Abstract
In the endoplasmic reticulum (ER), nascent membrane and secreted proteins that are misfolded are retrotranslocated into the cytosol and degraded by the proteasome. For most ER-associated degradation (ERAD) substrates, ubiquitylation is essential for both their retrotranslocation and degradation. Yeast Doa10 is a polytopic membrane ubiquitin ligase (E3) that along with its cognate ubiquitin-conjugating enzymes (E2s), Ubc7 and the C-terminally membrane-anchored Ubc6, makes a major contribution to ER-associated degradation. Ubc6 is also a substrate of Doa10. One highly conserved Doa10 element, the uncharacterized ∼130-residue TEB4-Doa10 domain, includes three transmembrane helices (TMs). We find that the first of these, TM5, includes an absolutely conserved ΦPΦXXG motif that is required for Doa10 function, as well as highly conserved negatively charged glutamate and aspartate residues. The conservative exchange of the TM5 glutamate to aspartate (doa10-E633D) results in complete stabilization of Ubc6 but has little if any effect on other substrates. Unexpectedly, mutating the glutamate to glutamine (doa10-E633Q) specifically accelerates Ubc6 degradation by ∼5-fold. Other substrates are weakly stabilized in doa10-E633Q cells, consistent with reduced Ubc6 levels. Notably, catalytically inactive ubc6-C87A is degraded in doa10-E633Q but not wild-type cells, but an active version of Ubc6 is required in trans. Fusion of the Ubc6 TM to a soluble protein yields a protein that is degraded in a doa10-E633Q-dependent manner, whereas fusion of the C-terminal TM from an unrelated protein does not. These results suggest that the TEB4-Doa10 domain regulates Doa10 association with the Ubc6 membrane anchor, thereby controlling the degradation rate of the E2.
Keywords: E3 Ubiquitin Ligase, Membrane Proteins, Protein Degradation, Ubiquitin-conjugating Enzyme (Ubc), Ubiquitylation, Doa10, Endoplasmic Reticulum-associated Degradation (ERAD), MARCH6, TD (TEB4-Doa10) Domain, TEB4
Introduction
Most secreted and integral membrane proteins in eukaryotes are initially translocated from the cytoplasm, usually co-translationally, across the ER3 membrane (1, 2) where they fold and assemble and are often proteolytically processed, glycosylated, or oxidatively modified. The ER contains specialized membrane translocation and protein modification machineries that execute these complex protein localization and processing reactions. These processes are not 100% accurate, so ER quality control mechanisms have evolved to identify misfolded, unassembled, or aberrantly modified proteins and either repair the errors or eliminate the abnormal proteins. The latter mechanisms fall under the category of ER-associated degradation (ERAD) (reviewed in Refs. 3–5).
ERAD substrates are retrotranslocated from the ER lumen or ER membrane to the cytoplasm and are degraded there by the proteasome (6–8). Substrate modification by polyubiquitin appears to be necessary for both efficient retrotranslocation and degradation in most cases (3). A series of enzymatic reactions catalyzed by three types of enzymes promote ubiquitin conjugation to substrates as follows: E1 ubiquitin-activating, E2 ubiquitin-conjugating, and E3 ubiquitin-ligating enzymes. The E3s are the primary factors determining substrate specificity, and they also stimulate transfer of ubiquitin from the E2 to the substrate. For ERAD, ubiquitin-substrate conjugation occurs at the cytoplasmic surface of the ER membrane (3, 4), and proteins are retrotranslocated through an as-yet unidentified conduit(s) across the membrane with the aid of different AAA+ ATPase-containing complexes, specifically the proteasome regulatory particle and/or the Cdc48/p97 homohexamer (9, 10).
Two integral membrane E3 ligases in the ER of the yeast Saccharomyces cerevisiae, Hrd1/Der3 and Doa10, account for most ER-associated proteasomal proteolysis (3, 4). Doa10 has a remarkably broad substrate range, being capable of recognizing both soluble nuclear and cytoplasmic substrates in addition to ER and nuclear envelope (NE) integral membrane proteins (11, 12). Doa10 orthologs are apparent in the great majority of fully sequenced eukaryotic genomes; among them is the human protein TEB4 (MARCH6/KIAA0597) (13–15). These proteins all contain a variant zinc-coordinating RING motif at their N termini called a RING-CH domain (13, 16). Doa10 is the only clear example of a RING-CH protein in S. cerevisiae, but the human genome encodes at least 11 different ones, most of which are also integral membrane proteins (the membrane-associated RING-CH or MARCH proteins) (14, 17). In addition to the more broadly distributed proteins with RING-CH motifs, all of the presumptive Doa10 orthologs are predicted to have at least 10 transmembrane helices (TMs), and all contain a conserved ∼130-residue internal element known as the TEB4-Doa10 (TD) domain (13).
For Doa10, the E2 requirements are unusual, with two distinct enzymes, Ubc6 and Ubc7, both required for efficient ubiquitylation of substrates (13, 18, 19). Ubc6 is bound to the ER membrane via a C-terminal transmembrane anchor (20), whereas the soluble Ubc7 enzyme localizes to the ER by binding the transmembrane Cue1 receptor protein, which also allosterically activates the E2 (21–23). Despite knowledge of the E2s and other cofactors implicated in Doa10-mediated ubiquitylation, our understanding of the activities of the Doa10 ubiquitylation complex and the interplay between its constituents is currently very limited. It is not known why Doa10 requires two separate E2s, nor is it understood in any detail how these E2s, or other associated factors such as the Cdc48 complex, interact with Doa10. Unlike Ubc7, which is a stable protein as long as it is bound to Cue1 (24), Ubc6 is constitutively turned over via the Doa10 pathway with a half-life of ∼55 min (13, 25). The physiological rationale behind this turnover is unclear. Doa10 was also proposed to have a direct role in substrate passage through the membrane bilayer to the cytosol (13), but this remains unproven.
Here, we describe a surprising link between an intramembrane charged residue in the first TM of the TD domain of Doa10 and the behavior of the Ubc6 E2, which has uncovered at least one function of the TD domain. When the highly conserved TM5 glutamate residue, Glu-633, was mutated to an aspartate (E633D), Ubc6 was strongly stabilized, but degradation of other Doa10 substrates was not detectably impaired. Conversely, mutation of Glu-633 to glutamine (E633Q) dramatically enhanced Ubc6 turnover with a concomitant weak impairment in the degradation of other substrates. This rapid Ubc6 degradation required Doa10 RING-CH function as well as the Ubc7 E2, which is also true of the normal, much slower degradation of Ubc6 in wild-type (WT) DOA10 cells. Most interestingly, accelerated Ubc6 degradation also depended on the catalytic activity of Ubc6 itself, but this activity could be provided by a trans copy of the E2, unlike in WT cells. Thus, Ubc6 is both an integral component of the Doa10 ubiquitylation machinery and a substrate of this same machinery. Doa10 may either form a ubiquitylation complex containing both Ubc7 and (multiple subunits of) Ubc6, or it may interact sequentially with the E2s.
We speculate that Doa10 has multiple binding sites for Ubc6, with one site (the “E2 site”) for ubiquitin transfer from the ubiquitin-Ubc6 thioester to a substrate and a nearby site (the “substrate site”) where substrates normally bind and become polyubiquitylated. In this model, Ubc6 access to the substrate site is normally inhibited by structural features of the TD domain that depend on Glu-633 in TM5; the barrier is reduced in the doa10-E633Q protein, leading to abnormally rapid Ubc6 degradation. The charged residue(s) in the TMs of the TD domain could stabilize the architecture of the Ubc6-binding site(s) of Doa10 and/or allosterically connect the two different binding sites.
EXPERIMENTAL PROCEDURES
Yeast and Bacterial Methods
Yeast-rich (YPD) and minimal (SD) media were prepared as described previously, and standard methods were used for genetic manipulation of yeast (26). Standard techniques were used for recombinant DNA work in Escherichia coli. Yeast strains and cultures were grown at 30 °C. For spot growth assays, cells were grown in minimal medium to log phase, serially diluted (5-fold steps) in water, and spotted onto different media. Plates were incubated at 30 °C for 2–4 days.
Strain and Plasmid Constructions
The yeast strains used in this study are listed in supplemental Table S1. The full-length DOA10 gene is not stably maintained in E. coli. Therefore, for mutagenesis of DOA10, yeast strains with a mutated chromosomal copy of DOA10 were generated using the two-step delitto perfetto method (27). In the first step, a counterselectable reporter cassette (CORE cassette) containing KlURA3 and kanMX6 cassettes was PCR-amplified from the plasmid pCORE (27) and integrated in the DOA10 locus after Doa10 ORF nucleotide 115 (for generation of the doa10-C39S allele) or 1914 (for generation of mutations in TM5). In the second step, the mutant doa10 allele was generated by replacing the CORE cassette by homologous recombination with an oligonucleotide duplex encoding the desired Doa10 mutation flanked with ∼45 bp of homologous sequence at both ends. Correct recombination to generate the desired doa10 allele was in each case verified by DNA sequencing.
