Abstract
OBJECTIVE
A better understanding of the molecular mechanisms underlying the development and progression of diabetic neuropathy (DN) is essential for the design of mechanism-based therapies. We examined changes in global gene expression to define pathways regulated by diabetes in peripheral nerve.
RESEARCH DESIGN AND METHODS
Microarray data for 24-week-old BKS db/db and db/+ mouse sciatic nerve were analyzed to define significantly differentially expressed genes (DEGs); DEGs were further analyzed to identify regulated biological processes and pathways. Expression profile clustering was performed to identify coexpressed DEGs. A set of coexpressed lipid metabolism genes was used for promoter sequence analysis.
RESULTS
Gene expression changes are consistent with structural changes of axonal degeneration. Pathways regulated in the db/db nerve include lipid metabolism, carbohydrate metabolism, energy metabolism, peroxisome proliferator–activated receptor signaling, apoptosis, and axon guidance. Promoter sequences of lipid metabolism–related genes exhibit evidence of coregulation of lipid metabolism and nervous system development genes.
CONCLUSIONS
Our data support existing hypotheses regarding hyperglycemia-mediated nerve damage in DN. Moreover, our analyses revealed a possible coregulation mechanism connecting hyperlipidemia and axonal degeneration.
According to statistics published by the American Diabetes Association in 2007, 23.6 million people, approximately 8% of the U.S. population, have diabetes and 1.6 million new cases are diagnosed every year. Type 2 diabetes is the most common type of diabetes, accounting for 90–95% of all cases, and is characterized by hyperglycemia, insulin resistance, and dyslipidemia. In addition, an estimated 57 million American adults exhibit impaired glucose tolerance, which frequently develops into type 2 diabetes (1).
The hyperglycemia that defines diabetes is a major factor in the development of micro- and macrovascular complications, which severely affect patients’ longevity and quality of life. Microvascular complications include retinopathy, nephropathy, and neuropathy. From 60 to 75% of diabetic patients develop diabetic neuropathy (DN), the most common and debilitating complication of diabetes (1). DN is the leading cause of nontraumatic lower-extremity amputations, and the annual cost of DN management is estimated to be more than $10 billion (2).
DN results from length-dependent axonal loss affecting distal portions of extremities first and progressing proximally in a stocking-glove pattern (3). Signs and symptoms of DN vary, and though both sensory and motor nerve fibers may be involved, sensory manifestations appear earlier and are more prevalent. Although development of DN correlates with the severity and duration of diabetes, there is a great variability in the onset and progression of symptoms. Some diabetic patients with poorly controlled hyperglycemia do not display signs and symptoms of DN, whereas others with good glycemic control develop DN, suggesting the involvement of factors besides hyperglycemia. We contend that a better understanding of the molecular mechanisms underlying the development and progression of DN is needed for early diagnosis and intervention as well as for understanding the failure of existing treatments. Identification of potential nonglycemic mechanisms is critical for better prediction of progression and for designing preventive therapies (4). For example, obesity and dyslipidemia have recently been identified as additional risk factors in the development of microvascular complications and may play an important role in the pathogenesis of DN (5–7).
Because of a mutation in the leptin receptor, BKS db/db mice are hyperphagic and obese, develop severe type 2 diabetes with marked hyperglycemia, and serve as an experimental model of type 2 diabetes. These mice develop diabetes by 4 weeks and DN by 8 weeks of age, with prolonged thermal latencies, slowed nerve conduction velocities (NCVs), and loss of intraepidermal nerve fiber (IENF) density (8,9). In addition, db/db mice exhibit high plasma triglyceride and lipoprotein levels and may serve as a model for diabetic dyslipidemia (10). Heterozygous (db/+) mice do not develop diabetes and are used as nondiabetic controls in experimental DN (11).
Using DNA microarrays, we compared global gene expression in the sciatic nerve of db/db mice with that of db/+ mice at 24 weeks of age (20 weeks of diabetes) to study changes in gene expression in peripheral nerves. Bioinformatic analysis of microarray gene expression data identifies regulated genes and biological processes and provides new insights into the underlying pathogenesis of the disease process (12).
RESEARCH DESIGN AND METHODS
Mice.
BKS.Cg-m+/+Leprdb/J mice (stock number 000642) were purchased from Jackson Laboratories (Bar Harbor, ME). Animals were maintained at the University of Michigan in a pathogen-free environment and cared for following the University of Michigan Committee on the Care and Use of Animals guidelines. Mice were given continuous access to food (Purina 5001, Purina Mills LLC, St. Louis, MO) and water.
Measurement of blood glucose, GHb, cholesterol, and triglyceride levels.
Fasting blood glucose levels were measured every 4 weeks. After a 6 h fast, tail blood was analyzed using a standard glucometer (One Touch Profile, LIFESCAN, Inc., Milpitas, CA). At 24 weeks, GHb was measured using the Helena Laboratories Test Kit, Glyco-Tek affinity column method (Helena Laboratories Corp., Beaumont, TX). Plasma samples from six mice per group (db/db and db/+) were pooled, and each pool was analyzed by fast-protein liquid chromatography. The fractions were assayed for cholesterol and triglyceride levels. These assays were performed as previously published (13).
Assessment of neuropathy.
Hind paw withdrawal time and tail flick time were recorded as measures of thermal latency (14). Sural and sciatic NCVs were recorded and IENF density in footpads of the mice was measured as previously described (8,15).
Tissue harvest and RNA preparation.
At 24 weeks of age, mice were killed by sodium pentobarbital overdose and tissues were harvested as previously described (8). The left sciatic nerve was dissected and stored in 30 μL RNAlater (Ambion, An Applied Biosystems Business, Austin, TX). Total RNA was isolated from sciatic nerve using a commercially available silica gel–based isolation protocol (RNeasy Mini Kit, QIAGEN Inc., Valencia, CA), including an on-column DNase digestion following the manufacturer’s protocol. RNA quantity and quality were measured by microfluid electrophoresis using the RNA 6000 Pico LabChip on a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
Affymetrix microarrays.
