Abstract
CD59 is a glycosylphosphatidylinositol-anchored protein that inhibits the assembly of the terminal complement membrane attack complex (MAC) pore, whereas Streptococcus intermedius intermedilysin (ILY), a pore forming cholesterol-dependent cytolysin (CDC), specifically binds to human CD59 (hCD59) to initiate the formation of its pore. The identification of the residues of ILY and hCD59 that form their binding interface revealed a remarkably deep correspondence between the hCD59 binding site for ILY and that for the MAC proteins C8α and C9. ILY disengages from hCD59 during the prepore to pore transition, suggesting that loss of this interaction is necessary to accommodate specific structural changes associated with this transition. Consistent with this scenario, mutants of hCD59 or ILY that increased the affinity of this interaction decreased the cytolytic activity by slowing the transition of the prepore to pore but not the assembly of the prepore oligomer. A signature motif was also identified in the hCD59 binding CDCs that revealed a new hCD59-binding member of the CDC family. Although the binding site on hCD59 for ILY, C8α, and C9 exhibits significant homology, no similarity exists in their binding sites for hCD59. Hence, ILY and the MAC proteins interact with common amino acids of hCD59 but lack detectable conservation in their binding sites for hCD59.
Keywords: Bacterial Toxins, Cholesterol, Complement, Membrane, Receptor Structure-Function
Introduction
The cholesterol-dependent cytolysins (CDCs)4 are a family of pore-forming toxins produced by a diverse group of Gram-positive pathogens. Recently crystal structures of the membrane attack complex/perforin proteins complement C8α, a C9-like protein from Photorhabdus luminescens, and mouse perforin (1–4) suggested that they are structurally and mechanistically related the CDCs (5–7) and may be ancient ancestors (8). Interestingly, this relationship between the CDCs and membrane attack complex/perforin proteins extends to other features of the complement system. Two members of the CDC family, intermedilysin (ILY) secreted by Streptococcus intermedius and vaginolysin (VLY) secreted by Gardnerella vaginalis, specifically bind to the human form of CD59 (hCD59), a glycosylphosphatidylinositol (GPI)-anchored terminal inhibitor of the mammalian complement membrane attack complex (MAC) (9), rather than cholesterol (10, 11). These CDCs bind to hCD59 to initiate the assembly of their oligomeric pore complex on the membrane of human cells (10), whereas the main function of CD59 is to block the assembly of the MAC pore on host cells, thereby protecting them from the lytic effects of activated complement MAC.
Domain 4 of the CDCs mediates its interaction with cholesterol-rich membranes (12–14). Most CDCs use cholesterol as their receptor and specifically recognize its 3-β-hydroxy group. Recently Farrand et al. (15) defined the cholesterol recognition motif as a threonine/leucine pair located in loop 1 that is conserved in all members of the CDC family, including ILY and VLY. However, ILY initiates its interaction with the cell by binding to hCD59 (10) rather than cholesterol. Receptor binding triggers domain 3 structural changes that lead to oligomerization and pore formation and also allows the cholesterol recognition motif to recognize and bind cholesterol, which initiates the membrane insertion of loops L1-L3. The insertion of L1-L3 is necessary to firmly anchor ILY to the membrane (15, 16) as ILY disengages from hCD59 during prepore to pore conversion (17). Hence, the cholesterol-dependent insertion of the L1-L3 loops is necessary to maintain its anchor to the membrane during this critical transition (15, 17).
Human CD59 is a 20-kDa GPI-anchored membrane protein that inhibits the formation of the complement MAC pore on host cells when complement is activated during infection (18). CD59 specifically binds to MAC components C8α and C9, thereby preventing their oligomerization into the MAC pore complex (18–20). An important feature of CD59 is its species selectivity, which is responsible for the homologous restriction of CD59 activity (21, 22). C8α and C9 binding to hCD59 has been linked to the variable region residues 40–58, which exhibits the least homology between species and is responsible for its species selective inhibition of complement (21–23). The same region of hCD59 has been shown to contribute to its species-specific interaction with ILY (10). We recently reported the interaction between non-lytic complexes of ILY and hCD59 abrogated the ability of CD59 to protect host cells from lysis by MAC, suggesting the binding sites on hCD59 for ILY and C8α and/or C9 overlap (17).
To better understand the interaction of ILY with its receptor, we performed a detailed analysis of the surface residues of hCD59 and those of domain 4 of ILY that contribute to this interaction. These studies revealed a significant correspondence in the hCD59 residues that contribute to ILY binding and its interaction with the MAC proteins. These similarities extended to a far deeper level than expected; ILY, C8α, and C9 all interact with common residues of hCD59. Their hCD59 binding sites, however, exhibit a remarkable absence of similarity. We further show that the residues of ILY that contribute to its binding site for hCD59 appear to be a signature motif for hCD59 binding CDCs.
MATERIALS AND METHODS
Antibodies, Plasmids, and Chemicals
The gene for ILY was cloned into pTrcHisA (Invitrogen) expression vector as described previously (24). The gene for hCD59 and its derivatives was cloned into pcDNA3.1 (+) as previously described (25). All chemicals and enzymes were obtained from Sigma, VWR, and Research Organics except where noted. All fluorescent probes were obtained from Molecular Probes (Invitrogen). Anti-hCD59 MEM-43 fluorescein isothiocyanate (FITC) conjugated was obtained from AbCam. Anti-hCD59 H19 conjugated to FITC was obtained from BD Pharmingen. Anti-HA conjugated to FITC was obtained from Sigma. Anti-hCD59 10G10 antibody was purified as previously described from a mouse B-cell myeloma (17). Rabbit antiserum to Chinese hamster ovary cells (CHO) cell membranes was previously prepared as described (26).
Generation ILY and hCD59 Mutants
The generation of amino acid substitutions in the genes for ILY and hCD59 was accomplished using PCR QuikChange mutagenesis (Stratagene). Most ILY mutants were generated in the monomer-locked version of ILY (ILYml) in which a free cysteine was substituted for Asp-280 as a site for modification with sulfhydryl-specific fluorescent probes (17) unless noted otherwise. The Oklahoma Medical Research Foundation Core DNA sequencing facility performed DNA sequence analysis of each mutant toxin plasmid.
Expression, Purification, and Fluorescent Labeling of ILY, Perfringolysin O (PFO), and Their Derivatives
The expression and purification of recombinant ILY and its derivatives and PFO from Escherichia coli were carried out as previously described (6, 24).
Fluorescent modification of ILY and its derivatives at cysteine-substituted Asp-280 with Alexa-Fluor 488 C5-maleimide (AF488; Molecular Probes) via the sulfhydryl was carried out as described previously (10, 27). Proteins were typically labeled at an efficiency of 60–100%. The dye-labeled protein samples were made 10% (v/v) in sterile glycerol, quick-frozen in liquid nitrogen, and stored at −80 °C.
