Abstract
The helicase and primase activities of the hexameric ring-shaped T7 gp4 protein reside in two separate domains connected by a linker region. This linker region is part of the subunit interface between monomers, and point mutations in this region have deleterious effects on the helicase functions. One such linker region mutant, A257T, is analogous to the A359T mutant of the homologous human mitochondrial DNA helicase Twinkle, which is linked to diseases such as progressive external opthalmoplegia. Electron microscopy studies show that A257T gp4 is normal in forming rings with dTTP, but the rings do not assemble efficiently on the DNA. Therefore, A257T, unlike the WT gp4, does not preassemble on the unwinding DNA substrate with dTTP without Mg(II), and its DNA unwinding activity in ensemble assays is slow and limited by the DNA loading rate. Single molecule assays measured a 45 times slower rate of A257T loading on DNA compared with WT gp4. Interestingly, once loaded, A257T has almost WT-like translocation and DNA unwinding activities. Strikingly, A257T preassembles stably on the DNA in the presence of T7 DNA polymerase, which restores the ensemble unwinding activity of A257T to ∼75% of WT, and the rescue does not require DNA synthesis. The DNA loading rate of A257T, however, remains slow even in the presence of the polymerase, which explains why A257T does not support T7 phage growth. Similar types of defects in the related human mitochondrial DNA helicase may be responsible for inefficient DNA replication leading to the disease states.
Keywords: ATPases, Bacteriophage, DNA-binding Protein, DNA Enzymes, DNA Helicase, DNA Polymerase, DNA Primase, DNA-Protein Interaction, DNA Replication, Mitochondrial Metabolism
Introduction
Helicases are molecular motors that are involved in almost all of the metabolic processes of the nucleic acids in the living cell. Helicases catalyzing genome replication and recombination assemble into six-membered ring-shaped proteins around DNA (1–7). Bacteriophage T7 encodes a ring-shaped helicase, the gp4 protein, which has served as a model system to understand the mechanisms of this class of helicases (3, 4, 7). The human mitochondrial DNA helicase Twinkle is homologous to T7 gp4, showing 46% overall sequence similarity (8).
T7 gp4 is a bifunctional helicase-primase protein with helicase conserved motifs in the C-terminal domain and the primase motifs in the N-terminal domain. The two domains of the T7 gp4 are structurally independent, but each enzyme depends on the other for optimal activity (9, 10). Crystal structures of T7 gp4 have shown that the primase and helicase domains of T7 gp4 are connected via a linker region made of a helix-loop region (Fig. 1A) (11–13). The linker region is an important element that forms the subunit interface for assembling the helicase domains into a compact ring. A helix and loop in the linker region of one subunit interact with three helices of a neighboring subunit to connect the six subunits of the hexamer in a head-to-tail fashion (Fig. 1B).
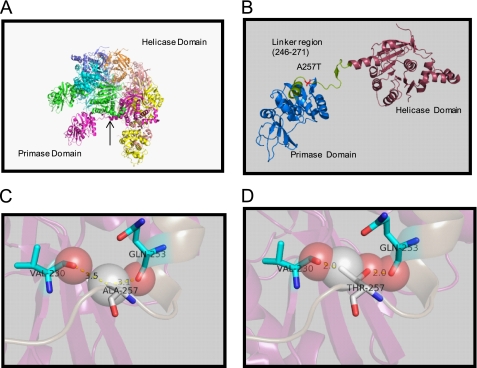
FIGURE 1.
Structure of T7 gp4 and location of the A257T mutation. A, T7 gp4 hexameric ring showing the arrangement of the helicase and primase domains (Protein Data Base ID code 1Q57). B, One subunit of the gp4 ring showing the 26-amino acid linker region between the C-terminal helicase and N-terminal primase domains. Ala257 is present in the linker region. C, close-up view of the Ala257 residue in relationship to proximal Gln253 and Val230. D, close-up view of A257T mutation showing clashes of Thr257 with the backbones of residues Gln253 and Val230.
Genetic screening has identified helicase-deficient but primase-proficient mutants that lie in the linker region (14, 15). Many of the disease-causing mutations in the human mitochondrial helicase Twinkle are also found in the highly homologous linker region (8, 16, 17). These mutations are linked to autosomal dominant progressive external opthalmoplegia, a human disease characterized by multiple mitochondrial DNA deletions (8, 18, 19). Individuals carrying this mutation suffer from early or late onset progressive external eye muscle weakness, neuropathy, depression, ataxia, and deafness.
One of the linker region mutations of the T7 gp4 is A257T, which was originally isolated from a genetic screen for helicase-deficient and primase-proficient mutants of gp4 (14, 15). An analogous mutation, A359T, is found in the human Twinkle protein (8). Twinkle contains a C-terminal helicase and N-terminal primase-like domain separated by a linker region that is highly homologous to the linker region of T7 gp4 (20). The similarity between T7 gp4 and human Twinkle indicates that the detailed characterization of the linker region mutant of T7 gp4 can provide important insights into the mechanisms that lead to mitochondrial deletions in humans. Biochemical studies of A257T or A257V have shown that the mutant protein is defective in DNA unwinding, and it was proposed that this might be due to its defect in translocating along single-stranded DNA (ssDNA)2 (15, 21). However, the precise mechanism by which A257T/A257V mutation causes this defect in translocation was not elucidated.
Here, we report an in-depth characterization of the biochemical and structural properties of the T7 gp4 A257T mutant to understand its helicase defect. Our studies show that A257T is not defective in translocation on ssDNA and DNA unwinding, but has a defect in DNA loading. We find that T7 DNA polymerase stabilizes the binding of A257T to the DNA. However, the assembly rate of A257T gp4 on the DNA remains critically slow, even in the presence of T7 DNA polymerase, which explains why it does not support T7 phage growth.