RNA isolated from 10 db/db and 9 db/+ mice was used for hybridization. Of the total RNA, 75 ng from each sample was amplified and biotin labeled using the Ovation Biotin-RNA Amplification and Labeling System (NuGEN Technologies Inc., San Carlos, CA) according to the manufacturer’s protocol. RNA amplification and hybridization was performed in two batches (batch 1: db/db [n = 6], db/+ [n = 5]; batch 2: db/db [n = 4], db/+ [n = 4]) by the University of Michigan Comprehensive Cancer Center Affymetrix and Microarray Core Facility using the GeneChip Mouse Genome 430 2.0 Array (Affymetrix, Santa Clara, CA). Hybridization intensities for the probe features on the chip were detected by laser scan. Image files were quantified using Affymetrix GeneChip software (MAS5) to generate CEL files.
Data analysis.
Body weight, blood glucose and GHb levels, plasma cholesterol and triglyceride levels, thermal latency measures, IENF density, and NCVs of the mice from the two groups were compared using a t test. Data for the mice corresponding to outlier microarrays (see below) were excluded from these analyses, leaving six db/db and seven db/+ animals.
The Affymetrix CEL files were processed using a local copy of GenePattern, a bioinformatics platform from the Broad Institute (16), and standard libraries from the BioConductor project (http://www.bioconductor.org) and the R project (http://www.r-project.org). Array quality assessment was performed using BioConductor Simpleaffy and AffyQCReport packages. Six arrays (two db/+ and four db/db) identified as outliers were removed from further analysis based on the array quality assessment.
Array intensities were normalized with the robust multichip average method, and probe set to gene mapping was done using the BrainArray Custom CDF version 12 (17). Log2-transformed data from the two batches of microarrays were adjusted for batch effect bias using distance weighted discrimination batch effect correction method (18). Out of 16,394 genes on the chip, 915 genes with intensity lower than background were removed from further analysis. Raw and processed microarray data are available in the National Center for Biotechnology Information Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo, accession number GSE27382).
Intensity-based moderated t test (19) was used to determine significant differential expression, and false discovery rate <0.05 was used as a threshold to obtain a list of differentially expressed genes (DEGs). Significantly overrepresented biological categories in the DEGs were identified using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (12) version 6.7, and relationships among the categories were visualized using the ConceptGen gene set relationship mapping tool (20). DEGs were clustered with Chinese Restaurant Clustering (CRC) (21), and functional enrichment analysis of DEGs from each of the clusters was performed using DAVID.
The Genomatix (http://www.genomatix.de) literature cocitation network analysis tool (BiblioSphere) was used to identify pathways and regulatory networks among the DEGs. Protein–protein interactions (PPIs) among the DEGs were identified using the Michigan Molecular Interaction (MiMI) database (http://mimi.ncibi.org) and visualized using Cytoscape (http://www.cytoscape.org). Twenty-two DEGs from one CRC cluster, all annotated with “lipid metabolism,” were selected for promoter sequence analysis. Genomatix FrameWorker database was used to search the promoter sequences for overrepresented transcription factor binding site (TFBS) motifs. A region of 1,000 base pair (bp) upstream and 100 bp downstream of a transcription start site was designated as a promoter region. Promoter sequences of only experimentally verified transcripts were included in the analysis. A distance of 1–500 bp and a distance variance of 100 bp were permitted between TFBSs.
Real time RT-PCR.
The expression levels of five DEGs (highly upregulated in the db/db samples) were measured by real time RT-PCR (qRT-PCR) in five db/db and six db/+ samples for technical validation of microarray data. The amount of RNA from each sample was first normalized to an endogenous reference (TATA box binding protein [Tbp]; ∆CT) and then relative to the control group (∆∆CT) and was expressed as 2−∆∆CT.
RESULTS
Diabetes, dyslipidemia, and neuropathy phenotyping.
Table 1 lists phenotyping parameters for the db/db and db/+ mice measured at 24 weeks of age. The db/db mice are significantly heavier and exhibit higher levels of fasting blood glucose and GHb than the db/+ mice at 24 weeks of age. Plasma cholesterol and triglyceride levels are significantly elevated in the db/db mice, confirming dyslipidemia in these mice. Thermal latency in the hind paw and tail is significantly increased in db/db mice, indicative of sensory loss. Both IENF density and sciatic motor nerve conduction velocity (SMNCV) are significantly lower in db/db mice. Increased thermal latency, loss of IENF density, and decreased SMNCV confirm the development of DN in the db/db mice at 24 weeks of age.
TABLE 1.
Phenotyping parameters for the db/db and db/+ mice
| Parameter | db/db (n = 6) | db/+ (n = 7) | P value |
|---|---|---|---|
| Body weight (g) | 51.8 ± 4.38 | 29.6 ± 1.64 | 1.79E-05 |
| Fasting blood glucose (mg/dL) | 507.7 ± 80.16 | 148.0 ± 33.98 | 1.57E-03 |
| GHb (%) | 12.3 ± 0.52 | 7.1 ± 0.56 | 3.06E-09 |
| Plasma cholesterol (mg/dL) | 107 ± 18.21 | 75.8 ± 11.89 | 6.60E-03 |
| Plasma triglyceride (mg/dL) | 104.2 ± 34.89 | 56.5 ± 9.47 | 1.91E-02 |
| Hind paw latency (s) | 4.0 ± 1.09 | 2.7 ± 0.64 | 3.66E-02 |
| Tail flick latency (s) | 5.6 ± 0.65 | 4.1 ± 1.00 | 8.73E-03 |
| IENF density (fibers/mm2) | 27.2 ± 2.84 | 39.8 ± 3.13 | 2.76E-05 |
| Sural NCV (m/s) | 15.7 ± 5.46 | 20.6 ± 5.03 | 1.24E-01 |
| SMNCV (m/s) | 30.2 ± 10.40 | 48.3 ± 4.23 | 6.26E-03 |
Diabetes, dyslipidemia, and neuropathy phenotyping parameters measured at 24 weeks of age (mean ± SD).
Differential gene expression.