Cells and Transfection
Chinese hamster ovary cells (CHO) were transfected with pcDNA3.1 (+) expression plasmid containing the gene for human CD59 (CHOhCD59) or its derivatives. The cells were maintained in F-12 Kaighn's medium (10% v/v fetal calf serum (FCS), 1% v/v Pen/Strep) (Invitrogen). As previously described (17), the first nine amino acids of the malaria epitope tag (NANPNANPNALG located between residues 2 and 3 of the mature human CD59 (25)), were replaced with the HA epitope from influenza (YPYDVPDYA) to create HA tagged hCD59 (HAhCD59). CHO cells were then transfected with this plasmid using the FuGENE 6 reagent (Roche Applied Science) as per the manufacturer's instructions. Positive clones were selected using anti-HA-FITC antibody (Sigma) by cell sorting (Influx cell sorter, University of Oklahoma Health Sciences Center Flow Cytometry Core Facility). Subsequent hCD59 mutants were generated in the HAhCD59 parental background using QuikChange site-directed mutagenesis. CHO lines expressing alanine-substituted hCD59 mutants for Glu-43, Asn-48, Asp-49, Thr-51, and Thre-52 were prepared as previously described (26). In all cases cells expressing similar levels of hCD59 and its derivatives were selected by flow cytometry using fluorescently tagged anti-HA epitope tag monoclonal antibody.
The conformational integrity of the CD59 mutants was determined by measuring the relative Kd of monoclonal antibodies for hCD59 and its derivatives that bound to different epitopes. This was achieved by incubating each mutant hCD59 with various concentrations of the FITC-conjugated anti-CD59 monoclonal antibodies. Briefly, the hCd59-expressing CHO cells were removed from the culture flask by gentle scraping in PBS supplemented with 5 mm EDTA. Cells (1 × 105) were incubated with monoclonal antibody FITC-MEM-43 (0–66.6 nm) or FITC-H19 (0–1000 nm) for 1 h at 4 °C (total volume 100 μl). Antibody binding was measured by fluorescence using flow cytometry to determine the geometric mean of the total bound fluorescent antibodies at each concentration of antibody. The relative Kd of the binding interaction was determined using Graphpad Prism to plot the geometric mean of the cell bound fluorescence intensity versus antibody concentration using the single site binding equation Y = BmaxX/Kd + X where X is the concentration of antibody, Y is the geometric mean of the fluorescence intensity, Bmax is the maximum binding sites, and the Kd is the dissociation constant.
Binding Affinity Analysis
Subconfluent CHOHAhCD59 cells and various mutants were detached with 5 mm EDTA in PBS, washed once with PBS, and resuspended in F-12 Ham's medium supplemented with 10% FCS. 50 μl of cells (1 × 105 cells) were added to 50 μl of various concentrations of AF488 labeled ILYml (170.9–0.167 nm) in PBS. Cells and toxin were incubated for 1 h at 37 °C and then transferred to 400 μl of PBS and read by flow cytometry. The Kd of the ILY-hCD59 interaction was calculated as described above for antibody binding except that the geometric mean and concentration of the bound fluorescently labeled ILYml was substituted for the monoclonal antibody.
Binding to human erythrocytes was performed in a manner similar to that described for the hCD59 binding assay with CHO cells with the following changes; 1 × 106 erythrocytes (RBC) were incubated with various concentrations (0.167–170.9 nm) of Alexa Fluor 488-labeled ILYml or its derivatives.
Cell Lysis Assays
Subconfluent CHOHAhCD59 cells and various mutants were detached with 5 mm EDTA in PBS, washed once with PBS, and resuspended in F-12 Ham's medium supplemented with 10% FCS. For lytic end point analysis, 50 μl of cells (1 × 105 cells) were added to 50 μl of PBS containing various concentrations of ILYwt (85.45–0.0835 nm) and incubated for 1 h at 37 °C to allow for completion of lysis. The cells were then transferred to tubes containing 400 μl of 5 μg/ml propidium iodide (PI) in PBS. Cell staining by PI was determined by flow cytometry. Percent lysed cells (geometric mean of cells stained with PI) were plotted versus toxin concentration using Prism to determine tissue culture lytic dose (TCLD50; amount of ILY needed to lyse 50% of tissue culture cells).
For kinetic assays of cell lysis, subconfluent CHOHAhCD59 cells or derivatives expressing mutated hCD59 were detached with 5 mm EDTA in PBS and washed once with PBS. Cells were resuspended in PBS + PI (5 μg/ml) at 2 × 105 cells/ml at room temperature. After an initial reading, a time course of cell lysis was performed by adding 214 nm ILYwt/1 × 105 cells, and the percent PI staining was monitored every 30 s by FLOW cytometry. Untreated cells were used as the negative control for lysis.
ILY Oligomerization Kinetics
Förster resonance energy transfer (FRET) between donor and acceptor dye-labeled ILY monomers was used to determine the kinetics of oligomerization of ILY on CHO cells expressing hCD59 or hCD59 mutants. Subconfluent CHOHAhCD59 and CHOHAhCD59D22A cells were detached with 5 mm EDTA in PBS, washed once with PBS, and resuspended in F-12 Ham's media supplemented with 10% FCS. 1 × 106 cells/ml were injected with an equilibrated solution of 170.9 nm prepore-locked ILY labeled with Alexa-Fluor 488 (ILYpp-AF488) (AF488; donor dye) and 170.9 nm unlabeled ILYpp or 170.9 nm ILYpp-Rho (tetramethylrhodamine; acceptor dye) in a total volume of 50 μl. Measurements of FL-1, the donor signal, were taken every second using a FACSCalibur flow cytometer (University of Oklahoma Health Sciences Center). Analysis of the data was performed using FLOWJO software (Treestar). Rate of oligomerization was calculated by the expression 1 − (DU − DA), where DU is the donor fluorescence in the absence of acceptor, and DA is the donor fluorescence in the presence of acceptor labeled toxin. The difference (DU − DA) reflects the total FRET at each time point. By calculating 1 − (DU − DA) for each sample taken at 30-s intervals, we derived the rate of oligomerization as determined by the rate of change in FRET between donor and acceptor labeled ILY.
Complement-mediated CHO Cell Lysis Assay
Assays were performed as previously described (26). Briefly, subconfluent CHO expressing human CD59 (CHOhCD59) cells were detached with 5 mm EDTA in PBS, washed once with PBS, and resuspended in F-12 Ham's media supplemented with 10% FCS (Invitrogen). 200 μl of cells (1 × 105 cells) were sensitized with rabbit anti-CHO membrane serum (5% final concentration) for 15 min on ice. Antibody-depleted normal human sera diluted in F-12 Ham's medium was then added to a final concentration of 10% (final volume 400 μl) to supply the complement proteins. After 60 min at 37 °C, cell viability was determined by adding PI (5 μg/ml) and measuring the proportion of PI-stained (dead) cells to total cells by FACSCalibur flow cytometer (Flow Cytometry Core Facility) and FLOWJO software (Treestar). Untreated cells were used as the negative control for lysis.
Hemolytic Activity
The hemolytic activity of each ILY mutant on human erythrocytes (RBC) was determined as described previously (6). The HD50 is defined as the concentration of toxin required to lyse 50% of the human erythrocytes under standard assay conditions.
For the antibody inhibition assay of lectinolysin (LLY)-mediated hemolysis of human erythrocytes, 100 nm of the H19, MEM43, or 10G10 anti-hCD59 monoclonal antibodies were incubated 30 min on ice with the human erythrocytes before the addition of toxin. LLY was then serially diluted and added to the erythrocytes and the HD50 determined as above. The inhibition of LLY-mediated hemolysis by prepore-locked ILY was carried in a similar manner except that 350 mm prepore-locked ILY was preincubated with the erythrocytes before the addition of LLY.