EXPERIMENTAL PROCEDURES
Reagents and Buffers
Chemical reagents were purchased from Sigma and JT Baker. Radionucleotides were purchased from PerkinElmer Life Science or GE Healthcare. Oligodeoxynucleotides were purchased from Integrated DNA Technology (IDT, Coralville, IA). 5′,6′-Fluorescein carboxyamide succinamidyle ester was purchased from Molecular Probes. All reactions were carried out at 25 °C in the T7 helicase buffer containing 50 mm Tris-Cl, pH 7.5, 40 mm NaCl, and 10% glycerol.
Proteins
WT gp4 (T7 gp4A′), A257T gp4, and T7 gp5 (exo-) were purified from recombinant Escherichia coli as described (22, 23). Protein concentrations were determined from its absorption at 280 nm and calculated extinction coefficients in 8 m guanidine hydrochloride.
DNA Substrates
Oligodeoxynucleotides purchased from IDT were PAGE-purified and electroeluted (Schleicher & Schuell Elutrap). DNA concentrations were determined from absorption at 260 nm and calculated extinction coefficients after digestion with snake venom phosphodiesterase I (24). The DNA strand was radiolabeled using [γ-32P]ATP and polynucleotide kinase. Labeled DNA was cleaned using Sephadex G-25 spin columns. Unwinding substrates were prepared by annealing together complementary DNA strands in the T7 helicase buffer. The 70-bp minicircle substrate was made using a DNA splint as reported (25, 26).
Ensemble Fluorescence-based DNA Unwinding
Fluorescence-based DNA unwinding experiments were performed in a stopped-flow instrument (Kin Tek Instruments, Austin, TX) (27). An equal volume of fluorescein-labeled 30-bp unwinding fork (20 nm), WT or A257T gp4 (200 nm hexamer), dTTP (4 mm), EDTA (6 mm), BSA (2 μm) in T7 helicase buffer from one syringe was rapidly mixed with MgCl2 (14 mm, and final free concentration of 2 mm), BSA (2 μm) with and without 3 μm dT90-DNA trap from a second syringe. The reaction mix was excited at 480 nm through a slit width of 2 mm (∼8-nm spectral bandwidth), and the increase in fluorescence was detected by monitoring light filtered through a long pass filter with a cutoff of 515 nm. The unwinding rates were determined from fitting as described (27).
In the DNA unwinding experiments involving mixed WT and A257T hexamers, the proteins were mixed in different molar proportions (40/0, 40/20, 40/40, 40/80, 40/200) in T7 helicase buffer and incubated on ice for 30 min, followed by the addition of EDTA, dTTP, BSA, and DNA and incubated for 10 min. The reaction was then carried out as indicated above.
Single Molecule DNA Unwinding
The single molecule DNA unwinding experiments were carried out as described previously (28) at 8-piconewton force using 2 mm dTTP. The raw initiation time under single molecule conditions was measured as the time period between rapid unzipping of ∼2000 bp of the double-stranded DNA (dsDNA) and when the helicase begins to unwind the dsDNA. This time periods were corrected for the time it takes to translocate on an average of 1000 nucleotides to determine the initiation time.
Electron Microscopy
WT or A257T gp4 (2.1 μm) was incubated at 25 °C for 20 min in 25 mm triethanolamine buffer, pH 7.2, with the nucleotides and DNA, applied to glow discharged grids, stained with 2% (w/v) uranyl acetate, and imaged under minimal dose conditions in a Tecnai T12 electron microscope, at a nominal magnification of ×30,000 and an accelerating voltage of 120 keV.
Minicircle Rolling Circle DNA Synthesis
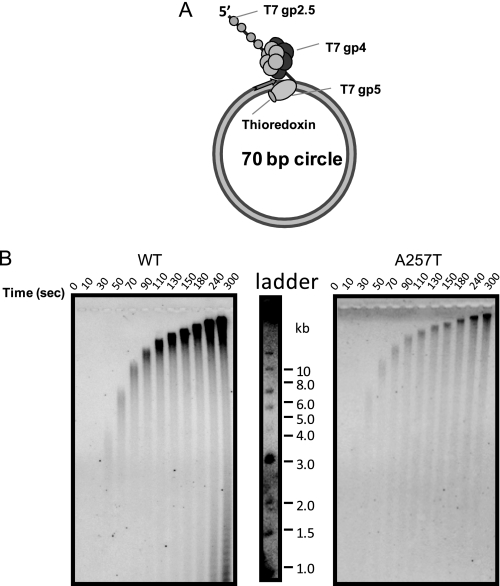
The rolling circle DNA synthesis of WT and A257T was carried out on the 70-bp minicircle substrate at 22 °C as reported (25).
Radiometric DNA Unwinding Assay
The rapid kinetics of dsDNA unwinding by WT and A257T was measured using the radiometric assay. WT or A257T (200 nm) was incubated for 30 min with 5 nm 40-bp replication fork (GC 20%), 5 mm EDTA, and 4 mm dTTP substrate in reaction buffer on ice in one syringe of the RQF-3 Quench-Flow apparatus (Kin Tek Instruments) and mixed with 13 mm MgCl2 and 3 μm dT90-DNA trap in the same buffer from a second syringe for 0.02 to 3 s and stopped by the addition 1.5 volume of 100 mm EDTA and 1% SDS. T7 DNA polymerase was preassembled by mixing gp5 and E. coli thioredoxin in 1:5 ratio at 22 °C for 5 min in reaction buffer containing freshly made 5 mm DTT. When present, T7 DNA polymerase (200 nm) was added to WT or A257T and DNA mix and further incubated at room temperature for 30 min. The quenched unwinding reaction with and without T7 DNA polymerase was loaded on a native 12% polyacrylamide gel for resolving the duplex and unwound DNA substrates, and the fraction of DNA unwound was determined and quantitated using the PhosphorImager (Molecular Dynamics, Sunnyvale, CA) instrument and ImageQuant5 software. The data were analyzed as described previously (29).
