There remains a critical need to better understand the underlying disease mechanisms responsible for diabetes complications in order to develop new and improved therapeutic strategies for these chronic conditions. These complications are broadly classified as microvascular, including neuropathy, nephropathy, and retinopathy, or macrovascular, including cardiovascular and peripheral vascular disease. The risk for developing complications is influenced by many factors including duration of diabetes and genetic factors. Current treatments have resulted in only a partial reduction in this risk, and the management of these conditions remains a major unmet need for those with diabetes. New insights have come from an unlikely ally, the worm C. elegans, in which research has identified a novel family of endogenous, small (∼22 nucleotides), single stranded, noncoding RNA molecules known as microRNAs (miRNAs) as developmental regulators (1,2). These molecules, only identified in humans in the last decade, modulate physiological and pathological processes by the posttranscriptional inhibition of gene expression (3). Many excellent reviews have been written dealing with biogenesis (4) through to their role in development and disease (5), although this work has generally focused on cancer and infection rather than diabetes and its complications (6).
The effect of miRNAs is via the incomplete binding of the “seed sequence” at the 5′ end of the miRNA to the complementary target site in the 3′ untranslated region (UTR) of the messenger RNA. Each miRNA species can inhibit the expression of many genes, while each mRNA can be potentially targeted by several miRNAs. Recent studies have demonstrated clear links between altered miRNA expression and certain diabetes complications, as described later in this article. These findings further highlight the complex nature of diabetes and the huge challenge in better understanding the various vascular complications associated with this disorder. Here we discuss some recent studies demonstrating the role of miRNAs in diabetes complications and at sites where complications occur, with the anticipation that these studies are likely to influence the treatment, diagnosis, and management of complications. We also explore the huge clinical challenge before us in attempting the targeted modulation of these rather promiscuous molecules in specific tissues in order to achieve better therapeutic outcomes.
miRNAS IN DIABETES COMPLICATIONS
There is a wealth of evidence on the diverse role of miRNAs in many biological processes, including proliferation, differentiation, apoptosis, and development. The list of diseases in which dysregulation of miRNAs has been implicated is constantly growing. Compared with cancer, far less is known about the role of miRNAs in diabetes and its complications. Several miRNAs have been identified as having a physiological role in tissues in which diabetes complications occur. Whether these miRNAs are involved in the damage that occurs in diabetes is yet to be established. The association between altered miRNA expression and the development and progression of the various diabetes complications implicates certain miRNAs in the development of diabetes-related injury in the heart, kidney, peripheral nerves, and retina. Given that each miRNA has the potential to regulate multiple genes, dysregulation of miRNAs would be certain to have impact on many biological processes that are of direct relevance to diabetes complications. As tempting as it is to have a blinkered approach to our favorite miRNA and target gene, it is naïve to ignore the incredible complexity of miRNA regulation and the depth of the downstream maze of targets, both direct and indirect. This complexity is highlighted later in this article, demonstrating how both increased and decreased expression of certain miRNAs are associated with diabetes complications. A summary of the miRNAs relevant to diabetes complications is found in Table 1.
TABLE 1.
Diabetes complications: miRNAs and their targets
| Complication | Known targets | Reference |
|---|---|---|
| Diabetic nephropathy miR-21 | PTEN | (29) unpublished* |
| miR-29 family | Collagens | (25) unpublished* |
| miR-192 | ZEB2, Col 1 | (16,18,22,23) |
| miR-93 | VEGF | (34) |
| miR-200 family | ZEB1, ZEB2, TGF-β2 | (18,19) |
| miR-216a | YB-1, PTEN | (15,26) |
| miR-377 | PAK1, MnSOD | (30) |
| Retinopathy | ||
| miR-93, miR-200, and miR-29 families? | VEGF | (34) indirect† |
| Atherosclerosis | ||
| miR-16 | COX-2 | (39) indirect† |
| miR-503? | CCNE1, cdc25A | (40) |
| Heart | ||
| miR-21 | SPRY1 | (50,51) |
| miR-29 | Collagens | (17) |
| miR-30? | CTGF | (48) indirect† |
| miR-1/133 | ERG, RhoA, Cdc42, WHSC2, HCN1, and HCN4 | (44–47) |
| miR-320 | VEGF, FGF, IGF-1, and IGF-1R | (46) |
*H.Y.L., unpublished data.
†Inferred from the reference.
CAN WE IDENTIFY PATIENTS AT RISK FOR DEVELOPING COMPLICATIONS?
Gene profiling technology has been used with a view to improve disease diagnosis and prognosis, especially in the area of cancer research. Over a third of diabetes patients develop serious complications. Can we use miRNA profiling to identify patients that go on to develop complications?
