Abstract
The protein α-synuclein has a central role in Parkinson disease, but the mechanism by which it contributes to neural degeneration remains unknown. We now show that the expression of α-synuclein in mammalian cells, including neurons in vitro and in vivo, causes the fragmentation of mitochondria. The effect is specific for synuclein, with more fragmentation by α- than β- or γ-isoforms, and it is not accompanied by changes in the morphology of other organelles or in mitochondrial membrane potential. However, mitochondrial fragmentation is eventually followed by a decline in respiration and neuronal death. The fragmentation does not require the mitochondrial fission protein Drp1 and involves a direct interaction of synuclein with mitochondrial membranes. In vitro, synuclein fragments artificial membranes containing the mitochondrial lipid cardiolipin, and this effect is specific for the small oligomeric forms of synuclein. α-Synuclein thus exerts a primary and direct effect on the morphology of an organelle long implicated in the pathogenesis of Parkinson disease.
Keywords: Membrane Bilayer, Mitochondria, Neurodegeneration, Parkinson Disease, Synuclein, Mitochondrial Dynamics
Introduction
Many observations have implicated mitochondria in the pathogenesis of PD.2 Mitochondria from the substantia nigra of affected patients show a selective reduction in the activity of respiratory chain complex I (1). Somatic mutations also accumulate with age and PD in the mitochondrial DNA of substantia nigra neurons (2). In addition, the neurotoxins MPTP and rotenone, which produce models of PD, both act by disrupting mitochondrial function. Genetic evidence further supports a primary role for mitochondria in the pathogenesis of PD. Mutations in parkin and the mitochondrial kinase PINK1 both cause autosomal recessive PD (3), and these genes appear required for the normal clearance of defective mitochondria by autophagy (4). However, the molecular mechanisms responsible for mitochondrial dysfunction in the much more common sporadic forms of PD have remained unclear.
Several observations suggest a central role for the protein α-synuclein in the pathogenesis of sporadic PD. Point mutations in synuclein produce a rare autosomal dominant form of PD (5–7), indicating a causative role for the protein. α-Synuclein also accumulates in the Lewy bodies and dystrophic neurites of essentially all patients with idiopathic PD (8), implicating the protein in sporadic as well as familial forms of the disease. Furthermore, duplication and particularly triplication of the SNCA (α-synuclein) gene cause a severe, highly penetrant form of PD (9, 10), indicating a dose-dependent pathogenic role for the wild type protein when overexpressed and suggesting that the accumulation of synuclein in sporadic PD is the primary pathogenic event. However, the mechanism by which α-synuclein causes PD remains poorly understood. Expressed in yeast and Drosophila, human α-synuclein produces severe toxicity (11–14), but these model organisms lack endogenous synuclein, and the overexpression of wild type synuclein in mammalian systems causes remarkably little if any consistent toxicity (15–18).
Although the mechanism by which α-synuclein causes PD remains poorly understood, circumstantial evidence has implicated mitochondria. Mice lacking α-synuclein show resistance to the mitochondrial neurotoxin MPTP (19) and a reduction in the mitochondrial lipid cardiolipin (20). Mice overexpressing a mutant form of synuclein also exhibit mitochondrial damage (21, 22). In addition, recent observations have begun to suggest a direct interaction of synuclein with mitochondria (21, 23–26). We have previously found that synuclein binds specifically to mitochondria rather than other organelles (27), and the amount of synuclein localized to the mitochondria of substantia nigra neurons increases dramatically in PD (24).
Does α-synuclein influence the behavior of mitochondria? We now find that in mammalian cells, including neurons, increased expression of synuclein produces mitochondrial fragmentation, and this effect precedes any loss of mitochondrial function. Surprisingly, the fragmentation does not require the mitochondrial fission protein Drp1. Rather, it involves a novel, direct effect of oligomeric synuclein on mitochondrial membranes.
EXPERIMENTAL PROCEDURES
Molecular Biology
All constructs used for transient transfection (except mRFP) were subcloned into the pCAGGS vector downstream of the chicken actin promoter (28). mRFP was subcloned into pGW1-CMV as described previously (29). A30P, A53T, and E46K mutations were introduced into the human α-synuclein cDNA by PCR. Azurite was subcloned from pCEP4-Azurite (Addgene), MitoDsRed2 from pDsRed2-mito, mitoGFP from pAcGFP1-Mito, and CFP from pECFP-N3 (Clontech) after introducing the L221K mutation to prevent dimerization (30). mCherry was fused to the N terminus of rat synaptophysin. Human HA-Drp1 and HA-K38A Drp1 were the generous gifts from A. van der Bliek (UCLA); Mfn1–10×Myc, Mfn1(K88T)-10×Myc, Mfn2–16×Myc, and Mfn2(K109A)-16×Myc were the generous gifts from D. Chan (California Institute of Technology), and huntingtin exon 1 fused to CFP was the generous gift from J. Shao and M. Diamond (University of California, San Francisco). Silencer select pre-designed RNAi s13204, s13205, s13206, and negative control 1 were obtained from Applied Biosystems.
