Abstract
The amount of available hypoxia-inducible factor (HIF)-1α has been considered to be largely a consequence of post-translational modification by multiple ubiquitin-proteasome pathways. However, the role of transcriptional regulation of HIF-1α is less certain, and the mechanisms of transcriptional regulation of HIF-1α require further investigation. Here we report that related transcriptional enhancer factor-1 (RTEF-1), a member of the TEF transcriptional factor family, transcriptionally regulates the HIF-1α gene under normoxic and hypoxic conditions. The expression of HIF-1α mRNA was decreased in endothelial cells in which RTEF-1 was knocked down with siRNA. Sequential deletional analysis of the HIF-1α promoter revealed that the MCAT-like element in the HIF-1α promoter was essential for HIF-1α transcription. Binding of RTEF-1 to the MCAT-like element was confirmed by ChIP. Treatment of endothelial cells with a HIF-1 inhibitor resulted in retardation of RTEF-1-induced proliferation and tube formation. Moreover, increased HIF-1α expression was observed in transgenic mice expressing RTEF-1 under the VE-cadherin promoter (VE-Cad/RTEF-1). VE-Cad/RTEF-1 mice subjected to hindlimb ischemia demonstrated increased levels of HIF-1α, accelerated recovery of blood flow, and increased capillary density compared with littermate controls. These results identify RTEF-1 as a regulator of HIF-1α transcription, which results in up-regulation of HIF-1α and acceleration of recovery from ischemia.
Keywords: Endothelium, Gene Regulation, Hypoxia, Transcription Factors, Transcription Promoter
Introduction
Related transcriptional enhancer factor-1 (RTEF-1),3 also known as TEAD4, is one of four vertebrate members of the TEF transcriptional factor family that share a highly conserved domain capable of binding the MCAT element CATN(T/C)(T/C) (1, 2) in the promoter region of genes expressed in cardiac (3), skeletal, and smooth muscle cells (4). RTEF-1 regulates skeletal muscle α-actin transcription in myofibroblasts by binding to the MCAT element-containing region within the α-actin promoter (5). In hypertrophic cardiac myocytes, RTEF-1 is a direct target of α1-adrenergic signaling and enhances the α1-adrenergic response of the skeletal muscle α-actin promoter through the MCAT element (6). Elimination of RTEF-1 expression in mice causes preimplantation lethality with defects in trophoblast stem cells, trophectoderm, and blastocoel cavities (7). Transgenic mice with cardiac-specific overexpression of RTEF-1 have progressive atrial arrhythmias and increased levels of dephosphorylated Cx43 at the cardiac gap junctions (8). However, the target genes of RTEF-1 in nonmuscle cells have not been fully defined. In vascular endothelial cells, expression of RTEF-1 can be induced by hypoxia (9, 10). The sequential deletion and site-directed mutational analyses of the VEGF promoter demonstrated that a GC-rich region containing four Sp1 (special protein 1) response elements was essential for RTEF-1 function in endothelial cells (9, 10), although this region does not contain hypoxia-responsive elements or MCAT-related elements. Because the patterns of MCAT-like elements are relatively diverse, it remains unclear exactly how RTEF-1 confers cell-specific expression in myocytes and how RTEF-1 acts on genes in other cell types. HIF-1α is a major hypoxia-induced transcriptional factor that regulates a number of genes involved in energy metabolism and vascular homeostasis during low oxygen conditions (11, 12). Previous studies have shown that HIF-1α is tightly controlled by multiple ubiquitin-proteasome pathways that primarily affect protein stability (13), which indicates that the amount of HIF-1α is determined by post-translational modification. However, HIF-1α expression can also be up-regulated under normoxic conditions in response to growth factors, thrombin, angiotensin II, cytokines, or insulin (14–16), suggesting that multiple mechanisms are involved in the regulation of HIF-1α.
Here we report that the transcription factor, RTEF-1, is involved in hypoxia-related gene regulation as a transcriptional activator of HIF-1α. We demonstrate that HIF-1α gene expression was decreased when RTEF-1 was knocked down in endothelial cells. Under hypoxic conditions, the HIF-1α gene can be maintained at a sustained level by RTEF-1. RTEF-1 increased HIF-1α gene expression through interacting with MCAT-like elements. In addition, RTEF-1 induced substantial cell proliferation and vascular structure remodeling in vitro and accelerated recovery from ischemia in vivo, which correlated with its function of increasing the expression of HIF-1α.