RESULTS
DNA Unwinding Activity of the gp4 A257T
The DNA unwinding activity of gp4 WT and A257T was measured by the real-time fluorescence assay (27). T7 gp4 WT or A257T (100 nm hexamer) was preincubated with the fluorescein-labeled fork DNA (10 nm) in the presence of dTTP (without Mg2+), and the reaction was initiated by rapid mixing with MgCl2 (Fig. 2A).
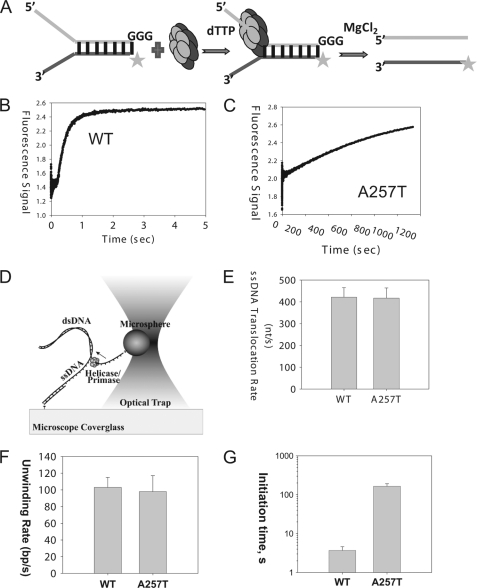
FIGURE 2.
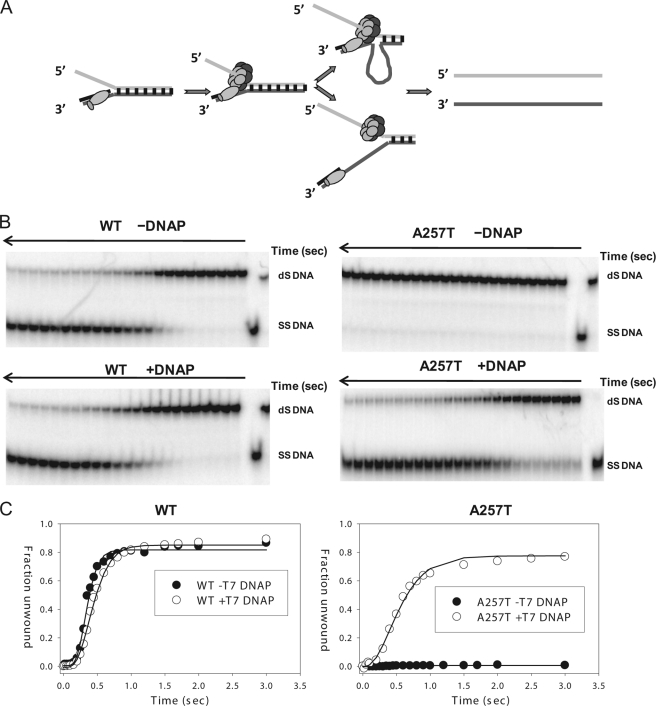
Helicase activities. A, scheme showing the ensemble unwinding assay. The ring-shaped hexameric helicase loads on the 5′-overhang and moves from the 5′-end to the 3′-end to unwind the fluorescein-labeled dsDNA fork substrate. T7 gp4 WT or A257T (100 nm) was preincubated with the fluorescent ds30 (23%) substrate (10 nm) and dTTP (2 mm) in one syringe of the stopped-flow instrument, and the unwinding reaction was initiated by addition of MgCl2 from a second syringe at 25 °C. B, unwinding kinetics of WT gp4. The lag kinetics was fit to the incomplete gamma function (29) to obtain a rate of ∼67 bp/s for the WT gp4. C, fluorescence signal resulting from unwinding of dsDNA into ssDNA by A257T plotted against time. The kinetics fit to a single exponential rate constant of 1.3 × 10−3 s−1. D, scheme of the single molecule optical trap setup (not to scale). A helicase/primase molecule separates the dsDNA, while a constant force is maintained on the DNA fork junction by adjusting the position of the glass coverslip relative to the optically trapped microsphere. The displacement of the trap relative the coverslip is utilized to determine the amount of DNA unwound (and subsequently the unwinding rate). E, comparison of ssDNA translocation rate of WT gp4 and A257T. Experiments were carried out at 0.3 nm WT and 30 nm A257T, 2 mm dTTP, 5 mm MgCl2. F, comparison of the unwinding rate WT gp4 and A257T. G, comparison of the initiation times of DNA unwinding by WT gp4 and A257T. The initiation times were experimentally determined at 5 nm hexamer, 2 mm dTTP, 5 mm MgCl2. The data represent average from >20 traces.
A time-dependent increase in fluorescence was observed in the WT gp4 reaction within 100 ms of reaction start (Fig. 2B). The fast kinetics indicates that WT gp4 is preassembled on the fork DNA and immediately initiates DNA unwinding upon addition of Mg(II) with rates of 67 bp/s at 25 °C. In the same time span, no fluorescence increase was observed in the A257T reaction; however, when the reaction was monitored for longer periods, we did observe a slow increase in fluorescence that indicated ∼1500-fold slower rate of unwinding by A257T compared with WT gp4 (Fig. 2C). As indicated previously, these experiments show that A257T has a defect in DNA unwinding (15, 21). However, the exact reason for the defect was not determined.