Plasmids encoding fusions of the Deg1 degron-coding sequence to the URA3 reporter have been described previously (11, 18). The p414MET25-Deg1-Vma12-KanMX plasmid was made by recombination in yeast between the PCR-amplified KanMX6 ORF from pFA6a-KanMX6 (28) and gapped p414MET25-Deg1-Vma12-PrA (11). An expression plasmid for internally HA-tagged Ubc6 (pRS416-Ubc6HA) was a gift from T. Sommer (Max-Delbrück-Center, Berlin) and was described previously (25). Plasmid pRS416-ubc6(C87A)HA was generated using the QuikChange protocol (Stratagene) with pRS416-Ubc6HA as template. Plasmid p414MET25-URA3-HA-Ubc6TM was generated in two steps. First, the URA3 ORF was PCR-amplified from pRS426 (29), adding SpeI and PstI sites to the 5′ and 3′ ends, respectively. Second, following digestion with SpeI and PstI, the URA3 insert was cloned into the same sites in p414MET25 (30) to yield p414MET25-URA3. The HA-Ubc6TM insert was generated by PCR amplification of the coding sequence for Ubc6TM (Ubc6 residues 213–250, which includes the membrane anchor plus 18 upstream residues) and adding a flanking sequence encoding HA and a PstI site at the 5′ end and a SalI site at the 3′ end. The HA-Ubc6TM insert was cloned into p414MET25-URA3 using the PstI and SalI sites to yield p414MET25-URA3-HA-Ubc6TM. To generate plasmid p414MET25-URA3-HA-Prm3TM, the sequence encoding Prm3 residues 92–133 was PCR-amplified from MHY500 genomic DNA, adding flanking BamHI and XhoI sites at the 5′ and 3′ ends, respectively. The BamHI/XhoI-digested PRM3 PCR fragment was ligated to BamHI/SalI-cut p414MET5-URA3-HA-Ubc6TM to yield p414MET25-URA3-HA-Prm3TM. All plasmids were verified by DNA sequencing.
Preparation of Cell Extracts and Immunoblotting
Cell extracts were prepared as described previously (31). Briefly, 2.5 A600 eq of logarithmically growing cells were lysed in β-mercaptoethanol/NaOH, and proteins were precipitated in 5% trichloroacetic acid. The pellet was resuspended in SDS gel-loading buffer. Proteins were separated by SDS-PAGE and electrotransferred onto PVDF membranes (Amersham Biosciences). Immunodetection was carried out with appropriate primary antibodies and horseradish peroxidase-conjugated secondary antibodies. Immunoreactive species were visualized using ECL reagents (Amersham Biosciences). The following antibodies were used: anti-HA mouse monoclonal antibody 16B12 (Covance); anti-Myc mouse monoclonal antibody 9E10 (Covance); anti-Pgk1 mouse monoclonal antibody 22C5 (Molecular Probes); anti-Doa10 antiserum from rabbit raised against a His6-tagged N-terminal 128-residue fragment of Doa10 (17), and a rabbit polyclonal anti-Ubc6 antiserum (gift from T. Sommer, Max-Delbrück Center, Berlin, Germany).
Co-immunoprecipitation Analysis
Logarithmically growing yeast cells (25 A600 eq) were harvested by centrifugation and resuspended in 0.5 ml of ice-cold extraction buffer (50 mm Tris-HCl, pH 7.5, with protease inhibitors PMSF (625 μm) and aprotinin (5 μg/ml)). All steps were done at 4 °C except where indicated. Following addition of 1 volume of glass beads, cells were disrupted by four cycles of vortexing (30-s pulse/30 s on ice). Lysates were diluted with 0.5 ml of extraction buffer and cleared by centrifugation for 5 min at 370 × g. The crude microsomal fraction was collected by centrifugation of the resulting supernatant at 16,000 × g for 10 min and was resuspended in 0.5 ml of resuspension buffer (RB) (50 mm Tris-HCl, pH 7.5, 200 mm NaAc, 10% glycerol with protease inhibitors (625 μm PMSF and 5 μg/ml each of aprotinin, leupeptin, and chymostatin; 10 μg/ml pepstatin; and 2 μg/ml antipain)). Membranes were solubilized by addition of digitonin to 1% with a 10-min incubation on ice. The supernatant after centrifugation (16,000 × g for 10 min) was diluted 1:1 with RB. An aliquot (5% of total) of the diluted supernatant was removed and served as input for immunoblot. Rabbit anti-Doa10 serum or mouse 9E10 anti-Myc antibody was added to the remaining supernatant and incubated overnight with constant agitation. Subsequently, protein A-Sepharose beads (for anti-Doa10) or protein G-Sepharose beads (for anti-Myc) equilibrated in RB were added and incubated for 2 h with gentle agitation. Beads were washed four times with 0.5 ml of RB without protease inhibitors, and bound proteins were eluted by addition of 50 μl of 2× sample buffer and incubation for 15 min at 37 °C before SDS-PAGE and immunoblotting.
Degradation Assays
Cycloheximide-chase/Western blot analysis was carried out as described previously (17). Briefly, cycloheximide (0.25 mg/ml) was added to logarithmically growing yeast cultures, and cell aliquots were removed at the indicated times after addition. Cells were pelleted, resuspended in cold STOP mix (0.5× SD, 10 mm NaN3), and stored on ice until all time points had been collected. Lysates were generated, and proteins were visualized by immunoblotting. Protein degradation rates were measured using a G:Box system (Syngene). Values for each time point were normalized using an anti-Pgk1 loading control.
Pulse-chase analysis was carried out as described previously (18). Deg1-βgal was immunoprecipitated with an anti-α2 antiserum (32). Radiolabeled Deg1-βgal was visualized and quantified using a PhosphorImager (GE Healthcare) and ImageQuant software, and half-lives were calculated from exponential curve fits using DeltaGraph 5.
Fluorescence Microscopy of Live Yeast Cells
For localization of GFP fusion proteins, cells (MHY3612, Doa10-GFP; MHY3693, doa10-E633Q-GFP) were grown to mid-log phase, washed twice, and resuspended in sterile water; 2.5 μl were spotted on a slide and sealed under a coverslip. GFP was excited using a 470 nm (40-nm bandwidth) bandpass filter and visualized with a 525 nm (50-nm bandwidth) bandpass filter on a Axioplan epi-fluorescence microscope (Carl Zeiss) equipped with a 100× plan-apochromat 1.4 NA objective lens. Pictures were taken on a Zeiss Axiocam camera using a Uniblitz shutter driver (model VMM-D1; Vincent Assocs., Rochester, NY) and the program Open Lab 3.1.5 (Improvision, Lexington, MA).
RESULTS
Sequence Analysis of the Doa10 Subfamily of RING-CH E3s
With the availability of complete genome sequences from multiple eukaryotes, we could explore both the broad distribution of Doa10 orthologs throughout the Eukarya and the extent of conservation among these proteins. All Doa10 orthologs were characterized by an N-terminal RING-CH domain, an internal conserved segment of ∼130 residues called the TD domain, and at least 10 predicted TMs (Fig. 1A) (13). The TD domain includes three TMs (17). Sequences that fit the above criteria for a Doa10 ortholog are found in species from all four of the five currently recognized eukaryotic “supergroups” for which fully sequenced genomes are available (Plantae, Excavates, Unikonts, and Chromalveolates). From this, it can be inferred that a Doa10-like protein was already present in the last eukaryotic common ancestor (33). On the other hand, Doa10 has been lost in certain lineages within the different supergroups. For instance, among the Plantae, a Doa10 ortholog was not detected in three fully sequenced green alga genomes, but it is present in land plants and red algae (Cyanidioschizon merolae) (Fig. 1B). In species that lack Doa10, its functions in ERAD and nuclear protein degradation might be assumed by other ubiquitin ligases, such as HRD1 (34).
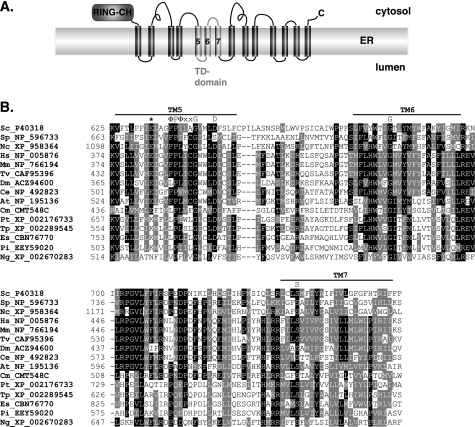
FIGURE 1.
TD domain of the Doa10 subfamily of RING-CH proteins. A, schematic of the yeast Doa10 ubiquitin ligase highlighting the N-terminal RING-CH and TD domains. B, sequence alignment (ClustalW) of the TD domains from a wide range of highly divergent eukaryotes. The three predicted TM segments are highlighted by horizontal bars. The conserved glutamate (Glu-633 in S. cerevisiae) is marked by an asterisk. The invariant TM5 ΦPΦXXG motif (where Φ is a hydrophobic residue), the highly conserved aspartate (Asp-646 in S. cerevisiae), and the strictly conserved glycine in TM6 and serine in TM7 are indicated. Sc, S. cerevisiae; Sp, Schizosaccharomyces pombe; Nc, Neurospora crassa; Hs, Homo sapiens; Mm, Mus musculus; Tv, Tetraodon nigroviridis; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; At, Arabidopsis thaliana; Cm, Cyanidioschyzon merolae; Pt, Phaeodactylum tricornutum; Tp, Thalassiosira pseudonana; Es, Ectocarpus siliculosus; Pi, Phytophthora infestans; and Ng, Naegleria gruberi. To simplify the figure, TD domain residues 504–529 from T. nigroviridis and residues 541–601 from N. gruberi were deleted.