EXPERIMENTAL PROCEDURES
Cell Culture and Hypoxia
Human umbilical vein endothelial cells (HUVECs, Lonza) and human aortic endothelial cells (Lonza) were cultured in EBM-2. Human microvascular endothelial cells (HMEC-1) (Center of Disease Control) were cultured in MCDB-131 (Invitrogen); HEK293 and 293T were cultured in Dulbecco's Modified Eagle Media (DMEM, Invitrogen). Hypoxic exposure was performed using a molecular incubator chamber (Billups-Rothenberg) flushed with 5% CO2, 95% N2. The concentration of oxygen (1–3%) was determined before and after incubation using an oxygen analyzer (Vascular Technology).
Gene and Protein Expression
RNA levels of HIF-1α were also measured by Quantitative real time PCR using the primers 5′-AAGTCTCGAGATGCAGCCA-3′ and 5′-AGGAAGCTTCGCTGTGTGTTTTG-3′. Protein levels were measured by Western blot with the antibodies: anti-RTEF-1 (Genemed Inc.); anti-HIF-1α, anti-VEGF-A, and anti-TEF-1 (BD Biosciences); anti-p65 and anti-p50 (Santa Cruz); and anti-DTEF-1 (a gift from Dr. Iain Farrance, University of Maryland).
Retroviral Transduction and siRNA Transfection
The coding sequence of RTEF-1 (NM_003213) obtained from a PXJ40/RTEF-1 construct (gifted from Dr. Alexandre Stewart, University of Ottawa) was subcloned into the pBMN-GFP vector (Orbigen). 293T cells were transfected with pBMN-GFP-RTEF-1, pMD-VSVG, pJK3, and pCMV-tat using polyethylenimine (Polysciences). The virus-containing medium was used to transduce HMEC-1, and stable transfectants were selected with puromycin (250 ng/ml). The RTEF-1 siRNA was synthesized by Qiagen (siRNA1, 5′-AAGCCGGAACGTCCCATGATG-3′; siRNA2, 5′-AAACCCAAGATGCTGTGTATT-3′). A siRNA not targeted to any human gene was used as a negative control.
Promoter Activity and ChIP
A DNA construct encompassing a fragment of the 5′ flanking region (−557 to +300) of the human HIF-1α gene (17) was a generous gift from Drs. Scot Ebbinghaus and Daekyu Sun (University of Arizona). HEK293 cells were transfected with the constructs using polyethylenimine (Polysciences), NFκB inhibitors, wedelolactone (Sigma), or SM7368 (Calbiochem) and incubated for 5 h prior to measurement of luciferase activity. Luciferase activity was determined using the dual luciferase assay system (Promega). The ChIP assay was performed according to the instructions of the ChIP-IT Express kit (Active Motif). PCRs were performed with the primer pairs (18) including HIF-1α: 5′-CACCCCCATCTCCTTTCTCT-3′ and 5′-GGGTTCCTCGAGATCCAATG-3′; and actin: 5′-TGCACTGTGCGGCGAAGC-3′ and 5′-TCGAGCCATAAAAGGCAA-3′.
Proliferation and Matrigel Assays
BrdUrd (10 μm) was added to HMEC-1 in each well of a 96-well plate. Cell proliferation was measured by an ELISA kit (Roche Applied Science) over 24 h at 4-h intervals. Matrigel analysis was performed by placing endothelial cells on growth factor-reduced Matrigel (BD Biosciences); images were photographed using a TS100 Nikon microscope after 3 and 6 h. HIF-1 inhibitor, YC-1, was purchased from Sigma. YC-1 (50 μm) was incubated with the Matrigel in the hypoxia chamber for 15 min before HMEC-1 cells were added and incubated for 6 h.
Immunofluorescence Analysis
The sections prepared from optimum cutting temperature compound (OCT, Tissue-Tek)-embedded tissue samples of quadriceps were used for immunofluorescence analysis. Briefly, the sections were fixed in 4% formaldehyde and incubated with blocking solution followed by incubation with anti-CD31 antibodies (Santa Cruz). The sections were mounted in mounting medium (Vector Laboratories) and viewed with a fluorescence microscope (Laica DM5000B).
Transgenic Mice and Hindlimb Ischemia
RTEF-1 transgenic mice were generated on the Friend Virus B-type (FVB) background at the Beth Israel Deaconess Medical Center Transgenic Core Facility using the mouse VE-cadherin promoter to drive endothelial-specific expression of human RTEF-1. The mice were maintained at the animal facility at Beth Israel Deaconess Medical Center. The institutional animal care and use committee approved all animal procedures. Hindlimb ischemia was induced by ligation of the femoral artery as described (19). Blood perfusion in ischemic hindlimbs was measured by laser Doppler image (MoorLDI) on intermittent days until day 30.
Statistical Analysis
Dunnett's multiple comparison test was used for comparison of band density in Northern blot analysis. One-way analysis of variance was used to assess significance in the hindlimb ischemia study. Two-tailed independent Student's t test was used to determine significance in all other assays. The p values were based on three individual experiments with standard deviation.
