Abstract
HIV-1 integrase (IN) orchestrates the integration of the reverse transcribed viral cDNA into the host cell genome and participates also in other steps of HIV-1 replication. Cellular and viral factors assist IN in performing its multiple functions, and post-translational modifications contribute to modulate its activities. Here, we show that HIV-1 IN is modified by SUMO proteins and that phylogenetically conserved SUMOylation consensus motifs represent major SUMO acceptor sites. Viruses harboring SUMOylation site IN mutants displayed a replication defect that was mapped during the early stages of infection, before integration but after reverse transcription. Because SUMOylation-defective IN mutants retained WT catalytic activity, we hypothesize that SUMOylation might regulate the affinity of IN for co-factors, contributing to efficient HIV-1 replication.
Keywords: HIV, Integrase, Post-translational Modification, Ubiquitination, Viral Replication, SUMOylation
Introduction
HIV-1 IN6 is a 288-amino acid protein consisting of three functionally independent domains. The N-terminal domain harbors a highly conserved HHCC zinc binding motif that contributes to IN multimerization and enzymatic activities. The central core domain contains the catalytic DDE motif that is conserved in all retroviral and retrotransposon INs and in certain bacterial transposases. The C-terminal domain is the least conserved among retroviral IN and binds DNA nonspecifically (for reviews, see Refs. 1 and 2). The best characterized activity of HIV-1 IN is the catalysis of integration, which is crucial for HIV-1 replication (3). This reaction can be reproduced in vitro in the presence of recombinant IN alone and synthetic DNA species mimicking the viral LTR ends and an acceptor substrate (4, 5). However, other components of the preintegration complex (PIC) contribute to the specificity and efficiency of integration in vivo (6–8). Independently of its enzymatic activity, HIV-1 IN plays additional roles during the viral life cycle that are still ill defined. Indeed, many catalytically active IN mutants have pleiotropic effects impairing reverse transcription and/or uncoating (9–17), PIC nuclear import (18, 19), and virion protein composition and/or morphology (16, 20, 21).
Post-translational modifications contribute to the regulation of IN activities. HIV-1 IN interacts with and is acetylated by both histone acetyltransferases p300 and GCN5 on C-terminal lysine residues (22, 23). Acetylation increases IN affinity for the viral cDNA, enhances its strand transfer activity in vitro, and might regulate the interaction between IN and cellular factors (24). However, the role of this modification during HIV-1 replication is still controversial (25). HIV-1 IN is also ubiquitinated and subsequently degraded by the proteasome (26–28). In the viral context, IN degradation seems to occur after integration and to be required for correct gap repair (29, 30) and viral gene expression (26). Recently, phosphorylation of HIV-1 IN by cellular JNK has also been proposed to modulate its stability and to be necessary for efficient integration (31).
SUMOylation consists of the covalent attachment of small ubiquitin-like modifier (SUMO) peptides to a Lys residue within the consensus motif (ΨKX(E/D), where Ψ is a large hydrophobic residue) of a substrate protein. SUMO conjugation is mediated by SUMO-specific E1-activating, E2-conjugating, and E3-ligating enzymes and is reversed by SUMO-specific proteases (reviewed in Ref. 32). In mammals, three major forms of SUMO proteins are expressed. SUMO-1 has ∼45% amino acid sequence homology to SUMO-2 and SUMO-3, which are 96% identical to each other. SUMO modification is implicated in numerous cellular processes, including signal transduction, protein stability and localization, transcriptional regulation, chromatin structure, and genome stability (32). It is also well established that viruses interfere with and/or hijack the cellular SUMOylation machinery to replicate (for reviews, see Refs. 33 and 34). Interactions between murine leukemia virus capsid (CA) protein and components of the SUMOylation pathway are required for proper execution of the early steps of replication after reverse transcription but before integration (35). SUMOylation events have also been implicated in the early phase of HIV-1 infection. Indeed, SUMO-2 and RanBP2 (Ran-binding protein 2), a SUMO E3 ligase, were identified in genome-wide screens for cell factors that promote HIV-1 reverse transcription and PIC nuclear import, respectively (36, 37). Interaction of HIV-1 or Mason-Pfitzer monkey virus Gag proteins with SUMO-1 and the E2-conjugating enzyme Ubc9 during the late phases of replication have also been reported and are probably involved in the production of fully infectious virions (38–41).
Here, we show that HIV-1 IN is SUMOylated and that three Lys residues, which are found within conserved consensus motifs, represent the major SUMO acceptor sites. In the viral context, mutation of SUMO acceptor residues in IN led to reduced infectivity and slower replication kinetics. Biogenesis, release, and reverse transcription steps of mutant HIV-1 particles were not affected. However, cells infected with viruses harboring SUMOylation-defective IN mutants showed a significant decrease in integration events compared with HIV-1WT-infected cells. Because SUMOylation-site IN mutants retained WT catalytic activity, we inferred that modification by SUMO might participate in the modulation of the HIV-1 IN interaction network by regulating its affinity for co-factors, which are required for the efficient execution of early events of HIV-1 replication.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
HeLa and 293T cells were grown in DMEM (Invitrogen). CEM-GFP cells (AIDS Reagent Program), a human T cell line harboring the GFP reporter gene under the control of HIV-1 LTR (42), were grown in RPMI (Invitrogen). Media were supplemented with 10% fetal calf serum (Invitrogen), 100 units/ml penicillin, and 100 μg/ml streptomycin. Antibodies used were as follows: mouse anti-IN (IN-2), mouse anti-His, rabbit anti-SUMO-1 (FL-101) (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)), rabbit anti-IN 756 and 757 (AIDS Reagent Program), rabbit anti-SUMO-2/3 (Zymed Laboratories Inc.), mouse anti-CA (Hybridolab), mouse anti-FLAG M2 (Sigma-Aldrich), mouse anti-HA 12CA5 (Roche Applied Science), and rabbit anti-acetyl-lysine (Abcam).
Plasmid Construction and Mutagenesis
psPAX2, pWPI, and pMD2.G (a gift from D. Trono); pNL4–3EnvFsGFP (a gift from D. Gabuzda), which contains a complete HIV-1 provirus with an env-inactivating mutation and enhanced GFP in the place of Nef (43); and INWT-FLAG, which encodes a codon-optimized IN gene harboring an ATG initiation codon and a C-terminal FLAG tag (44), have been described previously. The cDNAs encoding WT IN, SUMO-1, -2, and -3, were amplified by PCR from IN-FLAG and YFP-SUMO-1, -2, and -3 (45) and were subcloned in frame with an N-terminal His6 tag into the pcDNA3.1(−) vector (Invitrogen), yielding His-INWT and His-SUMO-1, -2, and -3. The cDNA encoding the C-terminal region of LEDGF/p75 (amino acids 325–530) was amplified by PCR from WT and D366N mutant HA-LEDGF/p75 (46). To produce IN mutants, changes were introduced by PCR into the IN sequence of the suitable plasmid using the Expand Long Templates PCR System (Roche Applied Science). The entire recombinant IN coding fragment was confirmed by sequencing and swapped for the corresponding WT fragment into the appropriate recipient vector.
Purification of Recombinant His-tagged IN and in Vitro SUMOylation Assay
Bacterially expressed His-tagged full-length IN, N-terminal (INΔN, encoding amino acids 50–288) or C-terminal (INΔC, encoding amino acids 1–213) truncation forms (47), and IN3KR mutant were purified as described previously (48). Next, recombinant His-tagged full-length or mutant IN proteins (200 nm) were used to perform an in vitro assay with the SUMOylation kit (BIOMOL) according to the manufacturer's instructions.
