Abstract
Heteropolymeric B-band lipopolysaccharide in Pseudomonas aeruginosa PAO1 is synthesized via the so-called Wzy-dependent pathway, requiring a functional Wzy for polymerization of O-antigen repeat units in the periplasm. Wzy is an integral inner membrane protein for which the detailed topology has been mapped in a recent investigation (Islam, S. T., Taylor, V. L., Qi, M., and Lam, J. S. (2010) mBio 1, e00189-10), revealing two principal periplasmic loops (PL), PL3 and PL5, each containing an RX10G motif. Despite considerable sequence conservation between the two loops, the isoelectric point for each peptide displayed marked differences, with PL3 exhibiting a net-positive charge and PL5 showing a net-negative charge. Data from site-directed mutagenesis of amino acids in each PL have led to the identification of several key Arg residues within the two RX10G motifs that are important for Wzy function, of which Arg176, Arg290, and Arg291 could not be functionally substituted with Lys. These observations support the proposed role of each PL in a catch-and-release mechanism for Wzy-mediated O-antigen polymerization.
Keywords: Antigen, Carbohydrate-binding protein, Lipopolysaccharide (LPS), Membrane biogenesis, Membrane proteins, Polysaccharide, O antigen, Polymerase, Topology, Wzy
Introduction
Lipopolysaccharide (LPS) is an important virulence factor in the opportunistic human pathogen Pseudomonas aeruginosa, exerting direct endotoxic effects (1), as well as impacting numerous other virulence traits including the export of type III secretion effectors (2) and the formation of biofilm architecture (3). LPS is composed of the proximal lipid A moiety, the core oligosaccharide linker, and the distal capping polysaccharide, of which P. aeruginosa synthesizes two glycoforms: one capped by homopolymeric A-band common polysaccharide and the other by heteropolymeric O-specific B-band O-antigen (O-Ag),3 the latter being the immunodominant cell surface antigen that differentiates this species into 20 serotypes (4).
B-band O-Ag biosynthesis in P. aeruginosa PAO1 follows the Wzy-dependent pathway, requiring (i) translocation of undecaprenyl pyrophosphate- (UndPP-) linked O-Ag repeat units from the cytoplasmic to the periplasmic leaflets of the inner membrane, (ii) polymerization of these units in the periplasm (iii) to preferred modal lengths, (iv) followed by ligation of the heteropolymer to lipid A-core. These processes are mediated by the integral inner membrane proteins Wzx (5), Wzy (6), Wzz1/Wzz2 (7), and WaaL (8), respectively, with their assembly roles mainly characterized via genetic evidence. However, their precise mechanisms of function remain poorly understood. O-Ag subunit polymerization by Wzy has recently been demonstrated in vitro (9), but characterization of the reaction mechanism has yet to take place.
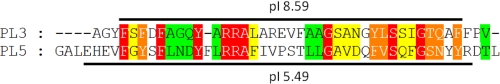
Recently, our group mapped the topology of Wzx, Wzy, and WaaL from P. aeruginosa PAO1; instead of relying on in silico topology prediction algorithm outputs to dictate the location of α-helical transmembrane segments (TMS), we used an unbiased experimental approach (10). This resulted in the identification of previously undetected motifs in the TMS, as well as in the cytoplasm and periplasm for each protein. Wzy was found to contain 14 TMS, with two large periplasmic loops (PL) of comparable size localized between TMS5–6 (PL3) and TMS9–10 (PL5), each containing an RX10G tract of amino acids (supplemental Fig. S1) (10). Alignment of the peptide sequences of the two loops via ClustalW2 revealed a high degree of conservation for structurally and/or functionally equivalent amino acid residues (see Fig. 1).
FIGURE 1.
Alignment of PL3 (Ala162–Val203) and PL5 (Gly272–Leu319) from Wzy of P. aeruginosa PAO1 via ClustalW2. Included are 3 residues from each flanking α-helical TMS (supplemental Fig. S1). Calculated pI values for both PL3-specific and PL5-specific polypeptides (specified by a black line) are indicated. Highlighted residues have been colored based on their conservation score (out of 10) as presented in Jalview (33). The color key is as follows: red, score = 10; orange, score = 9; yellow, score = 8; green, score = 7. Both loop regions contain identical and structurally equivalent amino acids, alluding to a common motif between the two. Adapted from Islam et al. (10).