RESULTS
RTEF-1 Regulates HIF-1α Expression in Endothelial Cells
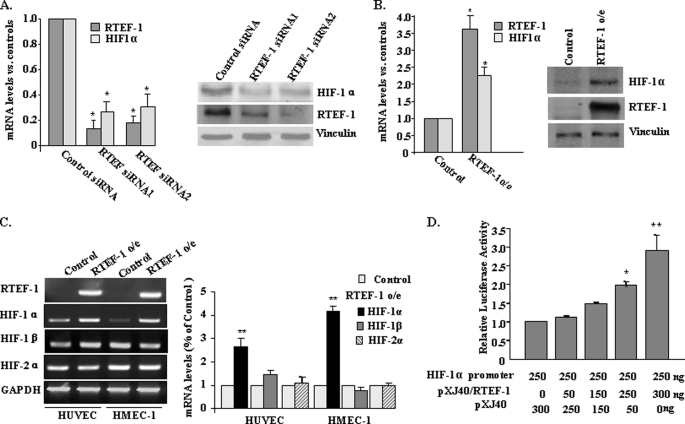
To explore the target genes of RTEF-1 in endothelial cells, DNA array analysis was performed in HUVEC transfected with RTEF-1 siRNA. HIF-1α was significantly down-regulated following RTEF-1 knockdown. Fig. 1A demonstrated that HIF-1α mRNA and protein levels were reduced after using two different sequences of RTEF-1 siRNA for knockdown of RTEF-1 in HUVEC by quantitative PCR and immunoblotting. To confirm this finding that HIF-1α is a potential target gene of RTEF-1, RTEF-1 was expressed in HMEC-1 by retroviral infection. HIF-1α mRNA and protein levels were significantly increased in RTEF-1/HMEC-1 (RTEF-1 o/e) (Fig. 1B and supplemental Fig. S1), whereas the mRNA levels of HIF-1β and HIF-2α were not affected by RTEF-1 in either HUVEC or HMEC-1 (Fig. 1C). To determine whether HIF-1α was regulated by RTEF-1 on a transcriptional level, a fragment of the HIF-1α promoter (17) was inserted into a luciferase reporter construct and cotransfected with RTEF-1 cDNA. The HIF-1α promoter exhibited an RTEF-1 dose-dependent increase in activity, exhibiting a maximum 2.9 ± 0.4-fold increase (Fig. 1D). Together, these data indicate that RTEF-1 up-regulates HIF-1α expression in endothelial cells.
FIGURE 1.
RTEF-1 is required for HIF-1α gene expression in endothelial cells. A, the expression of HIF-1α mRNA (left panel) and protein (right panel) was assessed after knockdown of RTEF-1 by its siRNA in HUVEC. B, quantitative PCR (left panel) and immunoblotting (right panel) confirmed that RTEF-1 was overexpressed in HMEC-1 and HIF-1α was up-regulated. HMEC-1 transfected with pBMN vector was used as control. C, semi-quantitative PCR for HIF-1α, HIF-1β, and HIF-2α in HUVEC and HMEC-1 transfected with RTEF-1 or control plasmid. Quantitative densitometry data are shown in the right panel. D, cotransfection of HEK293 cells with HIF-1α promoter construct (250 ng) and various combinations of PXJ40 (vector) and RTEF-1 (550 ng of total DNA). Luciferase activity was determined. The data are from three independent experiments (means ± S.D.). *, p < 0.05; **, p < 0.01.
RTEF-1 Regulates HIF-1α Transcription by Binding to an MCAT-like Element
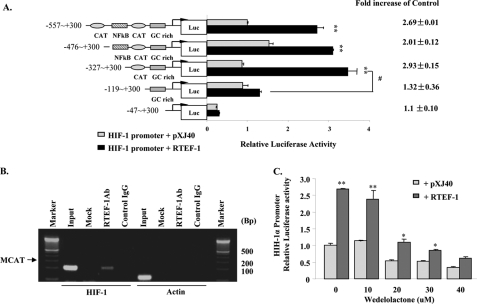
Sequence analyses of the HIF-1α gene have identified two MCAT-like sequences (5′-CATTACA-3′, −486 to −480; 5′-CATTGGA-3′, −192 to −186) in the 5′-UTR, an NFκB binding site (−216 to −207) (18, 20), and a quadruplex structure formed by a polypurine/polypyrimidine tract (−104 to −84) that was reported to regulate basal HIF-1α expression (17). Additionally, alignment analysis showed that an MCAT-like sequence was located 16 base pairs downstream of the NFκB binding site in the human HIF-1α promoter and was conserved in mouse and bovine (supplemental Fig. S2). To identify the specific regulatory element in the HIF-1α promoter bound by RTEF-1, a series of truncations of the HIF-1α promoter were cotransfected with RTEF-1 cDNA. RTEF-1 induced an increase in HIF-α promoter activity that was significantly attenuated by deletion of the MCAT-like sequence (Fig. 2A). The physical interaction of RTEF-1 with the HIF-1α promoter was examined by ChIP in RTEF-1 o/e endothelial cells. As shown in Fig. 2B, a 172-bp DNA fragment (−342 to −171) including MCAT-like elements was detected in RTEF-1 immunoprecipitated chromatin.
