Abstract
A number of human malignancies exhibit sustained stimulation, mutation, or gene amplification of the receptor tyrosine kinase human mesenchymal-epithelial transition factor (c-Met). ARQ 197 is a clinically advanced, selective, orally bioavailable, and well tolerated c-Met inhibitor, currently in Phase 3 clinical testing in non-small cell lung cancer patients. Herein, we describe the molecular and structural basis by which ARQ 197 selectively targets c-Met. Through our analysis we reveal a previously undisclosed, novel inhibitory mechanism that utilizes distinct regulatory elements of the c-Met kinase. The structure of ARQ 197 in complex with the c-Met kinase domain shows that the inhibitor binds a conformation that is distinct from published kinase structures. ARQ 197 inhibits c-Met autophosphorylation and is highly selective for the inactive or unphosphorylated form of c-Met. Through our analysis of the interplay between the regulatory and catalytic residues of c-Met, and by comparison between the autoinhibited canonical conformation of c-Met bound by ARQ 197 to previously described kinase domains of type III receptor tyrosine kinases, we believe this to be the basis of a powerful new in silico approach for the design of similar inhibitors for other protein kinases of therapeutic interest.
Keywords: Drug Design, Kinetics, Protein Phosphorylation, Protein Structure, Receptor Tyrosine Kinase, X-ray Crystallography, c-Met, Kinase Autoinhibition, Noncompetitive, Small Molecule Protein Inhibitor
Introduction
The human mesenchymal-epithelial transition factor (c-Met)2 pathway is one of the most frequently dysregulated pathways in human cancer (1, 2). An activated c-Met signaling pathway promotes tumor cell growth, survival, migration, and invasion, as well as tumor angiogenesis and metastasis. The aberrant activation of c-Met in many human cancers is due to met gene amplification, transcriptional up-regulation, point mutations, or ligand-mediated (hepatocyte growth factor) autocrine or paracrine stimulation (3, 4). As a result, c-Met has attracted considerable attention as a potential target for therapeutic intervention in oncology (3, 5). Small molecule c-Met inhibitors and therapeutic monoclonal antibodies that inhibit c-Met activity have exhibited anti-tumor activity in preclinical models (6). ARQ 197 (Fig. 1A) is a low molecular weight, orally bioavailable, selective inhibitor of c-Met (7, 8). It has been shown to arrest c-Met-dependent downstream signaling by disrupting both constitutive and ligand-mediated c-Met phosphorylation. ARQ 197 inhibits c-Met activation across a range of human tumor cell lines and shows anti-tumor activity in several human tumor xenografts (7). In clinical studies to date, ARQ 197 has been well tolerated and has yielded encouraging clinical responses including prolonged stable disease across a range of human tumors either alone or in combination with other agents (9, 10). In a subset of patients with non-small cell lung cancer, met gene amplification is associated with both de novo and acquired resistance to pharmacologic EGFR inhibition (11). Recent results from a randomized Phase II trial of ARQ 197 in combination with erlotinib demonstrated a 66% improvement in median progression-free survival in patients with advanced refractory non-small cell lung cancer when compared with patients treated with erlotinib alone (9).
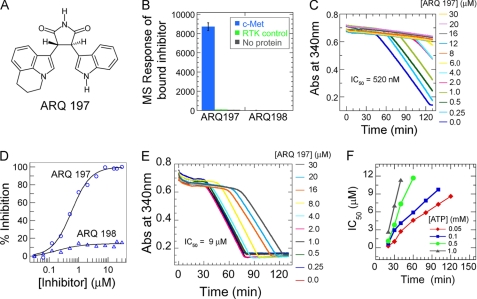
FIGURE 1.
Analysis of the binding of ARQ 197 to the unphosphorylated c-Met and its inhibitory activity. A, chemical structure of ARQ 197. B, the relative affinity of the two enantiomers (ARQ 197 and ARQ 198, 20 μm) to the inactive c-Met (14 μm) as measured by indirect affinity mass spectrometry. Selectivity of the binding was probed by comparison of the binding to inactive FGFR2 (RTK control). C, inhibition of c-Met autophosphorylation by ARQ 197. The enzyme progress curve for c-Met autophosphorylation was monitored by a continuous spectrophotometric assay at various inhibitor concentrations (shown on right side). The reaction was followed at 0.1 mm ATP, the inhibition was observed by shifts in induction time followed by a decrease in absorption. The rate of activation was calculated from the slopes within 20 min of the induction time (lag phase) of autophosphorylation of c-Met without inhibitor. Fitting of the c-Met inhibition data at various ARQ 197 concentrations gave an IC50 value of 520 nm. D, ARQ 197 enantioselectively inhibits c-Met kinase activity. The inhibitory activity of these two compounds was measured in the c-Met autophosphorylation reaction. Although ARQ 197 displayed a potency of 520 nm, ARQ 198 showed only a very weak inhibition when measured up to a 40 μm concentration. E, inhibition of the autophosphorylation reaction by ARQ 197 is dependent on c-Met activation state. The potency of inhibition (IC50) was measured at various concentrations of ATP. The progress curve is shown for c-Met inhibition with various concentrations of ARQ 197 at 1 mm ATP, and F, the IC50 values were calculated from the slopes of the curves derived at constant time points after the first lag phase of the control (no inhibitor). Similarly, the IC50 values were also calculated at a segment of the 10-min time intervals at each ATP concentration across the length of the progress curve.
ARQ 197 was initially identified as being of potential therapeutic interest in cell-based systems. Using classical enzyme kinetics analyses, ARQ 197 was subsequently characterized as non-ATP competitive (7). Because ARQ 197 is not competitive with ATP and retains its potency in cells, we sought to characterize the molecular basis for this behavior by using biophysical, biochemical, and structural studies.
Our results demonstrate that ARQ 197 recognizes the canonical autoinhibited conformation of c-Met and selectively inhibits the inactive, unphosphorylated form of this kinase. These findings advance our understanding of how c-Met signaling can be inhibited by small molecules. Furthermore, ARQ 197 represents a novel class of kinase inhibitor, and by understanding its mode of inhibition, it is predicted that inhibitors of other kinases of interest that target the inactive or autoinhibited form can be discovered.