One study observed increased expression of several miRNAs in the diabetic GK rat model, including increased expression of the miR-29 family in muscle, fat, and liver, correlating with insulin resistance (7). Another study profiled miRNA expression in skeletal muscle of the spontaneous diabetic GK rat and observed the downregulation of seven and overexpression of two miRNAs, with miR-24 showing the highest fold change (8). Kloting et al. (9) examined miRNA expression profiles in omental and subcutaneous fat deposits in control subjects and newly diagnosed diabetic patients. Interestingly, similar expression patterns were observed in both fat deposits for each patient, but differences were observed between patients based on parameters of obesity and metabolism. Zampetaki et al. (10) profiled miRNAs in the plasma of 800 patients in a prospective study of type 2 diabetic patients and established a diabetes signature based on ten miRNAs. They made similar observations in hyperglycemic leptin obese mice. Interestingly, they observed the decreased expression of six miRNAs several years prior to the onset of type 2 diabetes. This decreased expression correlated with subclinical peripheral artery disease, suggesting that such a signature may be used to identify high risk individuals prone to getting diabetes complications. Studies such as this are very important and have the potential to make important contributions toward earlier identification of high risk individuals and better management of the early disease. Further advances in profiling techniques using readily accessible biological fluids will lead to good advances in this important area.
The identification of single nucleotide polymorphisms (SNPs) in certain miRNAs (11) and, in particular, in miR-29 in type 1 diabetes is interesting given the potential role of miR-29 in renal fibrosis (see below). Whether these SNPs alter the regulation of miRNA transcription/stability and whether they are relevant on a broader scale in diabetes complications will need to be clarified.
A real advance in this area would be the confirmation of the previously identified miRNA signature (10) and further development to identify prediabetes in patients before they develop overt disease. Equally important would be the identification of a signature for the subset of diabetes patients that go on to develop complications.
miRNAS IN DIABETIC NEPHROPATHY
Diabetic nephropathy (DN) is the leading cause of kidney failure, affecting the glomerulus and ultimately leading to progressive kidney scarring. Fibrosis is characterized by the excess accumulation and deposition of extracellular matrix (ECM), the predominant proteins being collagens, but also fibronectin, fibrillin and elastin. ECM is synthesized by many cell types but the principal effector cells in the kidney are myofibroblasts, arising from renal tubular cells via epithelial to mesenchymal transition (EMT) and endothelial cells via endo-MT, bone marrow derived cells and resident interstitial fibroblasts (12). Blockade of specific steps of EMT dramatically reduces fibrotic lesions in models of kidney fibrosis, including diabetes, highlighting the role of some aspects of the EMT pathway in nephropathy (12).
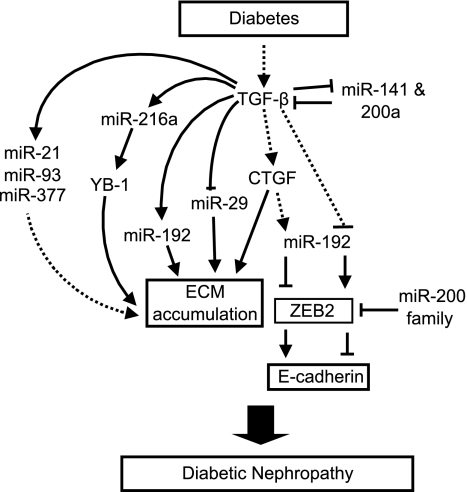
Recent attention has turned to the role of miRNAs in renal disease where Dicer knockout experiments have established the general importance of miRNA in normal renal development and function (13). Other studies have provided evidence linking prosclerotic factors and miRNAs to fibrosis (14–19) and have provided the much needed impetus in this area with the promise of new approaches for the treatment of fibrosis. Figure 1 shows the main miRNAs that have been reported to play a role in diabetic nephropathy and are outlined below.
FIG. 1.
Summarized here are the miRNAs known to have a role in the development and progression of diabetic nephropathy. A number of miRNAs have been identified as contributing to diabetes complications and in particular in diabetic nephropathy. Several of the miRNAs shown here have been directly connected to this disease via specific targets, while the specific details of others are yet to be clearly established. In the case of miR-192, the dual published roles are shown.
miR-192.