As expected, the most conserved regions in all the identified Doa10 orthologs were the RING-CH and TD domains (Fig. 1B). A striking feature of the TD alignment is the presence in TM5 (S. cerevisiae TM numbering) of two highly conserved charged residues as well as strictly conserved proline and glycine residues. In addition, TM6 has a fully conserved glycine and TM7 a strictly conserved serine; the latter residue has been associated with kinking of transmembrane helices from analysis of known membrane protein structures (35). In TM5, the absolutely conserved proline and glycine residues are part of a ΦPΦXXG motif (where Φ is a hydrophobic residue). Such a motif has not been described previously but might help create a specific bend or interruption in the TM5 helix (see “Discussion”).
Charged and polar residues are only infrequently encountered in the membrane interior (36) and often form favorable contacts with residues of other proteins or other TMs of the same protein within the lipid bilayer (37). There are two highly conserved negatively charged residues in TM5. One is an aspartate (yeast Asp-646) that is broadly conserved, and the other is a glutamate (yeast Glu-633) that is absolutely conserved in the Unikonts and Plantae (Fig. 1B, first 10 species in alignment). By contrast, a lysine is found instead of glutamate at this position in all the fully sequenced stramenopiles (Phaeodactylum tricornutum, Thalassiosira pseudonana, Ectocarpus siliculosus, and Phytophthora infestans), which represent a major branch within the Chromalveolates. In land plants, a second Doa10-related protein can be identified, and it also has a lysine at this position, unlike its paralog, which has a glutamate (Fig. 1B, At). The charged side chains at these positions could mediate specific interactions with substrates or cofactors or could be important for achieving a specific structure of Doa10 within the membrane (or both).
ΦPΦXXG Motif in TM5 of Doa10 Is Important for Function
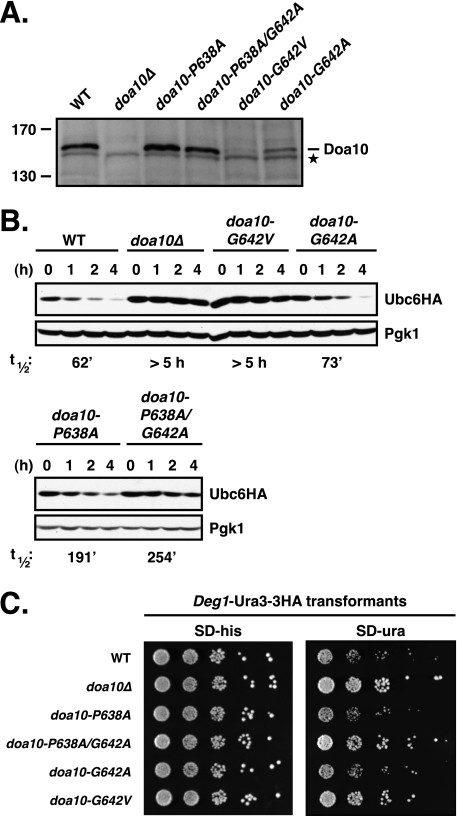
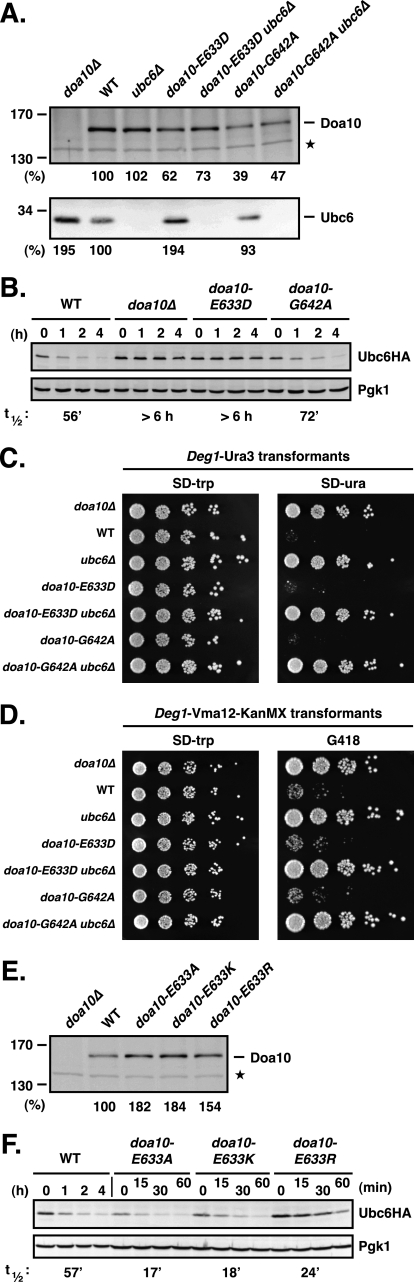
We focused our mutagenesis studies on TM5 because of its highly unusual sequence. First we asked whether the ΦPΦXXG motif in TM5 was indeed important for Doa10 function. To this end, we replaced the proline and glycine residues individually or together with either alanine or valine. Levels of the doa10-P638A and the P638A/G642A proteins expressed from the natural chromosomal locus were comparable with that of WT Doa10, whereas the levels of G642A and G642V were significantly reduced (to ∼40% and <5% of WT levels) (Fig. 2A). We tested the activity of the ΦPΦXXG motif mutants toward Ubc6, one of two cognate E2 enzymes of Doa10 (Fig. 2B). The C-terminally membrane-anchored Ubc6 protein is normally ubiquitylated and degraded relatively slowly by a mechanism that depends on Doa10, Ubc7, as well as the active site of Ubc6 itself (13, 25). Consistent with previous results, we found that a functional HA-tagged version of Ubc6 (20) had a half-life of ∼50–60 min in wild-type (WT) cells (Fig. 2B) and was strongly stabilized in cells lacking Doa10 (t½ >5 h). Ubc6 was only slightly stabilized in doa10-G642A-expressing cells (t½ = 73 min) but was significantly more stable in the other doa10 ΦPΦXXG mutant cells.
FIGURE 2.
ΦPΦXXG motif in TM5 of Doa10 is important for Doa10 function. A, steady state levels of individual ΦPΦXXG motif mutants. The mutant alleles were expressed from the natural chromosomal locus, and lysates were prepared and analyzed by immunoblotting with a Doa10-specific polyclonal rabbit antiserum. Asterisk, a cross-reacting yeast protein that allows comparison of protein loading. B, degradation of Ubc6HA in ΦPΦXXG motif mutant cells. Following addition of cycloheximide, aliquots of cells were taken at the indicated times, and lysates were analyzed by anti-HA and anti-Pgk1 immunoblotting. The ΦPΦXXG mutants were expressed from the chromosomal DOA10 locus. Ubc6HA was expressed from a CEN plasmid in the presence of endogenous Ubc6. Approximate half-lives, listed below the panel, were determined as described under “Experimental Procedures.” C, degradation of the soluble substrate Deg1-Ura3–3HA in ΦPΦXXG motif mutant cells inferred from growth assays. 5-Fold serial dilutions of the indicated strains that had been transformed with a HIS3-marked Deg1-Ura3–3HA expression plasmid were spotted onto minimal plates lacking histidine (SD−his) or uracil (SD−ura) and incubated at 30 °C for 2.5 days (−his) and 4 days (−ura), respectively.
We also evaluated the activity of each mutant toward the model Doa10 substrate Deg1-Ura3–3HA using a degradation-sensitive growth assay (Fig. 2C). The Deg1 degradation signal is within the first 67 residues of the yeast MATα2 transcriptional repressor and is recognized by the Doa10 pathway (13, 18). WT cells expressing Deg1-Ura3–3HA as the only source of Ura3 enzyme, which is necessary for uracil biosynthesis, grow poorly on media lacking uracil due to rapid degradation of the fusion protein. Deletion of DOA10 greatly stimulated growth (Fig. 2C). Growth rates of the TM5 mutants on plates lacking uracil generally correlated with the degree of Ubc6HA protein stabilization (Fig. 2, B and C). Thus, mutation of both Pro-638 and Gly-642 in the ΦPΦXXG motif strongly reduced activity toward Ubc6 and Deg1-Ura3–3HA. These data show that the highly conserved TM5 ΦPΦXXG motif is indeed critical for Doa10 function.