EXPERIMENTAL PROCEDURES
Expression and Purification of Unphosphorylated Form of c-Met
cDNA of full-length c-Met purchased from Origen Technologies was used as a template for PCR amplification. The cloning and expression protocol was adopted from that previously reported (12). The DNA fragment encoding the kinase domain (1038–1346) was inserted into Novagen vector pET28a between the NcoI and SalI sites. The primers were designed to contain a His6 tag to the N terminus. To express unphosphorylated c-Met kinase protein, a tyrosine phosphatase PTP1B-(1–283) was sequentially ligated into the construct between the SalI and NotI sites. A second ribosome binding site was incorporated in the PTP1B primer after the SalI site. The resulting construct produced N-terminal His-tagged fusion kinase protein expressed in Codon-Plus BL21 (DE3)RIL Escherichia coli cells (Stratagene). Cells were grown in 2× YT broth (MP Biomedicals), cultured up to 0.8 absorbance units at 600 nm at 25 °C, and induced with 0.25 mm of isopropyl 1-thio-β-d-galactopyranoside overnight at 12 °C. The co-expressed protein was purified by metal chelation chromatography followed by anion and cation exchange columns. In brief, cell pellets from 9 liters of culture medium was suspended in 50 mm Tris, pH 8.5, 150 mm NaCl, 10% glycerol, 25 mm imidazole, pH 8.5, 1 mm PMSF. The cell suspension was lysed by sonication and 0.5% Triton X-100 was added to the lysate before centrifugation. A clear supernatant was obtained by centrifugation at 50,000 × g for 45 min and was passed onto the nickel-nitrilotriacetic acid column beads (Invitrogen) at 4 °C. The column was washed with a high salt buffer (25 mm Tris, pH 8.5, 0.5 m NaCl, 25 mm imidazole), and the protein was eluted with 300 mm imidazole, pH 8.5, 100 mm NaCl, and 7.5% glycerol. Following concentration using Amicon ultrafiltration centrifugal tubes (30 kDa molecular mass cutoff), the protein was dialyzed in a buffer containing 25 mm Tris, pH 8.5, 10% glycerol, 0.1% 2-mercaptoethanol for 5 h at 4 °C. The dialyzed protein was purified using QFF-ion exchange cartridge (GE Healthcare) and eluted using buffer containing a salt gradient of 0–0.3 m NaCl. The c-Met protein was further purified using size exclusion chromatography on a Superdex 200 column and eluted with 25 mm Tris-HCl, pH 8.5, 100 mm NaCl, 10% glycerol, and 0.1% 2-mercaptoethanol. The c-Met protein was concentrated to 15 mg/ml and stored at −80 °C. The resulting c-Met preparations were analyzed for their degree of phosphorylation by mass spectrometry and confirmed as fully unphosphorylated.
Indirect Affinity Mass Spectrometry Assay
The relative affinity of ARQ 197, and its c-Met inactive enantiomer ARQ 198, for the unphosphorylated c-Met protein and for a control type III RTK kinase domain (FGFR2) was measured by indirect affinity mass spectrometry (13). Briefly, binding mixtures were 25 μl in volume and contained 14 μm protein, 20 μm inhibitor in 25 mm Tris-HCl, pH 7.5, 100 mm NaCl, 0.1% 2-mercaptoethanol, and a 2% final DMSO concentration. Protein and inhibitors were incubated for 1 h at room temperature, and cooled briefly on ice prior to the separation step. Protein-bound inhibitor was separated from the unbound inhibitor by fast centrifugation at 4 °C through a size exclusion gel in a 96-well plate format. A control containing 20 μm inhibitor in buffer was used to verify that no inhibitor passed through the gel in the absence of protein carrier. Eluent from the size exclusion column (∼25 μl) was mixed with 30 μl of internal standard in DMSO:H2O (1:1) and analyzed for the presence of small molecular weight inhibitor by LC/MS. Sample was injected onto a Waters 2795 HPLC equipped with an Atlantis C18 column (2.1 × 30 mm, 3 μm) and separated at 0.5 ml/min by a water/acetonitrile gradient with 0.1% formic acid as modifier starting from 10% and reaching 90% acetonitrile in 5 min, and re-equilibrating in 10% acetonitrile for another 3 min. HPLC flow was directed to a triple quadrupole mass spectrometer equipped with an ESI probe, which was operated in single ion reaction monitoring mode, with the following parameters; capillary voltage 3.5 V, cone voltage 20 V, source temperature 120 °C, desolvation temperature 450 °C, dwell time of 0.5 s, and unit mass resolution on each quadrupole.
Continuous Spectrophotometric Assay
Kinase Autophosphorylation Assays
Kinase activity was monitored using a continuous spectrophotometric assay as described previously (14). In this assay, the consumption of ATP is coupled via the pyruvate kinase:lactate dehydrogenase enzyme pair to the oxidation of NADH, which is monitored as the decrease in absorption at 340 nm. Reaction mixtures contained 25 mm Tris, pH 8.0, 100 mm NaCl, 10 mm MgCl2, 1 mm phosphoenolpyruvate, 0.28 mm NADH, 89 units/ml of pyruvate kinase, 124 units/ml of lactate dehydrogenase, and 2% DMSO. The enzyme and various concentrations of ARQ 197 were incubated for 30 min at 4 °C. The ADP-coupling reaction was initiated by the addition of ATP to the assay mixtures containing enzyme and ARQ 197 complex in a total reaction volume of 50 μl. The assay was carried out in 384-well plates, and the rate of absorbance decrease at 340 nm was monitored using a Tecan Safire II instrument at 30 °C. For IC50 determinations, the autophosphorylation reaction was carried out at 1 μm enzyme concentration and 0.1 mm ATP, and the linear slope of the biphasic curve after the lag phase was used to calculate IC50 values. The effect of ATP concentration on the kinetics of c-Met autophosphorylation was monitored by measuring the enzyme activity at various ATP concentrations at a fixed c-Met concentration of 1 μm.
Substrate Phosphorylation Assays
The substrate phosphorylation reaction was measured with 0.5 μm c-Met, 50 μm Pyk2 peptide (AGAGSIESDIYAEIPDETC), 0.1 mm ATP, and 10 mm MgCl2. The enzyme inhibitory activity of unphosphorylated c-Met (0.5 μm) was followed by adding previously incubated enzyme and ARQ 197 complex to an assay mixture containing ADP-coupled reaction mixtures with substrate peptide. The assay was initiated by the addition of 0.1 mm ATP and the decrease in absorbance was measured at 340 nm. The activated form of c-Met was prepared by preincubating c-Met (10 μm) with 0.5 mm ATP and 10 mm MgCl2 for 1 h at 25 °C. The extent of c-Met phosphorylation was assessed by MS analysis. For enzyme inhibition assays, the fully phosphorylated c-Met was diluted with buffer (25 mm Tris, pH 8.5, 100 mm NaCl, and 0.1% 2-mercaptoethanol) to a final concentration of 0.5 μm and incubated with various concentrations of ARQ 197 at 4 °C. The ADP-coupling reaction was initiated by the addition of 0.5 mm ATP to assay mixtures containing enzyme (0.5 μm final concentrations) and ARQ 197 complex and 50 μm Pyk2 peptide. The reaction was monitored by following the decrease in absorbance in a microplate reader at 30 °C.
c-Met Autophosphorylation Monitored by Mass Spectrometry
c-Met (1 μm) was incubated with ARQ 197 (5 and 20 μm) in the presence of ATP (0.1 and 1 mm) in 25 mm Tris buffer, pH 7.5, containing 2 mm DTT, 5% glycerol, and 2% DMSO. Reactions were stopped at different time intervals by addition of EDTA. The progress of c-Met autophosphorylation was measured by monitoring the increase of the mass of the intact protein using a Q-TOF mass spectrometer. Aliquots of the phosphorylation reaction were separated on a BEH C18 column (1 × 50 mm, 1.7 μm, Waters) using an acetonitrile/H2O gradient with 0.1% formic acid as modifier, and the eluent was directed to a Q-TOF Premier mass spectrometer equipped with the ESI probe. MS spectra were deconvoluted by the MaxEnt1 software package (Masslynx, Waters).