Figure 1 outlines the steps in the transcriptional analysis of the microarray data. Using intensity-based moderated t test false discovery rate <0.05 as a threshold, we identified 4,017 DEGs; 2,122 genes were upregulated and 1,895 genes were downregulated in the db/db relative to the db/+ samples. Microarray data were validated using qRT-PCR for 5 selected DEGs, which demonstrated high correlation between microarray and qRT-PCR expression levels (Supplementary Table 1).
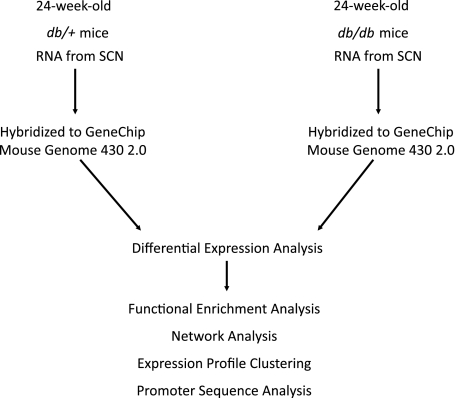
FIG. 1.
Transcriptional data analysis workflow. RNA samples obtained from the sciatic nerve (SCN) of db/db and db/+ mice were hybridized to Affymetrix GeneChip microarrays. Data quantified from scanned microarrays were analyzed to identify DEGs. Functional enrichment analysis of DEGs was performed to identify overrepresented biological categories and pathways. Network analysis identified functionally related subgroups of DEGs. Expression profile clustering identified subgroups of coexpressed DEGs. Promoter sequence analysis identified overrepresented TFBS modules in the promoter sequences of coexpressed genes.
Functional enrichment.
Enrichment analysis identifies functional categories that have a higher representation in the regulated genes and, thus, are likely to be active in the experimental condition. Table 2 lists Gene Ontology (GO) terms overrepresented in DEGs identified using DAVID. Overrepresented biological processes with a higher number of upregulated genes are cell cycle, lipid metabolic process, lipid transport, carbohydrate metabolic process, response to stress, and apoptosis. Biological processes with a majority of genes downregulated are axonogenesis and cell adhesion. Overrepresented cellular component terms are mitochondrion and axon, with a majority of mitochondrial genes upregulated and a majority of axonal genes downregulated.
TABLE 2.
Overrepresented biological annotation terms
| Category | Term | Number of genes (up/down) | Fold enrichment | BH P value |
|---|---|---|---|---|
| GO_BP | Cell cycle | 191 (126/65) | 1.45 | 2.10E-05 |
| GO_BP | Lipid metabolic process | 172 (119/53) | 1.47 | 2.40E-05 |
| GO_BP | Vesicle-mediated transport | 126 (67/59) | 1.55 | 3.20E-05 |
| GO_BP | Response to stress | 200 (129/71) | 1.34 | 9.36E-04 |
| GO_BP | Cell adhesion | 164 (70/94) | 1.36 | 2.14E-03 |
| GO_BP | Axonogenesis | 52 (7/45) | 1.72 | 5.55E-03 |
| GO_BP | Apoptosis | 160 (99/61) | 1.31 | 1.29E-02 |
| GO_BP | Lipid transport | 30 (22/8) | 1.82 | 2.27E-02 |
| GO_BP | Carbohydrate metabolic process | 100 (76/24) | 1.2 | 5.24E-02 |
| GO_CC | Axon | 33 (6/27) | 1.95 | 2.31E-02 |
| GO_CC | Mitochondrion | 227 (175/52) | 1.91 | 5.24E-02 |
Functional terms significantly overrepresented in the DEGs are listed. BH P value, Benjamini-Hochberg corrected P value of enrichment in DAVID; GO_BP, GO biological process; GO_CC, GO cellular component.
Network analysis.
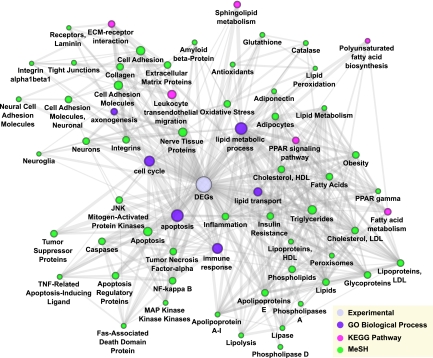
Gene network analysis is useful for exploring both published and novel gene associations that may be involved in the process being studied. ConceptGen is a gene set relation mapping tool that identifies relationships (significant overlap) between an experimental gene set and curated gene sets in sources such as GO terms, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and Medical Subject Heading (MeSH) terms (20). ConceptGen was used to analyze 717 DEGs annotated with the GO biological processes shown in Table 1. The mouse gene identifiers were converted by ConceptGen Homology conversion tool to 695 human homologs. Relationship among selected concepts (GO biological processes, KEGG pathways, and MeSH terms) associated with the gene set were visualized using ConceptGen network graph tool (Fig. 2). KEGG pathways with significant overlap with the DEG set include fatty acid metabolism, peroxisome proliferator–activated receptor (PPAR) signaling, and extracellular matrix–receptor interaction. Concepts related to the DEG set clustered in three groups: lipid metabolism, apoptosis, and axonogenesis (Fig. 2).
FIG. 2.
ConceptGen network graph of overrepresented concepts visualized in Cytoscape. Network depicting relationships and overlap among significantly overrepresented concepts (GO biological processes, KEGG pathways, and MeSH terms) in the DEGs. Node color represents concept type, node size represents number of genes in the concept, and thickness of the edge represents gene overlap between concepts.