Changing the Conserved Glu-633 to Gln in TM5 Causes Rapid Degradation of Ubc6
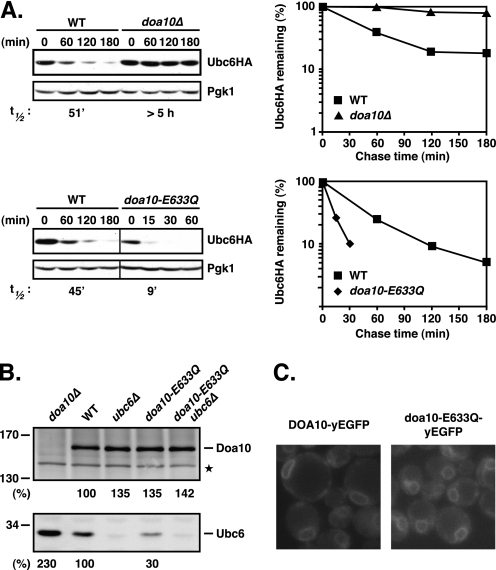
To test whether the negatively charged Glu-633 residue in TM5 was necessary for proper Doa10 function, we mutated the chromosomal copy of DOA10 so that it instead encoded glutamine. Unexpectedly, the Ubc6 E2 enzyme became extremely unstable in the doa10-E633Q mutant (Fig. 3A). Ubc6HA was degraded at least 5-fold faster in doa10-E633Q (t½ ∼9 min) than in WT cells. The accelerated degradation of Ubc6HA was associated with a substantial drop in Ubc6HA steady state levels, indicating the absence of a compensatory increase in Ubc6 synthesis rates (Fig. 3A; twice the amount of lysate was loaded in the doa10-E633Q lanes to facilitate protein detection). The enhanced degradation of Ubc6HA was not due to the HA tag or plasmid-based expression as we observed similar effects on the steady state levels of endogenous untagged Ubc6 protein (Fig. 3B). Moreover, the WT Doa10 and the doa10-E633Q proteins were expressed at comparable levels with or without Ubc6 present (Fig. 3B) and showed a similar localization to the ER and nuclear envelope (Fig. 3C). Therefore, the dramatically enhanced degradation of Ubc6 in doa10-E633Q cells is unlikely to be due to any gross perturbation in Doa10 structure or localization.
FIGURE 3.
E633Q mutation in TM5 of Doa10 strongly enhances degradation of Ubc6. A, degradation of Ubc6HA in WT, doa10Δ, and doa10-E633Q cells. Left panels, cycloheximide chase/immunoblot was done as in Fig. 2B. Note: twice as much lysate was loaded for the doa10-E633Q samples compared with WT. Right panels, quantitation of Ubc6HA degradation kinetics from cycloheximide/Western chase data. Values were normalized to Pgk1 controls. B, steady state levels of WT Doa10 and doa10-E633Q in the presence and absence of Ubc6 and corresponding Ubc6 steady state levels. The doa10-E633Q mutant was expressed from the genomic locus. Lysates were prepared, separated on a 5–15% SDS-PAGE gradient gel, and analyzed by immunoblotting with a Doa10- or Ubc6-specific antiserum (as indicated). The asterisk denotes a cross-reactive yeast protein used for normalization. Quantitation of Doa10 and Ubc6 steady state levels was carried out as described under “Experimental Procedures.” Doa10 and Ubc6 levels in WT were set to 100%. C, subcellular localization of Doa10-yEGFP and doa10-E633Q-yEGFP proteins in live cells. GFP fluorescence of both chromosomally expressed proteins localizes to the ER/NE.
Degradation of Soluble and Membrane Substrates Is Weakly Impaired in doa10-E633Q Cells
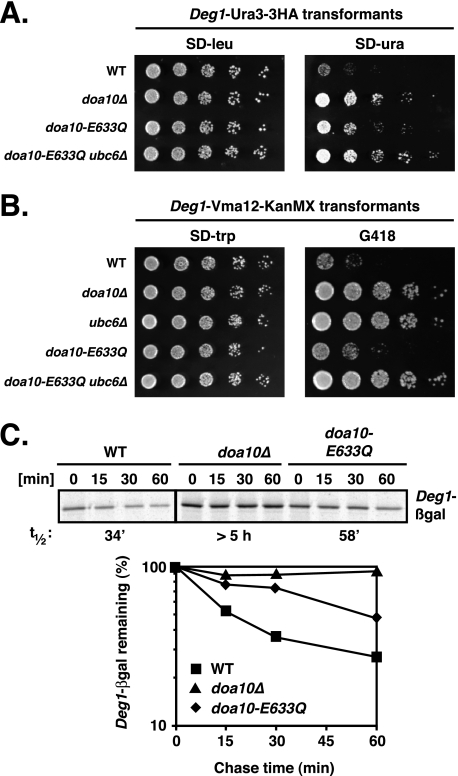
Other Doa10 substrates might also be degraded more rapidly in the doa10-E633Q strain, or conversely, the reduced levels of Ubc6 in this mutant might impair the ubiquitylation and degradation of these substrates. To investigate this, we determined the degradation rates of different model substrates in doa10-E633Q cells using degradation-sensitive growth assays. First we tested the activity of doa10-E633Q against the soluble substrate Deg1-Ura3–3HA (Fig. 4A). In doa10-E633Q cells, a modest increase in growth on plates lacking uracil was observed, suggesting a mild defect in Deg1-Ura3–3HA degradation. Doa10 also targets integral membrane protein substrates. We generated a novel membrane reporter substrate by fusing Deg1 to Vma12, an ER membrane protein with two TMs, followed by a KanMX sequence, which confers resistance to the antibiotic G418 (geneticin). As shown in Fig. 4B, WT cells expressing Deg1-Vma12-KanMX grew very poorly on plates containing G418, whereas doa10Δ and ubc6Δ cells grew well. Mutant doa10-E633Q cells transformed with the reporter grew slightly better on G418 than WT cells, suggesting a weak impairment of Deg1-Vma12-KanMX degradation; this degradation still required Ubc6. Hence, the doa10-E633Q allele does not appear to accelerate degradation of other substrates of Doa10 but instead weakly stabilizes them.
FIGURE 4.
Degradation of both soluble and membrane Doa10 substrates is slightly slower in doa10-E633Q cells. A, decreased degradation of the soluble substrate Deg1-Ura3–3HA in doa10-E633Q cells inferred from growth assays. Serial dilutions of the indicated strains that had been transformed with a LEU2-marked Deg1-Ura3–3HA expression plasmid were spotted onto minimal plates lacking leucine (SD−leu) or uracil (SD−ura) and incubated for 3 days at 30 °C. B, decreased degradation of the membrane substrate Deg1-Vma12-KanMX. Serial dilutions of the indicated strains that had been transformed with a TRP1-marked Deg1-Vma12-KanMX expression plasmid were spotted onto minimal plates lacking tryptophan (SD−trp) or on YPD plates with 300 μg/ml G418 and incubated for 2 days. C, degradation of the soluble protein Deg1-βgal. Degradation rates of Deg1-βgal were determined by [35S]Met pulse-chase analysis in WT, doa10-E633Q, and doa10Δ cells expressing Deg1-βgal from the chromosome. Deg1-βgal was immunoprecipitated with an anti-βgal antibody. Lower panel, quantitation of Deg1-βgal pulse-chase data.
For a more quantitative readout of doa10-E633Q activity toward Doa10 substrates, we determined the half-life of Deg1-β-galactosidase (Deg1-βgal) (13, 18) by pulse-chase analysis (Fig. 4C). Deg1-βgal had a half-life of ∼34 min in WT cells and was stabilized at least 10-fold in doa10Δ cells (t½ >5 h), comparable with what is seen in a ubc6Δ mutant (18). Consistent with what was seen in the growth assays, Deg1-βgal was weakly stabilized (∼2-fold) in doa10-E633Q cells (t½ ∼58 min). The slight increase in half-life is most likely due to the 3–4-fold reduction in Ubc6 enzyme levels in doa10-E633Q cells (Fig. 3B).
We conclude from these data that doa10-E633Q is still functional and able to work with the Ubc6 E2 to target Doa10 substrates for degradation. Accelerated degradation therefore appears to be specific to Ubc6 in the doa10-E633Q mutant.
Divergent Effects of Different Glu-633 Mutations
We also changed Glu-633 to several other amino acids to determine their effects on Doa10, particularly its role in Ubc6 degradation. The doa10-E633D mutation, which preserved the negative charge but reduced the length of the side chain by one methy-lene group, resulted in an ∼40% drop in E3 protein level compared with WT Doa10 (Fig. 5A). Despite causing only a modest reduction in Doa10 levels, this mutation severely impaired degradation of Ubc6HA (Fig. 5B); this correlated with an increase in endogenous Ubc6 to levels similar to those in doa10Δ cells (Fig. 5A). Stabilization of Ubc6 in doa10-E633D cells was not due simply to the slightly reduced levels of the E3 as Ubc6 is degraded with near-WT kinetics in doa10-G642A cells, in which the level of E3 protein is reduced even further (by ∼60% compared with WT) (Fig. 5, A and B). Therefore, the effect on Ubc6 degradation of changing Glu-633 to an Asp residue was opposite to that of changing it to a Gln residue, causing strong stabilization of the E2 in the former case and strong destabilization in the latter.
FIGURE 5.