The role of miR-192 remains controversial and highlights the complex nature of miRNA research. Kato et al. (16) observed increased renal expression of miR-192 in streptozotocin (STZ)-induced diabetes and in the db/db mouse and demonstrated that tranforming growth factor (TGF-β1) upregulated miR-192 in mesangial cells (MCs). miR-192 repressed the translation of ZEB2, a transcriptional repressor that binds to the E-box in the collagen 1a2 (col1a2) gene (16). They proposed that miR-192 repressed ZEB2 and resulted in increased col1a2 expression in vitro and contributed to increased collagen deposition in vivo. These data suggest a role for miR-192 in the development of the matrix accumulation observed in DN. Interestingly, miR-192 and a number of other miRNAs have also been correlated with disease severity and progression in patients with IgA nephropathy (20) and hypertensive nephrosclerosis (21), suggesting that miR-192 may also play a role in other forms of renal disease. More recently, a tight association was observed between upregulation of miR-192 in the fibrotic kidney and TGF-β1/Smad signaling (22) with deletion of Smad7 resulting in increased miR-192 expression in the rat subtotal nephrectomy model. Importantly, this study also revealed the regulatory mechanisms by which TGF-β1 regulates miR-192 expression via the Smad3 but not the Smad2 signaling pathway. Indeed, overexpression of the miR-192 mimic promoted whereas knockdown of miR-192 inhibited TGF-β1–induced collagen expression.
In contrast to the above, other reports suggest the relationship between miR-192 and renal fibrosis may be more complicated. Krupa et al. (23) identified two miRNAs in human renal biopsies, the expression of which differed by more than twofold between progressors and nonprogressors with respect to DN, the greatest change occurring in miR-192 which was significantly lower in patients with advanced DN, correlating with tubulointerstitial fibrosis and low glomerular filtration rate. They also reported, in contrast to the Kato et al. (16) study in MCs, that TGF-β1 decreased expression of miR-192 in cultured proximal tubular cells (PTCs) (23). These investigators concluded that a decrease in miR-192 is associated with increased renal fibrosis in vivo. Our own work in this area confirms the view that loss of miR-192 is associated with renal fibrosis (18). TGF-β1 treatment induced decreased expression of miR-192 and miR-215, which target ZEB2, along with changes typical of EMT, including increased fibrogenesis and decreased E-cadherin expression in PTCs. Interestingly, connective tissue growth factor (CTGF) treatment also resulted in fibrogenesis but caused the induction of miR-192/215 and, consequently, decreased ZEB2 and increased E-cadherin. Ectopic expression of miR-192/215 resulted in increased E-cadherin by targeting ZEB2, but had no impact on fibrogenesis. Thus, our work demonstrates a clear link between miRNA-192/215 and ZEB2 in TGF-β1/CTGF-mediated changes in E-cadherin expression, independent of fibrosis (18).
The contrasting findings above highlight the complex nature of miRNA research. Some of the differences may relate to models and/or experimental conditions; however, one often overlooked explanation is that some effects of miRNAs and inhibitors are likely to be indirect in nature. Our understanding of how miRNAs function is continually evolving, with recent evidence demonstrating that miRNAs can also regulate gene expression via deadenylation and altering message stability, and by modulation of transcription. Bioinformatics using pathway analysis will be needed to better understand the cross-talk between factors that drive many downstream processes and how those processes ultimately impact the expression of individual genes.
The miR-200 family.
The next family of miRNAs to become a hot area of research in DN was the miR-200 family with researchers quickly making the obvious link between miR-200 and EMT. The pioneering work of Gregory et al. (14) established the role of the miR-200 family as regulators of the epithelial phenotype primarily by targeting ZEB1/2, the transcriptional repressors of E-cadherin. These observations established the important link between miRNA, the prosclerotic TGF-β pathways, and EMT in fibrosis. Both TGF-β1 and TGF-β2 decreased the expression of the miR-200 family in renal cells (18,19). This decrease is accompanied by an increase in expression of TGF-β2, the 3′ UTR of this prosclerotic cytokine being a target for miR-200a. This work demonstrated a feedback loop by which increased TGF-β2 causes decreased miR-200a, which in turn relieves repression of TGF-β2 expression, and hence promotes fibrosis. Interestingly, we also observed that miR-200a could decrease Smad3 activity and attenuate TGF-β–induced ECM protein synthesis, however whether this effect was direct was not determined (19). Identifying and deciphering the complex regulation of multiple targets and pathways impacted on by each miRNA remains a challenge, both in terms of understanding and using this knowledge for clinical benefit.
miR-29.
Recent reports have linked miR-29 to fibrosis, especially in renal disease. miR-29a has been shown to be important in the regulation of a number of collagens in a model of hypertensive renal disease (24) and miR-29b in the kidneys of salt-sensitive rats (24). Qin et al. (25) found that miR-29 was downregulated in the fibrotic kidney of obstructive nephropathy and was negatively regulated by TGF-β1 via the Smad3, but not the Smad2, signaling pathway. Indeed, Smad3 physically bound the miR-29 promoter and repressed miR-29 expression, thereby promoting collagen matrix expression. Our own unpublished work has also demonstrated significantly decreased expression of miR-29 family members in STZ-induced diabetic mice and rats. More importantly, the finding of inhibition of progressive renal fibrosis in obstructive nephropathy by ultrasound-microbubble–mediated miR-29b gene transfer demonstrated that miR-29 may be a novel therapeutic molecule for renal fibrosis (25).
miR-216a.