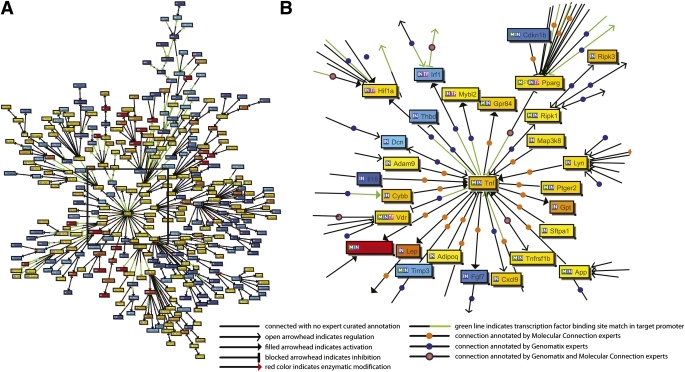
BiblioSphere generates a network of genes based on their cocitation in PubMed abstracts and identifies functionally related subnetworks based on known regulatory relationships and pathway annotation. Figure 3A illustrates a BiblioSphere network of DEGs centered on tumor necrosis factor-α (TNF-α) with gene–gene connections restricted to expert-curated connections. TNF-α was chosen as the central node because of its importance in type 2 diabetes and lipid metabolism (22). Central nodes of subnetworks and associated pathways include Hifna (apoptosis), Serpine1 (p53 signaling), App (axonogenesis), Lyn (Fc ε RI signaling), and Pparg (PPAR signaling and fatty acid oxidation) (Fig. 3B).
FIG. 3.
BiblioSphere cocitation network of DEGs. Nodes represent genes, node color represents fold change (red/yellow: upregulation; blue: downregulation). The network is centered at TNF-α. Only shortest paths to the central node are shown for clarity. A: Network of DEGs. B: Enlarged view of the part of the network identified in A illustrates connections between the central node and other highly connected nodes in the network. Highly connected nodes in the network include Pparg, App, Serpine1, Hif1a, Irf1, Vdr, Lyn, and Ripk1.
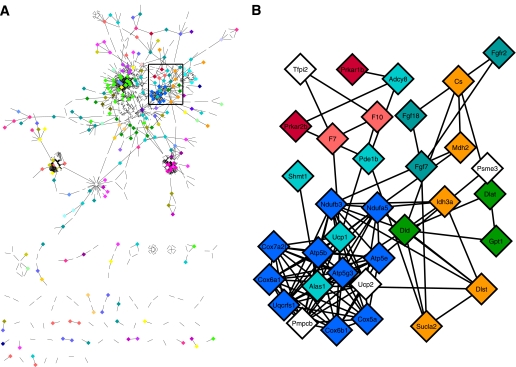
PPI network of DEGs was constructed using Cytoscape MiMI plug-in (23). Subnetworks were identified based on pathway annotation of genes in MiMI (Fig. 4). Regulated pathways identified by PPI network are carbohydrate metabolism (glycolysis and tricarboxylic acid cycle [TCA]), energy metabolism (oxidative phosphorylation), lipid metabolism (fatty acid metabolism and glycerolipid metabolism), glutathione metabolism, cell development (cell cycle, apoptosis, axon guidance, and neurotrophin signaling), cell communication (cell adhesion and extracellular matrix–receptor interaction), and signal transduction pathways (PPAR signaling, transforming growth factor-β [TGF-β] signaling, adipocytokine signaling, Wnt signaling, mitogen-activated protein kinase [MAPK] signaling, ErbB signaling, and mammalian target of rapamycin [mTOR] signaling).
FIG. 4.
MiMI PPI network of DEGs visualized in Cytoscape. Nodes represent genes; edges represent PPIs. A: Entire network. B: Enlarged view of part of the network identified in A. Node color represents pathway annotation of genes (for example, blue: oxidative phoshorylation; orange: TCA; red: apoptosis).
Supplementary Table 2 lists DEGs in the pathways regulated in the db/db nerve. The majority of DEGs in the glycolysis, TCA, oxidative phosphorylation, fatty acid metabolism, glycerolipid metabolism, mitochondrial fatty acid elongation, lipid transport, adipocytokine signaling, PPAR signaling, and apoptosis pathways are upregulated, whereas most of the axonogenesis-related genes are downregulated in db/db nerve.
Expression profile clustering and promoter sequence analysis.
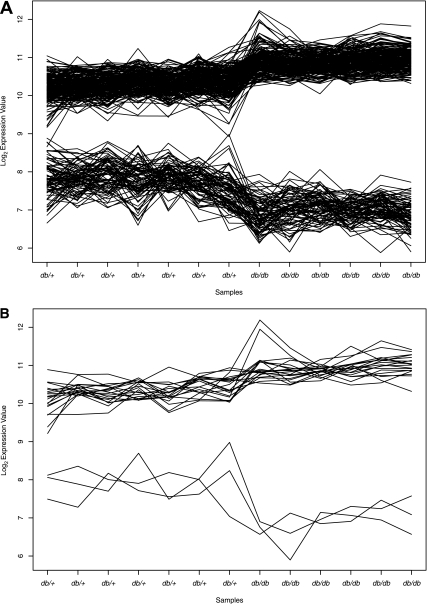
Clustering of gene expression profiles identifies potentially coregulated and functionally related subgroups of genes. CRC, a model-based Bayesian clustering tool, grouped the DEGs into 18 clusters based on positive as well as negative correlation among their expression profiles. We performed enrichment analysis for gene sets from individual clusters to identify overrepresented functional categories (Supplementary Table 3). Figure 5A illustrates a cluster of 275 DEGs enriched in “mitochondrion,” “lipid metabolic process,” and “axonogenesis.” Gene regulatory sequence elements are organized into motifs of TFBSs known as promoter modules, possibly suggesting coordinated regulation by multiple transcription factors (TFs) (24). We used Genomatix FrameWorker tool to analyze 41 promoter sequences of 22 genes annotated with “lipid metabolism” from the above cluster (Fig. 5B) to look for significantly overrepresented TFBS motifs. Five of these genes are also annotated with “mitochondrion.” Table 3 lists the top five most significantly overrepresented promoter modules consisting of three vertebrate TF matrix families (groups of functionally similar TFs) (25). The HOXF family TFs, the HBOX family TF Meox1, the LHXF family TFs, and the HOMF family TF Msx1 are downregulated, whereas the ETSF family TFs are upregulated in the db/db nerve (Supplementary Table 4).
FIG. 5.
Gene expression profiles generated by CRC. Log2 expression levels in seven db/+ and six db/db samples. A: Expression profile cluster of 275 DEGs enriched in “mitochondrion,” “lipid metabolic process,” and “axonogenesis.” B: Expression profiles of 22 “lipid metabolism” genes selected for promoter sequence analysis.
TABLE 3.