Substrate Phosphorylation Monitored by Mass Spectrometry
In one set of experiments, unphosphorylated c-Met (0.5 μm) was preincubated with ARQ 197 (20 μm) for 1 h. Pyk2 peptide (40 μm) and ATP (0.1 mm) were added to initiate the phosphorylation reaction. The reaction was stopped at various time intervals by addition of EDTA, and aliquots were analyzed using an Acquity/Q-TOF system. The phosphorylation time course of the Pyk2 peptide was monitored by the increase of the peptide mass due to the phosphate addition. In a second set of experiments, c-Met (0.5 μm) was preactivated by incubation with ATP (0.5 mm) for 1 h. Activated c-Met was treated with ARQ 197 (20 μm) for another 1 h, followed by addition of Pyk2 peptide (40 μm) and ATP (0.5 mm). Pyk2 phosphorylation was measured as described above.
Crystallization and Structure Determination of c-Met·ARQ197 Complex
For co-crystallization, unphosphorylated c-Met was diluted to 10 mg/ml with a buffer of 25 mm Tris, pH 8.5, 100 mm NaCl, 5% glycerol, 0.1% 2-mercaptoethanol, and ARQ 197 was added from a 50 mm (100% DMSO) stock solution to a final concentration of 1 mm. Thin needle crystals were obtained at 4 °C by vapor diffusion using 13% ethanol, 12% ethylene glycol, 100 mm imidazole, pH 8.5. Microseeding was employed to generate single diffraction quality crystals. Crystals were flash frozen in liquid nitrogen in the presence of well solution supplemented with 30% ethylene glycol. The diffraction data were collected at NSLS X29 beamline synchrotron x-ray source at 100 K. The crystals were indexed to the triclinic space group with two molecules in the asymmetric unit and a solvent content of 45%. The raw data were processed and integrated using MOSFLM, and the intensities were sorted and merged with SCALA. The structure was solved by molecular replacement using the program Phaser as implemented in CCP4 (15) using a search model of the apo-c-Met kinase crystal structure (PDB code 2G15). The critical structural elements, the A-loop (residues 1222–1247) and the glycine-rich loop (residues 1086–1091) were deleted from the search model. After a few initial rounds of automated model building and refinement using Arp/wARP (16), the structure was refined using iterative cycles of model building in COOT, followed by simulated annealing using CNX, restrained refinement using REFMAC (Table 1). Molecular graphics were rendered with PyMol (17).
TABLE 1.
Data collection and refinement statistics
| Data collection | |
|---|---|
| Resolution, Å | 50–1.94 |
| Space group | P1 |
| Unit cell parameters | |
| a, Å | 53.470 |
| b, Å | 58.670 |
| c, Å | 64.960 |
| α, ° | 88.410 |
| β, ° | 68.100 |
| γ, ° | 85.520 |
| Content of the asymmetric unit | 2 |
| No. of measured reflections | 508,319 |
| No. of unique reflections | 52,006 (1662)a |
| Data redundancy | 3.8 (3.8) |
| Data completeness, % | 94.5 (96.4) |
| Rsym | 0.064 (0.309) |
| I/σ | 10.4 |
| Refinement | |
| Rfactor/Rfree | 20.3/25.4 |
| Root mean square deviationsb | |
| Bond length, Å | 0.009 |
| Bond angle, ° | 1.429 |
| B factor, Å2 | 14.9 |
a Numbers in parentheses refer to the highest resolution shell.
b Rsym = Σ|Iavg − Ij|/Σ Ij. Rfactor = Σ|Fo − Fc|/ΣFo, where Fo and Fc are observed and calculated structure factors, respectively. Rfree was calculated from a randomly chosen 5% of reflections excluded from the refinement, and Rfactor was calculated from the remaining 95% of reflections. The root mean square deviation values are the rms deviation from ideal geometry.
RESULTS
ARQ 197 Shows Enantioselective Binding to the Unphosphorylated c-Met
We began our studies by measuring the binding of ARQ 197 and its c-Met inactive enantiomer ARQ 198 to the unphosphorylated form of c-Met. The c-Met kinase domain was expressed and purified in an unphosphorylated state as reported previously (18) and the phosphorylation status was confirmed by mass spectrometry. The low aqueous solubility and strong UV absorption properties of the two enantiomers precluded the use of traditional binding assays using SPR, ITC, or fluorescence techniques to obtain quantitative affinity measurements. Instead, the relative binding affinity of the two enantiomers was measured by indirect-affinity mass spectrometry (19). The specificity of binding was tested by including an additional inactive receptor tyrosine kinase (FGFR2) as control (Fig. 1B). ARQ 197 bound to the inactive c-Met protein and did not bind to FGFR2. In contrast, the enantiomer ARQ 198 did not bind to either protein.
ARQ 197 Inhibits the Catalytic Activity of c-Met
To elucidate the mechanism by which ARQ 197 inhibits c-Met, we measured the inhibition of both autophosphorylation and substrate phosphorylation reactions. A continuous spectrophotometric kinase assay was used to measure the kinase activity of the catalytic domain of c-Met. To determine the IC50 for the inhibition of c-Met autophosphorylation, the enzyme was incubated with varying concentrations of ARQ 197 and activity was measured in the presence of 100 μm ATP. The progression of the autophosphorylation was found to be biphasic, indicating that the activation of c-Met is a bimolecular trans-phosphorylation reaction (Fig. 1C). As the concentration of ARQ 197 increased so did the lag phase preceding the autophosphorylation reaction. Following the lag phase, inhibition of the initial rate (the first 20 min) of autophosphorylation gave an IC50 value of 548 ± 120 nm. However, once the autophosphorylation reaction had been allowed to continue for 1 h, the IC50 value increased by 10-fold, to ∼5 μm. Under similar experimental conditions, ARQ 198 did not inhibit c-Met activity at concentrations up to 40 μm (Fig. 1D). Consistent with weak binding to c-Met, as shown by the indirect affinity mass spectrometry experiment, these results show ARQ 198 to be c-Met inactive.
The effect of ATP concentration on the inhibition of c-Met autophosphorylation by ARQ 197 was also evaluated. A rapid decrease in the lag phase of c-Met autophosphorylation with increasing ATP concentration was observed as was a rapid increase in IC50 values with the progress of the reaction (Fig. 1, E and F). The IC50 values determined for c-Met inhibition by ARQ 197 within 20 min of the start of the autophosphorylation reaction, following the lag phase, increased non-linearly with increasing ATP concentration. At an ATP concentration close to the Km of c-Met (0.1 mm), the IC50 value was 550 nm. This increased to 9 μm at a saturating ATP concentration of 0.5 mm.
To better understand the ATP-dependent inhibition of c-Met activation by ARQ 197, levels of phosphorylation of residues Tyr1234 and Tyr1235 of the A-loop were measured by mass spectrometry. Phosphorylation of c-Met in the absence or presence of ARQ 197 was determined either with 0.1 or 1 mm ATP, and the mass of c-Met was measured at various time intervals (Fig. 2, A and B, and supplemental Figs. S1, A and B, and S2, A–D). As reported for other RTKs, activation of c-Met was sequential. The sequence of these phosphorylation events was determined by mass spectrometric analysis of the phosphopeptides (supplemental Fig. S3–S5). The A-loop Tyr1234 was the first residue to get phosphorylated followed by the adjacent Tyr1235. The order of phosphorylation remained the same in the presence of ARQ 197. Consistent with the spectrophotometric experiments, ARQ 197 inhibited c-Met autophosphorylation in a time- and ATP concentration-dependent manner. Preincubation of the enzyme with increasing concentrations of ARQ 197 increased the length of the c-Met lag phase and significantly inhibited autophosphorylation of the first tyrosine residue in a concentration-dependent manner (Fig. 2B and supplemental Fig. S3A). Phosphorylation of the Tyr1235 residue was also dramatically inhibited by ARQ 197. A similar trend was observed at 1 mm ATP concentration, although the overall rate of c-Met activation was increased (supplemental Fig. S1, A and B). In the presence of ARQ 197 the lag phase was increased over that measured in the absence of ARQ 197.