His-tagged Protein Purification on Nickel-NTA Beads in Denaturing Conditions
293T cells (3 × 106) were seeded into 10-cm dishes and transfected 24 h later using a calcium phosphate precipitation technique with plasmids encoding FLAG-tagged WT or mutant IN proteins and vectors expressing Ubc9 and His-SUMO-1, -2, or -3 or an appropriate empty vector. After 40 h, cells were lysed under denaturing conditions in buffer A (6 m guanidium HCl, 0.1 m Na2HPO4/NaH2PO4, 10 mm imidazole, pH 8.0) and sonicated (10 cycles, 40-s pulse, 15-s pause with the BioruptorTM (Diagenode)). Cell lysates were incubated with nickel-NTA-agarose beads (Qiagen) (3 h, room temperature) and next extensively washed with decreasing amounts of guanidium HCl. Bound proteins were eluted by boiling in Laemmli buffer with 200 mm imidazole and resolved by SDS-PAGE. Tagged proteins were probed for by Western blot.
Analysis of HIV-1 IN Protein Localization and Half-life
For indirect immunostaining, HeLa cells were grown on glass coverslips and transfected with Polyfect reagent (Qiagen) according to the manufacturer's instructions. After 48 h, cells were fixed with phosphate-buffered saline (PBS), 4% paraformaldehyde (10 min, 4 °C), permeabilized with ice-cold methanol (5 min, 4 °C), and incubated with primary antibodies overnight at 4 °C, followed by corresponding secondary antibodies conjugated to Alexa-Fluor488 (Jackson ImmunoResearch Laboratories, Inc.). Images were acquired on a laser-scanning confocal microscope (LSM510 Meta; Carl Zeiss) equipped with an Axiovert 200 M inverted microscope, using a Plan Apo 63/1.4 numerical aperture oil immersion objective.
For fractionation experiments, 293T cells expressing FLAG-tagged WT, 3KR, or 3EQ IN were lysed in buffer C (10 mm Tris-Cl, pH 7.4, 0.15 m NaCl, 1% CHAPS, EDTA-free complete protease inhibitors (Roche Applied Science)) (30 min, 4 °C). Supernatant (cytosol) and pellet (nuclei) were separated by centrifugation (top speed, 5 min, 4 °C). Nuclear content was extracted in buffer N (buffer C with 0.85 m NaCl final) (30 min, 4 °C).
For IN stability studies, cycloheximide (100 μg/ml; Sigma) or MG132 (5 μm; Calbiochem) were added to the culture medium 24 h after transfection. Next, cells were lysed in buffer N. Total protein content was measured with a Bradford assay (Sigma). Proteins (25 μg/lane) were resolved by SDS-PAGE and detected by Western blot.
Virus Stock Production and Infectivity Assay
Single-round viruses were produced by co-transfection of 293T cells using a standard calcium phosphate precipitation technique with a plasmid encoding WT or mutant HIV-1-packaging DNA (psPAX2) and the genomic transfer vector encoding GFP (pWPI) or the pNL4–3EnvFsGFP vector and an expression vector for the glycoprotein G of vesicular stomatitis virus (VSVg) (pMD2.G). Replication-competent viruses were produced by transfecting the pNL4-3 plasmid that encodes a complete HIV-1 infectious provirus. Supernatants were collected 40 h post-transfection, clarified by low speed centrifugation, filtered through 0.45-μm pore size filters, and treated with 10 units/ml Turbo DNase (Ambion) (1 h, room temperature). Viral particles were concentrated by ultracentrifugation (24,000 rpm, 1 h 30 min, 4 °C) using a SW32 rotor (Beckman) on a 20% sucrose cushion. All viral stocks were normalized for the p24CA antigen content, as determined by ELISA (Zeptometrix) and used to infect target cells (6 × 104 293T or 1 × 106 CEM-GFP cells). After 48 h, the percentage of GFP-expressing cells was measured by flow cytometry on a FACSCalibur flow cytometer with CellQuest software (BD Biosciences).
Western Blot Analysis of Viral Proteins
Viral proteins associated with virions or with infected cells were analyzed by Western blot with anti-CA and anti-IN antibodies. For quantification of virion-associated CA and IN proteins, secondary antibodies coupled with IRDye near infrared dyes (IRDye800CW and IRDye680LT, Science Tec) were used. Proteins were visualized on an Odyssey infrared imager and quantified with Odyssey software (LI-COR Biosciences).
Real-time PCR Analysis
Total genomic DNA was extracted using a blood and body fluid kit (Qiagen) from 293T cells (5 × 105) infected with single-round viruses. Full-length reverse transcripts, integrated HIV-1 DNA, and 2-LTR circles were quantified using a previously described protocol (49). Parallel infections with heat-inactivated HIV-1WT viruses were performed to control for residual levels of plasmid DNA that may have resisted DNase treatment. Viral RNA was extracted with the RNeasy Mini kit (Qiagen) and amplified with the HIV-1 real time RT-PCR kit (BioEvolution). Real-time PCR and RT-PCR were performed on a Lightcycler 1.0 (Roche Applied Science).
Vpr-integrase Complementation
Viral stocks generated by co-transfecting 293T cells with pNLX.Luc(R-Env−), pRL2P-Vpr-IN WT or mutant, and pNLXE7 were used to infect Jurkat cells (2 × 106 cells/ml, 5 × 105 reverse transcriptase cpm), as described (50). Cells were harvested 48 h after infection and lysed in passive lysis buffer (Promega). Frozen and thawed lysates were clarified by centrifugation (18,730 × g, 15 min, 4 °C), and supernatants were analyzed for luciferase activity in duplicate using the Promega luciferase assay system, an EG&G Berthold Microplate LB 96V luminometer, and a Microlite 1 flat bottom microtiter plate (Thermo Labsystems). Luciferase activity was normalized to the protein concentration as determined by the Bio-Rad protein assay kit (Bio-Rad) and corrected for background levels from lysates of cells infected with Env-negative controls.
Immunoprecipitation
293T cells were co-transfected with plasmids encoding FLAG-tagged WT or 3KR or 3EQ IN and WT or D366N HA-LEDGF/p75Cter. The immunoprecipitation assay was performed as described (46). Briefly, precleared cell extracts were incubated with HA.11 affinity matrix (Covance) (3 h, 4 °C). Following extensive washing, bound proteins were eluted in Laemmli buffer. Cell extracts and immunoprecipitates were analyzed by Western blot.
Statistical Analyses
Pairwise comparison between groups was performed using Student's t test. p < 0.05 was set as a threshold for statistical significance.
RESULTS
HIV-1 IN Is Covalently Modified by the Three SUMO Paralogues in Vitro
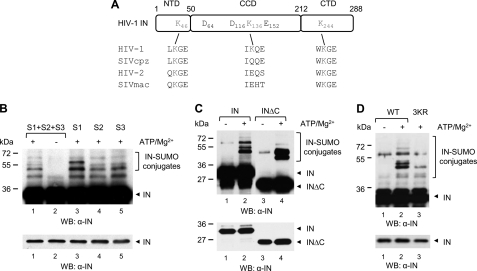
By analyzing HIV-1 IN sequences, we identified three Lys residues at positions 46, 136, and 244 (HXB2 numbering scheme) within canonical SUMOylation consensus motifs, which represent potential sites for modification by SUMO (SUMOplot; SUMO sp 2.0 (51)). SUMO consensus motifs harboring Lys46 and Lys244 are conserved in HIV-2, SIVcpz, and SIVmac (Fig. 1A and supplemental Fig. S1), whereas the SUMO consensus harboring K136 is found in about one-third of HIV-1 strains (52).
FIGURE 1.