One reason for the defect might be that the mutant does not translocate along ssDNA. We therefore used a single molecule assay to measure the translocation rate of T7 gp4 on ssDNA. In this assay, one strand of a 4.1-kb length dsDNA molecule is attached to an optically trapped microsphere and the other strand is attached to the surface of a glass coverslip (30, 31) (Fig. 2D). By pulling on the ends of the two strands, ∼400 bp of dsDNA is rapidly unzipped by mechanical movement of the coverslip relative to the trapped microsphere, creating ssDNA loading region for freely diffusing T7 gp4 hexamers to bind. Following binding of the gp4 to the ssDNA loading region, several hundred bp of dsDNA are quickly unzipped in front of the helicase, allowing the helicase to translocate on ∼500 nucleotides of ssDNA without working against the DNA fork junction (28). The translocation rate is then determined by measuring the time for the helicase to travel across the unzipped ssDNA region. Using such experiments, we found that A257T can translocate on ssDNA, and the translocation rate of A257T is nearly the same as WT gp4 (Fig. 2E).
To determine whether A257T can unwind dsDNA, we used the same single molecule assay described above. T7 gp4 was allowed to reach the unwinding fork junction, and the unwinding reaction was monitored by measuring the increase in the length of ssDNA region. To our surprise, we found that the A257T is capable of unwinding a kilobase pair length of dsDNA with rates that are only a little slower than WT gp4 (Fig. 2F). The only defect we observed was that we needed higher A257T concentrations to detect enough unwinding molecules. WT gp4 at 0.3 nm hexamer concentration initiated DNA unwinding with an average initiation time of ∼60 s. To get a comparable initiation time (∼60 s) with A257T, the concentration of A257T had to be increased to 30 nm. These results indicate that contrary to what was concluded from previous ensemble reactions (15, 21), A257T is not defective in ssDNA translocation and DNA unwinding. The requirement of higher A257T protein indicates a defect in initiation; hence, we designed single molecule experiments to measure the initiation times to unwinding.
To measure the initiation time, about 2000 bp of the dsDNA was mechanically unzipped rapidly to create ssDNA region for the helicase to bind. Then, the time between mechanical unzipping of DNA and the beginning of the dsDNA unwinding reaction was measured. After correcting for the time it takes to translocate on the ssDNA (using the ssDNA translocation rates determined from Fig. 2E), we determined that at 5 nm hexamer, 2 mm dTTP, and 5 mm MgCl2, it takes on an average 3.7 s for the gp4 WT to initiate unwinding, and it takes A257T about 165 s to initiate unwinding at the same 5 nm concentration (Fig. 2G). These results indicate that A257T is 45 times slower at initiation compared with WT gp4. However, once it is bound to DNA, A257T unwinds with almost a WT rate. At 5 nm, we do not expect to have multiple A257T hexamers unwinding DNA because the initiation time of 165 s on an average is much longer than its unwinding time of <30 s.
A defect in assembling around DNA can explain the slow rate of unwinding by A257T detected in the ensemble assays (Fig. 2C). However, in the ensemble assays preincubation of A257T should have bypassed any slow assembly steps. WT gp4 preassembles on the fork DNA with dTTP without Mg(II), and in this manner we can bypass the assembly steps and synchronize the unwinding reaction, which allows us to measure the intrinsic unwinding rate. But this was not observed with A257T. This indicates that A257T does not preassemble on the fork DNA in the presence of dTTP without Mg(II). This is consistent with the filter binding assays that showed very little or no binding of A257T on ssDNA in the presence of dTTP without Mg(II) (supplemental Fig. 1). However, once Mg(II) is added, A257T must load on the DNA, as we observe DNA unwinding, but the loading rate must be slow, as measured by the single molecule assays above. Thus, the observed rate of DNA unwinding by A257T is slow under ensemble conditions because it is limited by the slow rate of DNA loading. The inefficient loading of A257T on the DNA also explains the lower DNA-stimulated Vmax of dTTPase (supplemental Fig. 2). The WT-like dTTP Km indicates that A257T is not defective in binding dTTP.
A257T Forms Rings
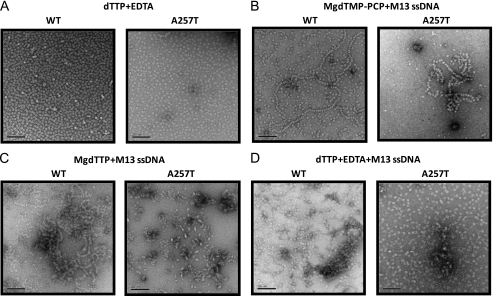
To determine whether the reason for not binding DNA in the presence of dTTP without Mg(II) is due to a defect in ring formation, we imaged WT and A257T gp4 proteins by electron microscopy (32, 33). Both WT and A257T gp4 assemble into ring-shaped structures in the presence of dTTP or dTMPPCP (Fig. 3A, supplemental Fig. 3), and the rings with dTMPPCP and MgCl2 bind to M13 ssDNA like beads on a string (Fig. 3B), consistent with the NC-DEAE binding assay with dTMPPCP (supplemental Fig. 1). In the presence of MgdTTP, WT gp4 binds to DNA like beads on a string, but A257T forms bullet-like structures, which is either an aggregate or a structure bound to DNA (Fig. 3C). With dTTP without Mg(II), the rings of A257T do not bind DNA (Fig. 3D). We conclude that A257T gp4 is normal in forming rings with dTTP; however, the rings do not bind DNA in the presence of dTTP without Mg(II).
FIGURE 3.
Electron microscopy studies of WT and A257T gp4. A, WT T7 gp4A or A257T 2.1 μm was incubated at 24 °C in triethanolamine buffer, pH 7.5, with 1.3 mm dTMPPCP, 2 mm magnesium acetate, 0.6 nm M13 ssDNA. B, WT or A257T was incubated at 24 °C with 2 mm dTTP, 7 mm MgCl2, 3 mm EDTA, 0.6 nm M13 ssDNA. C, WT or A257T was incubated with 2 mm dTTP and 3 mm EDTA. D, WT or A257T was incubated with 2 mm dTTP, 3 mm EDTA, and 0.6 nm M13 ssDNA.