To further explore the functional significance of this finding, we analyzed the charge nature of both PL3 and PL5. Significant differences in the isoelectric point (pI) of each loop were observed despite the high degree of sequence conservation. Furthermore, we created site-directed mutants of conserved charged residues between PL3 and PL5, subsequently identifying key Arg residues required for the O-Ag subunit polymerization reaction; certain of these residues highlighted the importance of the guanidinium functional group of Arg in O-Ag polymerization as Lys-substituted variants simply maintaining an analogous charge at the same position were unable to restore function. This is the first investigation to examine the function of a Wzy protein within the context of a rigorously defined topological structure and in so doing provides evidence for the “catch-and-release” mechanism for Wzy function we have previously proposed (10) and herein refined.
EXPERIMENTAL PROCEDURES
DNA Manipulations
QuikChange (Agilent) site-directed mutagenesis was performed on a previously created template encoding Wzy fused with a C-terminal His8-tagged green fluorescent protein (GFP) moiety (Wzy-GFP-His8). This fusion tag does not affect wild type O-Ag polymerase function and provides the additional advantage of displaying GFP fluorescence upon expression induction (10). This fusion construct was contained in an arabinose-inducible pHERD26T vector, selected by LB supplementation with 90 μg/ml tetracycline (11). Plasmid DNA was isolated with a GenElute miniprep kit (Sigma). PCR amplifications for mutagenesis were carried out using KOD Hot Start DNA polymerase (Novagen). The oligonucleotide primer sequences used to create the various mutants are available upon request.
Complementation Analysis
The in vivo function of each mutant construct was assayed by using the mutant genes to complement B-band LPS deficiency in a P. aeruginosa PAO1 wzy chromosomal knock-out mutant created by our group; this mutant was previously demonstrated to be deficient in B-band LPS production while maintaining the production of A-band LPS (6). Analysis of the Wzy coding sequence from P. aeruginosa PAO1 has revealed the presence of numerous rare codons, with many immediately downstream of the start codon (6, 12). This characteristic likely contributes to the low expression levels of the protein (13). To avoid dose-dependent overexpression artifacts of a given Wzy mutant, cells containing the various constructs were grown to stationary phase by overnight incubation at 37 °C, shaking in tetracycline-supplemented LB medium without expression induction. The potential for overexpression artifacts was monitored through the measurement of GFP fluorescence for each of the overnight samples processed for LPS analysis; these levels did not register above the background fluorescence detected for the pHERD26T empty vector control, indicating that all mutant constructs were also expressed at background plasmid levels (data not shown). LPS was extracted from bacteria after the complementation experiment and analyzed by SDS-PAGE and Western immunoblotting as described previously (10, 14) with anti-B-band O-Ag antibody MF15-4 (15).
Membrane Localization Quantitation
Cultures of P. aeruginosa PAO1 Δwzy (25 ml) expressing various Wzy-GFP-His8 mutants were grown overnight with 0.1% l-arabinose, equilibrated to A600 of 1.5, and sedimented at 4000 × g. To minimize mechanical loss of samples during processing, equilibrated cell pellets were permeabilized by using the vortex and resuspending the cell materials in 3 ml of B-PER II protein extraction reagent (Thermo Scientific) with 2× Complete protease inhibitor mixture lacking EDTA (Roche Applied Science) followed by shaking incubation at 30 °C (260 rpm, 30 min). Sample volume was increased by the addition of 2 ml 20 mm Tris-HCl, pH 7.5, with 2× Complete protease inhibitor lacking EDTA, after which crude debris and inclusion bodies were sedimented at 12,000 × g in a Beckman super-speed centrifuge (30 min). The resultant supernatant was sedimented at 120,000 × g in a Beckman ultracentrifuge (1 h) to obtain the membrane pellet, which was subsequently resuspended directly in the ultracentrifuge tube in 130 μl of 1× SDS-PAGE sample buffer using a Dounce homogenizer plunger. Samples were incubated at 37 °C for 30 min, with 25 μl resolved on a 12% SDS-PAGE gel; the full-range ECL Plex Fluorescent Rainbow Marker (GE Healthcare) protein standard was used. In-gel fluorescence scans were obtained using a Typhoon 9410 imager and ImageQuant TL software (GE Healthcare). The Blue2(488), Green(532), and Red(633) excitation lasers as well as the 526SP, 580BP, and 670BP emission filters were used to detect GFP, Cy3, and Cy5 fluorescence, respectively, after which final densitometry analysis of full-length Wzy-GFP-His8 fluorescence was performed in quadruplicate using the ImageJ software.