Kato et al. (26) have proposed miR-216a as another potential contributor to DN. miR-216a was upregulated by TGF-β1 in MCs, resulting in increased col1a2 expression. This involved an interaction between the col1a2 E-box and a complex of a number of molecules, one of which was shown to be regulated by the RNA-binding protein YB-1 (26). Kato et al. (26) postulated that increased miR-216a regulates the expression of Col1a2 by targeting YB-1 expression. Fraser et al. (27) demonstrated that YB-1 is a regulator of TGF-β1 mRNA translation in PTCs. Therefore, repression of YB-1 by miR-216a may also significantly contribute to fibrogenesis through increased TGF-β1. miR-216a and miR-217 have also been implicated by targeting the repression of the phosphatase and tensin homolog (PTEN) tumor suppressor gene, which results in activation of AKT, a key driver of DN (15,28). This example demonstrates the indirect and complex pathways by which miRNAs can affect important processes in cell physiology and biochemistry. One needs to add to this scenario the promiscuous nature of miRNAs and the multiple miRNA regulatory sites found on mRNAs to fully appreciate the tremendous complexity of miRNA regulation of cellular processes and disease.
miR-21.
Decreased expression of miR-21 has been observed early in DN, while ectopic expression of miR-21 inhibits MC proliferation under conditions of elevated glucose (29). Ectopic expression in vivo resulted in decreased albuminuria in diabetic db/db mice (29). PTEN is a target of and is decreased by miR-21, leading to increased p-Akt in MCs and db/db mice. How the opposing actions of decreased miR-21 (29) and increased miR-216a (15,28), which both target PTEN, contribute to renal fibrosis will need to be clarified. The relative contribution of each miRNA and the overall balance of miRNA expression needs to be established in order to make sense of the published data. Once again the contrasting experimental findings highlight the complex role of miRNAs in disease. We have recently observed that miR-21 expression was significantly upregulated in a mouse model of obstructive nephropathy and was also regulated by the TGF-β/Smad3 signaling pathway in vivo and in vitro. The findings that overexpression of miR-21 enhanced but that knockdown of miR-21 blocked renal fibrosis in vitro in response to TGF-β1 and in vivo in a mouse model of obstructive nephropathy suggests that miR-21 may also be an attractive target for the prevention of renal fibrosis under certain conditions (unpublished data, P.K., B.W., R.M.C., and H.Y.L.).
miR-377.
miR-377 was found to be elevated in spontaneous and STZ-induced models of DN (30). This study also found miR-377 to be elevated in MCs following TGF-β1 treatment in high glucose conditions. Ectopic expression of miR-377 in MCs resulted in increased fibronectin expression as well as other genes that contribute to DN. Fibronectin is not a direct target of miR-377, however miR-377 targets the expression of PAK1 and MnSOD, which indirectly lead to elevated fibronectin expression (30) and hence contribute to DN. The overall contribution of miR-377 to DN remains to be determined.
miR-93.
Vascular endothelial growth factor (VEGF) has been implicated in microvascular complications and is considered to be a significant player in diabetic retinopathy and nephropathy where it is often upregulated (31,32). Anti-VEGF treatment in animal models of diabetes showed significant improvement in kidney function (33). Long et al. (34) recently identified miR-93 to be a “signature miRNA” in hyperglycemic conditions both in vitro and in vivo, and that VEGF-A was a target of miR-93. They reported that hyperglycemia led to reduced miR-93 expression by downregulating the promoter of the host gene MCM7 in which miR-93 resides. The authors demonstrated that increased miR-93 expression prevented the hyperglycemia-induced expression of VEGF and its downstream signaling. Conversely, targeting miR-93 in vivo with morpholino oligomers induced VEGF expression (34).
DIABETIC RETINOPATHY
There are currently no studies linking miRNAs to diabetic retinopathy (DR). It is generally believed that the hyperglycemic environment and elevated expression of several growth factors are key mediators of DR. Of particular importance is VEGF which has been shown to be elevated in DR, resulting in pathogenic changes to the retinal structure, including neovascularization (32,35). Anti-VEGF therapy appears to be promising, although the findings of large trials are awaited to confirm these early findings (36). Given that VEGF is a predicted target of several miRNAs, including those that have already been implicated in other diabetes complications (miR-200b/c, miR-29a/b/c, and miR-93), it would be interesting to determine whether these miRNAs may also be relevant in DR.