HIV-1 IN is modified by the three SUMO paralogues in vitro on conserved Lys residues. A, schematic representation of IN protein. Amino acid sequence alignment of candidate SUMOylation motifs of HIV-1 (HXB2 reference sequence), SIVcpz, HIV-2rod, and SIVmac are shown. DDE, active site residues. B, purified recombinant His-IN (200 nm) was incubated with purified SUMO-specific E1 and E2 enzymes, and the three SUMO proteins (S1, S2, and S3) were added simultaneously or individually. The reaction was conducted in the presence or in the absence of ATP/Mg2+, as indicated. Modified and unmodified forms of IN were visualized with an anti-IN antibody. Incubation of purified His-tagged full-length and C-terminal truncation mutant IN (INΔC) (C) or WT and 3KR mutant IN proteins (D) with the three SUMO proteins simultaneously was performed as in B. The lower panels from B–D show shorter exposure times. WB, Western blot.
The extent of conservation of these motifs prompted us to assess whether HIV-1 IN is post-translationally modified by SUMO proteins. With this aim, we performed an in vitro SUMOylation assay. Slow migrating bands reactive to an antibody against IN were observed when purified recombinant IN bearing an N-terminal His6 tag was incubated with components of the SUMOylation machinery in the presence but not in the absence of ATP (Fig. 1, B and C, lanes 1 and 2). The appearance of several high molecular weight species indicates the addition of multiple SUMO moieties to IN. Of note, similar levels of modification but slightly different patterns of conjugation by SUMO-1 or SUMO-2/3 were observed when IN was incubated in the presence of each SUMO protein separately (Fig. 1B, lanes 3–5).
Because putative SUMO attachment sites localize to each IN functional domain, we examined the modification of IN mutants in which the N-terminal or the C-terminal region was deleted. SUMOylated species were observed when INΔC was subject to the in vitro SUMOylation reaction (Fig. 1C), whereas modified forms of INΔN could not be detected, probably due to technical limits (absence of the epitope or low affinity of the antibodies used) (data not shown).
To confirm that sites identified in silico are SUMOylated, we simultaneously replaced Lys residues at positions 46, 136, and 244 by Arg, an amino acid with similar positive charge that cannot be modified by SUMO, into the His-IN plasmid. The resulting triple IN mutant (IN3KR) was then tested in the in vitro SUMOylation assay. Under these settings, conjugation of SUMO to IN3KR was dramatically decreased compared with INWT (Fig. 1D). Altogether, these results show that HIV-1 IN is modified with similar efficiency, but with slightly different specificity, by the three SUMO paralogues on candidate consensus sites in vitro.
SUMOylation of HIV-1 IN upon Expression in the Cell
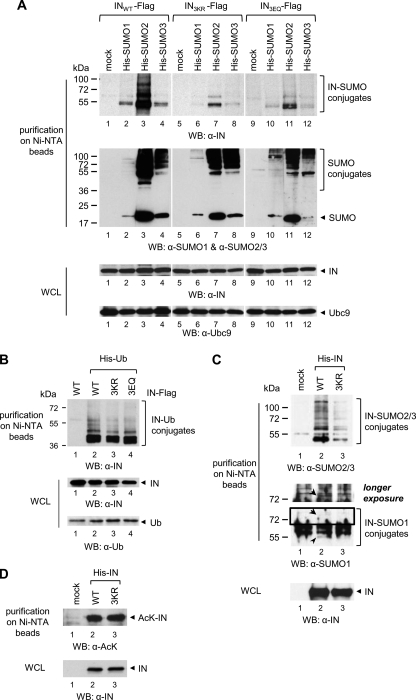
Because we found that HIV-1 IN is covalently modified by SUMO proteins in vitro, we next assessed SUMO conjugation in a cellular context. With this aim, 293T cells were co-transfected with a plasmid encoding HIV-1 IN bearing a C-terminal FLAG tag (INWT-FLAG) and expression vectors for Ubc9 and for N-terminal His-tagged SUMO-1, -2 or -3 or the appropriate control empty vector. Forty hours after transfection, purification by immobilized metal ion affinity chromatography in highly denaturing conditions was performed to ensure that only molecules covalently linked to IN were recovered and that the activity of SUMO proteases was blocked. Although IN expression levels were comparable in all samples (Fig. 2A, bottom, lanes 1–4), modified species reactive to an anti-IN antibody and migrating at the size expected for IN conjugated to one (∼36 + 12 = 48 kDa) or several SUMO moieties, were observed when SUMO-1, -2, or -3 was expressed but not in the control (Fig. 2A, top, lanes 1–4). Because Lys-to-Arg changes at positions 46, 136, and 244 lead to a drastic reduction of IN SUMOylation in vitro, the same mutations were introduced into the INWT-FLAG plasmid. Candidate SUMOylation sites were disrupted either individually or in various combinations, and modified IN derivatives were analyzed as described above. Single and double mutant IN proteins displayed SUMOylation profiles analogous to INWT (data not shown) (supplemental Fig. S2). As expected, the enrichment of modified forms of IN3KR, which was expressed at levels similar to INWT, was considerably diminished regardless of the SUMO protein expressed (Fig. 2A, compare lanes 5–8 with lanes 1–4). We also generated an IN mutant in which the Glu residues at positions 48, 138, and 246 were substituted by Gln, yielding the IN3EQ mutant. Indeed, the acidic amino acid (Glu/Asp) at +2 of the SUMOylation consensus motifs is indispensable for SUMO conjugation to Lys (53). In agreement with the results obtained with IN3KR, the SUMO-modified species of IN3EQ were much less abundant compared with that of INWT (Fig. 2A, compare lanes 9–12 with lanes 1–4).
FIGURE 2.
HIV-1 IN is SUMOylated by overexpressed or endogenous SUMO proteins in the cell. A, 293T cells were co-transfected with plasmids encoding FLAG-tagged INWT, or IN3KR, or IN3EQ and Ubc9; and His-tagged SUMO-1, or SUMO-2, or SUMO-3 or an empty vector (mock) and, 40 h later, were lysed in denaturing conditions followed by purification on nickel-NTA beads. B, 293T cells co-expressing WT or 3KR or 3EQ IN-FLAG and His-tagged ubiquitin were treated as in A. C, 293T cells expressing His-INWT or His-IN3R were treated as in A, and modification by endogenous SUMO-1 or SUMO-2/3 was assessed. A longer exposure time of the inset enclosed in the black square is shown (middle). The arrows indicate SUMO-1-conjugated IN forms. D, acetylated forms of His-INWT or His-IN3R were analyzed following purification as in A. Proteins expressed in the cells or enriched on nickel-NTA beads in A–D were visualized by Western blot with the indicated antibodies. WB, Western blot; WCL, whole cell lysate.
To evaluate whether disruption of IN SUMOylation motifs affected other post-translational modifications targeting Lys residues, FLAG-tagged WT or mutant IN proteins were expressed together with His-tagged ubiquitin in 293T cells, and purification in denaturing conditions was performed. Both IN3KR and IN3EQ displayed ubiquitination profiles similar to INWT, demonstrating that Lys-to-Arg or Glu-to-Gln changes specifically impaired SUMOylation but not ubiquitination (Fig. 2B).
Finally, we analyzed conjugation of endogenous SUMO proteins to ectopically expressed IN. Thus, we generated plasmids encoding WT or 3KR IN with an N-terminal His6 tag that were used to transfect 293T cells, followed by affinity purification in denaturing conditions. Post-translational derivatives of IN enriched on nickel-NTA beads were detected with an anti-SUMO-2/3 antibody in the presence, but not in the absence, of INWT expression (Fig. 2C, top, lane 2). Under the same settings, weak but specific bands were also detected with an antibody against SUMO-1 (Fig. 2C, middle, lane 2). However, although IN3KR was expressed at levels similar to INWT (Fig. 2C, bottom, compare lanes 2 and 3), corresponding SUMO-1- and SUMO-2/3-modified forms were significantly reduced (Fig. 2C, top and middle). Analysis of acetylation and ubiquitination of WT and 3KR IN under the same settings showed that both proteins were modified to a similar extent (Fig. 2D) (data not shown).