RESULTS
PL3 and PL5 Possess Different Charge Characteristics Despite High Sequence Equivalence
Upon calculation of the pI for PL3 and PL5 of Wzy using the ExPASy proteomics server, strikingly distinct results were obtained. The pI values for PL3 and PL5 were 8.59 and 5.49, respectively (Fig. 1). Even when the two loops were compared by accounting for only the conserved residues, pI values of 9.99 and 5.96 were calculated, respectively (Fig. 1, highlighted residues). As the pH of the periplasm has been demonstrated to match that of the extracellular environment (16), at a physiological pH of ∼7.4, this would indicate a significant net-positive charge on PL3 and a net-negative charge on PL5 (Fig. 1) despite the high level of sequence equivalence between the two.
This compositional equivalence was previously demonstrated to confer similar helical secondary structure propensity patterning between PL3 and PL5 (10). Additionally, construction of knowledge-based tertiary structure models of these highly conserved amino acid sequences using the I-TASSER platform (17) revealed strikingly similar three-dimensional folding models for PL3 and PL5. Each of these structures contained two antiparallel helices of similar length with a flexible intervening loop region (supplemental Fig. S2).
RX10G Motifs in PL3 and PL5 Contain Key Arg Residues Important for Wzy Function
Based on the experimentally derived topology map of Wzy (supplemental Fig. S1), site-directed mutagenesis was performed to replace specific residues within PL3 and PL5 with alanine to detect those that are important for polymerase function. Each mutant construct was assayed for its ability to complement a P. aeruginosa PAO1 wzy knock-out mutant and restore the synthesis of B-band LPS. Constructs containing changes to amino acid residues not essential for function should positively complement the wzy mutant and restore B-band production; in contrast, constructs containing changes to amino acid residues essential for function should not be able to complement Wzy deficiency.
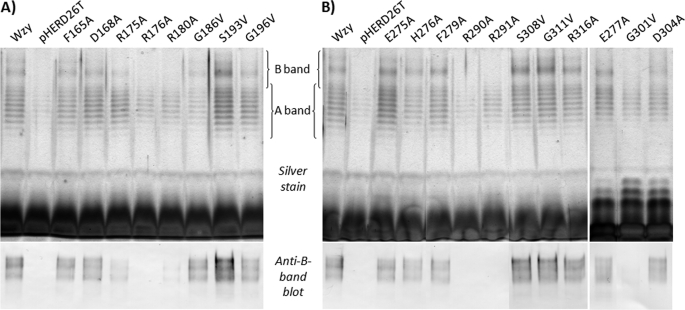
Individual substitution of the Arg residues in PL3 (positions 175, 176, and 180) to Ala adversely affected the ability of Wzy to restore B-band LPS biosynthesis (Fig. 2A). Incidentally, these positively charged residues correspond to the 1st, 2nd, and 6th amino acids contained within the RX10G tract of PL3, respectively (supplemental Fig. S1). Complete loss of B-band O-Ag was observed for Wzy with the R176A substitution, whereas only trace amounts were detected for substitutions R175A and R180A (Fig. 2A), indicating the importance of these amino acids in Wzy function. Substitutions of F165A and D168A located at the base of PL3 (supplemental Fig. S1) did not affect the function of the protein (Fig. 2A), suggesting that these residues do not form part of the tertiary structure that contributes to O-Ag subunit polymerization.
FIGURE 2.
SDS-PAGE and Western immunoblotting analyses of LPS from cells of P. aeruginosa PAO1 Δwzy complemented with various mutant Wzy constructs. In both A and B, the top portion is a silver-stained SDS-PAGE gel, and the bottom portion is a Western blot probed with B-band LPS-specific monoclonal antibody MF15-4. A, PL3 mutants. B, PL5 mutants. The results shown are reproducible and representative of three independent sample sets.