DIABETIC ATHEROSCLEROSIS
Advanced glycation end products (AGEs) act through the receptor for AGEs (RAGE), activating inflammatory pathways. One of the endogenous physiological ligands of RAGE is S100b, a member of the S100/calgranulin family (37), and interaction of these ligands with RAGE is critical in inflammation, including in diabetic atherosclerosis (38). Shanmugam et al. (39) recently reported that expression and binding of miR-16 to the COX-2 mRNA 3′UTR in THP-1 monocytic cells was inhibited by the binding of S100b to the RAGE receptor. In this case, the binding of S100b displaced the nuclear heterogeneous nuclear ribonucleoprotein K such that it translocates to the cytoplasm, to interact with the COX-2 3′UTR, to interfere with the binding of miR-16. The complexity of such interactions serves to remind us of the various layers of regulation that need to be understood before our knowledge of miRNAs can be used to produce better therapeutic outcomes in diabetes complications. More recent work has identified increased miR-503 as an important contributor to diabetes-induced impairment of endothelial function and angiogenesis by directly targeting CCNE1 and cdc25A (40). Whether this miRNA plays a role in diabetic atherosclerosis remains to be established.
HEART
miR-133.
Diabetic cardiomyopathy, although not fully characterized, typically displays hypertrophy of the heart and contractile dysfunction, which eventually leads to heart failure. Much attention has focused on miR-133, since it is abundantly expressed in heart and is a regulator of myogenesis (41). In the diabetic context, miR-133 has been postulated to be a significant contributor to cardiac pathology. The role of miR-133 in the diabetic heart is very interesting as it appears to have a dual role depending on whether expression is increased or decreased. How the balance between the two opposing actions of miR-133 works in diabetes remains an intriguing mystery.
Elevated cardiac expression of miR-133 is dependent on the serum response factor (SRF), which is elevated in the diabetic heart (42). Knockdown of SRF prevents increased expression of miR-133 (42), and cardiac specific dysregulation of SRF expression results in cardiac hypertrophy and other cardiac pathologies similar to those observed in the early stages of congestive heart failure. One consequence of increased cardiac expression of miR-133 is a perturbation to cardiac conductivity (42). An important electrical disturbance that is seen in diabetic patients is the prolongation of the QT interval, which represents the duration for ventricular depolarization and repolarization of myocytes. This cycle is controlled by the flow of ion currents through K+ channels, which when perturbed result in what is known as the long QT syndrome (LQTS) (42), potentially a life threatening disorder. The rapid delayed rectifier K+ current (IKr) is regulated by the human ERG gene, which is downregulated in cardiac myocytes in diabetes, resulting in prolonged repolarization and therefore long QT syndrome (43). The 3′UTR of the ERG mRNA is a target of miR-133 (42).
Decreased cardiac expression of miR-133 has also been associated with cardiac hypertrophy (44). In this condition, a thickening of the heart muscle results in smaller ventricular size and reduced cardiac output, a phenomenon observed with hypertension, the prevalence of which is increased in subjects with diabetes. Consistent with these observations, overexpression of miR-133 inhibits cardiac hypertrophy whereas inhibition of miR-133 induces more pronounced hypertrophy (44). Indeed, Feng et al. (45) have demonstrated decreased miR-133a in the diabetic mouse heart. Furthermore, in vitro exposure of cardiomyocytes to high glucose resulted in reduced miR-133a and more hypertrophic changes. The genes that are dysregulated in this case are RhoA, Cdc42 and WHSC2, all of which are known to be important in cardiac hypertrophy (44).
miR-320.
Recent work by Wang et al. (46) identified elevated miR-320 as a potential mediator of several angiogenic factors in diabetic myocardial microvascular endothelial cells as compared with control cells. The factors regulated by this miRNA include VEGF, fibroblast growth factor, IGF-1, and IGF-1R—all of which are particularly relevant to diabetic cardiomyopathy. Increased expression of miR-320 and consequently decreased expression of IGF-1 and IGF-1R are likely to play a role in the impaired angiogenesis observed in diabetes.
miR-29.
The work of van Rooij et al. (17) demonstrated a role for miR-29 in human and mouse hearts during myocardial infarction. Ectopic expression of miR-29b was shown to decrease collagen expression in cardiac fibroblasts. Conversely, miR-29 inhibitors delivered by tail vein injection were able to decrease collagen expression not only in the heart, but also in the liver and kidney. Whether dysregulation of the miR-29 family members occurs in the diabetic heart as in the kidney remains to be established.
miR-30.