Altogether, these results confirm that HIV-1 IN is modified by the SUMO paralogues in vivo and that conserved Lys residues at positions 46, 136, and 244 are the principal sites of SUMOylation. Moreover, these data indicate that candidate SUMOylation sites are not targeted by ubiquitination or acetylation.
Impairment of HIV-1 IN SUMOylation Does Not Affect Its Subcellular Localization or Stability
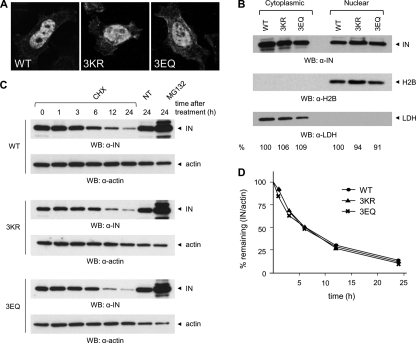
SUMOylation has been shown to regulate numerous cellular processes, including protein localization and stability (32). First, the involvement of SUMO conjugation in HIV-1 IN subcellular distribution was studied by expressing FLAG-tagged WT or mutant IN proteins in HeLa cells, followed by immunofluorescence and confocal microscopy analysis. As already established, INWT was enriched in the nucleus of transfected cells (Fig. 3A). Likewise, IN3KR and IN3EQ concentrated in the nucleus (Fig. 3A). A similar distribution was also observed upon analysis of the steady state nucleocytoplasmic partitioning of FLAG-tagged INWT, IN3KR, or IN3EQ expressed in 293T cells by cell fractionation followed by Western blot with an anti-IN antibody. The proper separation of the nuclear and the cytoplasmic fraction was confirmed by immunoblotting with antibodies against the cytoplasmic protein LDH or the nuclear histone H2B, showing no detectable cross-contamination (Fig. 3B). Second, to investigate the involvement of SUMOylation on IN stability, we studied the half-life of INWT, IN3KR, and IN3EQ overexpressed in 293T cells by carrying out a chase analysis upon treatment with the protein synthesis inhibitor cycloheximide. Although actin levels were not affected by the addition of the drug to the culture medium, INWT protein levels decreased over time, with an approximate half-life of 6 h (Fig. 3, C and D). Different experimental settings (ectopic expression of IN from a codon-optimized gene versus stable expression in a cell line) might account for discrepancies between this and earlier results, which reported a shorter life span for HIV-1 IN (26, 27). Similar decay kinetics was also determined for IN3KR and IN3EQ (Fig. 3, C and D). In agreement with previous reports (26–28), treatment with MG132, an inhibitor of the ubiquitin-proteasome system, prevented the degradation of both WT and mutant IN proteins (Fig. 3C), implicating that their degradation occurred mainly through the proteasomal pathway. Based on these results, we concluded that SUMOylation of Lys46, Lys136, and Lys244 significantly influences neither the subcellular distribution of HIV-1 IN nor the kinetics of its proteasome-dependent degradation.
FIGURE 3.
Subcellular distribution and stability of HIV-1 IN are not affected by mutation of SUMOylation consensus motifs. A, HeLa cells expressing WT or 3KR or 3EQ IN-FLAG were fixed with 4% PFA and then stained with an antibody anti-FLAG, followed by an Alexa488-conjugated secondary antibody. Cells were visualized on a confocal microscope, using a ×63 magnification. Representative images are shown. B, 24 h after transfection, 293T cells expressing FLAG-tagged INWT, IN3R, or IN3Q were subject to cell fractionation followed by Western blot with anti-IN. Purity of the fractions was verified with a nuclear marker, H2B, and a cytoplasmic marker, LDH. Following quantification of ECL signals with ImageJ, the distribution of WT or mutant IN proteins in the nucleus and the cytoplasm was determined by dividing the intensity of IN signals for the corresponding H2B or LDH signals. The values for INWT were arbitrarily set to 100. C, 24 h after transfection, 293T cells expressing FLAG-tagged INWT, IN3KR, or IN3EQ were treated with cycloheximide (CHX) or MG132 or left untreated (NT). At the indicated times, cells were lysed, and total proteins (25 μg/lane) were separated by SDS-PAGE followed by Western blot with anti-IN or anti-actin antibodies. D, ECL signals from Western blots of B were quantified using ImageJ, and values were plotted as the percentage of the signal at t = 0 (given an arbitrary value of 100%) remaining at the indicated time points. Results shown are representative of three independent experiments. WB, Western blot.
Mutation of Key Residues of IN SUMOylation Consensus Motifs Impairs HIV-1 Infectivity
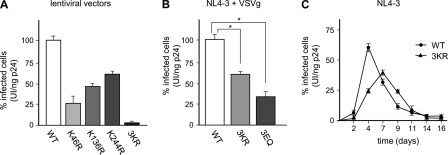
Having established that HIV-1 IN is SUMOylated in vivo, outside the viral context, we next addressed the role of SUMO conjugation to IN during viral replication. Identified SUMO acceptor Lys residues were mutated either individually or simultaneously to Arg in an HIV-1-derived packaging construct. Lentiviral vectors, prepared by transfection of 293T cells with WT or mutant HIV-1-packaging DNA, a vector expressing the VSVg envelope, and a genomic transfer vector encoding GFP, were harvested after 48 h and concentrated by ultracentrifugation through a sucrose cushion. Next, viral preparations of equal p24CA content were used to challenge 293T cells, and the infectivity of WT and mutant viral particles was determined by measuring the amount of GFP-expressing cells by flow cytometry 48 h postinfection. Although vectors harboring different single-point mutations of SUMO-acceptor K sites displayed moderate reductions in infectivity (between 25 and 59% WT levels), an HIV-1-derived viral vector harboring IN3KR was 3% as infectious as WT (Fig. 4A). Although they can fully recapitulate the early phase of infection, lentiviral vectors lack both the ability to perform the late stages of HIV-1 replication and viral accessory proteins present in their parental counterpart, which are mostly dispensable for replication in vitro but essential for viral pathogenesis in vivo. Thus, we analyzed the effect of the disruption of the SUMOylation sites within IN also on the infectivity of authentic HIV-1 viruses. With this aim, Lys-to-Arg substitutions were introduced in a plasmid encoding full-length HIV-1 proviral DNA with a frameshift in Env and expressing the GFP reporter gene in the place of Nef (NL4–3EnvFsGFP). Mutant virions harboring an IN protein bearing Glu-to-Gln changes at identified SUMOylation consensus motifs were also generated to specifically disrupt SUMOylation without affecting other post-translational modification that targets Lys residues. Single-round viruses were produced and used in infection experiments as described before. As shown in Fig. 4B, HIV-1 harboring IN3KR or IN3EQ was about 57 and 33% as infectious as HIV-1WT, respectively. The contribution of nonstructural viral proteins, absent from the lentivector background, might account for the differential viral infectivity of viruses bearing SUMOylation-deficient IN proteins in these settings. Finally, to study the outcome of the impairment of IN SUMOylation on HIV-1 replication following entry via receptor-mediated fusion at the plasma membrane, Lys-to-Arg changes were introduced into the NL4-3 molecular clone of HIV-1. Human T-lymphoid CEM-GFP cells were infected with equal p24CA amounts of HIV-1WT or HIV-13KR, and the percentage of GFP-positive cells was monitored by flow cytometry over time. Under these experimental conditions, the infectivity of HIV-13KR was 32 and 41% that of HIV-1WT at 48 and 96 h postinfection, respectively (Fig. 4C). Additionally, HIV-13KR replicated with a delay of ∼3 days compared with HIV-1WT, reaching peak growth at 7 days postinfection. Altogether, these findings indicate that SUMO-modified Lys46, Lys136, and Lys244 play a role during HIV-1 replication.