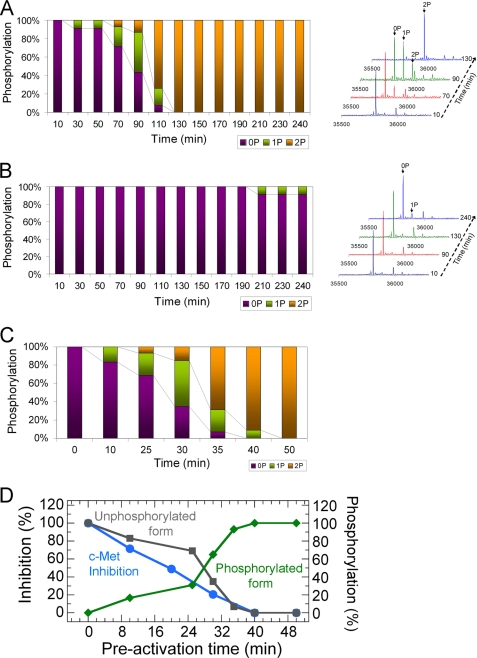
FIGURE 2.
Activation state-dependent inhibition of c-Met by ARQ 197. c-Met autophosphorylation in the presence and absence of ARQ 197 was measured by mass spectrometry (0P, no phosphorylation; 1P, phosphorylation on a single site; and 2P, phosphorylation on two sites). A, 1 μm c-Met, 0.1 mm ATP in the absence of inhibitor; B, 1 μm c-Met, 20 μm ARQ 197, 0.1 mm ATP. C, preactivation of c-Met (5 μm) with 0.1 mm ATP for different lengths of time as monitored by mass spectrometry. D, inhibition of preactivated forms of c-Met (as analyzed in C) by ARQ 197 monitored by continuous spectrometry. The assay mixtures contained final concentrations of 1 μm c-Met (preactivated forms and the control that was not preactivated), 0.1 mm ATP, and 30 μm ARQ 197. The percent inhibition of c-Met by ARQ 197 was calculated for each phosphorylated species with respect to the control, and the inhibition data were plotted against the phosphorylation levels measured by mass spectrometry as shown in C.
ARQ 197 Preferentially Inhibits the Inactive Form of c-Met
To further probe loss of the inhibitory potency of ARQ 197 as a function of time and ATP concentration in the autophosphorylation reaction we investigated the effect of c-Met activation upon inhibition by ARQ 197. A range of preactivated forms of c-Met were generated by preincubation with ATP activation (0.1 mm) for different lengths of time. The degree of phosphorylation was determined by mass spectrometry (Fig. 2C) under similar conditions. The resulting individually phosphorylated species of c-Met were tested in the autophosphorylation inhibition assay and the inhibitory activity of ARQ 197 was measured at a saturating concentration (30 μm). The decrease in inhibition by ARQ 197 from the unphosphorylated form (set at 100% inhibition) to the doubly phosphorylated form is shown in Fig. 2D. The loss of inhibitory activity of ARQ 197 correlated well with the decrease in the amount of unphosphorylated species. These results indicate that ARQ 197 preferentially targets the inactive form of c-Met to exert its inhibitory activity. This is discussed later in the context of the co-crystallographic results and the conformational changes that take place upon ARQ 197 binding to c-Met.
Previous studies have indicated that the kinase domain of c-Met is catalytically highly active, once c-Met autophosphorylation has been initiated, its catalytic efficiency increases rapidly, converting it to a fully active form in a time- and ATP-dependent manner (18, 20). The striking decrease in the lag phase in the autophosphorylation reaction in response to increasing ATP concentration further indicates the rapid depletion of the inactive form of c-Met.
ARQ 197 Inhibits Substrate Phosphorylation by Selectively Inhibiting c-Met Autophosphorylation
The inhibitory activity of ARQ 197 was evaluated by measuring the inhibition of substrate phosphorylation. Unphosphorylated c-Met was preincubated with varying concentrations of ARQ 197 and the substrate phosphorylation reaction was measured at a saturating concentration of synthetic Pyk2 peptide at 0.1 mm ATP. From the initial rates of peptide phosphorylation an IC50 value of 480 ± 100 nm was obtained, which is consistent with the potency observed for the inhibition of autophosphorylation (Fig. 3A). At 1 mm ATP, the IC50 value for the inhibition of substrate phosphorylation by ARQ 197 was increased by 15-fold, which confirmed the results observed in its ATP-dependent loss of inhibition of the autophosphorylation reaction.
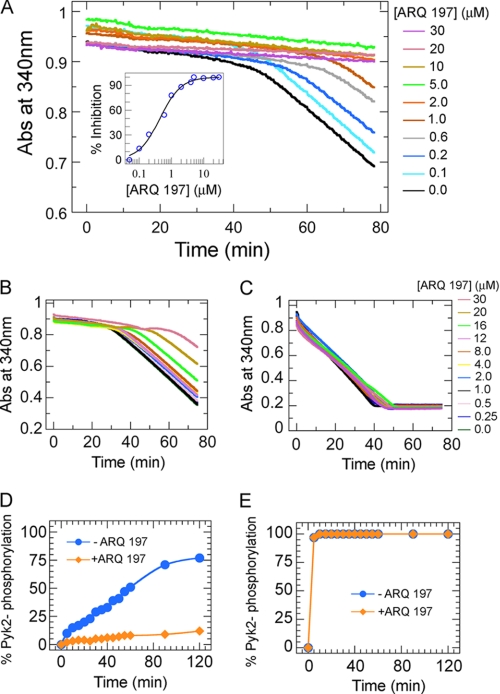
FIGURE 3.
ARQ 197 inhibits c-Met catalyzed substrate phosphorylation and preferentially targets the inactive form of c-Met. A, c-Met was preincubated at the indicated concentrations of ARQ 197 and the reaction was initiated by adding 0.1 mm ATP and 40 μm Pyk2 peptide. The progress of phosphorylation inhibition was monitored by following the absorbance change at 340 nm. The slopes of the curves at the indicated ARQ 197 concentrations were used to calculate the IC50 value (inset). The active form of c-Met (10 μm) was generated in the presence of 0.5 mm ATP, the resulting enzyme was diluted 20-fold (0.5 μm final concentration) with assay mixtures containing increasing concentration of ARQ 197. The reaction was initiated by the addition of 0.5 mm ATP and the inhibition of Pyk2 peptide phosphorylation followed. In a similar experiment c-Met (0.5 μm) not pretreated with ATP was used as a control. B, the control (unphosphorylated c-Met) showed ARQ 197 concentration-dependent inhibition. C, the phosphorylated c-Met showed no detectable change in enzyme activity with increasing concentrations of ARQ 197. The concentrations of ARQ 197 used in both experiments are shown on the right-hand side of the panel. D, the inhibition of substrate phosphorylation by ARQ 197 was monitored by mass spectrometry by following the progress of phosphorylation of 40 μm Pyk2 peptide with 0.5 μm inactive c-Met and 0.1 mm ATP in the presence (diamonds) or absence (circles) of 20 μm ARQ 197. E, same as D but starting with preactivated c-Met.