E633D mutation in TM5 of Doa10 strongly decreases degradation of Ubc6 but has no detectable effect on other substrates. A, comparison of Doa10, doa10-E633D, and doa10-G642A protein levels in the presence and absence of Ubc6 and the corresponding Ubc6 steady state levels. Immunoblot analysis and quantitation was done as in Fig. 3B. Doa10 and Ubc6 levels in WT were set to 100%. Asterisk, a cross-reacting yeast protein used to compare protein loading. B, conservative doa10-E633D substitution leads to stabilization of Ubc6HA. Cycloheximide chase/anti-HA immunoblot analysis was done as in Fig. 2B. C, degradation of the soluble substrate Deg1-Ura3 in doa10-E633D cells (and doa10-G642A cells) inferred from growth assays. Degradation of Deg1-Ura3 is dependent on the presence of Ubc6. Serial dilutions of the indicated strains that had been transformed with a TRP1-marked Deg1-Ura3 expression plasmid were spotted onto minimal plates lacking tryptophan (SD−trp) or uracil (SD−ura) and incubated for 2.5 days at 30 °C. D, degradation of the membrane substrate Deg1-Vma12-KanMX in doa10-E633D cells (as well as doa10-G642A cells) inferred from growth assays. Degradation of Deg1-Vma12-KanMX is dependent on the presence of Ubc6. Serial dilutions of the indicated strains that had been transformed with a TRP1-marked Deg1-Vma12-KanMX6 expression plasmid were spotted onto a minimal plate lacking tryptophan (SD−trp) or on a YPD plate supplemented with 300 μg/ml G418 and incubated for 2 days at 30 °C. E, steady state levels of Doa10 and different doa10-E633 substitutions (Ala, Lys, or Arg). Immunoblot analysis and quantitation was done as in Fig. 3B. F, doa10-E633A, doa10-E633K, or doa10-E633R mutations all accelerate Ubc6HA degradation. Cycloheximide chase/anti-HA immunoblot analysis was done as in Fig. 2B.
To investigate whether the doa10-E633D substitution affected the degradation of substrates other than Ubc6, we again examined model substrates using the degradation-sensitive growth assays described earlier. Degradation of the soluble substrate Deg1-Ura3 was unaffected based on growth on media lacking uracil (Fig. 5C). This degradation still depended on Ubc6. Notably, doa10-G642A cells also showed apparent full activity toward Deg1-Ura3 despite having strongly reduced levels of the E3 ligase (Fig. 5, A and C). Mutant doa10-E633D cells expressing the Deg1-Vma12-KanMX membrane reporter grew as poorly on G418 as WT cells, suggesting rapid degradation of this substrate as well (Fig. 5D).
In summary, the mutant doa10-E633D ligase is inactive toward Ubc6 but retains strong activity toward both the soluble Deg1-Ura3 and membrane Deg1-Vma12-KanMX substrates. This activity is dependent on Ubc6. Therefore, degradation of Ubc6 is not essential for Doa10 activity toward other substrates.
Strains with Doa10-Glu-633 changed to alanine, lysine, or arginine were also evaluated. Each substitution resulted in a modest increase in Doa10 steady state levels, and Ubc6 degradation was accelerated for each mutant (Fig. 5, E and F). The doa10-E633R allele enhanced Ubc6 degradation ∼2-fold, whereas the doa10-E633A and -E633K mutations accelerated Ubc6 degradation ∼3-fold (Fig. 5F). This last result is particularly striking because in some distant species, orthologs of yeast Doa10 have a lysine at this position (Fig. 1B); this implies either that Doa10 in these species has co-evolved complementary changes elsewhere in the protein or in interacting factors (possibly Ubc6) or that these orthologs do not function with Ubc6.
Mutagenesis of Additional TM5 Residues
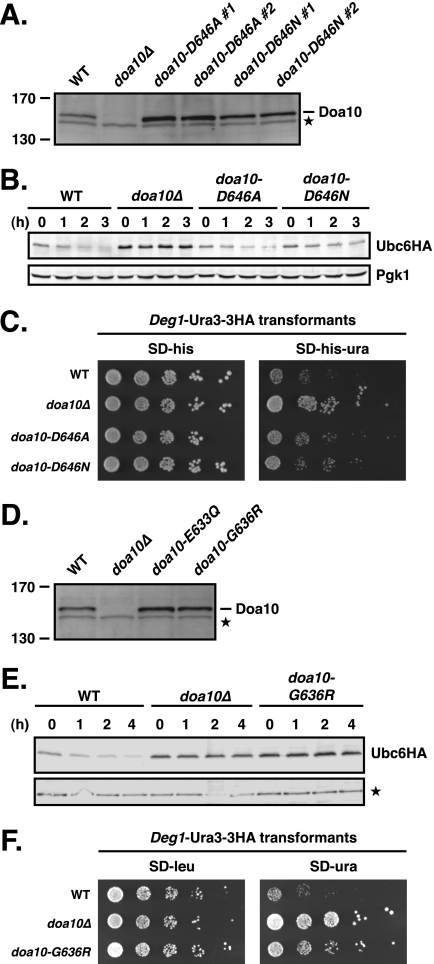
Mutation of the other conserved negatively charged residue in Doa10 TM5, Asp-646, to Ala or Asn led to at least partial stabilization of Ubc6 and other Doa10 substrates (Fig. 6, A–C, and data not shown). Therefore, these substitutions of Asp-646 (which also led to moderately enhanced levels of Doa10; Fig. 6A) caused reduced, rather than enhanced, rates of Ubc6 degradation, in contrast to the analogous mutations in Glu-633. We also mutated the other glycine in TM5, the nonconserved Gly-636. In doa10-G636R cells, both Ubc6 and Deg1-Ura3–3HA were significantly stabilized despite moderately elevated Doa10 levels (Fig. 6, D–F). Hence, perturbation of TM5 structure or membrane insertion by the G636R mutation had a general negative effect on Doa10 activity.
FIGURE 6.
Characterization of doa10-D646A, doa10-D646N, and doa10-G636R mutants. A, steady state levels of doa10-N646A and doa10–646N proteins. Lysates were prepared as in Fig. 2A (two independent strains of each doa10-D646 mutant were tested in parallel). Asterisk, a cross-reacting yeast protein that allowed comparison of protein loading. B, degradation of Ubc6HA in doa10-D646A and doa10-G646N cells. Cycloheximide chase/immunoblot was done as in Fig. 2B. C, degradation of the soluble substrate Deg1-Ura3–3HA in doa10-D646A and doa10-D646N cells inferred from growth assays. Serial dilutions of the indicated strains that had been transformed with a HIS3-marked Deg1-Ura3–3HA expression plasmid were spotted onto minimal plates lacking histidine (SD−his) or both histidine and uracil (SD−his−ura) and incubated for 3 days. D, comparison of steady state levels of WT Doa10, doa10-E633Q, and doa10-G636R proteins. Lysates were prepared as in Fig. 2A. Asterisk, a cross-reacting yeast protein that allowed comparison of protein loading. E, degradation of Ubc6HA in doa10-G636R cells. Cycloheximide chase/immunoblot was done as in Fig. 2B with the exception that a protein cross-reacting with the anti-HA antibody served as loading control. F, degradation defect for the soluble substrate Deg1-Ura3–3HA in doa10-G636R cells inferred from growth assays. Serial dilutions of the indicated strains that had been transformed with a LEU2-marked Deg1-Ura3–3HA expression plasmid were spotted onto SD−leu or SD−ura plates.
Collectively, these data indicate that the Doa10 TM5 structure is extremely sensitive to mutation and demonstrate a range of effects of mutations within TM5 on Doa10 activity and Ubc6 proteolysis. Most remarkably, they show that Ubc6 degradation rate is intimately linked to the identity of the side chain of residue 633 in this transmembrane helix. Because the doa10-E633Q substitution is the most conservative change that led to accelerated Ubc6 proteolysis, we focused our remaining analysis on this mutant.
Ubc6 Degradation Mediated by doa10-E633Q Requires Its E3 Ligase Activity and Ubc7
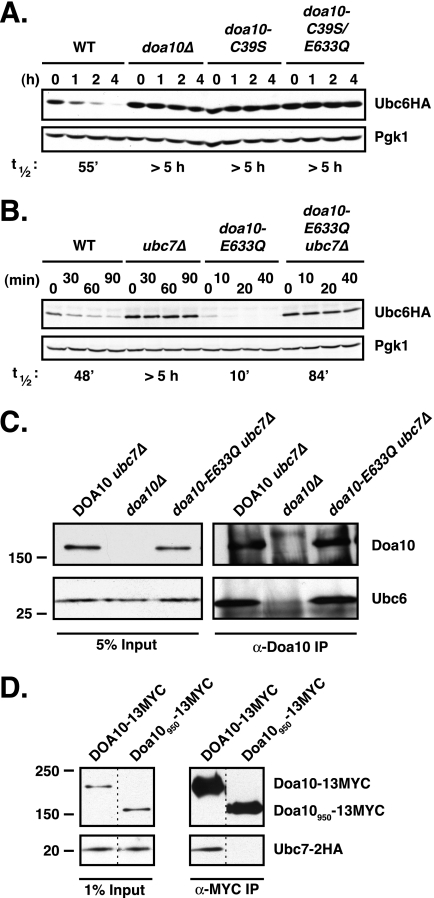
The moderate rate of Ubc6 degradation in WT cells depends on both the Doa10 E3 ligase and the Ubc7 E2 (13, 25). We asked whether the far more rapid degradation of Ubc6 in doa10-E633Q cells depended on the same activities. A second point mutation (encoding C39S) was engineered into the chromosomal doa10-E633Q allele to inactivate the RING-CH domain and thereby inactivate E3 ligase activity (13). WT and doa10-E633Q proteins with the C39S mutation were expressed at levels similar to their counterparts containing the intact RING domain (data not shown). Introduction of the C39S mutation into the RING-CH of doa10-E633Q strongly stabilized Ubc6HA (Fig. 7A; t½ >5 h). This degree of stabilization was similar to that observed with the doa10-C39S single mutant or complete deletion of Doa10. Ubc6HA was also strongly stabilized (>8-fold) in a doa10-E633Q ubc7Δ double mutant (Fig. 7B). Therefore, the rapid Ubc6 degradation caused by the doa10-E633Q substitution depends on both a functional RING-CH domain in Doa10 and the presence of Ubc7, as had been found for the slower turnover of Ubc6 in cells expressing Doa10 without this TM5 mutation. This suggests that the same mechanism of ubiquitin-dependent degradation is involved in both cases, but it becomes much more efficient in doa10-E633Q cells.