FIGURE 4.
HIV-1 virions harboring SUMOylation site mutant IN display reduced infectivity. A, equivalent p24CA amounts (5 ng) of VSVg-pseudotyped lentiviral vectors harboring WT or SUMOylation site mutant IN and encoding GFP were used to infect 293T cells. Infectivity of each viral stock, expressed as the percentage of GFP-positive cells at 48 h postinfection, was measured by flow cytometry. The infectivity of WT viruses was arbitrarily set to 100. B, the infectivity of equal p24CA amounts of VSVg-pseudotyped HIV-1WT, HIV-13KR, or HIV-13EQ (5 ng) was determined as described in A. *, p < 0.05 (paired t test). C, for spreading infection experiments, CEM-GFP cells were infected with equal amounts of HIV-1WT or HIV-13KR (20 ng of p24CA, an approximate multiplicity of infection of 0.15). Mock infections were performed using heat-inactivated WT viruses. Plotted values in A–C represent the mean ± S.D. (error bars) from three independent experiments.
SUMOylation Site Mutant HIV-1 Is Impaired at an Early Step(s) of Replication Preceding Integration
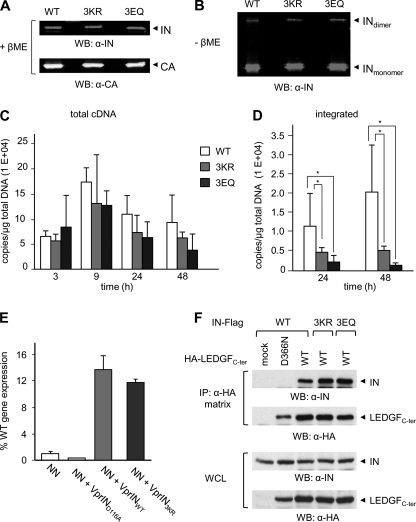
We observed that simultaneous disruption of IN SUMOylation consensus motifs correlated with an impairment of HIV-1 infectivity. To establish what stage(s) of HIV-1 replication cycle was affected, we first asked whether the defects observed could be accounted by improper viral particle assembly, composition, and/or release. Similar amounts of WT and mutant progeny virions were obtained following transfection of 293T cells with NL4–3EnvFsGFP plasmid encoding INWT, IN3KR, or IN3EQ, in independent production experiments, as determined by p24CA ELISA (Table 1). The expression levels and patterns of protease-mediated cleavage of the Gag precursor were analyzed and found to be similar for WT and mutant HIV-1-producing cells (data not shown). Analysis of HIV-1WT, HIV-13KR, and HIV-13EQ protein content by Western blot using near infrared dye-conjugated antibodies followed by quantification of the corresponding emission signals on an Odyssey infrared imaging system showed that WT and mutant viral particles displayed comparable amounts of IN protein relative to CA (Fig. 5A and Table 1). Additionally, two bands reactive to an anti-IN antibody, with the apparent molecular mass of IN monomer and dimer molecules, were detected in all samples by analysis performed under non-reducing conditions (without β-mercaptoethanol), as described previously (54). Quantification of the intensity of corresponding bands showed that HIV-13KR and HIV-13EQ displayed an IN dimer/monomer ratio comparable with that of HIV-1WT (Fig. 5B and Table 1). Finally, HIV-1WT, HIV-13KR, and HIV-13EQ virions packaged similar amounts of viral genomic RNA, as established by real-time RT-PCR on total RNA extracted from viral stocks of equivalent p24 content (Table 1). These results demonstrated that viral particle biogenesis, release, and maturation were mostly unaffected by the mutation of SUMO acceptor sites within IN.
TABLE 1.
Analysis of WT and SUMOylation site mutant virus release and composition
Results are the mean of three independent experiments.
| Efficiency of viral particle releasea | Virion composition |
|||
|---|---|---|---|---|
| IN/CAb | Dimer/monomerc | Genomic RNAd | ||
| % | % | % | ||
| WT | 100 | 100 | 100 | 161 ± 4 × 104 |
| 3KR | 79 ± 11 | 110 | 93 | 218 ± 10 × 104 |
| 3EQ | 86 ± 11 | 120 | 85 | 304 ± 31 × 104 |
a The efficiency of viral particle release was established by p24CA ELISA on the supernatant of virus-producing cells. The values corresponding to WT viruses are arbitrarily set to 100.
b Percentage of emission signals corresponding to virus-associated IN and CA measured on an Odyssey infrared imaging system.
c Percentage of emission signals corresponding to virus-associated monomeric and dimeric IN, measured as for IN/CA.
d Average copies of viral genomic RNA/ng of p24CA.
FIGURE 5.
HIV-1 harboring SUMOylation-defective IN is impaired in provirus formation but retains WT protein composition, catalytic activity, and LEDGF/p75-binding. Proteins contained in HIV-1WT or HIV-13KR or HIV-13EQ viral preparations (100 or 300 ng of p24CA) were separated by SDS-PAGE in reducing (with β-mercaptoethanol (+βME)) (A) or non-reducing (without β-mercaptoethanol (−βME)) (B) conditions and were revealed by Western blot with anti-CA or anti-IN antibodies. Emission signal intensities relative to CA and IN were quantified by laser scanning of corresponding bands on an Odyssey infrared imaging system. C, 293T cells were infected with the indicated single-round viruses (100 ng of p24CA, corresponding to a multiplicity of infection of 0.5 for HIV-1WT), and at the indicated time points, late reverse transcripts were quantified by real-time PCR and normalized for total DNA content. Signals detected in parallel infections with VSVg-minus viruses were subtracted from envelope-mediated infections to correct for input plasmid DNA carry over. D, integrated proviruses were quantified by Alu-PCR at 24 and 48 h postinfection. Results shown in A and B represent the mean ± S.D. of two independent experiments performed in duplicates. *, p < 0.05 (paired t test). E, analysis of the ability of WT, D116A or 3KR IN expressed in trans as Vpr fusion proteins to rescue the replication defect of integration-defective virions (NN), expressed as a percentage of WT HIV-1NLX.Luc(R−) activity (56). F, lysates from 293T cells expressing FLAG-tagged INWT, IN3KR, or IN3EQ and HA-tagged WT or D366N mutant LEDGFCter were immunoprecipitated with an HA affinity matrix. Bound proteins were analyzed by Western blot with anti-FLAG and anti-HA antibodies. WCL, whole cell lysate.
Because mutation of IN SUMOylation sites did not have an impact on the late events of the HIV-1 life cycle, we inferred that an early replication step might be affected. To address this question, we analyzed viral cDNA synthesis in cells infected with equivalent p24CA amounts of VSVg-pseudotyped single-round virions harboring INWT, IN3KR, or IN3EQ by real-time PCR. Although similar amounts of late reverse transcripts were synthesized over time (Fig. 5C), the levels of integrated proviruses at 24 and 48 h postinfection were significantly diminished in cells infected with HIV-13KR or HIV-13EQ as compared with HIV-1WT-infected cells (Fig. 5D). We also monitored the formation of 2-LTR circles, which are generally used as a marker of PIC nuclear import (reviewed in Ref. 55), and found that the number of copies of 2-LTR circles formed upon infection with HIV-1 harboring WT or mutant IN proteins was similar (data not shown).