Cell Culture and Morphologic Analysis
Spontaneously immortalized mouse embryonic fibroblasts were derived by serial passage (over 30 times) of mouse embryonic fibroblasts from E10.5 embryos as described previously (31). HeLa cells and immortalized mouse embryonic fibroblasts were transiently transfected by electroporation (Amaxa) and COS cells by FuGENE HD. In other studies, stable HeLa cell lines expressing mitoGFP were used to identify mitochondria. One to 2 days after transfection, healthy cells with similar levels of azurite fluorescence were imaged live in Tyrode's medium (in mm: 127 NaCl, 10 HEPES-NaOH, pH 7.4, 30 glucose, 2.5 KCl, 2 CaCl2, 2 MgCl2) with a 100× oil objective on a Zeiss LSM 510 confocal microscope. The images were then randomized and the prevailing mitochondrial morphology in each transfected cell was classified blind to the DNA transfected as tubular, fragmented, or intermediate. Cells classified as having tubular mitochondria contain almost entirely mitochondria with length/width (axis) ratios >10, as fragmented those containing mitochondria with axis ratio <3, and as intermediate those containing both tubular and fragmented mitochondria. Midbrain neurons were prepared from E14 embryos as described previously (32), transfected by electroporation at the time of plating, and imaged at 14–17 days in vitro. Length, width, axis ratio, perimeter, area, and number of distinct mitochondrial fragments were calculated from live neurons using Metamorph (Universal Imaging).
For survival studies using the automated microscope, primary hippocampal neurons were prepared from embryonic day 20–21 timed pregnant rats (Charles River Laboratories), cultured in Neurobasal-A with B27 (Invitrogen) for 5 days, and transfected with calcium phosphate as described previously (33).
Immunocytochemistry
Cell were fixed for 30 min in media containing 4% paraformaldehyde and immunostained in phosphate-buffered saline (PBS) containing 5% fetal bovine serum (FBS) and 0.2% Triton X-100. α-Synuclein was detected using either a mouse antibody to human α-synuclein (15G7, Axxora) or rabbit antibody to all the synuclein isoforms (AB5464, Chemicon), endoplasmic reticulum with a monoclonal antibody to the KDEL receptor (SPA-827, StressGen), peroxisomes with a rabbit polyclonal antibody to PMP70 (P0497, Sigma), cytochrome c with a mouse antibody (556432, BD Biosciences), the Golgi complex with a mouse antibody to GM130 (610822, BD Biosciences), lysosomes with a mouse antibody to CD107a (Lamp-1, 555798, BD Biosciences), and the microtubule cytoskeleton with an antibody to α-tubulin (Oncogene Science).
Analysis of Mitochondrial Function
Membrane Potential
Cells were treated for 1 h with tetramethylrhodamine methyl ester (TMRM) (1 nm) and imaged in Tyrode's medium containing TMRM. In selected experiments, cells were subsequently depolarized using 2.5 μm FCCP. For fluorescence-activated cell sorting (FACS), the cells were incubated 1 h with TMRM in the presence or absence of 5 μm FCCP, harvested in PBS containing 0.5% FBS, and sorted on an LSR-II (BD Biosciences), with GFP excited by a 20-milliwatt blue solid state 488 nm laser and TMRM by a 150-milliwatt green 532 nm laser.
Superoxide Levels
Live cells were exposed acutely to hydroethidium (3.2 μm), and the relative superoxide levels determined by the initial rate of increase in ethidium fluorescence, fit by linear regression as described previously (32).
Respiration
750,000 COS cells were added to an Oxygraph2 respirometer (Oroboros Instruments) in 2.1 ml, and oxygen consumption was measured after the sequential addition of 10 μm glutamate and 2.5 μm malate, 2 μg/ml oligomycin, 1 μm FCCP, and then 0.5 μm rotenone.
COS Cell Survival
16 h after transfection, COS cells were trypsinized and replated in 96-well plates. At 24, 48, 72, and 96 h after transfection, the cells were treated with 1 μm calcein green (to assess live cells) and either immediately with 5 μm ethidium (to assess dead cells) or after 30 min of incubation with 70% methanol (to assess total cells), and the fluorescence was quantified using a 96-well fluorescent plate reader.