Site-directed mutations R290A and R291A of Wzy, both within the RX10G motif of PL5, resulted in a complete abrogation of B-band LPS biosynthesis in the complementation assay (Fig. 2B). This is consistent with the aforementioned site-directed mutations made in PL3. These 2 residues correspond to the 1st and 2nd amino acids within the PL5 RX10G motif (supplemental Fig. S1) and align with Arg175 and Arg176 from PL3 (Fig. 1). Substitution of F279A in PL5, a residue in an analogous position to Phe165 of PL3 (Fig. 1), did not affect Wzy function, similar to substitutions E275A, H276A, E277A, and R316A (Fig. 2B) located near the PL5-membrane interface (supplemental Fig. S1). The D304A change in PL5 did not affect restoration of B-band LPS biosynthesis in the complementation assay, suggesting that Asp304 may not be involved in the tertiary structure environment of Wzy required for function (Fig. 2B). This observation is similar to the aforementioned PL3 changes, F165A and D168A.
The presence of the above mentioned substitutions of Arg resulting in a loss of Wzy function did not prevent membrane insertion of the respective mutant constructs as levels of GFP fluorescence comparable with the native construct were detected from membrane fractions isolated from cultures expressing each mutant construct (supplemental Fig. S3). This was a necessary verification as alteration of positive charge in loop domains can potentially affect membrane insertion of TMS in multispanning inner membrane proteins (18). Furthermore, generation of three-dimensional folding structures using the I-TASSER platform for variants of PL3 and PL5 corresponding to the above mentioned Arg mutants with demonstrated functional deficiencies were obtained (supplemental Fig. S4); these were shown to be nearly identical to those of the native sequences (supplemental Fig. S2), indicating similar folding propensities.
The conserved Gly and Ser residues between PL3 and PL5 (Gly186/Ser193/Gly196 and Gly301/Ser308/Gly311, respectively) (Fig. 1) were substituted with Val, prompted by a report of a functionally important Gly residue from Wzy of Francisella tularensis LVS (19). Of the six mutants screened, only Gly301 in PL5 was found to be important for function; all other Gly and Ser substitutions did not display any discernable deficiencies in B-band LPS production (Fig. 2).
Guanidinium Functional Groups of Arg Residues in PL3 and PL5 RX10G Motifs Are Required for Wzy Function
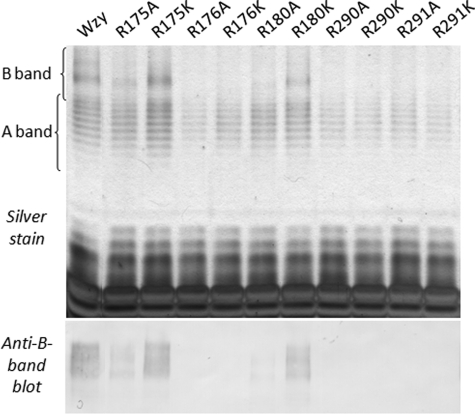
The overall importance of positive charge at the various RX10G Arg positions in PL3 and PL5 was examined through their respective substitutions with Lys. Following B-band LPS analysis, only the R175K and R180K substitutions in PL3 were able to restore Wzy function to native levels (Fig. 3). Substitutions R176K, R290K, and R291K were all unable to restore Wzy function (Fig. 3), yielding B-band LPS profiles equivalent to those seen for the respective Ala substitutions (Fig. 2). These results highlighted the essential nature of positive charge for PL3 function, as well as the importance of the various Arg-specific guanidinium side chains in PL3 and PL5 function.
FIGURE 3.
SDS-PAGE and Western immunoblotting analyses of LPS from cells of P. aeruginosa PAO1 Δwzy complemented with Lys substitutions of functionally important RX10G motif Arg residues from PL3 and PL5 of Wzy. The top panel is a silver-stained SDS-PAGE gel, and the bottom panel is a Western blot probed with B-band LPS-specific monoclonal antibody MF15-4. The results shown are reproducible and representative of three independent sample sets.
DISCUSSION
The Wzy-dependent pathway is required for the synthesis of various virulence-associated polysaccharides in Gram-negative bacteria. Thus far, it has been implicated in the biogenesis of LPS capped with heteropolymeric O-Ag, as well as in the synthesis of enterobacterial common antigen and various capsular polysaccharides (20). However, although the polymerase function of Wzy has recently been demonstrated in vitro (9), the mechanism of O-Ag subunit polymerization has yet to be discovered.
As it is difficult to obtain three-dimensional structural data of inner membrane proteins via standard crystallographic approaches, topology mapping of Wzy proteins has been attempted to uncover domains that may contribute to the function of the protein (10, 21, 22). However, of the three published topologies for Wzy proteins, two relied on topology prediction algorithms to first specify the locations of TMS followed by attempts to experimentally validate the prediction (21, 22). The most recent Wzy topology investigation, published by our group, was able to experimentally determine the topology of the protein without relying on initial in silico algorithm analysis (10). As such, we were able to obtain an unbiased map, and in so doing, we were able to uncover previously unidentified characteristics of the protein (10). Those properties led to a proposed mechanism of Wzy function, which we have further characterized and supported in the current investigation.