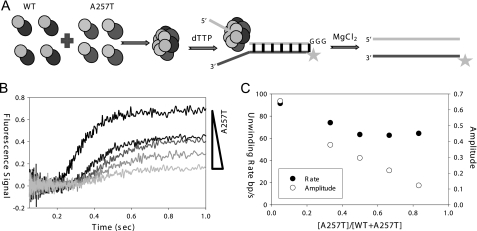
DNA Unwinding Activity of Mixed WT and A257T gp4
To determine whether mixed hexamers of WT gp4 and A257T are active in DNA loading, we measured the unwinding amplitude, which is a measure of the fraction of productive helicase bound to the forked substrate. WT gp4 at 20 nm was mixed with increasing amounts of A257T (10–100 nm) (Fig. 4A), and the unwinding kinetics was measured using the fluorescence-based assay (Fig. 4B). The data were analyzed to determine the unwinding rates and amplitudes. The unwinding rate of the mixed hexamers was 70% of the WT level and remained at that level even at a high A257T:WT ratio of 5:1 (Fig. 4C). The unwinding amplitude, on the other hand, decreased linearly with increasing fraction of A257T over the entire sampled region and predicted a close to zero amplitude for A257T alone. This indicates that the mixed A257T-WT hexamers are active in DNA unwinding; however, as the number A257T subunits increase in the mixed hexamers, the rings become increasingly defective in productive assembly on the DNA.
FIGURE 4.
DNA unwinding by mixed A257T-WT hexamers. A, WT gp4 (20 nm) was preincubated with various concentrations of A257T (10–100 nm) to make mixed hexamers, which were loaded on the fluorescein-labeled ds30 (23%) forked unwinding substrate with dTTP. The unwinding kinetics was measured by the stopped-flow fluorescence unwinding assay. B, the unwinding time traces show the decrease in amplitude with increasing fraction of A257T in the mixed hexamers. The kinetics was fit to the incomplete gamma function (29) to obtain the unwinding rates and amplitudes. C, the unwinding rate (filled circles) and amplitude (open circles) are plotted against a fraction of A257T.
A257T gp4 Supports DNA Replication
Previous studies had indicated that A257T supports DNA synthesis, but the mechanism of this rescue was not determined (15). We examined this reaction in more detail here with the minicircle replication assay (25, 34). A257T or WT gp4 was preincubated with T7 DNA polymerase on the minicircle fork DNA in the presence of dNTPs without MgCl2 (Fig. 5A). Reactions were initiated by adding MgCl2. Both WT and A257T supported the synthesis of kilobase pair size DNA products from rolling circle DNA synthesis (Fig. 5B). The only difference was the lower yield of DNA products in the A257T reactions, consistent with the lower efficiency of A257T assembly on the DNA.
FIGURE 5.
Rolling circle DNA synthesis by WT and A257T gp4 in the presence of T7 DNA polymerase. A, schematic shows the 70-bp minicircle replication fork. B, WT or A257T gp4 at 100 nm was incubated with 100 nm T7 DNA polymerase (T7 gp5-thioredoxin) and minicircle DNA (50 nm), and DNA synthesis was measured using [α-32P]dGTP in the presence of all dNTPs. The DNA products are resolved by electrophoresis on a 1% alkaline-agarose gel. Images show a time course of increasing kilobase pair length of DNA synthesis by WT and A257T in presence of T7 DNA polymerase.
T7 DNA Polymerase Rescues the Helicase Defect of A257T
There are two ways by which T7 DNA polymerase can rescue the activity of A257T gp4: (a) it helps load or stabilize the mutant helicase on the DNA, and/or (b) it “pushes” the helicase or prevents helicase slippage via its DNA synthesis activity. We did not observe any helicase slippage in the single molecule unwinding assays. However, we cannot eliminate the possibility that the rescue is because DNA polymerase pushes the helicase via DNA synthesis.
To distinguish between possibilities (a) and (b), the 40-bp fork DNA with 5′-ssDNA overhang for helicase loading and 3′-primer-template region for polymerase binding was incubated with the helicase and dTTP without Mg(II) in the absence and in the presence of T7 DNA polymerase for at least 30 min, and then reactions were initiated with MgCl2 (Fig. 6A). The 5′-strand was radiolabeled to monitor its conversion to ssDNA by the gel assay (27, 29, 35). We did not add the other three dNTPs required for DNA synthesis. Without the remaining dNTPs, the DNA polymerase incorporates one dTTP but cannot elongate the primer any further by DNA synthesis.
FIGURE 6.
T7 DNA polymerase rescues the DNA unwinding defect of A257T gp4. A, T7 DNAP and WT or A257T gp4 were assembled on the radiolabeled ds40 (20%) replication fork with dTTP. Unwinding reaction was initiated with MgCl2. DNA unwinding proceeds either by both proteins moving together, forming a loop-like structure, or helicase moving alone. B, native polyacrylamide gels show the unwinding kinetics of WT and A257T gp4 in the presence and absence of T7 DNAP. C, unwinding kinetics by WT and A257T in the presence (open circles) and in the absence of T7 DNAP (filled circles) is shown. WT gp4 unwinds the forked substrate in the absence of T7 DNAP at ∼82 bp/s in the presence of T7 DNAP at ∼65 bp/s. The unwinding rate of A257T gp4 in the presence of T7 DNAP is ∼50 bp/s.