The inhibitory activity of ARQ 197 on the active form of c-Met was evaluated. Fully activated c-Met was prepared by treatment of the unphosphorylated c-Met with 0.5 mm ATP for 1 h at room temperature and the progress of c-Met phosphorylation was measured by continuous spectrophotometry. Both the phosphorylated and unphosphorylated (control) c-Met were pretreated with increasing concentrations of ARQ 197 and the inhibition was measured at an identical substrate and enzyme concentration for both forms of kinase supplemented with 0.5 mm ATP. Although the control, unphosphorylated c-Met displayed ARQ 197 concentration-dependent inhibition of substrate phosphorylation (Fig. 3B), the phosphorylated c-Met under similar assay conditions showed no detectable inhibition up to 30 μm ARQ 197 (Fig. 3C). Thus ARQ 197 does not inhibit the active (phosphorylated) form of c-Met because phosphorylation of exogenous peptide substrate is not affected. Additionally, the phosphorylation state of the substrate Pyk2 peptide was determined directly by mass spectrometry. As shown in Fig. 3D, preincubation of ARQ 197 with unphosphorylated c-Met resulted in efficient inhibition of phosphorylation of the Pyk2 peptide. However, when ARQ 197 was preincubated with activated c-Met, the effect of the inhibitor on substrate phosphorylation was negligible (Fig. 3E and supplemental Fig. S8), confirming that ARQ 197 is highly selective for the inactive form of c-Met.
Structural Basis for the Inhibition of c-Met by ARQ 197
To determine how ARQ 197 achieves both high specificity for the inactive form of c-Met and selectivity across the kinome in conventional kinase assays (7), we solved the crystal structure of unphosphorylated c-Met in complex with ARQ 197. The structure consists of two identical copies of c-Met·ARQ 197 complexed in an asymmetric unit. ARQ 197 binds inactive c-Met between the N- and C-lobes and occupies the ATP-binding cleft (Fig. 4A). The five-membered pyrrolidine-2,5-dione ring system of ARQ 197 that connects the two aromatic rings of the tricyclic and indole ring makes canonical H-bond hinge interactions with the carbonyl and amide N-H groups of the backbone amide of Met1160 and carbonyl of Pro1158 (Fig. 4B). The indole ring is in close proximity to the adenine binding region projecting into the solvent area. The nitrogen of the indole ring makes a water-mediated hydrogen bond interaction with the backbone carbonyl of Lys1161 near the hinge. The periphery of the hydrophobic ring contacts Gly1085 of the glycine-rich loop β-strand. The tricyclic ring is perpendicular to the pyrrolidine-2,5-dione ring and is bound deep inside the hydrophobic pocket where it occupies the region between the ribose ring sugar and the α-phosphate groups of ATP.
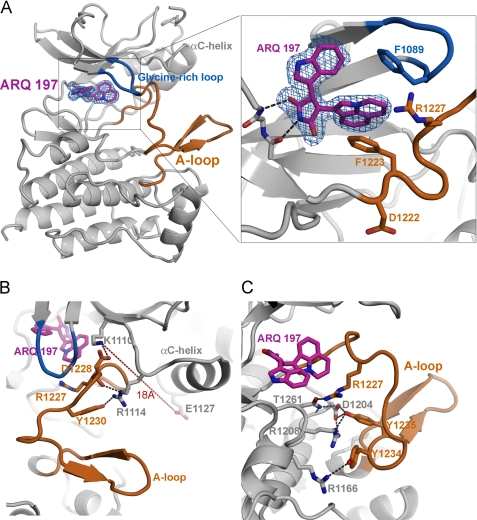
FIGURE 4.
The co-crystal structure of ARQ 197-bound c-Met kinase domain. A, the ribbon representation of c-Met is shown with ARQ 197 (magenta sticks). The conformations of the activation loop (orange) and the P-loop (marine blue) are highlighted. The bidentate hinge hydrogen bond interactions of ARQ 197 with c-Met are shown as dashed lines. The electron density map (2Fo − Fc contoured at 1.2 σ) for the inhibitor is shown as a mesh in marine blue. The tricyclic ring of ARQ 197 directly interacts with the A-loop and P-loop phenylalanines by inducing “hydrophobic collapse” through a π-π interaction. This hydrophobic interaction is also facilitated by the alkyl side chain of Arg1227. The Phe1089 residue of the glycine-rich loop makes a downward movement and the Phe1223 residue of the DFG motif adopts an out conformation. B, the hydrophobic interaction with ARQ 197 results in the insertion of the A-loop segment between the ATP-binding cleft and αC-helix leading to disruption of the ion-pair interaction between the catalytic residues followed by an outward movement of the αC-helix by 18 Å. The Lys1110 residue twists and makes both a hydrophobic interaction with ARQ 197 and an ion-pair interaction with the Asp1228 residue. This interaction is further stabilized by hydrogen bond interactions between Arg1114, Arg1227, and Tyr1230 residues. C, the Arg1227 directly couples the hydrophobic interaction of ARQ 197 with the hydrogen-bond network interactions of A-loop tyrosine residues (1235 and 1234) and the catalytic loop residues (Asp1204, Thr1261, and Arg1208).
ARQ 197 Recognizes a Novel Hydrophobic Pocket in Unphosphorylated c-Met
ARQ 197 binds to a previously uncharacterized hydrophobic pocket that accommodates the non-polar tricyclic ring in the ATP-binding cleft (Figs. 4A and 5A) with a high degree of complementarity. An ordered conformation of the glycine-rich loop folds down, and the aromatic ring of Phe1089 stacks on the upper plane of the tricyclic ring of ARQ 197. The lower plane of this ring forms stacking interactions with the phenylalanine of the DFG motif. Together the phenylalanine residues from the glycine-rich loop and DFG motif interact with the tricyclic group to form a “hydrophobic sandwich.” The non-polar alkyl side chain of Arg1227 of the A-loop covers, and provides an additional hydrophobic interaction with the tricyclic moiety. Likewise, the alkyl side chain of the catalytic Lys1110 caps the hydrophobic pocket.
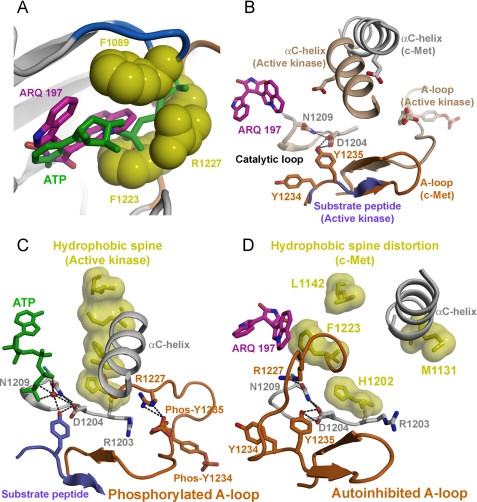
FIGURE 5.