CTGF expression is important in the heart where expression levels correlate with collagen levels and fibrosis. Liu et al. (47) reported that miR-133 knockout mice develop severe cardiac fibrosis. Both miR-30 and miR-133 target CTGF for repression (48) and may be relevant since elevated CTGF levels are seen in the context of diabetes, including within the heart (49).
miR-21.
Enhanced expression of miR-21 in the failing heart is selectively expressed in cardiac fibroblasts (50) and is increased in association with elevated extracellular signal-regulated kinase (ERK) signaling by downregulating the ERK inhibitor, Spry1. The ultimate result of activated ERK in cardiac fibroblasts is elevated collagen expression. In vivo silencing of miR-21 caused decreased ERK kinase activity and reduced interstitial fibrosis. Increased expression of miR-21 is found in the infarct zone in cardiac fibroblasts (51). Despite the importance of miR-21 in DN, it remains to be determined how relevant this miRNA is to complications in the diabetic heart.
WHAT DOES THE FUTURE HOLD?
Despite considerable progress in further understanding the link between miRNAs, development, physiology, and disease, what is becoming clearer is the ever-increasing complexity of the interactions between miRNAs and mediators of signaling pathways that are relevant to diabetes and particularly to its complications. It is becoming clearer that a number of factors need to be taken into account with miRNA research and that the study of individual miRNAs, which are interesting in their own right, should not be interpreted in isolation. While a number of miRNAs have been associated with diabetes complications (Table 1), the challenge remains to establish the role of these miRNAs in clinical disease and to identify the regulatory mechanisms or pathways related to miRNA expression in the pathogenesis of the various disorders. The development of new genetic miRNA mouse models will greatly facilitate advances in this exciting area and help to address some of the unanswered questions regarding the regulation of signaling pathways that emanate from many of the in vitro observations. There is also much commercial interest in the development of technologies to block or enhance miRNA expression in vivo as a new approach to the management of human disease, including the treatment of diabetes complications.
Two significant challenges remain a barrier to taking advantage of the knowledge currently gained in experimental studies to the next stage where they can be used clinically.
In recent times, the focus has moved toward a number of specific miRNAs that are emerging as potential therapeutic targets either by restoring expression or inhibition. Chemical advances in the area of delivery of miRNA inhibitors including cholesterol modifications and locked nucleic acid–modified anti-miRNAs have enabled efficient delivery of these molecules to various tissues (52,53), the stability of the inhibitors ranging up to 10 weeks in vivo following a single injection (17). However, in terms of in vivo delivery of miRNA mimics, huge challenges remain because mimics have a very short half-life and chemical modification alters their biological properties. Much of the literature describing such studies with mimics needs to be carefully evaluated. Some options being investigated include viral mediated delivery of mimics. This approach has been used in different models with a moderate degree of success (44,54–56).
A second obstacle relates to the targeting of specific tissues. Most approaches to date have involved nonspecific systemic delivery. The use of adeno-associated viral serotypes can improve specific delivery to some tissues (57). More recent attention has turned to the development of virally delivered tissue specific promoter driven miRNA expression. The development of therapeutic strategies to effectively deliver the miRNAs or inhibitors into the diseased tissues is urgently needed.
Despite the challenges for miRNAs as either a therapy or a target for new therapies, the use of miRNAs holds much promise as a new treatment, since these molecules are naturally occurring endogenous regulators of cell processes that are often dysregulated in disease. With improved understanding of the mechanisms whereby miRNAs repress gene expression and the development of more selective and stable mimics and inhibitors of miRNAs, this could lead to better approaches to address the “gene expression defects” that occur in various diseases, including diabetes complications. Where dysregulation of specific miRNA expression is linked to disease, the development of better targeted delivery systems is likely to lead to tailored treatment of patients. With the emergence of specific miRNAs that are common to a number of complications such as mir-29, which is implicated in organ fibrosis, the development of a more generalized approach to treat complications with the administration of a small cocktail of miRNAs or inhibitors becomes feasible. Indeed, profiling experiments to identify patterns of miRNA dysregulation that may be common to several complications would be a useful first step in this direction.
ACKNOWLEDGMENTS
This study was supported by a Centre Grant from the Juvenile Diabetes Research Foundation, the National Health and Medical Research Council of Australia (NHMRC526663), and the Research Grants Council of Hong Kong (RGC GRF 768207 and CUHK5/CRF09).
No potential conflicts of interest relevant to this article were reported.
P.K. researched data and wrote the manuscript. B.W. researched data and reviewed and edited the manuscript. R.M.C. researched data and reviewed and edited the manuscript. H.Y.L. researched data and reviewed and edited the manuscript.
Footnotes
See accompanying perspective, p. 1825.