Chymotrypsin Digestion of ILY and Its Derivatives
The folding of all ILY mutants was assayed by chymotrypsin digest. A 1 mg/ml stock of chymotrypsin (Thermo Scientific) was made in Tris buffer (20 mm Tris-HCl, pH 8.0, 10 mm CaCl2). 10 μg of each ILY mutant was incubated with chymotrypsin that ranged from a 1:1 molar ratio to 128:1 molar ratio (ILY:chymotrypsin) for 45 min at 37 °C in a final volume of 50 μl. The reaction was stopped by the addition of SDS loading buffer and boiling the sample for 5 min. The digested samples were then loaded onto a 4–20% SDS-PAGE gradient gel and Coomassie-stained for visualization. The peptide cleavage pattern of each mutant was compared with that for wild type ILY.
Receptor Blot
Receptor blots with ILY were performed as previously described (10).
Protein Docking
The availability of crystal structures for both ILY (28) and CD59 (29) and biological data herein allowed for computational docking to predict the structure of the ILY-hCD59 complex. We chose the 3D-dock suite Version 3.0 (30, 31) because it conveniently allows docking solutions to be filtered based on the residues shown to be involved in the protein-protein interaction. We have previously determined the crystal structure of ILY (PDB code 1S3R) (28). Of several structures of hCD59 available from the PDB, we selected 2UWR, a 1.34 Å resolution x-ray structure (21). The 3D-dock suite incorporates FTDock version 2.0, RPScore, and Multidock (30, 31). FTDock performs a global scan of translational and rotational space for possible positions of the two molecules, with the larger one being fixed and then applying surface complimentarity and electrostatic potential filters. RPScore ranks the possible docking solutions based on the empirical pair potential matrix derived from 90 non-homologous interfaces found in the Protein Data Bank (32). The 3D-dock suite also allows filtering of the docking solutions using biological information to reduce the number of possible complexes either before scoring with RPScore or after it. Using Multidock, the interface between the two proteins at the atomic level was then refined by iterative refinement of side chain orientations, rigid body refinement, and energy minimization.
The crystal structure of ILY contained two copies of the toxin in the asymmetric unit (referred to as molecule A and molecule B). Both copies are representative of possible ILY conformations, with several degrees of difference in the angular position of domains 1, 2, and 3 relative to domain 4. Thus, both copies of ILY were used in the docking to avoid any potential bias. The whole molecule of ILY was subjected to the docking even though it has been established that only domain 4 is involved in CD59 recognition (10). There are no significantly different side chain conformations on the surface of domain 4 between the two copies of ILY in the crystal structure with the exception of two loops at the end of the domain, i.e. residues 441–449 and 485–497 (the CDC tryptophan-rich motif). The largest differences in side chain conformations between the two copies of ILY in domain 4 are observed for residues Trp-491, Trp-492, Glu-492, Arg-495, and Leu-496.
The global docking performed using FTDock produced ∼10,000 possible solutions, and this was reduced to around 5000 hits by applying interaction filters. A docking solution passed the interaction filters if at least one of the following conditions was fulfilled: Tyr-434 of ILY is in contact with hCD59; Tyr-436 of ILY is in contact with hCD59; Arg-451 of ILY is in contact with hCD59; Phe-42 of hCD59 is in contact with ILY; Phe-47 of hCD59 is in contact with ILY; Tyr-62 of hCD59 is in contact with ILY. The filtered list of solutions was then scored with RPScore. The top three docking solutions obtained with molecule A of ILY had the pair potential scores of 8.11, 6.34, and 6.08, whereas the RPScores for molecule B were much lower (5.920, 5.83, and 5.8). The top-ranked solution obtained with molecule A had the highest protein-protein interface complimentarity score among these six docking solutions, and all of the residues in ILY and hCD59 shown experimentally to contribute to binding are involved in the protein-protein interaction. In addition, this solution also has the hCD59 molecule oriented in such a way that a GPI anchor attached at the C-terminal Asn-77 is in the vicinity of the tryptophan-rich loop of ILY, which is also expected to be embedded into the membrane. Of the top six ranked solutions for the ILY-hCD59 complex (i.e. three solutions for each ILY molecule), only the highest ranked solution for ILY molecule A was consistent with all of the experimental data; for example, the second ranked solution for ILY molecule A did not predict any of the ILY residues determined to be important for hCD59 recognition. The highest ranked solution for the ILY-hCD59 complex underwent further structural refinement using Multidock followed by energy minimization and simulated annealing in the program CNS (33).
RESULTS
Characterization of hCD59 Alanine Mutants
The surface residues of hCD59 were scanned by alanine substitution to identify the ILY binding site on hCD59. Alanine substitutions were made for surface-exposed residues within and around the variable region as well as residues involved in MAC inhibition (26) that were not in the variable region. In addition, residues within the CD59 epitope sites for monoclonal antibodies H19 (Tyr-61) and MEM43 (Arg-53 and Asp-23) were mutated to alanine because ILY had been shown to prevent these antibodies from binding (17). In total, 19 alanine substitutions were made at the following positions in hCD59: Asp-22, Phe-23, Lys-38, Lys-41, Phe-42, Glu-43, His-44, Asn-46, Phe-47, Asn-48, Asp-49, Thr-51, Thr-52, Arg-53, Glu-56, Asn-57, Tyr-61, Tyr-62, and Lys-65. The surface area of hCD59 covered by these substitutions is shown in Fig. 1, except for those residues shown below to be misfolded after alanine substitution.
FIGURE 1.
Human CD59 structure. Shown are representations of the NMR structure of the glycosylated form of the human CD59 molecule (34). Shown in a red ribbon representation is the variable region previously shown to contribute to ILY binding (10). Shown in yellow space-filled atoms are the positions of the disulfide forming cysteines and in the cyan space-filled atoms is the N-glycosylation at Asn-16. Shown in green space-filled atoms is Asn-77, which is attached to the GPI anchor. The residues examined in this study are shown in a transparent blue surface in the left figure and solid blue surface in the right figure (180° rotation). The structural representations were generated in VMD (40).
Human CD59 contains 10 cysteines involved in 5 disulfide bridges that are critical for maintaining its three-dimensional conformation (35). Mutations that affect disulfide bridge formation in hCD59 result in low expression, loss of function, misfolding of CD59, and loss of all antibody epitopes (22, 23, 26, 35). Studies that have mapped regions of functional importance on CD59 used monoclonal antibodies to detect conformational disruptions of the CD59 structure. Therefore, we used monoclonal antibody binding of MEM43 and H19 to assess the conformational integrity of the hCD59 mutants expressed on CHO cells. The epitopes for MEM43 and H19 do not overlap and are within the regions of hCD59 targeted in these studies. Mutants were not further characterized if both antibodies did not bind, as it was likely that the mutant was misfolded. If, however, the alanine substitution was in or near the epitope for one of the antibodies and disrupted its binding but binding of the other antibody was not affected then that mutant was further characterized for ILY binding and activity. The affinity of the antibody-hCD59 interaction for each mutant was compared with that derived for native hCD59 (Table 1) (23, 26). In most cases the affinity of the interaction of each antibody changed less than 2-fold compared with wild type hCD59. Also, the alanine mutations at Asp-22, Phe-23, Lys-38, Lys-41, Phe-42, Glu-43, His-44, Asn-48, Asp-49, Thr-51, Thr-52, and Lys-65 had been previously qualified by Huang et al. (26) using a larger repertoire of monoclonal antibodies. Mutants Y61A, N46A, D22G and D22N exhibited loss of binding of both antibodies and were not further characterized (Table 1). It should also be noted that hCD59 is not known to form higher order quaternary complexes with itself (i.e. dimers, trimers, etc.), so it is unlikely that a mutation would have disrupted a higher order structure that contained the ILY binding site. Also, ILY has been show to bind directly to hCD59 by receptor blot (10), so no other proteins are necessary for ILY binding to hCD59 that may have been displaced by any mutation.