Top five most significantly overrepresented TFBS modules
| TFBS module | Strand | Common to sequences (%) | P value |
|---|---|---|---|
| HOMF_HOMF_ETSF | + − + | 51 | 6.58E-07 |
| HOXF_LHXF_HOXF | − − + | 58 | 6.22E-04 |
| HOXF_HBOX_HBOX | − − + | 56 | 1.47E-03 |
| HBOX_LHXF_HOXF | − − + | 51 | 2.01E-03 |
| OCT1_LHXF_HOXF | + − + | 51 | 2.41E-03 |
Five promoter modules consisting of three elements were significantly overrepresented in the promoter sequences of lipid metabolism–related genes. Module matrix families, strand orientation, percent of promoter sequences containing the module, and P values of enrichment are shown. The P value is the probability of finding the module in a set of randomly selected promoters and is determined by scanning a background promoter sequence set of 5,000 human promoters.
DISCUSSION
How the metabolic dysregulation caused by insulin resistance leads to local nerve fiber damage in DN is unclear. In this study, we performed global gene expression profiling of sciatic nerve from 24-week-old db/db mice, which exhibit hyperglycemia, dyslipidemia, and DN typical of type 2 diabetes. We detected alterations in the expression of genes involved in carbohydrate and lipid metabolism, lipid transport, stress responses, and apoptosis. Furthermore, promoter sequences of coexpressed lipid metabolism–related DEGs exhibited significantly overrepresented TFBS motifs and are likely to share a common regulatory mechanism relevant to the development of DN.
Morphological and electrophysiological changes in the BKS db/db mice closely mimic the changes observed in DN patients (8,9,26–28). However, the db/db mouse model does not fully duplicate human DN pathology; not all structural changes observed in human DN are seen in the db/db mouse (27,29). Duration of diabetes in mice is likely not long enough for the development of severe long-term degeneration associated with human DN. The development of hyperglycemia and DN in the BKS db/db mice are considered to be consequences of hyperphagia and insulin resistance resulting from impaired leptin signaling in the hypothalamus (8). However, the role of leptin signaling in peripheral nerve is not understood, and whether leptin receptor mutation and obesity have a compounding effect on the development of DN in these mice is not known.
Schwann cells (SC) are major contributors to the mRNA in the sciatic nerve biopsies, with a small contribution coming from axons, epineural fibroblasts, adipocytes, and vascular endothelial cells (30). Our analyses revealed that genes encoding myelin structural proteins (P0, Pmp22, Connexin-32, Mag, E-Cadherin, and Periaxin), myelin structural lipid synthesis genes (Plp, Elovl6, and Apolipoprotein D), as well as Sox10, a TF required for myelination, are downregulated in db/db nerve. Segmental demyelination has been observed in human DN (31,32); mouse studies, however, do not report evidence of demyelination (27,29). Downregulation of myelin protein-encoding genes and myelin lipid synthesis genes may represent SC abnormalities preceding the structural changes.
The TFs c-Jun and Krox-24, and other genes expressed by denervated SC to promote axonal regeneration (GAP-43, Ncam, and L1) (33), are downregulated, as are positive regulators of axonogenesis (Cdh4, Nefl, and Mapt) and axon guidance pathway genes (Netrin G1 and Fyn), suggesting a lack of axonal regeneration. These changes are consistent with the axonal degeneration observed in morphological studies in the db/db mice (27,29).
Loss of neurotrophic signal in DN may impair nerve generation and cause dying back of axons (3,34). Neurotrophin Ngf; NT-3 receptor TrkC; Gdnf receptors Gfra1, Gfra3, and Gfra4; neuregulin receptor Erbb; and neurotrophin signaling pathway genes (Akt3 and Mapk10) are downregulated, indicating impaired neurotrophic support in the db/db nerve. Downregulation of Ngf, TrkC, and GFRα1 is consistent with the results of studies in diabetic rats (35–37). Other neurotrophins are not differentially regulated in our data. Some studies in rats with streptozotocin-induced diabetes report reduced expression of Bdnf, NT-3, NT-4/5, Igf-I, Igf-II, Cntf, Gdnf, and neurotrophin receptors p75LNGFR and TrkB approximately 12 weeks after the induction of diabetes (35–40), whereas others report no change in NT-3 expression in rat sciatic nerve (41). The difference in the animal models used in the studies (rat model of type 1 diabetes vs. mouse model of type 2 diabetes) may explain this discrepancy.
The SC marker S100β as well as antiapoptotic genes (Mapk8ip1 and TGF-β2) are downregulated, whereas proapoptotic genes (Fas, TNF-α, Casp8, and Brca1) are upregulated in the db/db samples (Supplementary Table 2). The downregulation of SC marker and antiapoptotic genes along with upregulation of proapoptotic genes suggests SC apoptosis in db/db nerve; however, no studies to date have demonstrated SC death in DN.
The causes of axonal and SC degradation in DN are not well understood, but several hypotheses have been developed in regard to the pathogenesis of the nerve injury (3,34). Our analyses indicate increased glucose, energy, and lipid metabolism in the db/db nerve and support the roles of hyperglycemia- and hyperlipidemia-induced oxidative stress, inflammatory response, and vascular ischemia in DN. In addition, our data indicate impaired axonal transport, neurotrophic signal, cell adhesion, and cell communication. In this study, we focused on hyperglycemia- and dyslipidemia-related gene expression changes in the nerve.
Hyperglycemia is a major factor in the development and progression of DN; current hypotheses suggest the effect of hyperglycemia is likely to be vascular, metabolic, or a combination of both (34,42). The metabolic hypothesis of axonal and SC damage in diabetes suggests that activation of glucose metabolism pathways in hyperglycemia results in oxidative stress. In our data, glycolysis, TCA, and oxidative phosphorylation genes are upregulated (Supplementary Table 2), suggesting activation of glucose and energy metabolism in the db/db nerve. Upregulation of oxidative phosphorylation genes and downregulation of mitochondrial H+ transporting ATP synthases in the db/db nerve suggest increased superoxide production (43). The mitochondrial proton carrier Ucp2, known to be upregulated in response to elevated reactive oxygen species (ROSs) (44), is highly upregulated; the oxidative stress-induced growth inhibitor Osgin1 is also upregulated in the db/db nerve. Upregulation of antioxidant genes (Sod2, Peroxiredoxins, and Catalase) suggests cellular response to increased ROS production. Edwards et al. (3) hypothesized that increased cellular oxidative stress results in activation of the Poly (ADP-ribose) polymerase (PARP) pathway, which in turn induces inflammatory responses in the nerve. Upregulation of PARP, inflammatory response, and MAPK signaling pathway genes in the db/db nerve support the PARP pathway–mediated inflammatory response hypothesis. Nuclear factor-κB (NF-κB) induced–inflammatory response is also implicated in demyelination and axon degeneration (3); however, NF-κB and genes regulated by NF-κB, such as iNOS, are not differentially regulated in our data.