Neuronal Survival
For the analysis of cell survival, images were taken at 24-h intervals using an automated microscope (29, 34), with image acquisition and analysis using ImagePro Plus 6.2 and with custom-designed programs. Transfected neurons were selected for analysis based on fluorescence intensity and morphology, including the presence of extended processes at the start of the experiment. Survival time was determined as the last time point at which the neuron was seen alive (supplemental Fig. S8). For statistical analysis, StatView software was used to construct Kaplan-Meier curves from the survival data. Survival functions were fitted to these curves and used to derive cumulative hazard (or risk of death) curves. Differences in cumulative risk of death curves were analyzed for statistical significance with the log-rank test, and each of the experiments was performed independently 2–4 times. The expression of α-synuclein was estimated by mRFP fluorescence intensity in the cell body. Images of mitochondria (visualized using mitoGFP) 48 h after transfection were randomized and classified as fragmented, intermediate, or more tubular blind to the genotype of transfection.
Fusion Assay
COS cells were cotransfected with azurite, the indicated combinations of α-synuclein, Drp1, or empty vector control and either mitoGFP or MitoDsRed to label mitochondria. One day later, the cells were trypsinized, and cells expressing the same plasmids and either mitoGFP or mitoDsRed were mixed and replated. On the 2nd day after transfection, the cells were preincubated with cycloheximide (50 μg/ml) for 30 min and then treated with polyethylene glycol 1500 (Roche Applied Science, catalog no. 13396000) for 1 min before washing and further incubation in media with cycloheximide. Cells were fixed in media containing 4% paraformaldehyde at 4, 6.5, and 9 h after polyethylene glycol treatment.
Preparation of Liposomes
Heart cardiolipin and synthetic dioleoylphosphatidylcholine (Avanti) were mixed in the ratios indicated and the chloroform evaporated under nitrogen. The resulting lipid film was dried under vacuum for 10 min and re-hydrated to a final concentration of 5 mm in 25 mm KCl, 2.5 mm magnesium acetate, 150 mm potassium gluconate, and 25 mm HEPES-KOH, pH 7.4 (cytosol buffer), for 30 min at room temperature. The resulting liposomes were subjected to five freeze/thaw cycles, passed 11 times through an extruder with a 1-μm pore (Avanti), stored in the dark at 4 °C under nitrogen, and used within 1 week.
Protein Expression
Synuclein
Recombinant human α-synuclein was expressed and purified as described (35). Purified protein was lyophilized and stored at −80 °C. Synuclein was resuspended in cytosol buffer and incubated 15 min on ice. The solution was then centrifuged at 184,000 × g for 15 min, and the supernatant was stored at 4 °C until use (typically within 5 days). Monomeric synuclein was isolated by size-exclusion chromatography through Sephadex G-100. To prepare intermediate oligomeric species, 140 mm protein was stirred at 600 rpm and 37 °C for 20 h and sedimented at 15,000 × g for 30 min, and the supernatant was separated by size-exclusion chromatography, with oligomer 1 fraction in the void volume. To prepare large oligomers and fibrils, agitation proceeded for 60 h, followed by centrifugation at 50,000 × g for 30 min to separate oligomer 2 fraction (in the supernatant, subsequently isolated by size-exclusion chromatography) from fibrils (pellet).
Huntingtin
GST-tagged monomeric mutant huntingtin (GST-Htt53Q) was a kind gift of Gregor Lotz and Paul Muchowski (Gladstone, University of California, San Francisco). Aggregation was initiated by cleavage of the GST tag with PreScission protease (Amersham Biosciences), and the mixture was shaken for 30 h at 700 rpm at 30 °C. Under these conditions, a mixture of oligomers and fibrils forms, as described previously (36).
Protein Analysis
Analytical Ultracentrifugation
The molecular weight of synuclein oligomers was estimated using equilibrium analytical ultracentrifugation, as described previously (37). Briefly 2 mg/ml purified synuclein was sedimented at three speeds (10,000, 14,000, and 20,000 rpm, AN-60 rotor) in a Beckman XL-1 ultracentrifuge for 16–20 h per speed at 20 °C, and protein concentration as a function of radius was determined by absorbance at 280 nm. Sedimentation curves were globally fit with Sedphit and Sedphat software (National Institutes of Health), using the monomer-dimer model with the dimer Kd value set to zero, and an extinction coefficient for synuclein of 5960 m−1 cm−1 (38).