We next assessed whether mutation of SUMOylation sites of HIV-1 IN affected its enzymatic activity. With that purpose, we tested the function of SUMOylation site mutant IN proteins in the viral context using a Vpr-IN complementation assay (56). WT or mutant IN proteins fused to Vpr were expressed together with an HIV-1 proviral vector encoding an active site IN mutant (HIV-1D64N/D116N). Vpr-mediated encapsidation of IN proteins, in which SUMOylated Lys residues were changed to Arg either individually or simultaneously, allowed recovery of viral infectivity comparable with that of Vpr-INWT (Fig. 5E) (data not shown), suggesting that mutation of SUMO acceptor sites did not significantly affect IN catalytic activity under these infection conditions.
Finally, we asked whether impairment of provirus formation upon infection with HIV-13KR or HIV-13EQ could be accounted for by loss of interaction with LEDGF/p75, an extensively studied IN-interacting protein and an essential chromatin-docking factor for HIV-1 PIC (29, 57–58). With this aim, 293T cells were co-transfected with an expression vector for FLAG-tagged INWT, or IN3KR, or IN3EQ and a plasmid encoding the C-terminal region (amino acids 325–530, encompassing the IN-binding domain) of LEDGF/p75 with an HA tag (HA-LEDGFC-ter) or the corresponding D366N mutant, which has lost the ability to bind IN (46) or the appropriate control empty vector. Immunoprecipitation assays performed 40 h after transfection showed that both IN3KR and IN3EQ were able to bind an LEDGF/p75C-ter in a manner similar to INWT (Fig. 5F). As expected, INWT was not enriched on HA matrix beads in the absence of WT LEDGFC-ter or in the presence of the D366N mutant (Fig. 5F). Altogether, these results demonstrated that impairment of IN SUMOylation by substitution of residues Lys46, Lys136, and Lys244 or Glu48, Glu138, and Glu246 affected neither IN catalytic activity nor LEDGF/p75 binding and correlates to an early replication defect occurring before proviral integration but after reverse transcription.
DISCUSSION
HIV-1 IN is the viral enzyme that orchestrates the integration of the viral cDNA into cellular genome, a key event of retroviral replication and the target of novel anti-HIV therapeutic agents. Numerous studies have contributed to quite an extensive understanding of the molecular basis of IN catalytic functions. On the contrary, although it is well established that integrity of IN structure and/or its interaction network is critical for optimal execution of various steps of HIV-1 life cycle other than integration (59), the mechanisms by which IN exerts these additional functions are presently still elusive. To address this issue, IN protein-protein interactions have been widely explored (reviewed in Ref. 60); however, information is scarce on IN post-translational modifications that represent a common, rapid, and generally reversible mechanism for fine tuning of protein activities.
We repeatedly observed that numerous bands were detected in an anti-IN immunoblot following affinity purification of HIV-1 IN under denaturing conditions from either cellular or viral extracts, suggesting that it undergoes a high degree of post-translational modification (data not shown). It has already been shown that IN is acetylated, ubiquitylated, and phosphorylated (22, 23, 25–29, 31). Because we had mapped putative sites of SUMO conjugation to Lys residues at positions 46, 136, and 244, located within phylogenetically conserved canonical SUMOylation consensus motifs (32), we asked whether HIV-1 IN is also SUMOylated. We identified these amino acids by comparative analysis of IN sequences, which showed that Lys46 and Lys244 are found within SUMOylation consensus motifs common to HIV-1, HIV-2, SIVmac, and SIVcpz (Fig. 1A and supplemental Fig. 1). Lys244 is also conserved among certain non-primate lentiviruses (61). Notably, analysis of naturally occurring variations within IN sequences of HIV-1 isolates showed that Lys46 and Lys244 are not polymorphic (62, 63). Conversely, Lys136 is conserved in about one-third of HIV-1 strains and displays both inter- and intrasubtype substitutions to residues that cannot be modified by SUMO with a frequency rate of >0.5% (62, 63). This extent of amino acid conservation suggests a role of these residues in preserved IN functions.
Analysis performed in vitro showed that recombinant purified HIV-1 IN is covalently coupled by SUMO proteins. Under these experimental conditions, similar efficiency of modification but slightly different patterns of conjugation by each paralogue were observed, pointing to the presence of both SUMO-1- and SUMO-2/3-specific attachment sites within IN. We further confirmed modification of IN by SUMO proteins in a cellular context, both in the presence of exogenous and endogenous expression levels of SUMO family members. In both cases, SUMO-2/3-conjugated IN species were more abundant than SUMO-1-conjugated forms, probably reflecting the higher availability of SUMO-2/3 compared with SUMO-1 (64).
Finally, the formation of numerous high molecular weight species is consistent with the addition of multiple SUMO moieties to IN. The attachment of single SUMO molecules to several Lys residues is supported by the fact that many SUMOylated IN species were detected when an in vitro reaction was conducted in the presence of SUMO-1 alone, which cannot form a poly-SUMO chain. However, the conjugation of polymeric SUMO chains to one Lys residue of IN under different experimental settings cannot be ruled out. In light of emerging evidence suggesting that SUMO proteins display both common and specific target protein preferences and play both redundant and non-redundant cellular functions, these results might underlie functional differences of SUMO-1 and SUMO-2/3 conjugation to IN.
Simultaneous substitution of key residues within the three canonical SUMOylation consensus motifs (either Lys or Glu) was required to significantly decrease the occurrence of SUMOylated, but not ubiquitinated or acetylated, IN species. However, IN SUMOylation was not abolished, indicating that Lys46, Lys136, and Lys244 represent the major, but not unique, SUMO acceptor sites. In addition to the ΨKX(E/D) consensus, the Lys residue within the reverse (E/D)XKΨ signature can also be modified by SUMO (65). Concordantly, Lys residues at positions 71 and 258, which are highly conserved among HIV-1 isolates, may represent additional sites of SUMOylation (supplemental Fig. S1). Disruption of preferred modified sites might also result in the transfer or the enhancement of SUMO conjugation to other Lys residues, as reported for other proteins (66, 67).
Mutation of identified SUMOylation sites did not significantly impact several IN properties, such as subcellular distribution and stability of recombinant HIV-1 IN in human cells or IN oligomerization within the virion. Moreover, SUMOylation-defective IN mutants retained LEDGF/p75 binding and were catalytically active, as demonstrated by their ability to rescue the replication defect of an IN active site mutant virus when expressed as Vpr fusion proteins in trans. On the basis of these observations, we deduced that Lys-to-Arg or Glu-to-Gln changes did not considerably alter IN structure. In agreement, analysis of available three-dimensional structures of HIV-1 IN (68, 69) showed that key residues of identified SUMOylation consensus motifs (Lys46 and Glu48, Lys136 and Glu138, and Lys244 and Glu246) are located at the surface of IN multimers and do not seem to be engaged in contacts between IN protomers, suggesting that SUMOylation of these sites would not directly affect HIV-1 IN oligomerization (supplemental Fig. S3, A and B).
In the viral context, mutation of SUMO acceptor sites correlated with decreased infectivity and slower replication kinetics compared with HIV-1WT, both in epithelial and T cell lines. Viruses harboring IN3KR displayed viral particle biogenesis and release similar to the WT counterpart, indicating that mutation of major SUMO acceptor sites did not affect late stages of the HIV-1 replication cycle. We next analyzed viral cDNA synthesis and provirus formation upon infection and found that SUMOylation site mutant viruses displayed a defect at the integration step, whereas reverse transcription and PIC nuclear import were mostly unaffected. Altogether, these results show that integrity of identified consensus sites for SUMOylation is required both for optimal SUMOylation levels of IN and efficient proviral integration. Either concomitant or consecutive modification of Lys residues at positions 46, 136, and 244 by SUMO might be required for a favorable outcome of HIV-1 replication. However, residual modification of noncanonical SUMOylation sites within IN might compensate for the lack of conjugation to major SUMO attachment sites and, thus, account for the moderate reduction of viral infectivity.