TABLE 1.
Relative binding affinity of anti-CD59 antibodies MEM43 and H19 to CHO cells expressing hCD59 mutants
The affinity of each monoclonal antibody for hCD59 was determined. Only those CD59 mutants that affected ILY binding or activity are listed. No detectable antibody binding is denoted as ND.
| Mutant CD59 | MEM43 Kd | H19 Kd |
|---|---|---|
| nm | nm | |
| WT | 5 ± 0.9 | 685 ± 151 |
| D22A | 4 ± 0.3 | 799 ± 83 |
| D22Ga | ND | ND |
| D22Na | ND | ND |
| D22R | 13 ± 9 | 355 ± 22 |
| D22Q | 3 ± 0.9 | 651 ± 105 |
| D22E | 5 ± 0.6 | 682 ± 175 |
| F23A | ND | 893 ± 167 |
| F42A | 3 ± 1 | 642 ± 231 |
| F42W | 2 ± 0.3 | 663 ± 44 |
| N46Aa | ND | ND |
| F47A | ND | 948 ± 153 |
| Y61Aa | ND | ND |
| Y62A | 4 ± 0.8 | ND |
| F23A/F42A | ND | 1029 ± 279 |
| F42A/F47A | ND | 1250 ± 647 |
| F42A/Y62A | 9 ± 4 | ND |
a Not further characterized.
CD59 Mutants Affect ILY Binding and Cytolytic Activity
Fluorescently labeled monomer locked ILY (ILYml) (17) was used to measure binding of ILY to the various hCD59 mutants expressed in CHO cells. Locking ILY into the monomer state minimized complications due to avidity effects of membrane oligomer formation and prevents cell lysis. Binding curves were established, and the relative Kd of the ILY-hCD59 interaction for each hCD59 mutant was determined and compared with wild type hCD59. Shown in Fig. 2A are the alanine substitutions that altered the affinity of the ILY-hCD59 interaction. Aromatic residues appear to play a dominant role in the interaction of hCD59 with ILY. Mutation of the aromatic residues Phe-23, Phe-42, Phe-47, and Tyr-62 all increased the Kd of ILY for hCD59. The substitution of alanine for Phe-42 or Phe-47 increased the Kd ∼2.5-fold, whereas alanine substitutions at Phe-23 and Tyr-62 increased the Kd 6-fold. Double mutants made with combinations of these mutants significantly increased the Kd of the binding interaction (Fig. 2A). Binding of ILY to the double mutants hCD59F42A·F47A and hCD59F42A·Y62A was not detected at the ILY concentrations used, and the hCD59F23A·F42A mutant increased the Kd almost 10-fold.
FIGURE 2.
Effect of hCD59 mutants on ILY binding affinity and cytolytic activity. A, CHO cells transfected with hCD59 mutants were incubated with various concentrations of fluorescently modified monomer-locked ILY (ILYml) for 1 h at 37 °C, and then binding was analyzed by flow cytometry to determined the level of bound ILYml from which the Kd was determined. Monomer-locked ILY was used to minimize the affects of avidity resulting from oligomerization of the ILY monomers on the membrane. B, CHO cells transfected with hCD59 mutants were also assayed for changes in their susceptibility to ILY cell lysis by determining the TCLD50 (tissue culture lethal dose of ILY required for 50% cell death). Cells were treated with various amounts of ILYwt for 1 h at 37 °C, and cell death was determined by propidium iodide staining and flow cytometry. The numbers above the bars indicate the -fold change in Kd or TCLD50 over wild type hCD59. Shown is the average of five independent analyses. ND, not detected, neither appreciable binding (B) nor cell lysis (A) was detected above background at the concentrations used.
CHO cells expressing mutant hCD59 were also assayed for their susceptibility to ILY-mediated lysis (Fig. 2B). The mutants that increased the Kd of the ILY-hCD59 interaction also decreased ILY cytolytic activity. Mutants in which the alanine-substituted Phe-42 was combined with the other alanine-substituted aromatic residues (Phe-23, Phe-47, or Tyr-62) were almost completely resistant to lysis by ILY, in agreement with the significantly decreased affinity of ILY for these mutants. The hCD59F42A mutant was unusual; its binding affinity was decreased only 2.5-fold, whereas the TCLD50 was increased 19-fold. This discrepancy suggests that Phe-42 may contribute to the activity of ILY in ways that are not confined to the binding interaction.
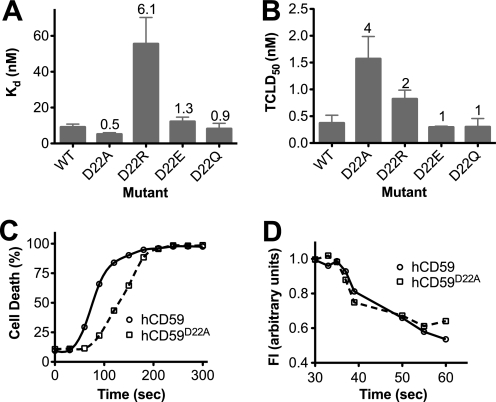
The alanine substitution for Asp-22 increased the affinity of ILY for CD59 by 2-fold (Fig. 2A). The same mutation was previously shown to increase the ability of hCD59 to protect cells against complement-mediated cell lysis about 2-fold, which was suggested to result from an increased affinity for complement proteins C8α and/or C9 (26). Substitution of either glutamate or glutamine for Asp-22 did not result in a significant change in the affinity of this interaction, whereas substitution with arginine increased the Kd of the interaction ∼6-fold (Fig. 3A). Although the alanine and arginine substitutions exhibited opposite effects on binding affinity, they both decreased the cytolytic activity (Fig. 3B). To understand this enigmatic result, we examined the kinetics of the cytolytic mechanism to identify the stage of the mechanism that was affected by the increased binding affinity.
FIGURE 3.
Effect of mutation of hCD59 Asp-22 on the ILY pore-forming mechanism. A, CHO cells transfected with hCD59 Asp-22 mutants were incubated with various concentrations of fluorescently labeled ILYml for 1 h at 37 °C to determine binding affinity by flow cytometry. B, CHO cells expressing the hCD59 Asp-22 substitutions were assayed for sensitivity to lysis by native ILY. The numbers above the bars indicated the -fold change in Kd or TCLD50 of ILY for each hCD59 Asp-22 mutant compared with wild type hCD59. C, the rate of cell lysis was determined for CHO cells expressing wild type hCD59 (solid lines) or hCD59D22A (dashed lines). Cells were incubated in media containing propidium iodide at 37 °C, and ILY was added to the cells at 30 s. Samples were taken and measured for PI uptake every 30 s by flow cytometry. D, the rate of oligomerization was determined by FRET between donor- and acceptor-labeled prepore-locked ILY (ILYpp). Prepore-locked ILY allowed the rate of oligomerization to be measured in the absence of cell lysis due to pore formation. Alexa488-labeled ILYpp (donor) was incubated with an equimolar amount of tetramethylrhodamine labeled ILYpp (acceptor) on CHO cells expressing wild type hCD59 (solid lines) or hCD59D22A (dashed lines). The rate of oligomerization was determined by monitoring the rate in change in donor fluorescence quenching due to FRET with acceptor-labeled ILY over time (see “Materials and Methods” for details). For C and D data are representative of 3–6 experiments. Experimental details for lytic and oligomerization assays are described in the “ILY Oligomerization Kinetics,” “Binding Affinity Analysis,“ and “Cell Lysis Assays” sections under “Materials and Methods.” FI, fluorescence intensity.
