOI] [PubMed] [Google Scholar]
- 30. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Al-Shahrour F., Minguez P., Vaquerizas J. M., Conde L., Dopazo J. (2005) Nucleic Acids Res. 33, W460–W464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang da W., Sherman B. T., Lempicki R. A. (2009) Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 33. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 34. Ho Sui S. J., Mortimer J. R., Arenillas D. J., Brumm J., Walsh C. J., Kennedy B. P., Wasserman W. W. (2005) Nucleic Acids Res. 33, 3154–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dichek H. L., Qian K., Agrawal N. (2004) J. Lipid Res. 45, 551–560 [DOI] [PubMed] [Google Scholar]
- 36. Li X., Syrovets T., Genze F., Pitterle K., Oberhuber A., Orend K. H., Simmet T. (2010) Arterioscler. Thromb. Vasc. Biol. 30, 582–590 [DOI] [PubMed] [Google Scholar]
- 37. Shi G. Y., Hau J. S., Wang S. J., Wu I. S., Chang B. I., Lin M. T., Chow Y. H., Chang W. C., Wing L. Y., Jen C. J., Wu H. L. (1992) J. Biol. Chem. 267, 19363–19368 [PubMed] [Google Scholar]
- 38. Larson M. G. (2008) Circulation 117, 115–121 [DOI] [PubMed] [Google Scholar]
- 39. Kunitake S. T., Chen G. C., Kung S. F., Schilling J. W., Hardman D. A., Kane J. P. (1990) Arteriosclerosis 10, 25–30 [DOI] [PubMed] [Google Scholar]
- 40. Lijnen H. R., Collen D. (1981) Thromb. Res. 24, 151–156 [DOI] [PubMed] [Google Scholar]
- 41. Ludwig A., Weber C. (2007) Thromb. Haemost. 97, 694–703 [PubMed] [Google Scholar]
- 42. Shimokawa H., Ito A., Fukumoto Y., Kadokami T., Nakaike R., Sakata M., Takayanagi T., Egashira K., Takeshita A. (1996) J. Clin. Invest. 97, 769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Véniant M. M., Withycombe S., Young S. G. (2001) Arterioscler. Thromb. Vasc. Biol. 21, 1567–1570 [DOI] [PubMed] [Google Scholar]
- 44. Getz G. S., Reardon C. A. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 242–249 [DOI] [PubMed] [Google Scholar]
- 45. Meir K. S., Leitersdorf E. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 1006–1014 [DOI] [PubMed] [Google Scholar]
- 46. Williams K. J., Tabas I. (1995) Arterioscler. Thromb. Vasc. Biol. 15, 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. May A. E., Schmidt R., Kanse S. M., Chavakis T., Stephens R. W., Schömig A., Preissner K. T., Neumann F. J. (2002) Blood 100, 3611–3617 [DOI] [PubMed] [Google Scholar]
- 48. Fuhrman B., Nitzan O., Karry R., Volkova N., Dumler I., Aviram M. (2007) Atherosclerosis 195, e108–116 [DOI] [PubMed] [Google Scholar]
- 49. Fuhrman B., Khateeb J., Shiner M., Nitzan O., Karry R., Volkova N., Aviram M. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 1361–1367 [DOI] [PubMed] [Google Scholar]
- 50. Piguet P. F., Vesin C., Donati Y., Tacchini-Cottier F., Belin D., Barazzone C. (1999) Circulation 99, 3315–3321 [DOI] [PubMed] [Google Scholar]
- 51. Ohwaki K., Bujo H., Jiang M., Yamazaki H., Schneider W. J., Saito Y. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 1050–1056 [DOI] [PubMed] [Google Scholar]
- 52. Gu J. M., Johns A., Morser J., Dole W. P., Greaves D. R., Deng G. G. (2005) J. Cell. Physiol. 204, 73–82 [DOI] [PubMed] [Google Scholar]
- 53. Johansson F., Kramer F., Barnhart S., Kanter J. E., Vaisar T., Merrill R. D., Geng L., Oka K., Chan L., Chait A., Heinecke J. W., Bornfeldt K. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2082–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McCormick M. M., Rahimi F., Bobryshev Y. V., Gaus K., Zreiqat H., Cai H., Lord R. S., Geczy C. L. (2005) J. Biol. Chem. 280, 41521–41529 [DOI] [PubMed] [Google Scholar]
- 55. Croce K., Gao H., Wang Y., Mooroka T., Sakuma M., Shi C., Sukhova G. K., Packard R. R., Hogg N., Libby P., Simon D. I. (2009) Circulation 120, 427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hermani A., De Servi B., Medunjanin S., Tessier P. A., Mayer D. (2006) Exp. Cell Res. 312, 184–197 [DOI] [PubMed] [Google Scholar]
- 57. Ryckman C., Vandal K., Rouleau P., Talbot M., Tessier P. A. (2003) J. Immunol. 170, 3233–3342 [DOI] [PubMed] [Google Scholar]
- 58. Newton R. A., Hogg N. (1998) J. Immunol. 160, 1427–1435 [PubMed] [Google Scholar]
- 59. Averill M. M., Barnhart S., Becker L., Li X., Heinecke J. W., Leboeuf R. C., Hamerman J. A., Sorg C., Kerkhoff C., Bornfeldt K. E. (2011) Circulation 123, 1216–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Viemann D., Barczyk K., Vogl T., Fischer U., Sunderkötter C., Schulze-Osthoff K., Roth J. (2007) Blood 109, 2453–2460 [DOI] [PubMed] [Google Scholar]
- 61. Ionita M. G., Vink A., Dijke I. E., Laman J. D., Peeters W., van der Kraak P. H., Moll F. L., de Vries J. P., Pasterkamp G., de Kleijn D. P. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 1220–1227 [DOI] [PubMed] [Google Scholar]
- 62. Yen T., Harrison C. A., Devery J. M., Leong S., Iismaa S. E., Yoshimura T., Geczy C. L. (1997) Blood 90, 4812–4821 [PubMed] [Google Scholar]
- 63. Higashimori M., Tatro J. B., Moore K. J., Mendelsohn M. E., Galper J. B., Beasley D. (2011) Arterioscler. Thromb. Vasc. Biol. 31, 50–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ehlermann P., Eggers K., Bierhaus A., Most P., Weichenhan D., Greten J., Nawroth P. P., Katus H. A., Remppis A. (2006) Cardiovasc. Diabetol. 5, 6–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lau W., Devery J. M., Geczy C. L. (1995) J. Clin. Invest. 95, 1957–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.