Interestingly, we obtained analogous results when studying HIV-13EQ, which bears an IN mutant in which Glu-to-Gln changes were introduced at identified SUMOylation consensus motifs to specifically impair SUMO conjugation but not other post-translational modifications that occur on Lys residues. The fact that viruses harboring either IN3KR or IN3EQ display an analogous phenotype supports a direct requirement for IN SUMOylation in the optimal execution of an event(s) following reverse transcription and nuclear entry but before integration. Thus, our results are agreement with data from recent genome-wide studies indicating that components of the SUMOylation pathway promote HIV-1 replication (36, 37).
To strengthen the role of IN SUMOylation during viral replication, we asked whether revertant viruses would emerge following passages of HIV-13KR-infected cells. Total genomic DNA was extracted at the peak of infection and used to amplify the IN fragment, which was then entirely sequenced. Our preliminary results show that one of 22 sequenced clones harbored an IN protein in which Arg residues at positions 136 and 244, but not 46, reverted simultaneously to Lys. This cloned harbored also a conservative Leu-to-Ile substitution at position 172.
We also attempted to establish the stage(s) of the HIV-1 life cycle during which IN SUMOylation could take place. We therefore generated 293T cell lines stably expressing His-SUMO proteins that were used to produce HIV-1 viral stocks. Bands migrating at the size expected for SUMO-conjugated IN or to Gag-Pol cleavage intermediates were detected following purification in denaturing conditions over nickel-NTA-agarose beads of virus-producing cells, indicating that SUMOylation occurs during the late phases of HIV-1 infection (supplemental Fig. S4). This result suggests also that SUMOylated IN might be incorporated within HIV-1 virions. However, SUMO-conjugated IN species were not detected upon lysis and purification of viral stocks (supplemental Fig. S4). The fact that each HIV-1 virion encapsidates an estimated 250 molecules of IN (70) and that only a small fraction of it might be SUMOylated at steady state could at least in part explain these observations. Moreover, SUMOylation is reversible, and efficient viral replication might rely on a dynamic process of conjugation-deconjugation of SUMO moieties to IN. We note also that HIV-1 p6 has been shown to be SUMOylated, but the incorporation of the SUMO-conjugated protein into virions has not been detected (38), despite the fact that p6 is about 20 times more abundant than IN.
Expression of components of the SUMOylation pathway, in particular SUMO-2 and the SUMO E3 ligase RanBP2, in target cells has recently been implicated in the completion of early events of HIV-1 infection (36, 37). We are currently addressing the role of a potential functional interaction between RanBP2 and HIV-1 IN, which could take place during PIC nuclear import. Preliminary experiments demonstrated also the existence of a physical and functional link between IN and SUMO E3 ligases of the PIAS family.7
In conclusion, we hypothesize that SUMOylation might modulate the affinity of IN for co-factors required for efficient viral replication, as recently reported for IN acetylation (24). Further studies will be required to clarify the molecular mechanisms by which the cellular SUMOylation pathway participates in the control of HIV-1 IN functions during the early steps of viral replication.
Supplementary Material
Acknowledgments
We thank D. Gabuzda, R. Niedenthal, F. Mammano, R. Hay, and D. Trono for reagents; P. Lesage, S. Basmaciogullari, V. Lallemand-Breitenbach, and M. Benkirane for discussion and critical reading of the manuscript; and F. Lacoste for informatics supplies. We thank N. Setterblad and C. Doliger at the Imagery and Cell Sorting Department of the Institut Universitaire d'Hématologie-Hôpital St-Louis IFR105 (supported by grants from the Conseil Regional d'Ile de France and the French Research Ministry) for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant AI052014 (to A. E.). This work was also supported by Sidaction and by Agence Nationale pour la Recherche sur le SIDA et les Hepatites Virales (ANRS).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
A. Zamborlini and G. Beauclair, unpublished results.
- IN
- integrase
- PIC
- preintegration complex
- SUMO
- small ubiquitin-like modifier
- CA
- capsid
- VSVg
- glycoprotein G of vesicular stomatitis virus.
REFERENCES
- 1. Craigie R. (2001) J. Biol. Chem. 276, 23213–23216 [DOI] [PubMed] [Google Scholar]
- 2. Esposito D., Craigie R. (1999) Adv. Virus. Res. 52, 319–333 [DOI] [PubMed] [Google Scholar]
- 3. Vandegraaff N., Engelman A. (2007) Expert Rev. Mol. Med. 9, 1–19 [DOI] [PubMed] [Google Scholar]
- 4. Craigie R., Fujiwara T., Bushman F. (1990) Cell 62, 829–837 [DOI] [PubMed] [Google Scholar]
- 5. Katz R. A., Merkel G., Kulkosky J., Leis J., Skalka A. M. (1990) Cell 63, 87–95 [DOI] [PubMed] [Google Scholar]
- 6. Carteau S., Gorelick R. J., Bushman F. D. (1999) J. Virol. 73, 6670–6679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hindmarsh P., Leis J. (1999) Adv. Virus. Res. 52, 397–410 [DOI] [PubMed] [Google Scholar]
- 8. Bowerman B., Brown P. O., Bishop J. M., Varmus H. E. (1989) Genes Dev. 3, 469–478 [DOI] [PubMed] [Google Scholar]
- 9. Hehl E. A., Joshi P., Kalpana G. V., Prasad V. R. (2004) J. Virol. 78, 5056–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tasara T., Maga G., Hottiger M. O., Hübscher U. (2001) FEBS Lett. 507, 39–44 [DOI] [PubMed] [Google Scholar]
- 11. Tsurutani N., Kubo M., Maeda Y., Ohashi T., Yamamoto N., Kannagi M., Masuda T. (2000) J. Virol. 74, 4795–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu X., Liu H., Xiao H., Conway J. A., Hehl E., Kalpana G. V., Prasad V., Kappes J. C. (1999) J. Virol. 73, 2126–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu K., Dobard C., Chow S. A. (2004) J. Virol. 78, 5045–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leavitt A. D., Robles G., Alesandro N., Varmus H. E. (1996) J. Virol. 70, 721–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Engelman A., Englund G., Orenstein J. M., Martin M. A., Craigie R. (1995) J. Virol. 69, 2729–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shin C. G., Taddeo B., Haseltine W. A., Farnet C. M. (1994) J. Virol. 68, 1633–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Briones M. S., Dobard C. W., Chow S. A. (2010) J. Virol. 84, 5181–5190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gallay P., Hope T., Chin D., Trono D. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9825–9830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ikeda T., Nishitsuji H., Zhou X., Nara N., Ohashi T., Kannagi M., Masuda T. (2004) J. Virol. 78, 11563–11573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bukovsky A., Göttlinger H. (1996) J. Virol. 70, 6820–6825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Quillent C., Borman A. M., Paulous S., Dauguet C., Clavel F. (1996) Virology 219, 29–36 [DOI] [PubMed] [Google Scholar]