ARQ 197 bound c-Met adopts the autoinhibitory conformation that is not compatible with ATP and substrate binding. This inactive conformation destabilizes the hydrophobic spine that prevents kinase activation. A, superposition of ARQ 197-bound c-Met with the AMP-PNP molecule present in the phosphorylated insulin receptor kinase structure (colored green). ARQ 197 interacts with c-Met by converting the polar “ATP-binding” cleft into an ATP-incompatible non-polar pocket. B, the autoinhibited “pseudo-substrate” conformation of the A-loop of ARQ 197-bound c-Met. The A-loop of c-Met adopts an anti-parallel β-strand hairpin conformation. Superposition of ARQ 197-bound c-Met with AMP-PNP and peptide-substrate bound insulin receptor kinase indicates that the A-loop mimics the peptide substrate. The Tyr1235 residue from the A-loop mimics the phosphorylated tyrosine by stabilizing the hydrogen bonding interactions with the catalytic loop residues. In the active state the Tyr1235 residue is phosphorylated and directly hydrogen bonds to the Arg1227 residue. C, comparison of the highly conserved regulatory hydrophobic spine interactions of the phosphorylated, AMP-PNP and substrate-bound insulin receptor kinase with the ARQ 197-bound c-Met structure. D, interaction of ARQ 197 with unphosphorylated c-Met prevents hydrophobic spine formation as one of these hydrophobic residues (Phe1223) directly participates in a non-polar interaction with ARQ 197 and the second hydrophobic residue of the spine makes a downward movement with the αC-helix.
ARQ 197 Stabilizes the DFG-out Conformation of c-Met and Disrupts the Interactions between the Key Catalytic Residues
In kinases, the DFG motif of the A-loop is an important determinant of activity, and the catalytically incompetent DFG-out conformation of the kinase has been the preferred conformation for designing inhibitors of inactive kinases (21). In the c-Met·ARQ 197 complex the kinase adopts the DFG-out conformation where Phe1223 sandwiches the tricyclic moiety of ARQ 197 and stabilizes the inactive conformation. The N-terminal region of this A-loop is further stabilized through the interaction of Arg1227 with the tricyclic group. Arg1114 from the β3-αC loop forms a hydrogen bond with the backbone Arg1227 and anchors the N-terminal region of the A-loop between ARQ 197 and the αC-helix. As a result of this insertion of part of the A-loop, the αC-helix moves away from ARQ 197 by a distance of 18 Å (Fig. 4B).
The αC helix, another key regulatory element in active kinase structures, is close to the ATP-binding pocket, which is crucial for maintaining the ion-pair interaction between the conserved catalytic Glu1127 and Lys1110 residues. The structural rearrangement imposed by ARQ 197 at the ATP-binding cleft breaks the ion-pair interaction between these catalytic residues and further stabilizes the inactive conformation of c-Met. Glu1127 swings out to the solvent accompanied by the αC-helix shift. The alkyl side chain of Lys1110 twists and participates in a hydrophobic interaction with the tricyclic moiety of ARQ 197, and the side chain -NH2 group forms a salt bridge with Asp1228 of the A-loop (Fig. 4B). A similar structural feature has been observed in several inactive kinases, including recently reported ATP-competitive c-Met·inhibitor complexes (22, 23), however, these inhibitors target the inactive c-Met conformations by different structural mechanisms. Importantly, the transformation of the highly polar-ATP binding region into an ATP-incompatible non-polar inhibitor binding pocket makes the c-Met·ARQ 197 complex a novel structure compared with previously described kinase-inhibitor structures (Fig. 5A).
The Substrate Binding Loop in the c-Met·ARQ 197 Complex Adopts the Canonical Autoinhibited Conformation
A centrally located A-loop in most kinases is an essential regulatory element for enzyme activity, and in the unphosphorylated state it adopts diverse conformations that are incompatible with substrate binding. In several inactive kinases, the activation loop forms a short anti-parallel β-strand and functions as a pseudosubstrate to prevent productive binding of ATP and/or peptide substrate (24). The A-loop of the c-Met·ARQ 197 complex exhibits such a canonical autoinhibited conformation (Figs. 4C and 5B). Tyr1234 of the A-loop is exposed and Tyr1235 is sequestered into the active site where it occupies the site of the substrate tyrosine residue. Tyr1235 is held by a hydrogen bond network formed by Asp1204 and Arg1208 of the catalytic loop (the strictly conserved residues across the type III RTKs) (25), and Tyr1234 is directed toward a positively charged region formed by the guanidinium groups of Arg1166 and Arg1170. The canonical autoinhibited conformation observed in the c-Met·ARQ 197 complex is distinct from the previously reported autoinhibited conformation of unphosphorylated apo-c-Met (12). Interestingly, c-Met adopts diverse inactive conformations bound to various c-Met inhibitors including nonspecific ATP-competitive kinase inhibitors (26). However, the canonical autoinhibitory conformation of the c-Met·ARQ 197 complex has some similarity with previously published inhibitor-bound c-Met structures (27) and the recently deposited structure (PDB ID 2WGJ) (supplemental Fig. S6, A and B), but they adopt distinct inhibitor binding pockets mediated by both polar and non-polar interactions typically observed in kinase-inhibitor structures (supplemental Fig. S6, C and D).
Although to date no published active c-Met structure is available, comparison of the c-Met·ARQ 197 complex with the active form of other type III RTKs (24, 28), and with phosphorylated insulin receptor bound to AMP-PNP and peptide substrate (29), suggests that the conserved Arg1227 residue plays a role in stabilization of the A-loop upon phosphorylation of c-Met through direct hydrogen bonding with phosphorylated Tyr1235 (Figs. 4B and 5B). Binding of ARQ 197 to c-Met induces a dramatic conformational change on Arg1227 through direct hydrophobic interaction of the alkyl side chain with the tricyclic moiety of ARQ 197. This interaction is further stabilized by a hydrogen bond between the amide carbonyl of Arg1227 and the guanidinium group of Arg1114. Disruption of a number of such conserved key electrostatic interactions in c-Met is necessary to stabilize the hydrophobic interaction with ARQ 197. Such structural alterations also impact the hydrophobic spine assembly, which is critical for kinase activation (30) as the hydrophobic interaction of ARQ 197 with Phe1223 (DFG-out conformation) followed by the movement of the αC-helix could prevent the hydrophobic spine assembly (Fig. 5, C and D). These interactions provide another level of autoinhibitory stabilization for c-Met.
ARQ 198 Does Not Fit the Structure of ARQ 197-bound c-Met
Docking experiments were performed to determine whether the biochemically inactive enantiomer ARQ 198 could be accommodated in the structure of c-Met bound to ARQ 197 in a similar binding mode. Either of the pyrrolidine-2,5-dione carbonyl groups can form the critical hydrogen bond with the hinge; however, in both cases ARQ 198 is not accommodated due to unacceptable steric contacts with the protein (supplemental Fig. S7).