Dynamic Light Scattering
Dynamic light scattering of protein was measured with a DynaPro molecular sizing instrument (Protein Solutions, Lakewood, NJ) in micro quartz cuvettes with 1.5-mm path length and 12-ml volume. The samples were filtered (100 nm pore size) before use, and measurements were collected at 10-s intervals for 2–5 min. Dynamic light scattering of liposomes in “cytosol buffer” was measured in a 384-well microplate (Corning Glass) using a DynaPro plate reader (Wyatt Technology).
Size-exclusion Chromatography
Size-exclusion chromatography was performed using either an AKTA Prime chromatographic system equipped with a Sephadex G-100 column (0.7 × 15 cm) or a Shimadzu LC-10AD liquid chromatography system with a Superdex 75 column (GE Healthcare). The columns were calibrated using a series of molecular weight standards: ribonuclease A (13.6 kDa), chymotrypsinogen A (25 kDa), ovalbumin (43 kDa), bovine serum albumin (65 kDa), and aldolase (158 kDa). The void volume was determined using blue dextran 2000 kDa.
Thioflavin T Fluorescence
Thioflavin T fluorescence measurements were performed in semimicro quartz cuvettes (Hellma) with a 1-cm excitation light path using a FluoroMax-3 spectrofluorometer. Spectra were recorded from 460 to 550 nm with excitation at 450 nm, increments of 1 nm, an integration time of 0.2 s, and 1-nm slits for both excitation and emission. The final concentration of thioflavin T was 20 mm, whereas protein concentrations were 100–200 nm.
Attenuated Total Reflectance Fourier Transform Infrared Spectroscopy
Data were collected on a Thermo-Nicolet Nexus 670 FTIR spectrometer equipped with MCT detector and out-of-compartment germanium trapezoidal internal reflectance element. The hydrated thin films were prepared by drying samples on the internal reflectance element under N2. Typically, 512 interferograms were co-added at 1 cm−1 resolution. Data analysis was performed using GRAMS32 (Galactic Industries). Secondary structure content was determined by curve-fitting deconvoluted spectra based on the second derivative and Fourier self-deconvolution to identify component band positions.
Production and Analysis of α-Synuclein Triple Knock-out Mice
Mice lacking α-, β-, and γ-synuclein (TKO mice) and littermate controls were produced by crossing α-β-double KO mice (The Jackson Laboratory) (39) to γ-synuclein KO mice (a generous gift of L. Lustig (40)).
Electron Microscopy
Cells
Eighteen hours after transfection, cells were sorted for GFP expression by FACS, and cells in the top quartile for fluorescence were plated onto aclar discs, cultured for an additional 6 h, fixed in 2.5% glutaraldehyde, and then processed for electron microscopy by staining in 0.2 m sodium cacodylate that contains 1% osmium tetroxide with 1.6% potassium ferricyanide. After dehydration in EtOH and embedding in resin, 60-nm sections were examined using a FEI Tecnai 12 transmission electron microscope.
Brain Sections
Six-month-old control and transgenic mice overexpressing α-synuclein from the mThy-1 promoter (Line 61) (41) and 5-month-old control and synuclein TKO mice were perfused, and the right hemibrain was postfixed in phosphate-buffered 4% paraformaldehyde, pH 7.4, at 4 °C for 48 h, as described previously. The hemibrains were then sectioned with a vibratome at 40 μm, postfixed in 1% glutaraldehyde, treated with osmium tetraoxide, embedded in Epon araldite, and the ventral midbrain sectioned with an ultramicrotome (Leica, Germany). Grids were analyzed with a Zeiss OM 10 electron microscope (42, 43), and serial electron micrographs obtained at ×5,000 and ×25,000. Approximately 100 mitochondria from the ventral midbrain were evaluated per animal to determine average diameter and distribution. For immunogold labeling, sections were mounted on nickel grids, etched, and incubated with a rabbit polyclonal antibody to α-synuclein (Millipore), followed by a secondary antibody conjugated to 10-nm Aurion ImmunoGold particles (1:50, Electron Microscopy Sciences, Fort Washington, PA) with silver enhancement. A total of 125 cells were analyzed per condition. Cells were randomly acquired from three grids, and electron micrographs were obtained at a magnification of ×25,000.
Liposomes
Liposomes were incubated in “cytosol” buffer, in the presence or absence of protein, for 5 min. A 2.5-μl sample was then applied to glow-discharged carbon-coated copper grids and stained twice in freshly prepared 0.75% aqueous uranyl formate (44). Samples were imaged using a Tecnai T12 (FEI, Netherlands) electron microscope equipped with a LaB6 filament and operated at an acceleration voltage of 120 kV. UCSF Tomo (45) was used for automatic image acquisition. Liposomes were selected at random, and imaged at ×26,000 with a defocus value of −5 μm on a Gatan 4k × 4k (Gatan, Pleasanton, Ca) CCD camera. The area of individual liposomes was measured using Metamorph software.