- 22. Cereseto A., Manganaro L., Gutierrez M. I., Terreni M., Fittipaldi A., Lusic M., Marcello A., Giacca M. (2005) EMBO J. 24, 3070–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Terreni M., Valentini P., Liverani V., Gutierrez M. I., Di Primio C., Di Fenza A., Tozzini V., Allouch A., Albanese A., Giacca M., Cereseto A. (2010) Retrovirology 7, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Allouch A., Cereseto A. (2009) Amino Acids, in press [DOI] [PubMed] [Google Scholar]
- 25. Topper M., Luo Y., Zhadina M., Mohammed K., Smith L., Muesing M. A. (2007) J. Virol. 81, 3012–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mousnier A., Kubat N., Massias-Simon A., Ségéral E., Rain J. C., Benarous R., Emiliani S., Dargemont C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13615–13620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mulder L. C., Muesing M. A. (2000) J. Biol. Chem. 275, 29749–29753 [DOI] [PubMed] [Google Scholar]
- 28. Devroe E., Engelman A., Silver P. A. (2003) J. Cell Sci. 116, 4401–4408 [DOI] [PubMed] [Google Scholar]
- 29. Emiliani S., Mousnier A., Busschots K., Maroun M., Van Maele B., Tempé D., Vandekerckhove L., Moisant F., Ben-Slama L., Witvrouw M., Christ F., Rain J. C., Dargemont C., Debyser Z., Benarous R. (2005) J. Biol. Chem. 280, 25517–25523 [DOI] [PubMed] [Google Scholar]
- 30. Yoder K. E., Bushman F. D. (2000) J. Virol. 74, 11191–11200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Manganaro L., Lusic M., Gutierrez M. I., Cereseto A., Del Sal G., Giacca M. (2010) Nat. Med. 16, 329–333 [DOI] [PubMed] [Google Scholar]
- 32. Geiss-Friedlander R., Melchior F. (2007) Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 33. Boggio R., Chiocca S. (2006) Curr. Opin. Microbiol. 9, 430–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wilson V. G., Rangasamy D. (2001) Virus Res. 81, 17–27 [DOI] [PubMed] [Google Scholar]
- 35. Yueh A., Leung J., Bhattacharyya S., Perrone L. A., de los Santos K., Pu S. Y., Goff S. P. (2006) J. Virol. 80, 342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. König R., Zhou Y., Elleder D., Diamond T. L., Bonamy G. M., Irelan J. T., Chiang C. Y., Tu B. P., De Jesus P. D., Lilley C. E., Seidel S., Opaluch A. M., Caldwell J. S., Weitzman M. D., Kuhen K. L., Bandyopadhyay S., Ideker T., Orth A. P., Miraglia L. J., Bushman F. D., Young J. A., Chanda S. K. (2008) Cell 135, 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brass A. L., Dykxhoorn D. M., Benita Y., Yan N., Engelman A., Xavier R. J., Lieberman J., Elledge S. J. (2008) Science 319, 921–926 [DOI] [PubMed] [Google Scholar]
- 38. Gurer C., Berthoux L., Luban J. (2005) J. Virol. 79, 910–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martinez N. W., Xue X., Berro R. G., Kreitzer G., Resh M. D. (2008) J. Virol. 82, 9937–9950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jaber T., Bohl C. R., Lewis G. L., Wood C., West J. T., Jr., Weldon R. A., Jr. (2009) J. Virol. 83, 10448–10459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weldon R. A., Jr., Sarkar P., Brown S. M., Weldon S. K. (2003) Virology 314, 62–73 [DOI] [PubMed] [Google Scholar]
- 42. Gervaix A., West D., Leoni L. M., Richman D. D., Wong-Staal F., Corbeil J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4653–4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. He J., Chen Y., Farzan M., Choe H., Ohagen A., Gartner S., Busciglio J., Yang X., Hofmann W., Newman W., Mackay C. R., Sodroski J., Gabuzda D. (1997) Nature 385, 645–649 [DOI] [PubMed] [Google Scholar]
- 44. Cherepanov P., Pluymers W., Claeys A., Proost P., De Clercq E., Debyser Z. (2000) FASEB J. 14, 1389–1399 [DOI] [PubMed] [Google Scholar]
- 45. Ayaydin F., Dasso M. (2004) Mol. Biol. Cell. 15, 5208–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cherepanov P., Sun Z. Y., Rahman S., Maertens G., Wagner G., Engelman A. (2005) Nat. Struct. Mol. Biol. 12, 526–532 [DOI] [PubMed] [Google Scholar]
- 47. Carayon K., Leh H., Henry E., Simon F., Mouscadet J. F., Deprez E. (2010) Nucleic Acids Res. 38, 3692–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Leh H., Brodin P., Bischerour J., Deprez E., Tauc P., Brochon J. C., LeCam E., Coulaud D., Auclair C., Mouscadet J. F. (2000) Biochemistry 39, 9285–9294 [DOI] [PubMed] [Google Scholar]
- 49. Brussel A., Sonigo P. (2004) J. Virol. 78, 11263–11271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lu R., Limón A., Ghory H. Z., Engelman A. (2005) J. Virol. 79, 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xue Y., Zhou F., Fu C., Xu Y., Yao X. (2006) Nucleic Acids Res. 34, W254–W257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kuiken C. L., Foley B., Freed E., Hahn B., Korber P. A., Marx F., McCutchan J. W., Wolinksy S. (2002) HIV Sequence Compendium 2002, Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM [Google Scholar]
- 53. Rodriguez M. S., Dargemont C., Hay R. T. (2001) J. Biol. Chem. 276, 12654–12659 [DOI] [PubMed] [Google Scholar]
- 54. Petit C., Schwartz O., Mammano F. (1999) J. Virol. 73, 5079–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Goff S. P. (2001) Fields Virology, 4th Ed., pp. 1871–1939, Lippincott Williams & Wilkins ed., Philadelphia [Google Scholar]
- 56. Liu H., Wu X., Xiao H., Conway J. A., Kappes J. C. (1997) J. Virol. 71, 7704–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Llano M., Saenz D. T., Meehan A., Wongthida P., Peretz M., Walker W. H., Teo W., Poeschla E. M. (2006) Science 314, 461–464 [DOI] [PubMed] [Google Scholar]
- 58. Shun M. C., Raghavendra N. K., Vandegraaff N., Daigle J. E., Hughes S., Kellam P., Cherepanov P., Engelman A. (2007) Genes Dev. 21, 1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Engelman A. (1999) Adv. Virus. Res. 52, 411–426 [DOI] [PubMed] [Google Scholar]
- 60. Al-Mawsawi L. Q., Neamati N. (2007) Trends Pharmacol. Sci. 28, 526–535 [DOI] [PubMed] [Google Scholar]
- 61. Cannon P. M., Byles E. D., Kingsman S. M., Kingsman A. J. (1996) J. Virol. 70, 651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ceccherini-Silberstein F., Malet I., D'Arrigo R., Antinori A., Marcelin A. G., Perno C. F. (2009) AIDS Rev. 11, 17–29 [PubMed] [Google Scholar]
- 63. Rhee S. Y., Liu T. F., Kiuchi M., Zioni R., Gifford R. J., Holmes S. P., Shafer R. W. (2008) Retrovirology 5, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Saitoh H., Hinchey J. (2000) J. Biol. Chem. 275, 6252–6258 [DOI] [PubMed] [Google Scholar]
- 65. Matic I., Schimmel J., Hendriks I. A., van Santen M. A., van de Rijke F., van Dam H., Gnad F., Mann M., Vertegaal A. C. (2010) Mol. Cell. 39, 641–652 [DOI] [PubMed] [Google Scholar]
- 66. Eladad S., Ye T. Z., Hu P., Leversha M., Beresten S., Matunis M. J., Ellis N. A. (2005) Hum. Mol. Genet. 14, 1351–1365 [DOI] [PubMed] [Google Scholar]
- 67. Lin D. Y., Huang Y. S., Jeng J. C., Kuo H. Y., Chang C. C., Chao T. T., Ho C. C., Chen Y. C., Lin T. P., Fang H. I., Hung C. C., Suen C. S., Hwang M. J., Chang K. S., Maul G. G., Shih H. M. (2006) Mol. Cell. 24, 341–354 [DOI] [PubMed] [Google Scholar]
- 68. Wang J. Y., Ling H., Yang W., Craigie R. (2001) EMBO J. 20, 7333–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen J. C., Krucinski J., Miercke L. J., Finer-Moore J. S., Tang A. H., Leavitt A. D., Stroud R. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Swanson C. M., Malim M. H. (2008) Cell 133, 742. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.