The vascular hypothesis of nerve damage in diabetes suggests that activation of glucose metabolism pathways causes functional and structural changes in the neuronal vasculature leading to endothelial hypoxia and axonal ischemia (45). Bradley et al. (46) noted thickening of perineurial cell basal lamina and increased endoneurial collagenization around SC in the sural nerve of DN patients. Activation of protein kinase C by hyperglycemia induces expression of vascular endothelial growth factor (VEGF)—an angiogenic gene and permeability factor—inhibits production of nitric oxide and alters Na+-K+-ATPase activity. Activation of glucose metabolism pathways induces TGF-β1 and Serpine1 expression, which results in endothelial fibrosis and thickening of vascular membranes (3). VEGF-C, TGF-β1, and Serpine1 are upregulated; Na+-K+-ATPases (Atp1b2 and Atp1b3) are downregulated in the db/db nerve. Upregulation of hypoxia-inducible factor Hif1a may indicate cellular response to ischemia (47). These changes in gene expression support the hypothesis of neuronal ischemia and hypoxia. Imbalance in expression of nitric oxide synthase (Nos) isoforms has been implicated in diabetic endothelial dysfunction (48); however, these are not differentially expressed in our data.
Dysregulation of lipid metabolism is implicated in peripheral neuropathy (7,30,33,49), but the mechanism of its effect is not well understood. We observed upregulation of genes expressed by adipocytes (Adiponectin, Perilipin, Lipin1, and Lpl), possibly due to increased amounts of adipose tissue in the epineurium of the db/db nerve. Verheijen et al. (30) demonstrated that epineurium of adult mouse sciatic nerve contains adipose-like tissue, and both endoneurium and epineurium express lipid metabolism genes; they suggested that adipocytokine signaling from endoneurial adipocytes may regulate lipid metabolism in SC and axons. Our analysis indicates upregulation of fatty acid metabolism, glycerolipid metabolism, lipid transport, PPAR signaling, and adipocytokine signaling genes in the db/db nerve (Supplementary Table 2).
Vincent et al. (7,50) hypothesized that increased production of ROS in hyperglycemia results in lipid peroxidation and that the formation of oxidized low density lipoproteins (oxLDL) contributes to the nerve injury in diabetic dyslipidemia. The apoptotic effect of oxLDL on vascular endothelial cells and neurons is mediated via oxLDL receptor (Olr1/LOX-1), which is induced by hyperglycemia, ROS, oxLDL, and TNF-α. In vitro studies demonstrate that binding of oxLDL to Olr1 leads to NADPH oxidase (Nox) activation, mitochondrial superoxide production, and neuronal oxidative stress in dorsal root ganglia of mice (50). oxLDL receptor Olr1 is highly overexpressed and Nox2 is upregulated in our data, supporting the hypothesis of oxLDL-induced nerve injury in dyslipidemia, possibly via activation of Nox. oxLDL also acts on macrophages via scavenger receptor (Scarb1/CD36) and triggers immune responses via MAPK and NF-κB signaling pathways (7). In our data, CD36 and MAPK signaling pathway genes (TNF-α, Il1a, and TGF-β1) are upregulated, supporting the hypothesis of CD36-mediated inflammatory response in the db/db nerve.
Coexpression of genes suggests shared mechanism of regulation. DEGs associated with “lipid metabolism,” “mitochondrion,” and “axonogenesis” clustered together based on their correlated expression pattern (Fig. 5A). Most of the lipid metabolism– and mitochondria-associated genes in the cluster are upregulated, whereas axonogenesis genes are downregulated. To investigate the potential coregulation mechanism, we analyzed promoter sequences of 22 DEGs from this cluster annotated with the term “lipid metabolism”; 5 of these genes are also annotated with “mitochondrion.”
It is interesting that TFs binding to the overrepresented TFBSs in promoter sequences of lipid metabolism genes are associated with neuron differentiation, axon guidance, and immune response, suggesting a possible common regulatory mechanism in lipid metabolism, stress response, and axonal degeneration. TFs annotated with “nervous system development”–related terms are downregulated in the db/db nerve. The HOXF family, the HBOX family, and the LHXF family TFs are annotated with “motor axon guidance” and “regulation of neuron differentiation.” HOMF family TF Msx1 is annotated with “negative regulation of vascular endothelial growth factor receptor signaling pathway” and “neuron differentiation.” “Immune response”–associated ETSF family TFs are upregulated in the db/db nerve (Supplementary Table 4). OCT1 family TF Oct-6, an SC TF important in myelination (30), is not among the DEGs.
In conclusion, gene expression changes suggest roles of multiple etiological factors in the development of DN. The changes are consistent with pathological characteristics of DN, such as axonal degeneration and potential loss of neurotrophic signal. Our findings support the role of hyperglycemia-induced oxidative stress and ischemia in nerve injury. Our results also support the hypothesis of oxidized lipid–mediated nerve injury and increased mitochondrial oxidative stress in dyslipidemia. In addition, our analyses revealed possible coregulation of lipid metabolism, stress response, and axonal degeneration genes and identified TFs that may modulate this coregulation. Further investigation into the signal mediated by these TFs is likely to provide more insight into dyslipidemia-induced nerve injury.