We recently showed that ILY disengages from its hCD59 receptor during prepore to pore conversion (17). This observation suggested that increasing the affinity of the ILY-hCD59 interaction would slow receptor disengagement, which would, therefore, slow the prepore to pore conversion and decrease the rate of cell lysis. If true, the combined binding and oligomerization kinetics would be unaffected (or slightly faster due to the higher affinity of the interaction), but the kinetics of lysis, reflecting pore formation, would be slowed. A slowed onset to ILY-mediated lysis was observed for cells expressing hCD59D22A (Fig. 3C). Next we examined the combined kinetics of binding and oligomerization by monitoring the rate of oligomerization by FRET between donor and acceptor labeled ILY. These data indicated that ILY binds and oligomerizes at similar rates on both hCD59WT- and hCD59D22A-expressing cells (Fig. 3D). Therefore, the lag in the rate of cytolysis does not correspond to a decrease in the rate of binding or oligomerization and can only result from a decreased rate of prepore to pore conversion. Hence, these data suggest that ILY is maintained in the prepore state for a longer period of time on cells expressing hCD59D22A than on cells expressing native receptor.
The Effect of hCD59 Mutants on Complement-mediated Lysis
Three of the five residues found to affect ILY binding and cytolytic activity (Asp-22, Phe-23, and Phe-42) had previously been shown to affect the complement inhibitory activity of CD59 (26). To confirm and extend these results, the complement susceptibility of CHO cells expressing each of the ILY binding mutants was examined. A clear correlation was observed between the effect of the mutations on complement protection and the affinity of the ILY-hCD59 interaction; mutants that were shown to increase or decrease the affinity of the ILY-hCD59 interaction correspondingly increased or decreased the complement protective activity of hCD59 (Fig. 4A). As shown in Fig. 3, the mutation of Asp-22 alone with different amino acids could increase or decrease the affinity of the ILY-hCD59 interaction. We found the effect on complement protection paralleled the effect on the affinity of the ILY-hCD59 interaction for alanine- and arginine-substituted Asp-22 (Fig. 4B); alanine substitution increased the affinity of the ILY-hCD59 interaction and increased protection of cells to complement-mediated lysis, whereas arginine substitution decreased the affinity of the ILY-hCD59 interaction and correspondingly decreased protection of cells to complement-mediated lysis.
FIGURE 4.
Residues of hCD59 necessary for ILY binding also contribute to its complement inhibitory function. CHO cells expressing the various hCD59 mutants were assayed for susceptibility to complement-mediated cell lysis. A, cells were sensitized with rabbit anti-CHO IgG antibody (26), human serum (10% v/v) was added as the source of complement components, and the cells were incubated for 45 min at 37 °C. B, as shown in Fig. 2, substitution of Asp-22 with different amino acids either increased or decreased binding affinity of ILY. The protective capacity of these mutants to MAC-mediated lysis is shown. Cell death was determined by PI staining and flow cytometry. Shown is the average of five independent analyses. Details are described under “Binding Affinity Analysis” under “Materials and Methods.”
The Gln and Glu mutations for Ala-22 did not result in any significant change in the binding affinity of the hCD59-ILY interaction or on the cytolytic activity of ILY (Fig. 3B). They did, however, increase the protection of CHO cells that expressed them to complement-mediated lysis, similar to the alanine substitution. The fact that these two mutants did not act consistently between ILY and human complement suggests that subtle differences exist in the binding interaction of hCD59 with ILY and human complement proteins C8α and C9.
Identification of Amino Acid Residues of ILY That Contribute to Binding of hCD59
Previous work showed that the hCD59 binding site resided within domain 4 of ILY (28, 36). Two CDCs have been identified, ILY and VLY, that bind specifically to hCD59 (10, 11). By comparing the primary structures of these CDCs to those of the cholesterol binding CDCs, we were able to identify four surface residues (Tyr-434, Tyr-436, Arg-451, and Ser-452) that were conserved in ILY and VLY but not the cholesterol binding CDCs (supplemental Fig. S1). Based on the crystal structure of ILY (28), these residues are exposed on the surface of domain 4 along two β-strands of the domain 4 β-sandwich that are predicted to face away from the pore (5–7, 37).
The close proximity of the Tyr-434 and Tyr-436 pair and the Arg-451 and Ser-452 pair prompted us to substitute each pair of mutants with alanine rather than generating single mutants in each residue. Double alanine substitution mutants were generated in the monomer-locked version of ILY (ILYml) background to minimize avidity effects on binding due to the formation of oligomeric complexes (17). These mutants were designated as ILYml·YY (alanine-substituted Tyr-434 and Tyr-436) and ILYml·RS (alanine-substituted Arg-451 and Ser-452). The mutations did not significantly alter the structure of ILY, as determined by chymotrypsin sensitivity or antibody binding, and each was well expressed in E. coli and was soluble (data not shown).
Giddings et al. (10) had previously used receptor blot analysis to demonstrate the interaction of ILY with hCD59 on CHAPS-extracted CD59 from human erythrocyte membranes. We used this technique to first determine, whether these mutations affected the interaction of ILY with hCD59. For these experiments we used human erythrocytes, which are a rich source of hCD59 (9). CHAPS-extracted erythrocyte membrane proteins were separated by SDS-PAGE and transferred to nitrocellulose paper. Replicate blots were probed with ILYml, ILYml·RS, or ILYml·YY. The ILYml·RS mutant exhibited a decrease in binding to hCD59 compared with ILYml, whereas no detectable interaction was seen when the blot was probed with ILYml·YY (Fig. 5A).
FIGURE 5.
ILY residues that contribute to the hCD59 binding site. A, blots of CHAPS-extracted human erythrocyte membrane proteins were probed with the ILY, ILYYY, and ILYRS (20 nm) or with the anti-CD59 monoclonal antibody H19. Each lane was loaded with the same amount of CHAPS-extracted membrane protein obtained from a single preparation of CHAPS-extracted human erythrocyte membrane proteins. B, human erythrocytes were incubated with fluorescently labeled ILYml or its derivatives, and changes in binding were determined by flow cytometry. Shown is the average of five independent analyses.
We next determined the Kd of these mutants for hCD59 on human erythrocytes relative to ILYml by flow cytometry using fluorescently tagged versions of ILYml and its derivatives. Both ILYml·YY and ILYml·RS mutants increased the Kd of ILY to 180 and 53 nm, respectively, compared with ILYml (Kd 4 nm) (Fig. 5B). When all four residues were mutated to alanines in the monomer-locked background (ILYml·YYRS), the decrease in ILY affinity for hCD59 was significantly more pronounced, exhibiting a Kd that was more than 130-fold higher than ILYml (Fig. 5B).
The hemolytic activity of these mutants was also compared with native ILY. Both ILYYY and ILYRS exhibited a decrease in their hemolytic activity as demonstrated by significantly increased HD50 values (Fig. 5B). Interestingly, the RS mutant exhibited a significantly greater effect on hemolytic activity than on binding, whereas the effect of the ILYYY mutant was much greater on affinity than on the cytolytic activity.