A257T gp4 without the polymerase does not unwind the fork DNA (Fig. 6B), consistent with the fluorescence-based unwinding assay. However, when A257T was preincubated with T7 DNA polymerase, it showed almost WT-like DNA unwinding activity (Fig. 6, B and C). WT gp4 unwinds the forked substrate in the absence of T7 DNA polymerase at ∼82 bp/s and in the presence of T7 DNA polymerase at ∼65 bp/s. The DNA polymerase in this case does not accelerate the DNA unwinding activity of WT as observed previously (36) because there is no DNA synthesis. In fact, it appears that a nonsynthesizing DNA polymerase slows the helicase. The unwinding rate of A257T gp4 in the presence of T7 DNA polymerase is ∼50 bp/s (Fig. 6C), which is ∼75% of WT. These results indicate that A257T can preassemble on the fork DNA in the presence of T7 DNA polymerase. The preassembled A257T on the DNA has almost WT-like unwinding activity, as observed in the single molecule unwinding assays (Fig. 2C).
How does T7 DNA polymerase help A257T preassemble on the fork DNA? It has been proposed that T7 DNA polymerase interacts with T7 gp4 via a basic loop that becomes exposed when T7 gp5 (DNA polymerase subunit of T7 DNA polymerase) interacts with thioredoxin (processivity factor). We therefore measured DNA unwinding by A257T without thioredoxin in the presence of T7 gp5 alone. We find that T7 gp5 alone was not able to rescue the helicase function of A257T (supplemental Fig. 4). These results indicate a specific mechanism involving the thioredoxin binding domain on gp5 in rescuing the helicase function of A257T.
Slow Assembly of A257T gp4 on the DNA
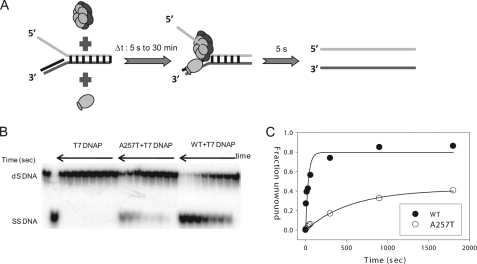
If A257T can unwind DNA to almost WT level in the presence of T7 DNA polymerase, then why does it not support phage growth? One reason could be that the loading of A257T on the DNA although rescued by T7 DNA polymerase is too slow to support T7 DNA synthesis. To measure the DNA loading rate in the presence of T7 DNA polymerase, we designed the following experiment (Fig. 7A): WT or A257T gp4 was incubated with T7 DNA polymerase and the replication fork substrate in the presence of dTTP without MgCl2, and this incubation time was controlled from 5 s to 30 min. Following controlled incubation, the unwinding activity was assayed within 5 s, which is enough time for the helicase to unwind the 40 bp duplex fully (see Fig. 6B). The unwinding activity here is a readout of the assembly process around DNA. The results showed that WT gp4 assembles on the fork at a rate of 0.03 s−1 and A257T assembles with a rate of 0.001 s−1 (Fig. 7, B and C), which is similar to the ensemble unwinding rate of A257T shown in Fig. 2F that we propose is limited by the DNA loading rate. Thus, T7 DNA polymerase does not speed up the DNA loading rate of A257T, but it aids in stabilizing the complex of A257T on the DNA, even when Mg(II) is absent. The intrinsically slow DNA loading rate of A257T explains why this mutant, although active in DNA unwinding, does not support phage growth.
FIGURE 7.
Slow binding of A257T to the replication fork. A, a mixture of WT or A257T gp4 (200 nm), T7 DNAP (200 nm), dTTP (4 mm), and radiolabeled ds40 (20%) replication fork substrate was incubated for various time intervals (Δt: 5 s to 30 min). A solution of MgCl2 and dT90 trap was then added to initiate the unwinding reaction, which was quenched in 5 s. The DNA substrate and the unwound ssDNA were resolved by native-PAGE. B, the image shows the extent of unwinding in 5 s in reactions containing T7 DNAP alone or T7 DNAP plus A257T or WT gp4 as a function of Δt. C, fraction of replication fork unwound as a function of Δt. The data were fit to single exponential equation to obtain the rate of helicase assembly in the presence of T7 DNAP: WT (filled circles) at 0.03 s−1 and A257T (open circles) at 0.001 s−1.
DISCUSSION
The studies in this paper show that the defect in the helicase function of the linker region mutant, A257T gp4, is caused by a defect in DNA loading rather than a defect in DNA unwinding, as previously thought. Both single molecule and ensemble assays indicate that A257T loads 30–45 times slower on DNA compared with WT gp4, but once it is assembled on the DNA into a stable complex, A257T has almost WT-like ssDNA translocation and unwinding activities.
The linker region between the N-terminal primase and the C-terminal helicase domain of T7 gp4 is an important element that forms the subunit interface for assembling the helicase domain into a compact ring (11–13). The helix and the loop of the linker region interact with three helices of the neighboring subunit to form a clasp of subunit interface (Fig. 1A). The primase domains are independent structural units emerging out of each subunit of the helicase ring, with their primase active sites pointing toward the neighboring subunit primase domain.
The residue Ala257 is located at the beginning in the loop of the linker region, close to the N-end, (Fig. 1B). Mutation of Ala257 to Val, Leu, Pro, Glu, Asp, Asn, Thr, His, or Tyr is lethal, and the mutant protein does not support T7 phage growth (21). These results suggest that the small size of Ala257 is important for its function. Because Ala257 lies closer to the primase domain than the helicase, it is unlikely that A257T mutation affects the folding of the helicase domain. This is consistent with the observation that A257T mutation does not affect ring formation, which occurs primarily through interactions between the helicase domains. Our modeling experiments using the crystal structure of the gp4 heptamer suggest that the position of Ala257 is such that any change to a bulky amino acid will produce a clash with the nearby amino acids (Fig. 1, C and D). The A257T change results in a clash with Gln253 and Val230, which both reside in the helix next to the linker region and in the primase domain. To accommodate the A257T change, therefore, the protein must undergo local structural changes. It appears that these local changes near the linker region affect the DNA loading mechanism of T7 gp4.