FIGURE 2.
RTEF-1 regulates HIF-1α transcription by interacting with an MCAT-like element. A, RTEF-1-activated HIF-1α promoter activity involves NFκB, MCAT, and a GC-rich area. Constructs with sequential deletion of the HIF-1α promoter were cotransfected with RTEF-1 (black bars) or pXJ40 (gray bars) into HEK293 cells. The results shown are the means ± S.D. of three independent experiments. **, p < 0.01; #, p < 0.05. B, ChIP assay showing coimmunoprecipitation of the HIF-1α promoter with anti-RTEF-1 antibody using chromatin isolated from RTEF-1 o/e HMEC-1 cells. The sequence containing the MCAT-like element was detected by PCR. PCR of the actin promoter was used to exclude nonspecific pulldown. C, failure of the IKK inhibitor, wedelolactone, to block HIF-1α promoter activity induced by RTEF-1. The induction ratio (RTEF-1/pXJ40) was unchanged in the presence of IKK inhibitor. The results are based on three experiments. The means ± S.D. are shown. *, p < 0.05; **, p < 0.01.
It is known that basal NFκB activity is required for HIF-1α protein accumulation under hypoxia (18). To clarify whether the NFκB element was also involved in RTEF-1-induced HIF-1α promoter activity, wedelolactone, an IκB kinase inhibitor, was incubated with bovine aortic ECs to block the activation of IKK before cotransfection of RTEF-1 and HIF-1α reporter. In the presence of wedelolactone, the basal activity of the HIF-1α promoter was decreased in a dose-dependent manner, whereas RTEF-1-induced HIF-1α promoter activity was decreased in parallel to a lesser extent (Fig. 2C). Another selective inhibitor of NFκB DNA binding, SM7368, showed a similar pattern and did not specifically block RTEF-1-induced HIF-1α promoter activity (supplemental Fig. S3). Together, these data suggested that NFκB was essential for basal HIF-1α promoter activity but was not required for RTEF-1-induced HIF-1α promoter activation.
RTEF-1 Is Required to Sustain HIF-1α Levels during Hypoxia in Endothelial Cells
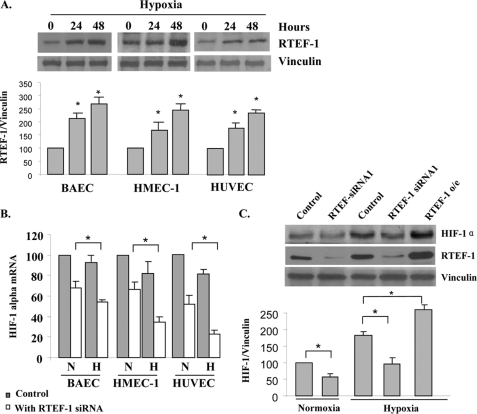
We next examined the role of RTEF-1 on HIF-1α during hypoxia. We confirmed our previous finding (9) that the expression of RTEF-1 was stimulated by hypoxia in endothelial cells including bovine aortic EC, HUVEC, and HEMC-1 (Fig. 3A). Induction of RTEF-1 expression was detectable by 1 h and continuously increased up to 72 h in response to hypoxia (supplemental Fig. S4A). By comparison, the other two members of the TEF protein family, TEF-1 and DTEF-1, did not respond to hypoxia (supplemental Fig. S4B). However, the expression of endogenous HIF-1α mRNA was either sustained or was slightly decreased under hypoxic conditions (Fig. 3B). In these same cells, when RTEF-1 was knocked down, HIF-1 mRNA levels were further attenuated under hypoxic conditions, indicating that RTEF-1 supported expression of HIF-1α mRNA during hypoxia. In addition, HIF-1 protein levels were significantly decreased when RTEF-1 was blocked by RTEF-1 siRNA in both normoxia and hypoxia (Fig. 3C and supplemental Fig. S5A). Thus, these results together suggest that hypoxia-induced RTEF-1 was required to maintain HIF-1α transcription during hypoxia in endothelial cells.
FIGURE 3.