Previously, it has been suggested that Wzy and WaaL proteins may share a similar mechanism of function as they are both involved in the addition of UndPP-linked O-Ag to a sugar acceptor (23). Conserved domain BLAST comparisons and alignment of WaaL amino acid sequences from Vibrio cholerae, Escherichia coli, and Salmonella enterica, as well as the Wzy amino acid sequence from Shigella flexneri, revealed a common sequence denoted as HX10G, containing a catalytically essential His residue predicted to be in the periplasm (23). Intriguingly, this specific motif was found to be absent from Wzy in P. aeruginosa PAO1 (10). The HX10G motif was found to be equally important for WaaL function in P. aeruginosa PAO1. However, a H303R site-directed mutant protein was fully functional. Therefore, substitution of His with Arg effectively created an RX10G tract (14). Based on this observation, we revisited the primary structure of Wzy, and within the context of the topology map, we localized RX10G motifs in each of PL3 and PL5 loops that align with each other (10). These RX10G motifs have now been shown to contain Arg residues that are essential for Wzy function in P. aeruginosa PAO1 (Fig. 2), but which are located in local environments that possess significantly different net charge characteristics (Fig. 1). Rather than a lack of membrane localization for these constructs (supplemental Fig. S3), the specific loss of key guanidinium functional groups contributed by Arg residues is a plausible reason for the abrogation of protein function as maintenance of a positive charge via Lys substitution was unable to preserve Wzy function in three of the five mutants (Fig. 3).
Consistent with the importance of Arg residues in the RX10G tracts of PL3 and PL5 described above, analyses of Protein Data Bank (PDB)-deposited structures solved in complex with bound substrate indicate that Arg is one of the two most widely observed residues present in the binding sites of carbohydrate- and sugar-binding proteins (24, 25). The guanidinium group itself of the Arg side chain has been shown to play a direct role in sugar binding as substitution of Arg with Lys, although maintaining the structure of a protein and a positive charge at a given location, may not be sufficient to maintain function (26). This is consistent with our findings for the Arg residues in PL3 and PL5 of Wzy (Fig. 3).
Intriguingly, based on existing structures in the PDB, UDP-glucuronate (co-crystallized in the structure of UDP-glucose dehydrogenase; PDB ID 2QG4) was indicated as the top-ranked ligand following I-TASSER binding site prediction for the R180A tertiary structure folding model (supplemental Fig. S4A), lending further support to the proposed sugar binding capacities of PL3 and PL5.
Recently, wzy from F. tularensis LVS (wzyFt) was identified by Kim et al. (19), who subjected the translated amino acid sequence (409 amino acids) of the protein to in silico topology prediction algorithm analysis, yielding a model containing 11 TMS. This low number of TMS is significantly different from the experimentally derived topological map of Wzy from P. aeruginosa PAO1 (438 amino acids), which showed that the protein contained 14 TMS (10) (supplemental Fig. S1). The authors' initial attempt to perform sequence alignment of Wzy proteins from various bacteria did not reveal conserved regions; however, restriction of the alignment to eight other γ Proteobacteria identified the conserved residue Gly323 (19). Based on the in silico-derived WzyFt topology map, these authors proposed that Gly323 was localized to the terminus of a second large PL. No effect on polymerization was observed upon Ala substitution. However, introduction of extraneous charges/polarities (via Glu, Arg, Asp, Gln, Ser, Tyr) or bulky/kink-inducing residues (via Leu, Pro) at that position yielded compromised WzyFt function. Consequently, the authors alluded to the potential importance of periplasmic Gly abundance in WzyFt function (19).