FIGURE 7.
E3 ligase activity of doa10-E633Q is required for the accelerated degradation of Ubc6. A, Ubc6HA is stabilized in the inactive doa10-C39S/E633Q double mutant. Cycloheximide-chase/immunoblot analysis of Ubc6HA stability was done as in Fig. 2B. B, rapid degradation of Ubc6HA in doa10-E633Q cells is dependent on Ubc7. C, comparable binding of Ubc6 to WT Doa10 and doa10-E633Q. Endogenous Doa10 or doa10-E633Q mutant proteins were immunoprecipitated from digitonin-solubilized microsomes with anti-Doa10 antibodies. Precipitates were analyzed by immunoblotting with anti-Doa10 or anti-Ubc6 antibodies. All panels are from same experiment. D, control showing that the isolation protocol used in Fig. 6C does not lead to nonspecific co-precipitation of proteins. Myc epitope-tagged WT Doa10 and a derivative, Doa10950, missing the last 369 residues, were precipitated with an anti-Myc antibody from digitonin-solubilized microsomes derived from cells expressing Ubc7–2HA. Precipitates were analyzed by immunoblotting with anti-Doa10 or anti-HA antibodies. Ubc7–2HA co-precipitation is only observed for the full-length E3. The strains expressing Ubc7–2HA and Doa10–13MYC (MHY3000) or the truncated Doa10950-13MYC (MHY3325) were made by Tommer Ravid (T. Ravid and M. Hochstrasser, unpublished data). The 1% input panel for Ubc7–2HA is from a longer film exposure than the other panels.
We asked whether changes in the binding of Ubc6 to the doa10-E633Q mutant protein might account for the observed accelerated degradation of Ubc6 (Fig. 7C). To ensure comparable cellular Ubc6 levels in the DOA10 and doa10-E633Q strains, the UBC7 gene was deleted. Doa10-specific antibodies co-precipitated similar amounts of endogenous Ubc6 from digitonin-solubilized microsomal extracts in the two strains. The solubilization protocol did not lead to nonspecific precipitation of membrane proteins inasmuch as we could demonstrate differential co-precipitation of Ubc7–2HA from cells expressing a WT epitope-tagged Doa10 compared with ones with a C-terminally truncated derivative analyzed under the same conditions (Fig. 7D). Therefore, the striking change in Ubc6 degradation in doa10-E633Q cells cannot be explained by a gross change in its binding affinity for the mutant E3.
Proteolytic Targeting of Ubc6 by a trans Copy of Ubc6 in doa10-E633Q Cells
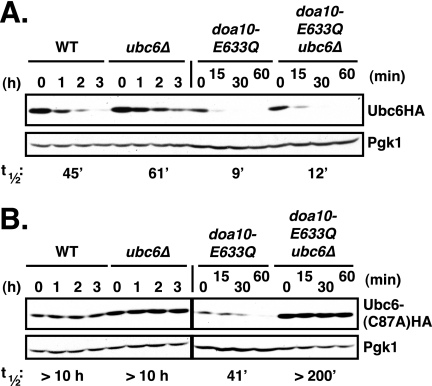
Degradation of Ubc6 in WT cells also depends on Ubc6 enzymatic activity, and the active site must be intact in the copy of the Ubc6 enzyme that is degraded, i.e. a catalytically inactive Ubc6 protein is stable even if a second, functional version of Ubc6 is expressed in the same cell (25). We tested whether the accelerated degradation of Ubc6 in doa10-E633Q cells was under similar constraints (Fig. 8). Consistent with previous findings (25), we observed that catalytically inactive Ubc6(C87A)HA protein was completely stable when ectopically expressed in DOA10 cells regardless of whether functional endogenous Ubc6 was present or not (in both cases t½ >10 h; Fig. 8B). In contrast, Ubc6(C87A)HA was readily degraded in doa10-E633Q cells, albeit at a decreased rate (t½ ∼41 min) compared with that of catalytically active Ubc6HA (t½ ∼9 min) (Fig. 8, A and B). Remarkably, the presence of Ubc6 E2 activity in trans was strictly required, as Ubc6(C87A)HA was fully stabilized in doa10-E633Q cells lacking endogenous Ubc6 (t½ >200 min). Therefore, a single amino acid exchange within TM5 of Doa10, E633Q, is sufficient to override the cis requirement for Ubc6 E2 activity for Ubc6 degradation but a requirement for an active trans copy of Ubc6 now becomes apparent.
FIGURE 8.
Catalytically inactive Ubc6 is degraded in doa10-E633Q but not DOA10 cells if functional Ubc6 is present in trans. A, Ubc6HA is degraded efficiently in cells lacking endogenous Ubc6. Ubc6HA was expressed from a low copy plasmid under control of its normal promotor. Analysis of Ubc6HA stability in WT, ubc6Δ, doa10-E633Q and doa10-E633Q ubc6Δ cells was done as in Fig. 2B. Twice as much lysate was loaded for doa10-E633Q and doa10-E633Q ubc6Δ samples compared with WT and ubc6Δ. B, catalytically inactive Ubc6(C87A)HA protein is degraded in doa10-E633Q cells but only if active Ubc6 is present in trans. The Ubc6 active site mutant Ubc6HA(C87A) was expressed from the UBC6 promoter on a low copy plasmid. Note: 2.5 times the amount of lysate was loaded for the doa10-E633Q samples relative to the others.
Notably, when active Ubc6HA was expressed in DOA10 cells lacking endogenous Ubc6 (ubc6Δ), Ubc6HA was degraded slightly slower than in WT cells (Fig. 8A). A similar difference was observed in doa10-E633Q cells with and without endogenous Ubc6 (Ubc6HA t½ = 9 versus 12 min, respectively). These differences could potentially reflect a stimulatory effect of Ubc6 in trans on Ubc6HA degradation even with WT Doa10 if Ubc6 levels become partially limiting or the tagged Ubc6HA has slightly lower activity than WT Ubc6 (or both). In any event, the ability of a trans copy of active Ubc6 to enable degradation of a catalytically inactive Ubc6 derivative in doa10-E633Q cells provides evidence that Ubc6 can function in a multimeric form, although active and inactive Ubc6 monomers need not be in direct contact (see “Discussion”).
Ubc6 Membrane Anchor Contributes to Degradation in doa10-E633Q Cells
The stimulation of Ubc6 degradation by doa10-E633Q raises the question of what Ubc6 feature(s) is responsible for this property. Ubc6 is an integral membrane protein that is held in the ER/NE membrane by a C-terminal anchor, with the N-terminal portion of the E2 facing the cytosol (20). This tail anchor (along with the preceding ∼30 residues) was previously shown to create a relatively short lived protein when appended to the C terminus of the normally stable, cytosolic Ubc4 E2 (25). However, the Ubc6 tail was not sufficient to trigger degradation of Ubc6 because its active-site cysteine was also needed (Fig. 8) (25); an E2 active site in the Ubc4-Ubc6TM fusion is provided by the Ubc4 catalytic domain.
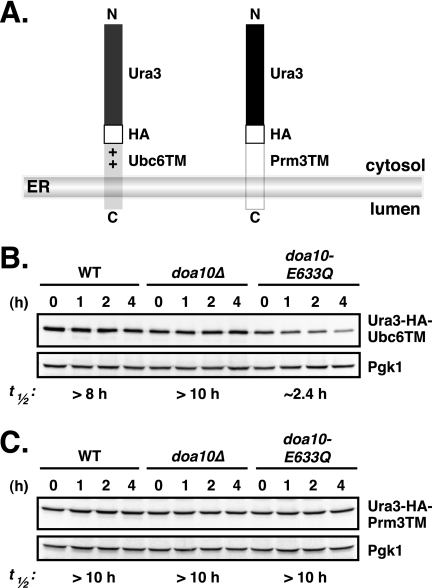
The requirement for a functional cis active site in Ubc6 for its degradation was relieved in doa10-E633Q cells (Fig. 8B). We therefore asked whether the Ubc6 C-tail would be sufficient to target an otherwise stable (non-E2) protein for degradation in the doa10-E633Q mutant. To this end, we generated a fusion between Ura3-HA and the C-tail membrane anchor of Ubc6, Ura3-HA-Ubc6TM (Fig. 9A), and determined its turnover in WT and doa10-E633Q cells (Fig. 9B). The model substrate was degraded only very slowly in WT and was completely stable in doa10Δ cells. In the doa10-E633Q mutant, Ura3-HA-Ubc6TM was degraded more rapidly, albeit still at a modest rate (t½ ∼2.4 h), indicating that the Ubc6TM was sufficient to destabilize an otherwise stable soluble protein.