RTEF-1 sustains HIF-1α levels during hypoxia in endothelial cells. A, Western blots analysis of RTEF-1 and vinculin expression under hypoxia in endothelial cells. B, quantitative real time PCR analysis of HIF-1α mRNA levels in endothelial cells with or without RTEF-1 knockdown with siRNA. N, normoxia; H, hypoxia. C, Western blot analysis of HIF-1α protein expression under hypoxic conditions after knockdown of RTEF-1 by siRNA in HUVEC when compared with that in normoxia and RTEF-1 o/e. The results are from three independent experiments (means ± S.D.). *, p < 0.05 indicates HIF-1 protein level in the cells of RTEF-1 siRNA or RTEF-1 o/e when compared with the controls under normoxic and hypoxic conditions.
RTEF-1 Increases Endothelial Proliferation and Aggregation via HIF-1 Induction in Hypoxia
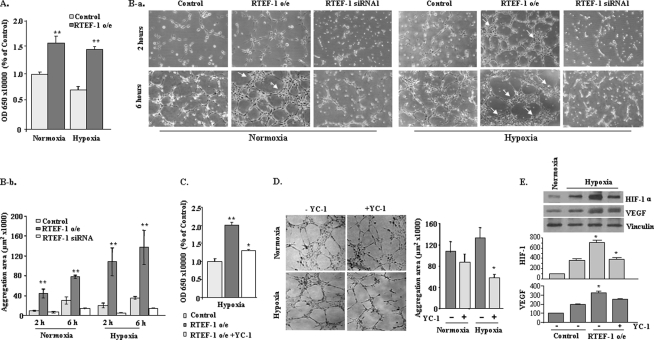
Because HIF-1α is a critical pro-angiogenesis factor during hypoxia, we explored the functional relevance of RTEF-1-regulated HIF-1α by analyzing cell proliferation and tube formation. In BrdUrd incorporation in HMEC-1 cells, RTEF-1 significantly increased the proliferation rate under both normoxic and hypoxic conditions (Fig. 4A). Matrigel tube formation assay revealed that RTEF-1 accelerated the rate of tube formation and demonstrated a progressive assembly of clusters in HMEC-1, whereas knockdown of RTEF-1 by siRNA caused the formation of disconnected tubes (Fig. 4B and supplemental Fig. S5B). Moreover, the RTEF-1-driven cell aggregation was enhanced by hypoxia (Fig. 4B and supplemental S5B). However, when YC-1, a blocker of HIF-1α at the post-transcriptional in hypoxia (21), was incubated with HMEC-1 cells under hypoxic conditions, we observed a significant reduction of proliferation (Fig. 4C). Similarly, in the Matrigel assay in the presence of YC-1, disconnected tube formation and less cluster area were observed under hypoxic conditions but had no significant effects under normoxic conditions (Fig. 4D). Further experiments were performed to verify whether blocking HIF-1 expression also impacted VEGF expression regulated by RTEF-1. As shown in Fig. 4E, the induction of VEGF expression was not completely attenuated when HIF-1 was blocked in RTEF cells during hypoxic conditions. Together, these data indicate that RTEF-1 accelerated endothelial cell proliferation and tube formation, which was in large part mediated by its transcriptional up-regulation of HIF-1α, although RTEF-1 targeted both VEGF and HIF-1.
FIGURE 4.
RTEF-1 increases cell proliferation and accelerates tube formation. A, BrdUrd incorporation assay in RTEF-1 o/e HMEC-1 cells in normoxia and hypoxia. **, p < 0.01. B, images (panel a) and quantification (panel b) demonstrate the Matrigel assay of HMEC-1 transfected with RTEF-1 and RTEF-1 siRNA at 2 and 6 h under normoxic and hypoxic conditions. C, BrdUrd incorporation assay of HMEC-1/RTEF-1 cells treated with 50 μm YC-1 for 15 min prior to hypoxia. D, Matrigel assay of HMEC-1/RTEF-1 cells treated with 50 μm YC-1 under normoxic and hypoxic conditions. E, Western blot analysis of expression of HIF and VEGF proteins in HMEC cells in the presence or absence of YC-1. The results are from three independent experiments (means ± S.D.). *, p < 0.05; **, p < 0.01.