REFERENCES
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843–854 [DOI] [PubMed] [Google Scholar]
- 2.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993;75:855–862 [DOI] [PubMed] [Google Scholar]
- 3.van Rooij E. The art of microRNA research. Circ Res 2011;108:219–234 [DOI] [PubMed] [Google Scholar]
- 4.Gregory RI, Chendrimada TP, Shiekhattar R. MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol Biol 2006;342:33–47 [DOI] [PubMed] [Google Scholar]
- 5.Lu M, Zhang Q, Deng M, et al. An analysis of human microRNA and disease associations. PLoS ONE 2008;3:e3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutvagner G. MicroRNAs and cancer: issue summary. Oncogene 2006;25:6154–6155 [Google Scholar]
- 7.He A, Zhu L, Gupta N, Chang Y, Fang F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol Endocrinol 2007;21:2785–2794 [DOI] [PubMed] [Google Scholar]
- 8.Huang B, Qin W, Zhao B, et al. MicroRNA expression profiling in diabetic GK rat model. Acta Biochim Biophys Sin (Shanghai) 2009;41:472–477 [DOI] [PubMed] [Google Scholar]
- 9.Klöting N, Berthold S, Kovacs P, et al. MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS ONE 2009;4:e4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zampetaki A, Kiechl S, Drozdov I, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 2010;107:810–817 [DOI] [PubMed] [Google Scholar]
- 11.Glinsky GV. An SNP-guided microRNA map of fifteen common human disorders identifies a consensus disease phenocode aiming at principal components of the nuclear import pathway. Cell Cycle 2008;7:2570–2583 [DOI] [PubMed] [Google Scholar]
- 12.Kalluri R. EMT: when epithelial cells decide to become mesenchymal-like cells. J Clin Invest 2009;119:1417–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li JY, Yong TY, Michael MZ, Gleadle JM. Review: the role of microRNAs in kidney disease. Nephrology (Carlton) 2010;15:599–608 [DOI] [PubMed] [Google Scholar]
- 14.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008;10:593–601 [DOI] [PubMed] [Google Scholar]
- 15.Kato M, Putta S, Wang M, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol 2009;11:881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato M, Zhang J, Wang M, et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA 2007;104:3432–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA 2008;105:13027–13032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang B, Herman-Edelstein M, Koh P, et al. E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-β. Diabetes 2010;59:1794–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B, Koh P, Winbanks C, et al. miR-200a Prevents renal fibrogenesis through repression of TGF-β2 expression. Diabetes 2011;60:280–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang G, Kwan BC, Lai FM, et al. Intrarenal expression of microRNAs in patients with IgA nephropathy. Lab Invest 2010;90:98–103 [DOI] [PubMed]
- 21.Wang G, Kwan BC, Lai FM, et al. Intrarenal expression of miRNAs in patients with hypertensive nephrosclerosis. Am J Hypertens 2010;23:78–84 [DOI] [PubMed]
- 22.Chung AC, Huang XR, Meng X, Lan HY. miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc Nephrol 2010;21:1317–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krupa A, Jenkins R, Luo DD, Lewis A, Phillips A, Fraser D. Loss of microRNA-192 promotes fibrogenesis in diabetic nephropathy. J Am Soc Nephrol 2010;21:438–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Taylor NE, Lu L, et al. Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension 2010;55:974–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin W, Chi-Kong A, Huang XR, Meng X, Lan HY. miR-29 Inhibits TGF-beta/Smad3-Mediated Renal Fibrosis In Vitro and In Vivo. In 43rd Annual Meeting of the American Society of Nephrology, Denver, Colorado, 2010, p. TH-FC062 [Google Scholar]
- 26.Kato M, Wang L, Putta S, et al. Post-transcriptional up-regulation of Tsc-22 by Ybx1, a target of miR-216a, mediates TGF-beta-induced collagen expression in kidney cells. J Biol Chem 2010;285:34004–34015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraser DJ, Phillips AO, Zhang X, et al. Y-box protein-1 controls transforming growth factor-beta1 translation in proximal tubular cells. Kidney Int 2008;73:724–732 [DOI] [PubMed] [Google Scholar]
- 28.Xin X, Chen S, Khan ZA, Chakrabarti S. Akt activation and augmented fibronectin production in hyperhexosemia. Am J Physiol Endocrinol Metab 2007;293:E1036–E1044 [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Peng H, Chen J, et al. MicroRNA-21 protects from mesangial cell proliferation induced by diabetic nephropathy in db/db mice. FEBS Lett 2009;583:2009–2014 [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Wang Y, Minto AW, et al. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J 2008;22:4126–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aiello LP, Avery RL, Arrigg PG, et al. Vascular endothelial growth factor in ocular fluid of patients with diabetic retinopathy and other retinal disorders. N Engl J Med 1994;331:1480–1487 [DOI] [PubMed] [Google Scholar]