Supplementary Material
ACKNOWLEDGMENTS
M.K. and E.L.F. have received National Institutes of Health grants U54-DA-021519 (National Center for Integrative Biomedical Informatics [NCIBI]) and R24-DK-082841-01. J.H. has received a University of Michigan Rackham Graduate School Predoctoral Fellowship. E.L.F. has received a grant from the Animal Models of Diabetic Complications Consortium (U01-DK-076160). This work was also supported by the Michigan Diabetes Research and Training Center, funded by a grant from the National Institute of Diabetes and Digestive and Kidney Diseases (DK-020572); the A. Alfred Taubman Medical Research Institute; and the Program for Neurology Research and Discovery, University of Michigan.
No potential conflicts of interest relevant to this article were reported.
M.P. researched data and wrote the manuscript. J.H. researched data and reviewed and edited the manuscript. Y.H. performed PCR validation. C.B., J.M.H., and S.S.O. conducted animal experiments. M.K. contributed to bioinformatic analysis design. E.L.F. designed and directed the study, contributed to discussion, and reviewed the manuscript.
Parts of this study were presented in abstract/poster form at the 18th Annual International Conference on Intelligent Systems for Molecular Biology, Boston, Massachusetts, 11–13 July 2010, and at the NCIBI 5th Annual Research Conference 2010 at the University of Michigan, Ann Arbor, Michigan, 20–21 April 2010.
The authors thank Kelli Sullivan, PHD, Department of Neurology, University of Michigan, for critical review of the manuscript.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db10-1541/-/DC1.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2007 national diabetes fact sheet [article online], 2007. Available from http://www.cdc.gov/diabetes/pubs/pdf/ndfs_2007.pdf Accessed 1 March 2010
- 2.Gordois A, Scuffham P, Shearer A, Oglesby A, Tobian JA. The health care costs of diabetic peripheral neuropathy in the US. Diabetes Care 2003;26:1790–1795 [DOI] [PubMed] [Google Scholar]
- 3.Edwards JL, Vincent AM, Cheng HT, Feldman EL. Diabetic neuropathy: mechanisms to management. Pharmacol Ther 2008;120:1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fioretto P, Dodson PM, Ziegler D, Rosenson RS. Residual microvascular risk in diabetes: unmet needs and future directions. Nat Rev Endocrinol 2010;6:19–25 [DOI] [PubMed] [Google Scholar]
- 5.Cameron NE, Eaton SE, Cotter MA, Tesfaye S. Vascular factors and metabolic interactions in the pathogenesis of diabetic neuropathy. Diabetologia 2001;44:1973–1988 [DOI] [PubMed] [Google Scholar]
- 6.Sugimoto K, Murakawa Y, Sima AA. Diabetic neuropathy—a continuing enigma. Diabetes Metab Res Rev 2000;16:408–433 [DOI] [PubMed] [Google Scholar]
- 7.Vincent AM, Hinder LM, Pop-Busui R, Feldman EL. Hyperlipidemia: a new therapeutic target for diabetic neuropathy. J Peripher Nerv Syst 2009;14:257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan KA, Hayes JM, Wiggin TD, et al. Mouse models of diabetic neuropathy. Neurobiol Dis 2007;28:276–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan KA, Lentz SI, Roberts JL, Jr, Feldman EL. Criteria for creating and assessing mouse models of diabetic neuropathy. Curr Drug Targets 2008;9:3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi K, Forte TM, Taniguchi S, Ishida BY, Oka K, Chan L. The db/db mouse, a model for diabetic dyslipidemia: molecular characterization and effects of Western diet feeding. Metabolism 2000;49:22–31 [DOI] [PubMed] [Google Scholar]
- 11.Sima AA, Robertson DM. Peripheral neuropathy in mutant diabetic mouse [C57BL/Ks (db/db)]. Acta Neuropathol 1978;41:85–89 [DOI] [PubMed] [Google Scholar]
- 12.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:3. [PubMed] [Google Scholar]
- 13.McMillen TS, Heinecke JW, LeBoeuf RC. Expression of human myeloperoxidase by macrophages promotes atherosclerosis in mice. Circulation 2005;111:2798–2804 [DOI] [PubMed] [Google Scholar]
- 14.Lee JH, Cox DJ, Mook DG, McCarty RC. Effect of hyperglycemia on pain threshold in alloxan-diabetic rats. Pain 1990;40:105–107 [DOI] [PubMed] [Google Scholar]
- 15.Christianson JA, Riekhof JT, Wright DE. Restorative effects of neurotrophin treatment on diabetes-induced cutaneous axon loss in mice. Exp Neurol 2003;179:188–199 [DOI] [PubMed] [Google Scholar]
- 16.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet 2006;38:500–501 [DOI] [PubMed] [Google Scholar]
- 17.Dai M, Wang P, Boyd AD, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 2005;33:e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benito M, Parker J, Du Q, et al. Adjustment of systematic microarray data biases. Bioinformatics 2004;20:105–114 [DOI] [PubMed] [Google Scholar]
- 19.Sartor MA, Tomlinson CR, Wesselkamper SC, Sivaganesan S, Leikauf GD, Medvedovic M. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics 2006;7:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sartor MA, Mahavisno V, Keshamouni VG, et al. ConceptGen: a gene set enrichment and gene set relation mapping tool. Bioinformatics 2010;26:456–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin ZS. Clustering microarray gene expression data using weighted Chinese restaurant process. Bioinformatics 2006;22:1988–1997 [DOI] [PubMed] [Google Scholar]
- 22.Moller DE. Potential role of TNF-alpha in the pathogenesis of insulin resistance and type 2 diabetes. Trends Endocrinol Metab 2000;11:212–217 [DOI] [PubMed] [Google Scholar]