Kim et al. (19) also randomly selected 17 charged residues for Ala substitution, and their results revealed that Asp177 was important for function of WzyFt, with this residue predicted to be located at the terminus of the first large predicted PL. Amino acids adjacent to Asp177 (Gly176, Gly178) and Gly323 (Tyr324, but not Ile322), all purportedly periplasmic, were subjected to Ala substitution, with no effect on protein function. Only upon similar charge perturbation (as described above for Gly323) was protein function compromised for Gly176 and Tyr324, whereas substitution of Gly178 showed no effect (19). Unfortunately, as the topology model of WzyFt was derived from in silico predictions with no experimental data obtained to validate the map, the subcellular localization of the residues mutated by Kim et al. (19) cannot be confirmed. Interestingly, the proposed model of WzyFt suggested that the C terminus of the protein is localized in the periplasm, yet the authors went on to create a full-length C-terminal fusion to GFP and were able to detect fluorescence (19). This is contradictory to published knowledge as GFP is widely used as a reporter for cytoplasmic residue localization due to its inability to fluoresce when expressed as part of a periplasmic fusion to a membrane protein (27, 28). As such, the proposed in silico algorithm-derived topological model for WzyFt clearly requires revision.
To test the catalytic importance of potentially similar residues in Wzy from P. aeruginosa PAO1, the mutually conserved residues from PL3 and PL5 (Fig. 1) were examined. However, because Gly, Ala, and Ser residues may be interchangeable in certain membrane protein motifs (29), a standard Ala-scanning approach was avoided. Instead, to minimize obvious tertiary structure disturbances while not introducing foreign charges at these locations, these 6 residues were individually subjected to steric hindrance mutagenesis by substitution with Val, resulting in the addition of two methyl groups (as compared with Ala substitution) (30). Interestingly, the only substitution that affected B-band LPS biosynthesis was G301V, the last position of the RX10G motif in PL5; all other substitutions showed no effect on the LPS phenotype (Fig. 2), indicating that these residues do not play a catalytic role in the polymerization of O-Ag by Wzy from P. aeruginosa PAO1.
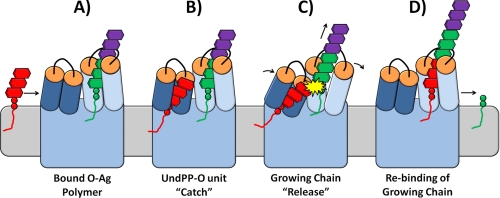
Based on the topological structure of Wzy from P. aeruginosa PAO1 (supplemental Fig. S1) (10), the presence of two similar sites of proposed substrate binding is initially consistent with the entirely conceptual ribosome-like model of O-Ag polymerization proposed by Bastin et al. (31). However, in addition to the high degree of PL3-PL5 sequence equivalence we have uncovered (Fig. 1), the above demonstrated importance of charged residues in PL3 and PL5, as well as the context of the local environments in which they are located (Figs. 1–3), are consistent with the mechanism of O-Ag polymerase function in P. aeruginosa PAO1 that we have previously proposed (10) and herein refined. Wzy of P. aeruginosa is required to interact with the negatively charged O-Ag units of this species (supplemental Fig. S5), linked to UndPP, in two different ways. As growth of O-Ag heteropolymers has been demonstrated to occur at the reducing terminus of the glycan chain (32), one mode of Wzy-substrate interaction would constitute retention of the growing O-Ag chain after it has been extended (Fig. 4A). The concurrent second interaction scenario would entail the capture and recruitment of a single UndPP-linked O-Ag subunit to be used for the next polymerization reaction (Fig. 4B). As PL3 and PL5 share such a high degree of physicochemical conservation (Fig. 1), it is intriguing to observe that although each contains identical key residues, the two periplasmic loops possess distinct net charge properties. Therefore, we propose that each functions in one of the two capacities described above.
FIGURE 4.
Proposed model for the catch-and-release mechanism of Wzy function in P. aeruginosa PAO1. PL3 (net-positive charge) is colored in dark blue. PL5 (net-negative charge) is colored in light blue. RX10G motifs on both PL3 and PL5 are colored in orange. A, polymerized O-Ag bound to PL5, mediated by its RX10G motif. B, catching (recruitment) of a new UndPP-linked O-Ag repeat by the positively charged PL3, mediated by its RX10G motif. C, transfer of the new UndPP-linked O-Ag repeat to the retention arm (PL5) of Wzy and polymerization at the reducing terminus of the growing chain, resulting in release of the growing chain to accommodate chain elongation. D, rebinding of the polymerized chain at the newly added O-Ag repeat unit via the RX10G motif.