RTEF-1 Regulates Angiogenesis and Blood Flow Recovery after Ischemia
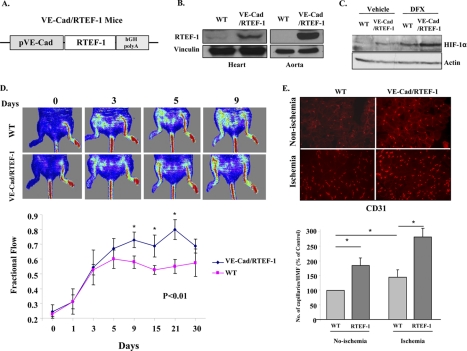
To achieve systemic expression of RTEF-1 in endothelial cells in vivo, transgenic mice were generated that expressed RTEF-1 under control of the VE-cadherin promoter (VE-Cad/RTEF-1) (Fig. 5A). Immunoblot analysis revealed that the RTEF-1 transgene was expressed in endothelial rich tissue, particularly in the heart and aorta (Fig. 5B). HIF-1α was significantly activated in the heart of the RTEF-1 transgenic mice (Fig. 5C). To determine whether RTEF-1 was involved in the normal vascular response to ischemia, RTEF-1 transgenic mice and wild-type littermates were subjected to ligation and ablation of the femoral artery. Blood perfusion in ischemic hind limbs was measured by Doppler analysis on intermittent days until day 30. At day 9, blood flow was restored to 75% in RTEF-1 transgenic mice but was restored to only 58% in littermate (control) mice (Fig. 5D). Immunohistochemical analysis indicated that VE-Cad/RTEF-1 mice had increased capillary formation in both ischemic and nonischemic quadriceps in comparison with WT mice (Fig. 5E).
FIGURE 5.
RTEF-1 regulates angiogenesis and blood flow recovery after ischemia in vivo. A, diagram of VE-cadherin promoter-driven human RTEF-1 transgene. B, immunoblot for RTEF-1 expression in the heart and aorta of endothelial cell-specific RTEF-1 transgenic mice (RTEF-1/TG). C, Western blot analysis of HIF-1α activation in RTEF-1/TG mice after intravenous injection of deferoxamine (DFX) to stabilize HIF-1α. D, RTEF-1/TG and wild-type mice were subjected to femoral artery ligation. Blood perfusion in ischemic hind limbs was measured by laser Doppler analysis on intermittent days until day 30. Sample images (upper panel) and quantification (lower panel) showing blood flow recovery at time points following ligation. *, significant increase of blood flow in the TG group when compared with the WT group from day 9 to day 21 (n = 5 for each group). E, sections of quadriceps from WT and RTEF-1/TG mice were stained for CD31 7 days after femoral artery ligation (n = 5; means ± S.D.). Top panel, images; bottom panel, quantification. *, p < 0.05.
DISCUSSION
Our present studies revealed an important role of RTEF-1 in vascular homeostasis. First, we demonstrated that HIF-1α is a target gene of RTEF-1, and RTEF-1 can maintain HIF-1α mRNA levels in hypoxic endothelial cells. Second, we identified a MCAT-like element in the HIF-1α promoter that directly binds RTEF-1. Finally, HIF-1α expression regulated by RTEF-1 resulted in accelerated recovery of blood flow after hindlimb ischemia in transgenic mice with endothelial-specific expression of RTEF-1.
The availability of HIF-1α in hypoxia-stimulated vascular growth has been considered to be largely the result of its post-translational regulation through the inhibition of O2-dependent prolyl hydroxylases that drive HIF-1α degradation under normoxia (13, 22). Recent reports that HIF-1α expression can also be induced under a normal oxygen environment suggest that more activation pathways are involved in HIF-1α regulation (14–16). We report here that RTEF-1 is required for maintaining HIF-1α mRNA expression in endothelial cells under both normoxic and hypoxic conditions. Although early studies demonstrated the induction of HIF-1α mRNA in experimental animals during development and hypoxia (23, 24), we have found that the HIF-1α mRNA level either showed no effect or was down-regulated in endothelial cells under hypoxic conditions. However, knockdown of RTEF-1 resulted in a further decrease in HIF-1α mRNA from hypoxia-attenuated levels. RTEF-1 was found to interact with the HIF-1α promoter and to maintain a sustainable level of transcription in response to hypoxia, which compensated for the reduced HIF-1α mRNA during hypoxia. Our results showed that transcriptional activation of the HIF-1α gene by RTEF-1, which precedes HIF-1α protein accumulation, was of critical importance under pathophysiologically relevant conditions ex vivo and in vivo.