DISCUSSION
ARQ 197 is an enantiospecific inhibitor of c-Met with selectivity across more than 200 kinases as shown in a conventional biochemical assay panel screen. Previous work showed ARQ 197 to inhibit the c-Met pathway in cell-based assays by selectively inhibiting c-Met activity (7). Additionally, it was characterized as enzymatically non-ATP competitive. The current study delineates the underlying mechanism of c-Met inhibition by ARQ 197 through the application of biochemical, biophysical, and structural studies. Collectively these data demonstrate that ARQ 197 strongly inhibits c-Met autoactivation by selectively targeting the inactive form of the kinase. ATP-dependent kinetic experiments further show that ARQ 197 does not inhibit substrate phosphorylation by activated c-Met kinase. However, ARQ 197 is highly effective in inhibiting substrate phosphorylation when preincubated with the inactive form of c-Met.
The observed loss in ARQ 197 potency to inhibit phosphorylated c-Met is likely due to the rapid shift in equilibrium from the inactive enzyme to the activated state. A study of the kinetics of activation of the catalytic domain of c-Met showed that the isolated kinase domain of c-Met is highly active even in the unphosphorylated state. The phosphorylation of c-Met has a profound effect on kinase activation and increases the catalytic efficiency (kcat/Km) by 160-fold (18). This dramatic increase in catalytic efficiency is attributed to the geometric progression of the autophosphorylation reaction resulting in a rapid conversion from the unactivated to the activated state.
The time- and ATP-dependent loss of ARQ 197 potency in c-Met inhibition is the result of c-Met activation and the concomitant conformational change from the inactive to the active conformation to which ARQ 197 does not bind. In cells, millimolar ATP concentrations are present, and yet ARQ 197 inhibits c-Met autophosphorylation with a potency equivalent to its biochemical potency measured at lower ATP concentrations. It is likely therefore that additional regulatory elements (including phosphatases) in cells assist in maintaining the inactive form (31, 32). This is especially likely in the case of c-Met because in the absence of such regulatory elements, the isolated kinase domain used in the current study in the in vitro experiments is rapidly activated by ATP. It is also possible that in cells given a longer time interval, sufficient inactive c-Met molecules become accessible for binding, which is ultimately detected as a pharmacodynamic shutdown of c-Met signaling (7). Additional studies will be required to delineate the complex interplay of such cellular regulatory elements to determine their effects on the inhibition of c-Met by ARQ 197.
The crystal structure of ARQ 197 in complex with the kinase domain of c-Met provides a structural rationale for the selectivity of ARQ 197 for the inactive form of c-Met. It reveals an unusual conformation not previously reported for any other protein kinase and suggests a potential mechanism for selectivity. The tricyclic group of ARQ 197 occupies a centrally located hydrophobic pocket and forms hydrophobic interactions with the phenylalanines of the glycine-rich loop and DFG motif. To form this pocket, the glycine-rich loop makes an inward movement, and the DFG motif flips to the “out” conformation. This interaction is further facilitated by the insertion of the N-terminal region of the A-loop between ARQ 197 and the αC-helix to cover the hydrophobic pocket with the alkyl side chain of Arg1227. As a result of the formation of this rigid hydrophobic network, the ion-pair interaction of the catalytic residues is disrupted, and there is a concomitant large displacement (18 Å) of the αC-helix. The dramatic transformation of a polar ATP-binding cleft into an ATP-incompatible non-polar pocket forces c-Met into an inactive conformation (Fig. 5A). Another striking feature of the c-Met·ARQ 197 complex is the position of the A-loop, which folds into a canonical autoinhibited conformation. Such an inactive conformation mimics the positioning of the protein substrate and occludes its binding site.
A survey of all available crystal structures of c-Met from the Protein Data Bank (26) in comparison with the c-Met·ARQ 197 complex shows that c-Met adopts diverse inactive conformations in the unphosphorylated state. The lack of an active c-Met structure limits structural comparisons, as even c-Met with its A-loop tyrosines mutated to mimic the active form (33) adopted a Src-like inactive conformation when co-crystallized with ATP (PDB code 3DKC). The majority of small molecule c-Met inhibitors described bind c-Met in an inactive conformation that involves a large movement of the αC-helix, often mediated by hydrophobic interactions with either the phenylalanine in a DFG-out conformation or the Tyr1230 of the A-loop. Although c-Met bound to these inhibitors adopts many structural features of an inactive conformation these inhibitors have been described as ATP-competitive (3, 34). The selectivity of these inhibitors for the inactive (un-activated) form of c-Met in biochemical assays is unclear. For example, AM7 (23, 35) binds c-Met in an inactive conformation but has been described as having only a 3-fold selectivity for the unphosphorylated form. A small difference in binding potency between the structurally distinct inactive and active conformations of c-Met suggests that this inhibitor interacts with c-Met in alternative binding modes.
Our studies have shown that ARQ 197 recognizes a novel inactive conformation. The association of ARQ 197 with hydrophobic residues of c-Met in an ATP-incompatible binding mode suggests that in the “quiescent state” c-Met favors a conformation similar to the autoinhibited conformations of apo-kinase structures such as c-Kit (36), Flt3 (37), and c-FMS (38). The non-polar ATP-binding cleft in these structures is not only incompatible with ATP binding, but the A-loop adopts a canonical pseudosubstrate autoinhibited conformation. Non-polar pockets in inactive kinases are routinely employed in the design of type II inhibitors, particularly the key regions of the “main pocket” (adenine binding region) and the “allosteric back pocket” (39, 40). Our analysis of the c-Met·ARQ 197 complex has identified clusters of hydrophobic residues that form distinct non-polar pockets that can be accessed by small molecule inhibitors such as ARQ 197. Given that the composition and conformation of these hydrophobic clusters are likely to be distinct for any given kinase in its autoinhibited conformation, it is possible that similar but kinase-specific pharmacophores can be identified for other kinases of therapeutic interest.
In summary, ARQ 197 is an example of a novel class of kinase inhibitor that binds to the inactive conformation of its target, c-Met. It is now possible to devise a systematic in silico design methodology that can be utilized for the design of similar inhibitors for other kinases and its application will be described elsewhere.
Supplementary Material
Acknowledgments
We thank Dr. Susan Taylor for critical reading of this manuscript. We also thank Drs. Chiang J. Li, former Chief Scientific Officer, and Yingwu Xu, former Senior Investigator, for their contributions to methodological refinement while at ArQule. The contribution of the scientific staff of ArQule and their helpful comments on the manuscript are also gratefully acknowledged.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Methods” and Figs. S1–S8.
The atomic coordinates and structure factors (code 3RHK) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- c-Met
- mesenchymal-epithelial transition factor
- RTK
- receptor tyrosine kinase
- DMSO
- dimethyl sulfoxide
- AMP-PNP
- 5′-adenylyl-β,γ-imidodiphosphate
- PDB
- Protein Data Bank.