Considering the significant net-positive charge on PL3, the positive charge dependence of Arg175 and Arg180, as well as the functional importance of the Arg176 guanidinium side chain in the RX10G motif (Figs. 2A and 3), it is conceivable that this periplasmic loop would function as the capture arm of Wzy; this would serve to “catch” incoming negatively charged O-Ag subunits (Fig. 4B), allowing for subsequent transfer to the retention site (Fig. 4C). The need for a consistent molecular framework with which to constantly recruit new O-Ag subunits is also reflected in the lack of variability in the tertiary structure modeling output based on PL3 amino acid sequence (supplemental Fig. S2A). Consequently, by virtue of its net-positive charge (Fig. 1) and important Arg residues (Figs. 2A and 3), this arm would provide sufficiently high capacity for Wzy to bind the negatively charged O-Ag subunits, as without initial recruitment (Fig. 4B), the subsequent polymerization reaction could not occur.
PL5, containing its own functionally important RX10G tract but with its more negatively charged local environment, would provide a tertiary structure motif conducive to a relatively transient interaction with an O-Ag subunit, capable of constant breakage and reformation. As such, PL5 would be a more likely candidate for the retention arm of Wzy, serving as a site required for constant binding and “release” of the growing O-Ag chain (Fig. 4, C and D), thus performing a more catalytic role. In this manner, we propose that the polymerized chain linked to UndPP would remain at the retention site (Fig. 4A) until the addition of a new UndPP-linked subunit from the recruitment arm was catalyzed (Fig. 4, B and C). This would result in loss of the UndPP formerly anchoring the growing chain at the retention site (Fig. 4D). Moreover, this would be more thermodynamically favorable as it would preclude the need for the back-and-forth-and-back-again transfer of the growing chain (with loss of the UndPP) from the retention site to the recruitment site and then back again to the retention site for each new subunit addition, as suggested by the concept developed by Bastin et al. (31). The demonstrated essential nature of the Arg290 and Arg291 side chains to the function of Wzy (Figs. 2B and 3), as well as their negatively charged local environment (Fig. 1), support the proposed release mechanism for PL5 with respect to its interaction with O-Ag subunits (Fig. 4C). This dynamic potential is also reflected in the more varied relative helix positions possible for the tertiary structure models of PL5 (supplemental Fig. S2B). Furthermore, PL5, which is located toward the latter half of the protein, is larger than PL3 (supplemental Fig. S1), and contains a GX3G (GASRight) helix-packing motif (29) in the downstream TMS; these are all analogous situations to the principal PL of WaaL from P. aeruginosa PAO1 required for transfer of O-Ag from UndPP to the core sugar acceptor linked to lipid A (10).
As polymerization of the O-Ag chain continues, the O-Ag becomes associated with either Wzz1 or Wzz2, the chain-length regulator proteins in P. aeruginosa PAO1 (7). This association appears to stabilize the interaction of the growing O-Ag chain with Wzy as removal of Wzz1 and Wzz2 function results in a complete loss of higher molecular weight B-band LPS and instead yields lower molecular weight banding patterns, indicative of significantly shorter lengths of heteropolymeric O-Ag chains ligated to lipid A-core (7).
To our knowledge, this investigation is the first to characterize Wzy polymerase function within the context of a rigorously validated and experimentally defined topological model of the protein. Considering the above mentioned results, taken together with the unbiased topology map for Wzy, the charge differences and functional importance of PL3 and PL5 would support a putative catch-and-release mechanism for the polymerization of O-Ag subunits, with both arms playing separate yet integral roles in the O-Ag assembly process.
Supplementary Material
This work was supported by operating grants from Cystic Fibrosis Canada and the Canadian Institutes of Health Research (CIHR) (Grant MOP-14687).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- O-Ag
- O-antigen
- TMS
- transmembrane segment(s)
- PL
- periplasmic loop
- UndPP
- undecaprenyl pyrophosphate.