Previous studies have suggested a role of the primase domain in DNA loading by the ring-opening mechanism (37, 38). Based on the DNA binding kinetics, it was proposed that ssDNA initially interacts with the primase domain, which chaperones the ssDNA into the central channel. It was proposed that the binding of the DNA to the primase domain causes the linker region clasp to weaken and to open the subunit interface like a lock washer, allowing the DNA to bind in the central channel. It has been shown that the A257T gp4 has primase activity; hence, the primase domain must be proficient in DNA binding (15, 21). However, local changes caused by the A257T mutation may restrict loading of the DNA into the central channel. The single molecule assays show that indeed A257T DNA loading rate is ∼45 times slower than WT gp4. The single molecule assay is sensitive to detect the small number of A257T molecules that load on the ssDNA, and hence it is able to observe the almost WT like unwinding and ssDNA translocation activities of A257T.
We did notice in the electron microscopy studies that A257T has a greater tendency to form aggregates in the presence of Mg2+. These aggregates appear as short filament type structures, which could be formed by altered interactions between subunits. It is possible that the local changes in the linker region alter interactions between the subunits, causing hydrophobic residues to be exposed that lead to protein aggregation. Perhaps these higher order oligomers or aggregates of A257T are refractory to DNA loading.
We find it curious that although both dTMPPCP and dTTP nucleotides promote ring formation, A257T binds DNA only in the presence of dTMPPCP, but not with dTTP. The decreased aggregation and increased rings around DNA with MgdTMPPCP as opposed to MgdTTP could be because dTMPPCP does not get hydrolyzed and interacts more tightly with the subunits than dTTP.
When A257T was preincubated with T7 DNA polymerase and fork DNA, the helicase could be stably assembled on the DNA such that it now displayed almost WT-like unwinding activity. This recovery was observed even in the absence of DNA synthesis. Hence, T7 DNA polymerase is able to restore the function of A257T gp4 by stabilizing the binding of the helicase on the replication fork.
T7 DNA polymerase interacts with T7 gp4 via the basic loops of the thioredoxin binding domain (39). The processivity factor thioredoxin binds to the thioredoxin binding domain of gp5 and causes exposure of the basic loops, which stimulate interaction between the helicase and polymerase. It appears that this interaction between the helicase and polymerase stabilizes A257T on the DNA. This is supported by our finding that T7 gp5 by itself (or thioredoxin by itself) does not stimulate the helicase activity of A257T. Although T7 DNA polymerase rescues the helicase function of A257T gp4, it does not accelerate the DNA loading rate. Measurement of the kinetics of protein assembly on the fork DNA showed that the loading time of A257T is about 30 times slower than of the WT gp4. Thus, it appears that T7 DNA polymerase rescues A257T by stabilizing the helicase complex on the DNA.
The slow assembly of A257T gp4 on the DNA explains why it is not a viable mutation and why it does not support phage growth. Helicase loading is a key step during the initiation of DNA replication, in addition to being required for processive DNA replication. Thus, any defect in helicase loading will lead to a defect in DNA replication. Although T7 DNA polymerase is able to rescue the DNA loading defect of A257T, the loading process is still inefficient to support the propagation of T7 phage, whose entire infection cycle lasts for 25–30 min at 30 °C (40).
The importance of studying the A257T mutation of T7 gp4 lies in the fact that an analogous mutation of A359T is naturally found in the human mitochondrial DNA helicase Twinkle (8). The Twinkle helicase contains a C-terminal helicase and N-terminal primase-like domain separated by a linker region that is highly homologous to the linker region of T7 gp4. Modeling studies suggest that the linker region of Twinkle plays a role similar to that of gp4 in connecting the helicase domains to form a hexameric ring (20). Many of the disease-causing Twinkle mutations are found within or near the linker region, including A359T.
The detailed studies of the T7 gp4 A257T mutant reported here potentially elucidate the defects of other helicases with similar conserved linker region. A biochemical study of A359T Twinkle has shown that the protein is not defective in oligomerization, but has reduced helicase and replication properties (20). If A359T Twinkle mutant cannot assemble efficiently on the DNA, it will hinder normal replication fork initiation and progression (41). A mutant defective in loading on DNA will also have problems with reinitiation of DNA replication following stalling or collapse of the replication fork. Mixed hexamers of T7 gp4 A257T and WT gp4 are more active in DNA loading even when 85% of the population was made of A257T. This is consistent with what has been observed with the A359T mutation in Twinkle. In the heterozygous form the symptoms are mild, but in the homozygous form of A359T, early onset autosomal dominant progressive external opthalmoplegia is observed.
Supplementary Material
Acknowledgment
We thank Dr. Vikas Nanda for help in structure modeling.
This work was supported, in whole or in part, by National Institutes of Health Grants GM55310 (to S. S. P.), GM035269 (to E. H. E.), and GM059849 (to M. D. W.). This work was also supported by National Science Foundation Grant MCB-0820293 (to M. D. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–4.
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA
- dTMPPCP
- deoxythymidine (β, γ, methylene) triphosphate.