FIGURE 9.
C-tail membrane anchor of Ubc6 promotes degradation of a model substrate in doa10-E633Q cells. A, schematic depiction of Ura3-HA-Ubc6TM and Ura3-HA-Prm3TM fusion proteins. The Ubc6TM segment consists of the C-terminal membrane anchor sequence of Ubc6 plus 18 residues preceding the TM, which is required for proper membrane insertion (38). The Prm3TM segment includes the C-terminal membrane anchor sequence of Prm3 plus the 13 residues preceding the TM. + denotes the positively charged nature of the stretch preceding the TM of Ubc6. B, cycloheximide-chase/anti-HA immunoblot analysis of Ura3-HA-Ubc6TM degradation in WT, doa10Δ, and doa10-E633Q cells. Pgk1 served as loading control. C, degradation of Ura3-HA-Prm3TM evaluated as in B.
We tested whether this property was specific to the Ubc6 C-tail anchor by generating a second fusion protein, this time between Ura3-HA and the last 42 residues of Prm3 (Fig. 9A). This segment of Prm3 included the C-tail membrane anchor plus the 13 preceding residues and was shown to be sufficient for ER membrane localization of a GFP reporter (38). Full-length Prm3 normally localizes to the outer NE, a subdomain of the ER (39). In contrast to the Ura3-HA-Ubc6TM protein, Ura3-HA-Prm3TM was equally stable in all three strains, WT, doa10Δ, and doa10-E633Q (t½ >10 h; Fig. 9C).
These results imply that doa10-E633Q (and presumably WT Doa10) recognizes specific features of the Ubc6 C-terminal membrane anchoring segment that are not shared by all C-terminally anchored proteins. The subtle alteration in TM5 of Doa10 is evidently changing the way Doa10 interacts with Ubc6, which is both a cofactor and substrate of the E3, thus making it much more prone to degradation via the Doa10-dependent proteolytic pathway.
DISCUSSION
Here we have analyzed the evolutionarily conserved but previously unstudied TD (TEB4-Doa10) domain of the S. cerevisiae Doa10 ubiquitin ligase. Phylogenomic comparisons among eukaryotes with fully sequenced genomes indicate that the TD domain is highly conserved. The three TMs within the domain are also well conserved, especially TM5. TM5 includes a glutamate (Glu-633) predicted to be positioned within the interior of the membrane bilayer, and Doa10 function is very sensitive to the identity of this residue. Most strikingly, replacement of the glutamate by glutamine at this position triggers abnormally rapid degradation of Ubc6, a transmembrane E2 that functions with Doa10. A major qualitative difference between the slow Ubc6 degradation that occurs normally and the rapid turnover in doa10-E633Q cells is that in the latter cells the active-site cysteine is no longer required in the Ubc6 molecule being degraded. Our data suggest that the polyubiquitylation of the rapidly degraded Ubc6 polypeptide involves ubiquitin chain assembly by Ubc7 and a second copy of Ubc6. Below, we discuss the mechanistic and functional implications of these findings.
Each of the three helices in the Doa10 TD domain has at least one absolutely conserved residue (Pro, Gly, or Ser) that is likely to help create deviations from ideal helical conformation, potentially favoring a specific structural architecture in the TD domain or increasing helical dynamics. Most intriguing is the ΦPΦXXG motif in TM5, which we find is important for Doa10 function (Fig. 2). No exact match to this consensus has previously been identified in transmembrane helices, but related motifs in integral membrane proteins of known structure suggest that the motif is linked to helix bending within the membrane. Proline is frequently found at kinks in transmembrane helices (40), and motifs containing proline, glycine, and serine often contribute to conformational flexibility (41, 42). In the transmembrane protein-conducting channel SecY, the sequence motif PΦXG is present at the end of TM1 (in most species), and a GΦXP motif is present in TM2; these contribute to a complex secondary structure, including a split of TM2 into two shorter helices (43). The kinked TM2 plays a crucial role in SecY function (43). By analogy, the unusual Doa10 TM5 may be part of a dynamic TD domain with a central role in Doa10-dependent transactions.
Interruption of transmembrane helices by a short nonhelical segment containing Pro, Gly, and/or Ser residues has also been observed in many classes of transporters, including amino acid antiporters (44), neurotransmitter-sodium symporters (45), and sodium-independent transporters (46). Interruption of helical structure exposes main chain carbonyl oxygen and nitrogen atoms for hydrogen bonding and ion coordination (45). It was also proposed that ion transport would be allosterically regulated, involving two substrate-binding sites where a well conserved charged residue coordinates a network of hydrogen bonds and/or salt bridges, allosterically connecting the two binding sites (46).
The idea that TM5 of Doa10 has a highly specific structure within the bilayer is consistent with the extreme sensitivity to mutation of the conserved Glu-633 residue in this helix. As charged membrane residues within TM helices are often critical determinants in helix-helix interactions in membrane proteins, it is plausible that this glutamate has a crucial role for Doa10 assembly and function (37, 46, 47). Changing Glu-633 to an aspartate, which maintains the negative charge but reduces the length of the side chain, strongly inhibits Doa10 activity toward its cognate E2 Ubc6, without strongly affecting activity toward other substrates. At the same time, degradation of the model substrates strictly depended on the presence of Ubc6, indicating that doa10-E633D and Ubc6 still productively interact in the degradation of these substrates. Hence, Ubc6 degradation does not seem to be a prerequisite for Doa10 function. Insofar as an aspartate has never been observed at this TM5 position in any Doa10 ortholog, some selective disadvantage is likely associated with this residue, possibly in the quality control of cellular Ubc6 or in Doa10 biogenesis.
In contrast, changing Glu-633 to a glutamine or several other amino acids has the surprising effect of greatly accelerating the degradation of Ubc6. Mutations elsewhere in TM5, such as Pro-638 and Gly-642 (in combination), as well as several hydrophobic residues (Fig. 6),4 all lead to loss of Doa10 activity. The fact that a P638A/G642A double mutant, but not either single mutant, was strongly impaired for Doa10 function may be related to the finding by Yohannan et al. (40) that a single substitution of a conserved intramembrane proline can often be tolerated because the kink is further stabilized by secondary mutations that have evolved elsewhere in the TM. Both single substitutions at Gly-642, G642A and G642V, caused a significant reduction in Doa10 levels, suggesting that flexibility at this residue in the ΦPΦXXG motif is important for Doa10 biogenesis or structural stability.
The accelerated Ubc6 degradation by doa10-E633Q shares hallmarks of the slower physiological Ubc6 degradation in WT cells; it is fully dependent on the RING-CH domain of Doa10 and on the Ubc7 E2. Ubc6 still functions as an E2 for doa10-E633Q despite its rapid degradation inasmuch as it continues to support nearly WT degradation rates of other Ubc6- and Doa10-dependent substrates. Taken together, these findings indicate that the doa10-E633Q protein functions much like its wild-type counterpart except for its behavior toward Ubc6.
What leads to the strongly enhanced degradation of Ubc6? In the simplest scenario, Ubc6 binding to doa10-E633Q might be tighter than to the WT E3, increasing the chance that Ubc6 will itself become ubiquitylated. However, co-immunoprecipitation experiments uncovered no obvious differences in binding of Ubc6 to doa10-E633Q and WT Doa10 (Fig. 7). We cannot completely rule out subtle changes in affinity that are not detectable by the co-immunoprecipitation assay. Alternatively, differences in Ubc6-doa10-E633Q binding geometry or dynamics may bring about the accelerated degradation of the E2. An important clue is undoubtedly the major qualitative change in the mechanism of Ubc6 degradation in doa10-E633Q relative to DOA10 cells, namely the dispensability of the active site in the Ubc6 polypeptide undergoing degradation (Fig. 8). A second, functional version of Ubc6 is absolutely required, however. (This latter requirement might also hold for DOA10 cells because we consistently observe a small decrease in Ubc6 degradation rate when a second copy of Ubc6 is deleted from the cell, possibly reflecting a dosage effect.) These observations strongly suggest that two Ubc6 molecules must interact for the doa10-E633Q-dependent ubiquitylation and degradation of Ubc6 in vivo. These could interact directly or via other components of the Doa10 complex.
Based on these considerations, we suggest a model in which two Ubc6 molecules bind at adjacent or overlapping sites in the Doa10 complex (supplemental Fig. S1). The first would be the site from which the Ubc6 would normally transfer its thioester-linked ubiquitin to substrate (the E2 site), while the second might be equivalent to or overlap a generally used site of substrate binding within Doa10 (the substrate site), although it could conceivably be a site normally used for E2 release or ubiquitin thioester transfer from E1. Transient movement of Ubc6 from the E2 site to the substrate site on Doa10, which might occur during normal ubiquitin chain assembly or only when another substrate is absent, could depend on Ubc6 thioester linkage to ubiquitin. If this cis-acting ubiquitin-dependent gating were bypassed in the doa10-E633Q mutant, the requirement for the Ubc6 active-site cysteine for its polyubiquitylation and degradation would be eliminated. Conversely, in the doa10-E633D ligase, which no longer allows Ubc6 degradation, access of Ubc6 to the substrate site might be constitutively hindered.