- 23.Gao J, Ade AS, Tarcea VG, et al. Integrating and annotating the interactome using the MiMI plugin for cytoscape. Bioinformatics 2009;25:137–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fessele S, Maier H, Zischek C, Nelson PJ, Werner T. Regulatory context is a crucial part of gene function. Trends Genet 2002;18:60–63 [DOI] [PubMed] [Google Scholar]
- 25.Cartharius K, Frech K, Grote K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 2005;21:2933–2942 [DOI] [PubMed] [Google Scholar]
- 26.Norido F, Canella R, Zanoni R, Gorio A. Development of diabetic neuropathy in the C57BL/Ks (db/db) mouse and its treatment with gangliosides. Exp Neurol 1984;83:221–232 [DOI] [PubMed] [Google Scholar]
- 27.Carson KA, Bossen EH, Hanker JS. Peripheral neuropathy in mouse hereditary diabetes mellitus. II. Ultrastructural correlates of degenerative and regenerative changes. Neuropathol Appl Neurobiol 1980;6:361–374 [DOI] [PubMed] [Google Scholar]
- 28.Sima AA, Robertson DM. Peripheral neuropathy in the diabetic mutant mouse. An ultrastructural study. Lab Invest 1979;40:627–632 [PubMed] [Google Scholar]
- 29.Robertson DM, Sima AA. Diabetic neuropathy in the mutant mouse [C57BL/ks(db/db)]: a morphometric study. Diabetes 1980;29:60–67 [DOI] [PubMed] [Google Scholar]
- 30.Verheijen MH, Chrast R, Burrola P, Lemke G. Local regulation of fat metabolism in peripheral nerves. Genes Dev 2003;17:2450–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malik RA, Veves A, Walker D, et al. Sural nerve fibre pathology in diabetic patients with mild neuropathy: relationship to pain, quantitative sensory testing and peripheral nerve electrophysiology. Acta Neuropathol 2001;101:367–374 [DOI] [PubMed] [Google Scholar]
- 32.Malik RA, Tesfaye S, Newrick PG, et al. Sural nerve pathology in diabetic patients with minimal but progressive neuropathy. Diabetologia 2005;48:578–585 [DOI] [PubMed] [Google Scholar]
- 33.Scherer SS. The biology and pathobiology of Schwann cells. Curr Opin Neurol 1997;10:386–397 [DOI] [PubMed] [Google Scholar]
- 34.Vincent AM, Feldman EL. New insights into the mechanisms of diabetic neuropathy. Rev Endocr Metab Disord 2004;5:227–236 [DOI] [PubMed] [Google Scholar]
- 35.Tomlinson DR, Fernyhough P, Diemel LT. Neurotrophins and peripheral neuropathy. Philos Trans R Soc Lond B Biol Sci 1996;351:455–462 [DOI] [PubMed] [Google Scholar]
- 36.Rodríguez-Peña A, Botana M, González M, Requejo F. Expression of neurotrophins and their receptors in sciatic nerve of experimentally diabetic rats. Neurosci Lett 1995;200:37–40 [DOI] [PubMed] [Google Scholar]
- 37.Liu GS, Shi JY, Lai CL, et al. Peripheral gene transfer of glial cell-derived neurotrophic factor ameliorates neuropathic deficits in diabetic rats. Hum Gene Ther 2009;20:715–727 [DOI] [PubMed] [Google Scholar]
- 38.Wuarin L, Guertin DM, Ishii DN. Early reduction in insulin-like growth factor gene expression in diabetic nerve. Exp Neurol 1994;130:106–114 [DOI] [PubMed] [Google Scholar]
- 39.Zhuang HX, Wuarin L, Fei ZJ, Ishii DN. Insulin-like growth factor (IGF) gene expression is reduced in neural tissues and liver from rats with non-insulin-dependent diabetes mellitus, and IGF treatment ameliorates diabetic neuropathy. J Pharmacol Exp Ther 1997;283:366–374 [PubMed] [Google Scholar]
- 40.Calcutt NA, Muir D, Powell HC, Mizisin AP. Reduced ciliary neuronotrophic factor-like activity in nerves from diabetic or galactose-fed rats. Brain Res 1992;575:320–324 [DOI] [PubMed] [Google Scholar]
- 41.Cai F, Tomlinson DR, Fernyhough P. Elevated expression of neurotrophin-3 mRNA in sensory nerve of streptozotocin-diabetic rats. Neurosci Lett 1999;263:81–84 [DOI] [PubMed] [Google Scholar]
- 42.Russell JW, Golovoy D, Vincent AM, et al. High glucose-induced oxidative stress and mitochondrial dysfunction in neurons. FASEB J 2002;16:1738–1748 [DOI] [PubMed] [Google Scholar]
- 43.Vincent AM, Russell JW, Low P, Feldman EL. Oxidative stress in the pathogenesis of diabetic neuropathy. Endocr Rev 2004;25:612–628 [DOI] [PubMed] [Google Scholar]
- 44.Chan SH, Wu CA, Wu KL, Ho YH, Chang AY, Chan JY. Transcriptional upregulation of mitochondrial uncoupling protein 2 protects against oxidative stress-associated neurogenic hypertension. Circ Res 2009;105:886–896 [DOI] [PubMed] [Google Scholar]
- 45.Horowitz SH. Diabetic neuropathy. Clin Orthop Relat Res 1993;(296):78–85 [PubMed] [Google Scholar]
- 46.Bradley JL, King RH, Muddle JR, Thomas PK. The extracellular matrix of peripheral nerve in diabetic polyneuropathy. Acta Neuropathol 2000;99:539–546 [DOI] [PubMed] [Google Scholar]
- 47.Probst-Cousin S, Neundörfer B, Heuss D. Microvasculopathic neuromuscular diseases: lessons from hypoxia-inducible factors. Neuromuscul Disord 2010;20:192–197 [DOI] [PubMed] [Google Scholar]
- 48.Zochodne DW, Verge VM, Cheng C, et al. Nitric oxide synthase activity and expression in experimental diabetic neuropathy. J Neuropathol Exp Neurol 2000;59:798–807 [DOI] [PubMed] [Google Scholar]
- 49.Tesfaye S, Stevens LK, Stephenson JM, et al. Prevalence of diabetic peripheral neuropathy and its relation to glycaemic control and potential risk factors: the EURODIAB IDDM Complications Study. Diabetologia 1996;39:1377–1384 [DOI] [PubMed] [Google Scholar]
- 50.Vincent AM, Hayes JM, McLean LL, Vivekanandan-Giri A, Pennathur S, Feldman EL. Dyslipidemia-induced neuropathy in mice: the role of oxLDL/LOX-1. Diabetes 2009;58:2376–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.