The protein docking analysis (see below) also predicted that Tyr-460 participated in the binding site. This mutant acted similarly to the hCD59D23A mutant; it also resulted in a modest increase in the Kd (2-fold) of the interaction, but it decreased the cytolytic activity 7-fold (Fig. 5B). Hence, mutations in either hCD59 or ILY that increase binding affinity decrease cytolytic activity.
Lectinolysin Is a CD59-binding CDC
Do ILY residues Tyr-434, Tyr-436, Arg-451, and Ser-452 represent a signature motif for hCD59-binding CDCs? These residues are also conserved in lectinolysin, a recently characterized member of the CDC family from Streptococcus mitis (38). To determine whether LLY was also a member of the hCD59 binding family of CDCs, we performed a hemolytic assay using human erythrocytes that had been preincubated with hCD59 monoclonal antibodies (H19 and 10G10), which we had previously shown to block ILY hemolytic activity and CD59 binding (10). Another monoclonal antibody to hCD59, MEM43, was also tested. LLY-mediated lysis of the erythrocytes was inhibited by all three antibodies (Fig. 6B) in a dose-dependent pattern that resembled that observed for ILY (Fig. 6A). The hemolytic activity of PFO, a CDC that uses cholesterol as its receptor, was not significantly altered by the monoclonal antibodies (Fig. 6C).
FIGURE 6.
Monoclonal antibodies to hCD59 and prepore-locked ILY inhibit ILY-mediated lysis of human erythrocytes. Human erythrocytes were either untreated or pretreated with monoclonal antibodies to huCD59 (H19, MEM43, and 10G10) and then treated with different amounts of ILY (A), LLY (B), or PFO (C), and the extent of hemolysis was determined by the amount of hemoglobin released at each concentration of toxin. In D we determined if monomer-locked ILY or PFO could inhibit LLY-mediated hemolysis by binding to their cognate receptors hCD59 or cholesterol, respectively, and blocking LLY-mediated hemolysis. Human erythrocytes were either untreated or preincubated with 350 nm prepore-locked ILY or PFO and then treated with various concentrations of active LLY. The extent of LLY-mediated hemolysis was then determined. Shown are representative data of three or more experiments.
We also examine the ability of prepore-locked versions of ILY and PFO to inhibit the hemolytic activity of LLY. Locking ILY in its prepore-locked complex maintains its interaction with hCD59, which is lost upon prepore to pore conversion (17). Furthermore, by locking both ILY and PFO in their non-hemolytic prepore-locked forms we could measure their ability to inhibit LLY-mediated hemolysis. In Fig. 6D we show that human erythrocytes preincubated with prepore-locked ILY, but not prepore-locked PFO, inhibits the hemolytic activity of LLY. Overall, these studies strongly suggest that LLY is a hCD59-binding CDC.
Protein Docking of the ILY-hCD59 Binding Sites
We used the information gained from the mapping studies to model the binding interface of the ILY-hCD59 interaction. The top ranked docking solution for the ILY-hCD59 complex obtained using 3D-dock suite is shown in Fig. 7. There are 16 residues involved in the interactions at the interface. The interface is rather polar, with a number of possible hydrogen bonds and salt bridges. The possible interactions involving the residues identified by mutagenesis are: a hydrogen bond between Tyr-434 of ILY via Arg-432 of ILY and main chain carbonyl O of Asn-77 of hCD59 as well as van der Waals interaction with Gln-74 and Tyr-62 of hCD59; a hydrogen bond between OH of Tyr-436 of ILY and OH of Tyr-62 of hCD59; a salt bridge between Arg-451 of ILY and Glu-43 of hCD59; hydrogen bonds between the main chain carbonyl O of Ser-452 in ILY and the main chain amide of Phe-47 or OH of Tyr-61 of hCD59; a hydrogen bond between the main chain amide of Ser-452 and OD1 of Asn-48 in hCD59 as well as van der Waals interaction with Phe-47 of CD59; a hydrogen bond between the OH of Ser-456 of ILY and the main chain amide of Lys-65 of hCD59 as well as van der Waals interaction between Ser-456 and Phe-42 of hCD59.
FIGURE 7.
Computer model of the interaction between ILY and hCD59. The molecular model of the ILY domain 4 (cyan) and hCD59 (magenta) interaction is shown. A, ILY residues involved in binding to hCD59 are depicted as cyan-colored sticks, and the hCD59 residues involved in ILY binding are shown as magenta-colored sticks. ILY loops L1-L3 are shown in green, and the tryptophan-rich undecapeptide is colored gray. hCD59 residue Asn-77 is represented as space fill (colored by atom type; carbon atoms are magenta, nitrogen atoms are blue, oxygen atoms are red, and hydrogen atoms are white) and is the site of attachment to the GPI membrane anchor. B, a 180° rotation about the y axis from the view shown in A. ILY residues Arg-451 and Ser-452 are located on opposite sides of the β-strand, whereas Tyr-434 and Tyr-436 are located on the same side of the β-strand.
Additional residues predicted to be involved in the interaction and possible contacts are evident from the proposed ILY-hCD59 complex. The side chain of Arg-453 of ILY is within hydrogen bond distance to the main chain carbonyl oxygens of Phe-42, Glu-43, Cys-45, and OE2 of Glu-43. Ser-454 of ILY is in van der Waals contact with Tyr-61 and Tyr-62. Asn-458 of ILY is in van der Waals contact with Tyr-62, and the main chain carbonyl O of Asn-458 is within hydrogen-bonding distance to the side chain of Gln-74. Gly-459 of ILY is in van der Waals contact with Gln-74 of hCD59. Tyr-460 of ILY is in van der Waals contact with disulfide Cys-64 to Cys-69, Phe-71, Glu-73, and Gln-74 of hCD59. The side chain and main chain amide of Asn-461 of ILY is within hydrogen bond distance of the side chain of Glu-73 of CD59. The protein docking data suggested that Tyr-460 on ILY might also interact with hCD59. We determined that the Tyr-460 to alanine mutation modestly increased the affinity between ILY and hCD59 (Kd = 2.7 nm) (Fig. 5B). Remarkably, this mutation decreased activity of ILY nearly 7-fold. This result is similar to that observed for the alanine substituted Asp-22 of hCD59, where we showed a similar phenomenon of increased binding leading to decreased activity (Figs. 2 and 3).
The protein docking data are generally consistent with our experimental findings and suggest that residues Phe-42, Phe-47, and Tyr-62 from hCD59 are predicted to be in direct contact with Tyr-434, Tyr-436, Arg-451, and Ser-452 of ILY. The proposed ILY-hCD59 complex can now be used to predict additional residues that may be involved in the protein-protein interaction but that are not necessarily conserved between the hCD59 binding CDCs.