Results of the present studies demonstrate that, in addition to muscle cells, endothelial cells also use MCAT-like elements for RTEF-1 transcriptional regulation of the HIF-1α gene. These results provide novel evidence indicating that transcriptional control mechanisms for the induction of genes may be similar between endothelial cells and muscle cells but do not rule out differences. For example, RTEF-1 regulated VEGF through the SP-1 element but not the MCAT element in endothelial cells (9). Transcriptional regulation of HIF-1α has been linked to activation of NFκB, which is associated with hypoxic responses in innate immunity and inflammation (18). A study using mice lacking IKK-β in different cell types demonstrated that NFκB is a critical transcriptional activator of HIF-1α, and basal NFκB activity is required for HIF-1α protein accumulation in the liver and brain of hypoxic animals (18). Our data showed that inhibition of NFκB did not block induction of HIF-1α promoter activity by RTEF-1, suggesting that RTEF-1 function is independent of NFκB activation. In addition to the NFκB binding site, other DNA elements such as a polypurine/polypyrimidine tract (−65 to −85) might participate in regulation of basal HIF-1α expression (17). Our results indicated that RTEF-1 bound directly to an MCAT-like element in the HIF-1α promoter. The absence of this element resulted in a loss of response to RTEF-1. Interestingly, deletion of this MCAT-like element did not further change the basal activity of the HIF-1α promoter. These findings suggest a novel transcriptional regulatory pattern for HIF-1α, in which a MCAT-like element is not required for basal expression of HIF-1α but mediates RTEF-1-induced up-regulation.
In a hypoxic environment, VEGF is up-regulated by activation of the HIF-1α pathway and enhances endothelial cell proliferation and tube formation (25). Recent studies in rodent models showed that IL-20 potently stimulates vessel tube formation (26). As an upstream regulator of HIF-1α, RTEF-1 promotes proangiogenic activity, suggesting the possibility of a regulatory cascade of growth factors and cytokines, including VEGF, under control of HIF-1α. It is interesting to note that the RTEF-induced proliferation rate was inhibited by ∼70% in endothelial cells by the HIF-1 inhibitor, YC-1, although blocking hypoxia-related HIF-1α stabilization did not completely decrease RTEF-1-induced VEGF expression. This is because RTEF-1 directly regulates VEGF (9, 10), as well as HIF-1, in endothelial cells. In addition to the fact that RTEF-1 accelerated endothelial network formation, an enlarged cluster area was observed in RTEF-1 o/e cells. Whether the proteins that are responsible for cytoskeleton organization or/and cell-cell connections are involved in endothelial cell structure formation is still unclear. Further investigations of the role of RTEF-1 in endothelial cell tube formation will help reveal the mechanisms of RTEF-1-driven cell aggregation.
In addition, we found increased blood flow restoration in VE-Cad/RTEF-1 transgenic mice during the recovery process after hindlimb ischemia. Increased expression of HIF-1α and its downstream angiogenic factor, VEGF, was observed in VE-Cad/RTEF-1 transgenic mice. A similar improvement of blood perfusion via support from endothelial cell organization, vessel maturation, and remodeling has been observed in vessels treated with IL-20 to stimulate collateral circulation (26). RTEF-1 may modulate angiogenesis by regulating its target gene, HIF-1α, to induce vascular sprouting and rearrangement of the cytoskeleton and promote vessel maturation.
In summary, our studies indicated that RTEF-1 is a positive regulator of HIF-1α transcription that acts by binding the MCAT-like elements in the HIF-1α promoter region in endothelial cells. By up-regulating HIF-1α transcription, RTEF-1 accelerated endothelial tube formation and enhanced cell aggregation in Matrigel models. In addition, accelerated ischemia recovery was observed in endothelial cell-specific RTEF-1 transgenic mice. Our observations suggest an important role for RTEF-1 in maintenance of endothelial homeostasis. Further understanding of the molecular basis of this regulation may lead to strategies for enhancing vascularization in ischemic diseases.
Supplementary Material
Acknowledgments
We thank Drs. S. W. Ebbinghaus and D. Sun (University of Arizona) for the human HIF-1α promoter construct, Dr. Iain Farrance (University of Maryland) for anti-DTEF-1 antibody, and Dr. Laura Benjamin (Beth Israel Deaconess Medical Center) for the VE-cadherin promoter.
This work was supported, in whole or in part, by National Institutes of Health Grant HLR01082837 (to J. L.). This work was also supported by the China Scholarship Council (to Y. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- RTEF-1
- related transcriptional enhancer factor-1
- HIF
- hypoxia-inducible factor
- NFκB
- nuclear factor κB
- HUVEC
- human umbilical vain endothelial cell
- BrdUrd
- bromodeoxyuridine
- HMEC
- human microvascular endothelial cell.