The (slow) doa10-E633Q-dependent degradation of Ura3-Ubc6TM suggests that the Ubc6 C-terminal membrane helix is sufficient for at least weak E3 interaction, presumably at the substrate site. The other Doa10-associated E2, Ubc7, interacts with the RING-CH motif, but stable in vivo binding requires downstream Doa10 sequences (Fig. 7D).5 No evidence to date has indicated that Ubc6 must also interact with the RING-CH, but this cannot be ruled out. The simplest model holds that Ubc6 and Ubc7 have distinct E2-binding sites on Doa10, and the two E2s function together to build a polyubiquitin chain on the substrate, which may sometimes be a second molecule of Ubc6.
A final question regarding our present results is whether Doa10-dependent Ubc6 binding and degradation are associated in any way with substrate retrotranslocation from the membrane. Like other transmembrane substrates, Ubc6 must be extracted from the bilayer for complete degradation (25). No “retrotranslocon” for Doa10 substrates has yet been identified experimentally. We previously proposed that Doa10 itself might contribute to a protein exit channel in the ER membrane (13). If true, this would likely involve the well conserved TD domain (17). The present findings establish TM5 of the TD domain as an unusual transmembrane element that is highly sensitive to mutation. It appears to have features that would be expected of a channel or transporter, such as structural flexibility, the ability to form intramembrane hydrogen bonds or salt bridges, and dynamic interactions with a transmembrane cofactor. Additional biochemical and biophysical experiments will be needed to address this and other mechanistic questions regarding this unique ubiquitin ligase complex.
Supplementary Material
Acknowledgments
We thank Hiroko Shimada-Kreft and V. J. Rubenstein for helpful discussions and comments on the manuscript. We are grateful to F. Storici for the pCORE plasmid and to T. Sommer for the pRS416-Ubc6HA plasmid and the anti-Ubc6 antibody. S. G. K. acknowledges Martin Scheffner for continuous support.
This work was supported, in whole or in part, by National Institutes of Health Grant GM046904 (to M. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
S. G. Kreft and M. Hochstrasser, unpublished data.
T. Ravid, S. G. Kreft, and M. Hochstrasser, unpublished data.
- ER
- endoplasmic reticulum
- ERAD
- endoplasmic reticulum-associated degradation
- E2
- ubiquitin-conjugating enzyme
- E3
- ubiquitin-protein ligase
- GFP
- green fluorescent protein
- HA
- hemagglutinin
- NE
- nuclear envelope
- Pgk1
- 3-phosphoglycerate kinase
- TD domain
- TEB4-Doa10 domain
- TM
- transmembrane helix.
REFERENCES
- 1. Blobel G., Dobberstein B. (1975) J. Cell Biol. 67, 835–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rapoport T. A. (2007) Nature 450, 663–669 [DOI] [PubMed] [Google Scholar]
- 3. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hirsch C., Gauss R., Horn S. C., Neuber O., Sommer T. (2009) Nature 458, 453–460 [DOI] [PubMed] [Google Scholar]
- 5. Hegde R. S., Ploegh H. L. (2010) Curr. Opin. Cell Biol. 22, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hiller M. M., Finger A., Schweiger M., Wolf D. H. (1996) Science 273, 1725–1728 [DOI] [PubMed] [Google Scholar]
- 7. Werner E. D., Brodsky J. L., McCracken A. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13797–13801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wiertz E. J., Jones T. R., Sun L., Bogyo M., Geuze H. J., Ploegh H. L. (1996) Cell 84, 769–779 [DOI] [PubMed] [Google Scholar]
- 9. Tsai B., Ye Y., Rapoport T. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 246–255 [DOI] [PubMed] [Google Scholar]
- 10. Bagola K., Mehnert M., Jarosch E., Sommer T. (2011) Biochim. Biophys. Acta 1808, 925–936 [DOI] [PubMed] [Google Scholar]
- 11. Ravid T., Kreft S. G., Hochstrasser M. (2006) EMBO J. 25, 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deng M., Hochstrasser M. (2006) Nature 443, 827–831 [DOI] [PubMed] [Google Scholar]
- 13. Swanson R., Locher M., Hochstrasser M. (2001) Genes Dev. 15, 2660–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bartee E., Mansouri M., Hovey Nerenberg B. T., Gouveia K., Früh K. (2004) J. Virol. 78, 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hassink G., Kikkert M., van Voorden S., Lee S. J., Spaapen R., van Laar T., Coleman C. S., Bartee E., Früh K., Chau V., Wiertz E. (2005) Biochem. J. 388, 647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dodd R. B., Allen M. D., Brown S. E., Sanderson C. M., Duncan L. M., Lehner P. J., Bycroft M., Read R. J. (2004) J. Biol. Chem. 279, 53840–53847 [DOI] [PubMed] [Google Scholar]
- 17. Kreft S. G., Wang L., Hochstrasser M. (2006) J. Biol. Chem. 281, 4646–4653 [DOI] [PubMed] [Google Scholar]
- 18. Chen P., Johnson P., Sommer T., Jentsch S., Hochstrasser M. (1993) Cell 74, 357–369 [DOI] [PubMed] [Google Scholar]
- 19. Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., Michaelis S. (2004) J. Biol. Chem. 279, 38369–38378 [DOI] [PubMed] [Google Scholar]
- 20. Sommer T., Jentsch S. (1993) Nature 365, 176–179 [DOI] [PubMed] [Google Scholar]
- 21. Biederer T., Volkwein C., Sommer T. (1997) Science 278, 1806–1809 [DOI] [PubMed] [Google Scholar]
- 22. Bazirgan O. A., Hampton R. Y. (2008) J. Biol. Chem. 283, 12797–12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kostova Z., Mariano J., Scholz S., Koenig C., Weissman A. M. (2009) J. Cell Sci. 122, 1374–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ravid T., Hochstrasser M. (2007) Nat. Cell Biol. 9, 422–427 [DOI] [PubMed] [Google Scholar]
- 25. Walter J., Urban J., Volkwein C., Sommer T. (2001) EMBO J. 20, 3124–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guthrie C., Fink G. R. (eds) (1991) Guide to Yeast Genetics and Molecular Biology, Vol. 194, Academic Press, New York [Google Scholar]
- 27. Storici F., Lewis L. K., Resnick M. A. (2001) Nat. Biotechnol. 19, 773–776 [DOI] [PubMed] [Google Scholar]
- 28. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 29. Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. (1992) Gene 110, 119–122 [DOI] [PubMed] [Google Scholar]
- 30. Mumberg D., Müller R., Funk M. (1994) Nucleic Acids Res. 22, 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Loayza D., Tam A., Schmidt W. K., Michaelis S. (1998) Mol. Biol. Cell 9, 2767–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hochstrasser M., Varshavsky A. (1990) Cell 61, 697–708 [DOI] [PubMed] [Google Scholar]
- 33. Koonin E. V. (2010) Cell 140, 606–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arteaga M. F., Wang L., Ravid T., Hochstrasser M., Canessa C. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11178–11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hall S. E., Roberts K., Vaidehi N. (2009) J. Mol. Graph. Model. 27, 944–950 [DOI] [PubMed] [Google Scholar]
- 36. Ulmschneider M. B., Sansom M. S., Di Nola A. (2005) Proteins 59, 252–265 [DOI] [PubMed] [Google Scholar]
- 37. Moore D. T., Berger B. W., DeGrado W. F. (2008) Structure 16, 991–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Beilharz T., Egan B., Silver P. A., Hofmann K., Lithgow T. (2003) J. Biol. Chem. 278, 8219–8223 [DOI] [PubMed] [Google Scholar]
- 39. Shen S., Tobery C. E., Rose M. D. (2009) Mol. Biol. Cell 20, 2438–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yohannan S., Faham S., Yang D., Whitelegge J. P., Bowie J. U. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 959–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jacob J., Duclohier H., Cafiso D. S. (1999) Biophys. J. 76, 1367–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chandrasekaran A., Ojeda A. M., Kolmakova N. G., Parsons S. M. (2006) J. Neurochem. 98, 1551–1559 [DOI] [PubMed] [Google Scholar]
- 43. Van den Berg B., Clemons W. M., Jr., Collinson I., Modis Y., Hartmann E., Harrison S. C., Rapoport T. A. (2004) Nature 427, 36–44 [DOI] [PubMed] [Google Scholar]
- 44. Gao X., Lu F., Zhou L., Dang S., Sun L., Li X., Wang J., Shi Y. (2009) Science 324, 1565–1568 [DOI] [PubMed] [Google Scholar]
- 45. Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E. (2005) Nature 437, 215–223 [DOI] [PubMed] [Google Scholar]
- 46. Schulze S., Köster S., Geldmacher U., Terwisscha van Scheltinga A. C., Kühlbrandt W. (2010) Nature 467, 233–236 [DOI] [PubMed] [Google Scholar]
- 47. Langosch D., Arkin I. T. (2009) Protein Sci. 18, 1343–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.