DISCUSSION
The ability of a subset of members of the CDC family to specifically use hCD59 as their receptor rather than cholesterol is an intriguing evolutionary change in receptor specificity, although a specific interaction with cholesterol is still required for successful pore formation. The primary function of CD59 function, to block MAC assembly on host cells, is homologously restricted, suggesting that the bacterial species expressing the hCD59 specific CDCs may have coevolved with humans. Our studies revealed a surprisingly deep correspondence of the ILY and MAC binding sites on hCD59. Mutation of residues in hCD59 that increased or decreased the affinity of its interaction with ILY correspondingly increased or decreased its protective effect to MAC-mediated lysis. This similarity in the hCD59 binding site, however, does not translate to homology in the binding sites of ILY, C8α, and C9. This shows that the interaction between these proteins and hCD59 is mediated by specific contacts to common residues on hCD59, but the structure of the binding site for hCD59 on its binding partners can vary considerably. We also identified four residues in the ILY binding interface that appear to constitute a signature motif for hCD59 binding CDCs. These residues are conserved between ILY and VLY, the two known hCD59 binding CDCs, but are not present in the large family of cholesterol binding CDCs. The motif is also present in LLY, shown herein to be the newest member of the hCD59 binding CDCs.
The hCD59 residues that contribute to its interaction with ILY also participate in its interaction with the MAC, demonstrating that there is significant correspondence in the two binding sites. Not all of the residues previously reported to contribute to the hCD59-MAC binding interface, however, were involved in the ILY-hCD59 binding interface. This observation suggests that ILY binds to a subset of the residues required for the interaction between hCD59 and the MAC. These residues, however, are sufficient to impart specificity for hCD59, similar to that seen for the homologous restriction of hCD59 inhibitory activity to the host MAC. Our model of the ILY-hCD59 interaction suggests that Phe-42, Phe-47, and Tyr-62 are located on the binding interface with ILY, and Asp-22 and Phe-23 are on the opposite face. This scenario is similar to that suggested by Huang et al. (26) for the binding sites for MAC proteins C8α and C9. They suggested that mutation of Asp-22 and Phe-23 indirectly affected the binding site structure of hCD59 for the MAC (26). Tyr-61 is also likely to participate in this interaction, as suggested by the molecular model of the ILY-hCD59 interaction, but was not tested since it appeared to be misfolded.
The binding phenotypes of hCD59 Asp-22 mutants are of special interest. Mutations at this site that increased or decreased the binding affinity for ILY only decreased the cytolytic activity of ILY. The fact that increasing the binding affinity decreased cytolytic activity supports our previous observation that showed ILY disengages from hCD59 during the prepore to pore transition (17). We had predicted that this disengagement was necessary to allow specific structural changes that were necessary for the insertion of the β-barrel pore. This implied that increasing binding affinity between ILY and hCD59 would slow the rate of hCD59 disengagement and, therefore, slow conversion of the prepore to pore. This is consistent with our observations herein that show the binding and oligomerization of ILY was largely unaffected, but pore formation was slowed. Similarly, alanine-substituted Tyr-460 of ILY also increased binding affinity about 2-fold yet decreased activity 7-fold, suggesting that it acted similarly to the hCD59 Asp-22 mutant, although secondary effects of the Tyr-460 mutation on ILY activity cannot be ruled out. Hence, the ILY affinity for hCD59 must be carefully balanced between maintaining binding rate and specificity but not so high that it significantly slows its disengagement from hCD59, which slows pore formation. This may also explain why ILY exhibits a reduced interface with hCD59 compared with the MAC proteins.
The phenotype of the alanine-substituted Phe-42 of hCD59 was also unusual; it had the smallest effect on the affinity of the hCD59-ILY interaction (2.5-fold decrease) but a disproportionally large effect on hemolytic activity (19-fold decrease). Similarly, the double alanine mutant for Arg-451 and Ser-452 of ILY also had a smaller effect on affinity (11-fold reduction) than the double alanine substitution of tyrosines 434 and 436 (39-fold reduction), yet it had a significantly larger effect on the cytolytic activity (24- versus 9-fold reduction, respectively). We have shown that hCD59 binding by ILY alone is sufficient to trigger the necessary structural changes for pore formation (15, 24). Our current studies suggest that Phe-42 of hCD59 and Arg-451 and Ser-452 of ILY may reside at a site within the binding interface that plays a greater role in triggering these structural changes than in binding affinity. Although we cannot rule out secondary effects of the Arg-451 and Ser-452 mutations on the ILY activity, the mutation of Phe-42 of hCD59 can only affect its interaction with ILY. Therefore, its disproportionally greater effect on lytic activity than on binding affinity likely results from the alteration of one or more specific contacts with ILY that are involved in triggering the structural changes necessary for pore formation. Hence, these data suggest that some interactions at the ILY-hCD59 interface contribute more to the binding strength of the interaction, whereas others play a greater role in triggering the structural changes necessary for pore formation. Conceptually this is not unlike an enzyme active site where residues may be dedicated to substrate recognition and binding and others to catalysis.
ILY and VLY were the only CDCs previously shown to use hCD59 as a receptor (10, 11). Our studies showed that the four conserved domain 4 residues Tyr-434, Tyr-436, Ser-451, and Arg-452 contribute to the binding interaction between ILY and hCD59. A fifth residue identified through the computer modeling of the ILY-hCD59 interaction, Tyr-460, also contributes to the binding interaction. The fact that Tyr-460 is not conserved among the hCD59 binding CDCs suggests that other domain 4 residues that are not strictly conserved are likely to participate in the binding interaction by forming direct contacts with hCD59 and/or by affecting the presentation of the conserved residues. These differences may modulate their binding affinity for hCD59 and therefore their pore-forming activity.
The ILY and VLY residues are also conserved in the recently characterized CDC, LLY, from S. mitis (38). Herein we showed that anti-hCD59 monoclonal antibodies that inhibited ILY hemolytic activity and binding to hCD59 (10) also inhibited LLY activity in a similar manner. We also showed that prepore-locked ILY, but not prepore-locked PFO, could effectively inhibit LLY hemolytic activity. These data suggest that LLY is the newest member of the hCD59 binding CDC family and that the Tyr-X-Tyr-X14-Ser-Arg motif is a signature motif for hCD59 binding CDCs.
Recently, Hughes et al. (39) proposed the hCD59 binding site on ILY was located in a 7-residue peptide in domain 4 near the conserved undecapeptide sequence. Their conclusions were based on similarity in a heptameric amino acid sequence in human C9 and ILY. The consensus sequence RXXYSKN is conserved between C9 residues 249–257 (RFSYSKN) and ILY (residues 495–501, RLIYSKN) (39). This site, however, is unlikely to directly participate in the interaction of ILY with hCD59 for several reasons. The C9 residues RFSYSKN have been recently shown not to participate in the CD59-MAC interaction (25). The change in the Kd was comparatively small in the ILY RLIYSKN → RLIYNRT mutant. and this sequence is poorly conserved in the hCD59 binding CDCs ILY, VLY, and LLY. Most problematic is that the NRT sequence that was used to replace the SKN sequence of ILY was derived from LLY, a CDC we show herein to be the newest member of the hCD59 binding CDCs. Hence, it appears unlikely that this sequence directly participates in the ILY-hCD59 interaction, although it is possible that its mutation could have indirectly perturbed the structure of the binding site described herein.
In conclusion, we have demonstrated that ILY and the MAC complement proteins C8α and C9 share a binding site on hCD59 but that the hCD59 binding sites on each protein do not exhibit any significant sequence similarity. Furthermore, we have identified residues conserved in the hCD59 binding CDCs that contribute to recognition and binding of hCD59, which appear to constitute a signature motif for hCD59 binding CDCs.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AI063444 (NIAID) (to R. K. T.) and AI47386 (to S. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- CDC
- cholesterol-dependent cytolysin
- GPI
- glycosylphosphatidylinositol
- MAC
- membrane attack complex
- hCD59
- human CD59
- ILY
- intermedilysin
- VLY
- vaginolysin
- LLY
- lectinolysin
- PFO
- perfringolysin O
- PI
- propidium iodide
- TCLD
- tissue culture lytic dose.