REFERENCES
- 1. Azakie A., Larkin S. B., Farrance I. K., Grenningloh G., Ordahl C. P. (1996) J. Biol. Chem. 271, 8260–8265 [DOI] [PubMed] [Google Scholar]
- 2. Farrance I. K., Mar J. H., Ordahl C. P. (1992) J. Biol. Chem. 267, 17234–17240 [PubMed] [Google Scholar]
- 3. Stewart A. F., Larkin S. B., Farrance I. K., Mar J. H., Hall D. E., Ordahl C. P. (1994) J. Biol. Chem. 269, 3147–3150 [PubMed] [Google Scholar]
- 4. Larkin S. B., Farrance I. K., Ordahl C. P. (1996) Mol. Cell. Biol. 16, 3742–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gan Q., Yoshida T., Li J., Owens G. K. (2007) Circ. Res. 101, 883–892 [DOI] [PubMed] [Google Scholar]
- 6. Ueyama T., Zhu C., Valenzuela Y. M., Suzow J. G., Stewart A. F. (2000) J. Biol. Chem. 275, 17476–17480 [DOI] [PubMed] [Google Scholar]
- 7. Yagi R., Kohn M. J., Karavanova I., Kaneko K. J., Vullhorst D., DePamphilis M. L., Buonanno A. (2007) Development 134, 3827–3836 [DOI] [PubMed] [Google Scholar]
- 8. Chen H. H., Baty C. J., Maeda T., Brooks S., Baker L. C., Ueyama T., Gursoy E., Saba S., Salama G., London B., Stewart A. F. (2004) Circulation 110, 2980–2987 [DOI] [PubMed] [Google Scholar]
- 9. Shie J. L., Wu G., Wu J., Liu F. F., Laham R. J., Oettgen P., Li J. (2004) J. Biol. Chem. 279, 25010–25016 [DOI] [PubMed] [Google Scholar]
- 10. Appukuttan B., McFarland T. J., Davies M. H., Atchaneeyasakul L. O., Zhang Y., Babra B., Pan Y., Rosenbaum J. T., Acott T., Powers M. R., Stout J. T. (2007) Invest. Ophthalmol. Vis. Sci. 48, 3775–3782 [DOI] [PubMed] [Google Scholar]
- 11. Iyer N. V., Kotch L. E., Agani F., Leung S. W., Laughner E., Wenger R. H., Gassmann M., Gearhart J. D., Lawler A. M., Yu A. Y., Semenza G. L. (1998) Genes Dev. 12, 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ryan H. E., Lo J., Johnson R. S. (1998) EMBO J. 17, 3005–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maxwell P. H., Wiesener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., Ratcliffe P. J. (1999) Nature 399, 271–275 [DOI] [PubMed] [Google Scholar]
- 14. Fukushima K., Murata M., Hachisuga M., Tsukimori K., Seki H., Takeda S., Asanoma K., Wake N. (2008) Placenta 29, 324–331 [DOI] [PubMed] [Google Scholar]
- 15. Richard D. E., Berra E., Pouyssegur J. (2000) J. Biol. Chem. 275, 26765–26771 [DOI] [PubMed] [Google Scholar]
- 16. Zelzer E., Levy Y., Kahana C., Shilo B. Z., Rubinstein M., Cohen B. (1998) EMBO J. 17, 5085–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. De Armond R., Wood S., Sun D., Hurley L. H., Ebbinghaus S. W. (2005) Biochemistry 44, 16341–16350 [DOI] [PubMed] [Google Scholar]
- 18. Rius J., Guma M., Schachtrup C., Akassoglou K., Zinkernagel A. S., Nizet V., Johnson R. S., Haddad G. G., Karin M. (2008) Nature. 453, 807–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Couffinhal T., Silver M., Zheng L. P., Kearney M., Witzenbichler B., Isner J. M. (1998) Am. J. Pathol. 152, 1667–1679 [PMC free article] [PubMed] [Google Scholar]
- 20. Belaiba R. S., Bonello S., Zähringer C., Schmidt S., Hess J., Kietzmann T., Görlach A. (2007) Mol. Biol. Cell 18, 4691–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yeo E. J., Chun Y. S., Cho Y. S., Kim J., Lee J. C., Kim M. S., Park J. W. (2003) J. Natl. Cancer Inst. 95, 516–525 [DOI] [PubMed] [Google Scholar]
- 22. Semenza G. L., Shimoda L. A., Prabhakar N. R. (2006) Novartis Found. Symp. 272, 2–8 [PubMed] [Google Scholar]
- 23. Jain S., Maltepe E., Lu M. M., Simon C., Bradfield C. A. (1998) Mech. Dev. 73, 117–123 [DOI] [PubMed] [Google Scholar]
- 24. Elson D. A., Ryan H. E., Snow J. W., Johnson R., Arbeit J. M. (2000) Cancer Res. 60, 6189–6195 [PubMed] [Google Scholar]
- 25. Pugh C. W., Ratcliffe P. J. (2003) Nat. Med. 9, 677–684 [DOI] [PubMed] [Google Scholar]
- 26. Tritsaris K., Myren M., Ditlev S. B., Hübschmann M. V., van der Blom I., Hansen A. J., Olsen U. B., Cao R., Zhang J., Jia T., Wahlberg E., Dissing S., Cao Y. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15364–15369 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.