REFERENCES
- 1. Ernst R. K., Hajjar A. M., Tsai J. H., Moskowitz S. M., Wilson C. B., Miller S. I. (2003) J. Endotoxin Res. 9, 395–400 [DOI] [PubMed] [Google Scholar]
- 2. Augustin D. K., Song Y., Baek M. S., Sawa Y., Singh G., Taylor B., Rubio-Mills A., Flanagan J. L., Wiener-Kronish J. P., Lynch S. V. (2007) J. Bacteriol. 189, 2203–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lau P. C., Lindhout T., Beveridge T. J., Dutcher J. R., Lam J. S. (2009) J. Bacteriol. 191, 6618–6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. King J. D., Kocíncová D., Westman E. L., Lam J. S. (2009) Innate Immun. 15, 261–312 [DOI] [PubMed] [Google Scholar]
- 5. Burrows L. L., Lam J. S. (1999) J. Bacteriol. 181, 973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Kievit T. R., Dasgupta T., Schweizer H., Lam J. S. (1995) Mol. Microbiol. 16, 565–574 [DOI] [PubMed] [Google Scholar]
- 7. Daniels C., Griffiths C., Cowles B., Lam J. S. (2002) Environ. Microbiol. 4, 883–897 [DOI] [PubMed] [Google Scholar]
- 8. Abeyrathne P. D., Daniels C., Poon K. K., Matewish M. J., Lam J. S. (2005) J. Bacteriol. 187, 3002–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Woodward R., Yi W., Li L., Zhao G., Eguchi H., Sridhar P. R., Guo H., Song J. K., Motari E., Cai L., Kelleher P., Liu X., Han W., Zhang W., Ding Y., Li M., Wang P. G. (2010) Nat. Chem. Biol. 6, 418–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Islam S. T., Taylor V. L., Qi M., Lam J. S. (2010) mBio 1, e00189-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qiu D., Damron F. H., Mima T., Schweizer H. P., Yu H. D. (2008) Appl. Environ. Microbiol. 74, 7422–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coyne M. J., Jr., Goldberg J. B. (1995) Gene 167, 81–86 [DOI] [PubMed] [Google Scholar]
- 13. Abeyrathne P. D., Lam J. S. (2007) Can. J. Microbiol. 53, 526–532 [DOI] [PubMed] [Google Scholar]
- 14. Abeyrathne P. D., Lam J. S. (2007) Mol. Microbiol. 65, 1345–1359 [DOI] [PubMed] [Google Scholar]
- 15. Lam J. S., MacDonald L. A., Lam M. Y., Duchesne L. G., Southam G. G. (1987) Infect. Immun. 55, 1051–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilks J. C., Slonczewski J. L. (2007) J. Bacteriol. 189, 5601–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roy A., Kucukural A., Zhang Y. (2010) Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gafvelin G., von Heijne G. (1994) Cell 77, 401–412 [DOI] [PubMed] [Google Scholar]
- 19. Kim T. H., Sebastian S., Pinkham J. T., Ross R. A., Blalock L. T., Kasper D. L. (2010) J. Biol. Chem. 285, 27839–27849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cuthbertson L., Mainprize I. L., Naismith J. H., Whitfield C. (2009) Microbiol. Mol. Biol. Rev. 73, 155–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Daniels C., Vindurampulle C., Morona R. (1998) Mol. Microbiol. 28, 1211–1222 [DOI] [PubMed] [Google Scholar]
- 22. Mazur A., Król J. E., Marczak M., Skorupska A. (2003) J. Bacteriol. 185, 2503–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schild S., Lamprecht A. K., Reidl J. (2005) J. Biol. Chem. 280, 25936–25947 [DOI] [PubMed] [Google Scholar]
- 24. Elumalai P., Rajasekaran M., Liu H. L., Chen C. (2010) Protoplasma 247, 13–24 [DOI] [PubMed] [Google Scholar]
- 25. Malik A., Ahmad S. (2007) BMC Struct. Biol. 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dahms N. M., Rose P. A., Molkentin J. D., Zhang Y., Brzycki M. A. (1993) J. Biol. Chem. 268, 5457–5463 [PubMed] [Google Scholar]
- 27. Feilmeier B. J., Iseminger G., Schroeder D., Webber H., Phillips G. J. (2000) J. Bacteriol. 182, 4068–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Drew D., Sjöstrand D., Nilsson J., Urbig T., Chin C. N., de Gier J. W., von Heijne G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2690–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Walters R. F., DeGrado W. F. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13658–13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Holst B., Zoffmann S., Elling C. E., Hjorth S. A., Schwartz T. W. (1998) Mol. Pharmacol. 53, 166–175 [DOI] [PubMed] [Google Scholar]
- 31. Bastin D. A., Stevenson G., Brown P. K., Haase A., Reeves P. R. (1993) Mol. Microbiol. 7, 725–734 [DOI] [PubMed] [Google Scholar]
- 32. Robbins P. W., Bray D., Dankert B. M., Wright A. (1967) Science 158, 1536–1542 [DOI] [PubMed] [Google Scholar]
- 33. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.