REFERENCES
- 1. Law R. H., Lukoyanova N., Voskoboinik I., Caradoc-Davies T. T., Baran K., Dunstone M. A., D'Angelo M. E., Orlova E. V., Coulibaly F., Verschoor S., Browne K. A., Ciccone A., Kuiper M. J., Bird P. I., Trapani J. A., Saibil H. R., Whisstock J. C. (2010) Nature 468, 447–451 [DOI] [PubMed] [Google Scholar]
- 2. Rosado C. J., Buckle A. M., Law R. H., Butcher R. E., Kan W. T., Bird C. H., Ung K., Browne K. A., Baran K., Bashtannyk-Puhalovich T. A., Faux N. G., Wong W., Porter C. J., Pike R. N., Ellisdon A. M., Pearce M. C., Bottomley S. P., Emsley J., Smith A. I., Rossjohn J., Hartland E. L., Voskoboinik I., Trapani J. A., Bird P. I., Dunstone M. A., Whisstock J. C. (2007) Science 317, 1548–1551 [DOI] [PubMed] [Google Scholar]
- 3. Hadders M. A., Beringer D. X., Gros P. (2007) Science 317, 1552–1554 [DOI] [PubMed] [Google Scholar]
- 4. Slade D. J., Lovelace L. L., Chruszcz M., Minor W., Lebioda L., Sodetz J. M. (2008) J. Mol. Biol. 379, 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shatursky O., Heuck A. P., Shepard L. A., Rossjohn J., Parker M. W., Johnson A. E., Tweten R. K. (1999) Cell 99, 293–299 [DOI] [PubMed] [Google Scholar]
- 6. Shepard L. A., Heuck A. P., Hamman B. D., Rossjohn J., Parker M. W., Ryan K. R., Johnson A. E., Tweten R. K. (1998) Biochemistry 37, 14563–14574 [DOI] [PubMed] [Google Scholar]
- 7. Rossjohn J., Feil S. C., McKinstry W. J., Tweten R. K., Parker M. W. (1997) Cell 89, 685–692 [DOI] [PubMed] [Google Scholar]
- 8. Rosado C. J., Kondos S., Bull T. E., Kuiper M. J., Law R. H., Buckle A. M., Voskoboinik I., Bird P. I., Trapani J. A., Whisstock J. C., Dunstone M. A. (2008) Cell Microbiol. 10, 1765–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rollins S. A., Sims P. J. (1990) J. Immunol. 144, 3478–3483 [PubMed] [Google Scholar]
- 10. Giddings K. S., Zhao J., Sims P. J., Tweten R. K. (2004) Nat. Struct. Mol. Biol. 11, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 11. Gelber S. E., Aguilar J. L., Lewis K. L., Ratner A. J. (2008) J. Bacteriol. 190, 3896–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohno-Iwashita Y., Iwamoto M., Mitsui K., Ando S., Nagai Y. (1988) Eur. J. Biochem. 176, 95–101 [DOI] [PubMed] [Google Scholar]
- 13. Tweten R. K. (1988) Infect. Immun. 56, 3228–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tweten R. K., Harris R. W., Sims P. J. (1991) J. Biol. Chem. 266, 12449–12454 [PubMed] [Google Scholar]
- 15. Farrand A. J., LaChapelle S., Hotze E. M., Johnson A. E., Tweten R. K. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 4341–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Soltani C. E., Hotze E. M., Johnson A. E., Tweten R. K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20226–20231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. LaChapelle S., Tweten R. K., Hotze E. M. (2009) J. Biol. Chem. 284, 12719–12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rollins S. A., Zhao J., Ninomiya H., Sims P. J. (1991) J. Immunol. 146, 2345–2351 [PubMed] [Google Scholar]
- 19. Farkas I., Baranyi L., Ishikawa Y., Okada N., Bohata C., Budai D., Fukuda A., Imai M., Okada H. (2002) J. Physiol. 539, 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hamilton K. K., Ji Z., Rollins S., Stewart B. H., Sims P. J. (1990) Blood 76, 2572–2577 [PubMed] [Google Scholar]
- 21. Zhao X. J., Zhao J., Zhou Q., Sims P. J. (1998) J. Biol. Chem. 273, 10665–10671 [DOI] [PubMed] [Google Scholar]
- 22. Yu J., Abagyan R., Dong S., Gilbert A., Nussenzweig V., Tomlinson S. (1997) J. Exp. Med. 185, 745–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bodian D. L., Davis S. J., Morgan B. P., Rushmere N. K. (1997) J. Exp. Med. 185, 507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Soltani C. E., Hotze E. M., Johnson A. E., Tweten R. K. (2007) J. Biol. Chem. 282, 15709–15716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang Y., Qiao F., Abagyan R., Hazard S., Tomlinson S. (2006) J. Biol. Chem. 281, 27398–27404 [DOI] [PubMed] [Google Scholar]
- 26. Huang Y., Smith C. A., Song H., Morgan B. P., Abagyan R., Tomlinson S. (2005) J. Biol. Chem. 280, 34073–34079 [DOI] [PubMed] [Google Scholar]
- 27. Giddings K. S., Johnson A. E., Tweten R. K. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11315–11320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Polekhina G., Giddings K. S., Tweten R. K., Parker M. W. (2005) Proc. Natl. Acad. Sci. 102, 600–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leath K. J., Johnson S., Roversi P., Hughes T. R., Smith R. A., Mackenzie L., Morgan B. P., Lea S. M. (2007) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 648–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gabb H. A., Jackson R. M., Sternberg M. J. (1997) J. Mol. Biol. 272, 106–120 [DOI] [PubMed] [Google Scholar]
- 31. Jackson R. M., Gabb H. A., Sternberg M. J. (1998) J. Mol. Biol. 276, 265–285 [DOI] [PubMed] [Google Scholar]
- 32. Moont G., Gabb H. A., Sternberg M. J. (1999) Proteins 35, 364–373 [PubMed] [Google Scholar]
- 33. Brunger A. T. (2007) Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 34. Fletcher C. M., Harrison R. A., Lachmann P. J., Neuhaus D. (1994) Structure 2, 185–199 [DOI] [PubMed] [Google Scholar]
- 35. Petranka J., Zhao J., Norris J., Tweedy N. B., Ware R. E., Sims P. J., Rosse W. F. (1996) Blood Cells Mol. Dis. 22, 281–296 [DOI] [PubMed] [Google Scholar]
- 36. Nagamune H., Ohkura K., Sukeno A., Cowan G., Mitchell T. J., Ito W., Ohnishi O., Hattori K., Yamato M., Hirota K., Miyake Y., Maeda T., Kourai H. (2004) Microbiol. Immunol. 48, 677–692 [DOI] [PubMed] [Google Scholar]
- 37. Czajkowsky D. M., Hotze E. M., Shao Z., Tweten R. K. (2004) EMBO J. 23, 3206–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Farrand S., Hotze E., Friese P., Hollingshead S. K., Smith D. F., Cummings R. D., Dale G. L., Tweten R. K. (2008) Biochemistry 47, 7097–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hughes T. R., Ross K. S., Cowan G. J., Sivasankar B., Harris C. L., Mitchell T. J., Morgan B. P. (2009) Mol. Immunol. 46, 1561–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.