REFERENCES
- 1. Christensen J. G., Burrows J., Salgia R. (2005) Cancer Lett. 225, 1–26 [DOI] [PubMed] [Google Scholar]
- 2. Danilkovitch-Miagkova A., Zbar B. (2002) J. Clin. Invest. 109, 863–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu X., Newton R. C., Scherle P. A. (2010) Trends Mol. Med. 16, 37–45 [DOI] [PubMed] [Google Scholar]
- 4. Martin T. A., Jiang W. G. (2010) Anticancer Agents Med. Chem. 10, 2–6 [DOI] [PubMed] [Google Scholar]
- 5. Eder J. P., Vande Woude G. F., Boerner S. A., LoRusso P. M. (2009) Clin. Cancer Res. 15, 2207–2214 [DOI] [PubMed] [Google Scholar]
- 6. Comoglio P. M., Giordano S., Trusolino L. (2008) Nat. Rev. Drug Discov. 7, 504–516 [DOI] [PubMed] [Google Scholar]
- 7. Munshi N., Jeay S., Li Y., Chen C. R., France D. S., Ashwell M. A., Hill J., Moussa M. M., Leggett D. S., Li C. J. (2010) Mol. Cancer Ther. 9, 1544–1553 [DOI] [PubMed] [Google Scholar]
- 8. Bagai R., Fan W., Ma P. C. (2010) IDrugs 13, 404–414 [PubMed] [Google Scholar]
- 9. Schiller J. H., Akerley W. L., Brugger W., Ferrari D., Garmey E. G., Gerber D. E., Orlov S. V., Ramlau R., Von Pawel J., Sequist L. V. (2010) J. Clin. Oncol. 28, 18s (suppl; abstr LBA 7502) [DOI] [PubMed] [Google Scholar]
- 10. Lee D. J., Ranganathan A., Xu B. (2010) Clin. Lung Cancer 11, 303–310 [Google Scholar]
- 11. Engelman J. A., Zejnullahu K., Mitsudomi T., Song Y., Hyland C., Park J. O., Lindeman N., Gale C. M., Zhao X., Christensen J., Kosaka T., Holmes A. J., Rogers A. M., Cappuzzo F., Mok T., Lee C., Johnson B. E., Cantley L. C., Jänne P. A. (2007) Science 316, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 12. Wang W., Marimuthu A., Tsai J., Kumar A., Krupka H. I., Zhang C., Powell B., Suzuki Y., Nguyen H., Tabrizizad M., Luu C., West B. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3563–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Muckenschnabel I., Falchetto R., Mayr L. M., Filipuzzi I. (2004) Anal. Biochem. 324, 241–249 [DOI] [PubMed] [Google Scholar]
- 14. Barker S. C., Kassel D. B., Weigl D., Huang X., Luther M. A., Knight W. B. (1995) Biochemistry 34, 14843–14851 [DOI] [PubMed] [Google Scholar]
- 15. CCP4 (1994) Acta Crystallogr. D Biol. Crystallogr. D50, 760–763 [DOI] [PubMed] [Google Scholar]
- 16. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Schrödinger, LLC, New York [Google Scholar]
- 18. Timofeevski S. L., McTigue M. A., Ryan K., Cui J., Zou H. Y., Zhu J. X., Chau F., Alton G., Karlicek S., Christensen J. G., Murray B. W. (2009) Biochemistry 48, 5339–5349 [DOI] [PubMed] [Google Scholar]
- 19. Whitehurst C. E., Annis D. A. (2008) Comb. Chem. High Throughput Screen. 11, 427–438 [DOI] [PubMed] [Google Scholar]
- 20. Sheth P. R., Hays J. L., Elferink L. A., Watowich S. J. (2008) Biochemistry 47, 4028–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kufareva I., Abagyan R. (2008) J. Med. Chem. 51, 7921–7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Albrecht B. K., Harmange J. C., Bauer D., Berry L., Bode C., Boezio A. A., Chen A., Choquette D., Dussault I., Fridrich C., Hirai S., Hoffman D., Larrow J. F., Kaplan-Lefko P., Lin J., Lohman J., Long A. M., Moriguchi J., O'Connor A., Potashman M. H., Reese M., Rex K., Siegmund A., Shah K., Shimanovich R., Springer S. K., Teffera Y., Yang Y., Zhang Y., Bellon S. F. (2008) J. Med. Chem. 51, 2879–2882 [DOI] [PubMed] [Google Scholar]
- 23. Bellon S. F., Kaplan-Lefko P., Yang Y., Zhang Y., Moriguchi J., Rex K., Johnson C. W., Rose P. E., Long A. M., O'Connor A. B., Gu Y., Coxon A., Kim T. S., Tasker A., Burgess T. L., Dussault I. (2008) J. Biol. Chem. 283, 2675–2683 [DOI] [PubMed] [Google Scholar]
- 24. Johnson L. N., Noble M. E., Owen D. J. (1996) Cell 85, 149–158 [DOI] [PubMed] [Google Scholar]
- 25. Reilly J. T. (2002) Br. J. Haematol. 116, 744–757 [DOI] [PubMed] [Google Scholar]
- 26. Asses Y., Leroux V., Tairi Kellou S., Dono R., Maina F., Maigret B. (2009) Chem. Biol. Drug Design. 74, 560–570 [DOI] [PubMed] [Google Scholar]
- 27. Porter J., Lumb S., Franklin R. J., Gascon-Simorte J. M., Calmiano M., Riche K. L., Lallemand B., Keyaerts J., Edwards H., Maloney A., Delgado J., King L., Foley A., Lecomte F., Reuberson J., Meier C., Batchelor M. (2009) Bioorg. Med. Chem. Lett. 19, 2780–2784 [DOI] [PubMed] [Google Scholar]
- 28. De Meyts P., Whittaker J. (2002) Nat. Rev. Drug Discov. 1, 769–783 [DOI] [PubMed] [Google Scholar]
- 29. Hubbard S. R. (1997) EMBO J. 16, 5572–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kornev A. P., Haste N. M., Taylor S. S., Eyck L. F. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17783–17788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hashigasako A., Machide M., Nakamura T., Matsumoto K., Nakamura T. (2004) J. Biol. Chem. 279, 26445–26452 [DOI] [PubMed] [Google Scholar]
- 32. Dikic I., Giordano S. (2003) Curr. Opin. Cell Biol. 15, 128–135 [DOI] [PubMed] [Google Scholar]
- 33. Schiering N., Knapp S., Marconi M., Flocco M. M., Cui J., Perego R., Rusconi L., Cristiani C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12654–12659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cañadas I., Rojo F., Arumí-Uría M., Rovira A., Albanell J., Arriola E. (2010) Clin. Transl. Oncol. 12, 253–260 [DOI] [PubMed] [Google Scholar]
- 35. Dussault I., Bellon S. F. (2008) Cell Cycle 7, 1157–1160 [DOI] [PubMed] [Google Scholar]
- 36. Mol C. D., Dougan D. R., Schneider T. R., Skene R. J., Kraus M. L., Scheibe D. N., Snell G. P., Zou H., Sang B. C., Wilson K. P. (2004) J. Biol. Chem. 279, 31655–31663 [DOI] [PubMed] [Google Scholar]
- 37. Griffith J., Black J., Faerman C., Swenson L., Wynn M., Lu F., Lippke J., Saxena K. (2004) Mol. Cell 13, 169–178 [DOI] [PubMed] [Google Scholar]
- 38. Walter M., Lucet I. S., Patel O., Broughton S. E., Bamert R., Williams N. K., Fantino E., Wilks A. F., Rossjohn J. (2007) J. Mol. Biol. 367, 839–847 [DOI] [PubMed] [Google Scholar]
- 39. Backes A. C., Zech B., Felber B., Klebl B., Muller G. (2008) Expert Opin. Drug Discov. 3, 1427–1449 [DOI] [PubMed] [Google Scholar]
- 40. Garuti L., Roberti M., Bottegoni G. (2010) Curr. Med. Chem. 17, 2804–2821 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.