REFERENCES
- 1. Matson S. W., Kaiser-Rogers K. A. (1990) Annu. Rev. Biochem. 59, 289–329 [DOI] [PubMed] [Google Scholar]
- 2. Lohman T. M., Bjornson K. P. (1996) Annu. Rev. Biochem. 65, 169–214 [DOI] [PubMed] [Google Scholar]
- 3. Patel S. S., Picha K. M. (2000) Annu. Rev. Biochem. 69, 651–697 [DOI] [PubMed] [Google Scholar]
- 4. Donmez I., Patel S. S. (2006) Nucleic Acids Res. 34, 4216–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singleton M. R., Dillingham M. S., Wigley D. B. (2007) Annu. Rev. Biochem. 76, 23–50 [DOI] [PubMed] [Google Scholar]
- 6. Pyle A. M. (2008) Annu. Rev. Biophys. 37, 317–336 [DOI] [PubMed] [Google Scholar]
- 7. Hamdan S. M., Richardson C. C. (2009) Annu. Rev. Biochem. 78, 205–243 [DOI] [PubMed] [Google Scholar]
- 8. Spelbrink J. N., Li F. Y., Tiranti V., Nikali K., Yuan Q. P., Tariq M., Wanrooij S., Garrido N., Comi G., Morandi L., Santoro L., Toscano A., Fabrizi G. M., Somer H., Croxen R., Beeson D., Poulton J., Suomalainen A., Jacobs H. T., Zeviani M., Larsson C. (2001) Nat. Genet. 28, 223–231 [DOI] [PubMed] [Google Scholar]
- 9. Bernstein J. A., Richardson C. C. (1989) J. Biol. Chem. 264, 13066–13073 [PubMed] [Google Scholar]
- 10. Mendelman L. V., Notarnicola S. M., Richardson C. C. (1993) J. Biol. Chem. 268, 27208–27213 [PubMed] [Google Scholar]
- 11. Singleton M. R., Sawaya M. R., Ellenberger T., Wigley D. B. (2000) Cell 101, 589–600 [DOI] [PubMed] [Google Scholar]
- 12. Sawaya M. R., Guo S., Tabor S., Richardson C. C., Ellenberger T. (1999) Cell 99, 167–177 [DOI] [PubMed] [Google Scholar]
- 13. Toth E. A., Li Y., Sawaya M. R., Cheng Y., Ellenberger T. (2003) Mol. Cell 12, 1113–1123 [DOI] [PubMed] [Google Scholar]
- 14. Rosenberg A. H., Griffin K., Washington M. T., Patel S. S., Studier F. W. (1996) J. Biol. Chem. 271, 26819–26824 [PubMed] [Google Scholar]
- 15. Washington M. T., Rosenberg A. H., Griffin K., Studier F. W., Patel S. S. (1996) J. Biol. Chem. 271, 26825–26834 [DOI] [PubMed] [Google Scholar]
- 16. Holmlund T., Farge G., Pande V., Korhonen J., Nilsson L., Falkenberg M. (2009) Biochim. Biophys. Acta 1792, 132–139 [DOI] [PubMed] [Google Scholar]
- 17. Matsushima Y., Kaguni L. S. (2009) Biochim. Biophys. Acta 1787, 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Copeland W. C. (2008) Annu. Rev. Med. 59, 131–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mao C. C., Holt I. J. (2009) Chang Gung Med. J. 32, 354–369 [PubMed] [Google Scholar]
- 20. Korhonen J. A., Pande V., Holmlund T., Farge G., Pham X. H., Nilsson L., Falkenberg M. (2008) J. Mol. Biol. 377, 691–705 [DOI] [PubMed] [Google Scholar]
- 21. Lee S. J., Richardson C. C. (2004) J. Biol. Chem. 279, 23384–23393 [DOI] [PubMed] [Google Scholar]
- 22. Patel S. S., Rosenberg A. H., Studier F. W., Johnson K. A. (1992) J. Biol. Chem. 267, 15013–15021 [PubMed] [Google Scholar]
- 23. Patel S. S., Wong I., Johnson K. A. (1991) Biochemistry 30, 511–525 [DOI] [PubMed] [Google Scholar]
- 24. Cavaluzzi M. J., Borer P. N. (2004) Nucleic Acids Res. 32, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pandey M., Syed S., Donmez I., Patel G., Ha T., Patel S. S. (2009) Nature 462, 940–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee J., Chastain P. D., 2nd, Kusakabe T., Griffith J. D., Richardson C. C. (1998) Mol. Cell 1, 1001–1010 [DOI] [PubMed] [Google Scholar]
- 27. Pandey M., Levin M. K., Patel S. S. (2010) Methods Mol. Biol. 587, 57–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johnson D. S., Bai L., Smith B. Y., Patel S. S., Wang M. D. (2007) Cell 129, 1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jeong Y. J., Levin M. K., Patel S. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7264–7269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koch S. J., Shundrovsky A., Jantzen B. C., Wang M. D. (2002) Biophys. J. 83, 1098–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shundrovsky A., Smith C. L., Lis J. T., Peterson C. L., Wang M. D. (2006) Nat. Struct. Mol. Biol. 13, 549–554 [DOI] [PubMed] [Google Scholar]
- 32. Egelman E. H., Yu X., Wild R., Hingorani M. M., Patel S. S. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 3869–3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu X., Hingorani M. M., Patel S. S., Egelman E. H. (1996) Nat. Struct. Biol. 3, 740–743 [DOI] [PubMed] [Google Scholar]
- 34. Lee J., Chastain P. D., 2nd, Griffith J. D., Richardson C. C. (2002) J. Mol. Biol. 316, 19–34 [DOI] [PubMed] [Google Scholar]
- 35. Donmez I., Rajagopal V., Jeong Y. J., Patel S. S. (2007) J. Biol. Chem. 282, 21116–21123 [DOI] [PubMed] [Google Scholar]
- 36. Stano N. M., Jeong Y. J., Donmez I., Tummalapalli P., Levin M. K., Patel S. S. (2005) Nature 435, 370–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ahnert P., Picha K. M., Patel S. S. (2000) EMBO J. 19, 3418–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Picha K. M., Ahnert P., Patel S. S. (2000) Biochemistry 39, 6401–6409 [DOI] [PubMed] [Google Scholar]
- 39. Ghosh S., Hamdan S. M., Cook T. E., Richardson C. C. (2008) J. Biol. Chem. 283, 32077–32084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Studier F. W. (1972) Science 176, 367–376 [DOI] [PubMed] [Google Scholar]
- 41. Goffart S., Cooper H. M., Tyynismaa H., Wanrooij S., Suomalainen A., Spelbrink J. N. (2009) Hum. Mol. Genet. 18, 328–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.