- 32.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature 2001;414:813–820 [DOI] [PubMed] [Google Scholar]
- 33.Flyvbjerg A, Dagnaes-Hansen F, De Vriese AS, Schrijvers BF, Tilton RG, Rasch R. Amelioration of long-term renal changes in obese type 2 diabetic mice by a neutralizing vascular endothelial growth factor antibody. Diabetes 2002;51:3090–3094 [DOI] [PubMed] [Google Scholar]
- 34.Long J, Wang Y, Wang W, Chang BH, Danesh FR. Identification of microRNA-93 as a novel regulator of vascular endothelial growth factor in hyperglycemic conditions. J Biol Chem 2010;285:23457–23465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan ZA, Farhangkhoee H, Chakrabarti S. Towards newer molecular targets for chronic diabetic complications. Curr Vasc Pharmacol 2006;4:45–57 [DOI] [PubMed] [Google Scholar]
- 36.Jardeleza MS, Miller JW. Review of anti-VEGF therapy in proliferative diabetic retinopathy. Semin Ophthalmol 2009;24:87–92 [DOI] [PubMed] [Google Scholar]
- 37.Hofmann MA, Drury S, Fu C, et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell 1999;97:889–901 [DOI] [PubMed] [Google Scholar]
- 38.Bucciarelli LG, Wendt T, Qu W, et al. RAGE blockade stabilizes established atherosclerosis in diabetic apolipoprotein E-null mice. Circulation 2002;106:2827–2835 [DOI] [PubMed] [Google Scholar]
- 39.Shanmugam N, Reddy MA, Natarajan R. Distinct roles of heterogeneous nuclear ribonuclear protein K and microRNA-16 in cyclooxygenase-2 RNA stability induced by S100b, a ligand of the receptor for advanced glycation end products. J Biol Chem 2008;283:36221–36233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caporali A, Meloni M, Völlenkle C, et al. Deregulation of microRNA-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation 2011;123:282–291 [DOI] [PubMed] [Google Scholar]
- 41.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 2005;436:214–220 [DOI] [PubMed] [Google Scholar]
- 42.Xiao J, Luo X, Lin H, et al. MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts. J Biol Chem 2007;282:12363–12367 [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Xiao J, Lin H, et al. Ionic mechanisms underlying abnormal QT prolongation and the associated arrhythmias in diabetic rabbits: a role of rapid delayed rectifier K+ current. Cell Physiol Biochem 2007;19:225–238 [DOI] [PubMed] [Google Scholar]
- 44.Carè A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med 2007;13:613–618 [DOI] [PubMed] [Google Scholar]
- 45.Feng B, Chen S, George B, Feng Q, Chakrabarti S. miR133a regulates cardiomyocyte hypertrophy in diabetes. Diabetes Metab Res Rev 2010;26:40–49 [DOI] [PubMed] [Google Scholar]
- 46.Wang XH, Qian RZ, Zhang W, Chen SF, Jin HM, Hu RM. MicroRNA-320 expression in myocardial microvascular endothelial cells and its relationship with insulin-like growth factor-1 in type 2 diabetic rats. Clin Exp Pharmacol Physiol 2009;36:181–188 [DOI] [PubMed] [Google Scholar]
- 47.Liu N, Bezprozvannaya S, Williams AH, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev 2008;22:3242–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duisters RF, Tijsen AJ, Schroen B, et al. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res 2009;104:170-178 [DOI] [PubMed]
- 49.Candido R, Forbes JM, Thomas MC, et al. A breaker of advanced glycation end products attenuates diabetes-induced myocardial structural changes. Circ Res 2003;92:785–792 [DOI] [PubMed] [Google Scholar]
- 50.Thum T, Gross C, Fiedler J, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008;456:980–984 [DOI] [PubMed] [Google Scholar]
- 51.Roy S, Khanna S, Hussain SR, et al. MicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovasc Res 2009;82:21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stenvang J, Silahtaroglu AN, Lindow M, Elmen J, Kauppinen S. The utility of LNA in microRNA-based cancer diagnostics and therapeutics. Semin Cancer Biol 2008;18:89–102 [DOI] [PubMed] [Google Scholar]
- 53.Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005;438:685–689 [DOI] [PubMed] [Google Scholar]
- 54.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA 2010;107:6328–6333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christensen M, Larsen LA, Kauppinen S, Schratt G. Recombinant adeno-associated virus-mediated microRNA delivery into the postnatal mouse brain reveals a role for miR-134 in dendritogenesis in vivo. Front Neural Circuits 2010;3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature 2009;457:426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther 2008;16:1073–1080 [DOI] [PubMed] [Google Scholar]