Protein
Samples were deposited on Formvar-coated 300 mesh copper grids and negatively stained with 1% aqueous uranyl acetate. Transmission electron micrographs were collected on a JEOL JEM-100B microscope operating with an accelerating voltage of 80 kV. Typical nominal magnifications were ×75,000.
RESULTS
α-Synuclein Specifically Disrupts Mitochondrial Morphology
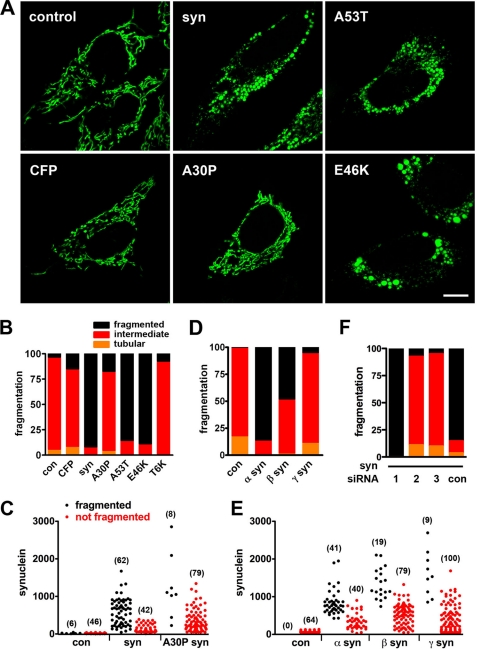
Considering the specific interaction of α-synuclein with mitochondrial membranes (27), we first assessed the potential effect of synuclein on mitochondrial morphology. Using HeLa cells because of their flat shape, large size, and dispersed tubular mitochondria, we cotransfected wild type human α-synuclein with a mitochondrially targeted enhanced green fluorescent protein (mitoGFP) and the blue fluorescent protein azurite as an independent reporter for transfection. The fluorescence of azurite correlates well with the overexpression of α-synuclein detected by immunostaining fixed cells (supplemental Fig. S1A), enabling us to identify live cells expressing synuclein in an unbiased manner solely on the basis of azurite expression. The mitochondrial morphology of azurite+ cells, assessed by cotransfection with mitoGFP and analyzed blind to the genotype of transfection, was then classified as tubular, fragmented, or intermediate. Relative to empty vector as control, the expression of α-synuclein produces a dramatic increase in the proportion of cells with fragmented mitochondria (p < 0.0001 by χ2 analysis), which typically cluster around the nucleus (Fig. 1, A and B). Quantitation confirms that synuclein reduces the length, perimeter, and area of mitochondria in transfected cells (supplemental Fig. S1B). Synuclein also increases mitochondrial width and hence reduces the axis ratio (length/width), producing shorter mitochondria with increased cross-sectional diameter. In addition, synuclein increases the mean number of distinct mitochondrial fragments per cell from 50.1 ± 5.9 to 161.6 ± 19.5 (p < 0.0001 by two-tailed t test, n = 14–17 cells per group, experiment repeated twice with similar results), thus excluding an effect on morphology independent of fission or fusion.
FIGURE 1.
Synuclein produces mitochondrial fragmentation in HeLa cells. A and B, HeLa cells were cotransfected with cDNAs encoding azurite (to identify transfected cells), mitoGFP (to identify mitochondria), and either empty vector control, CFP, wild type α-synuclein (syn), A30P, A53T, or E46K α-synuclein. 48 h after transfection, cells were selected on the basis of azurite fluorescence and imaged live; the images were randomized, and mitochondrial morphology was classified as fragmented, tubular, or intermediate blind to the genotype of transfection. Scale bar indicates 10 μm. The bar graph (B) shows the percentage of cells in each group. Wild type, A53T, and E46K α-synuclein produce mitochondrial fragmentation not observed in control (con), CFP, A30P, and T6K (replacement of Thr-22, Thr-33, Thr-44, Thr-59, Thr-81, and Thr-92 with lysines) synuclein, p < 0.0001 by χ2 analysis. n = 33–41 cells per group from two independent transfections. C, cells transfected as above were also fixed and immunostained for α-synuclein, and those with a range of synuclein levels (selected blind to mitochondrial morphology) were imaged and classified in terms of mitochondrial morphology. The number of cells in each group is indicated in parentheses. D and E, effects of α-, β-, and γ-synuclein on mitochondrial morphology were assessed the same way in live HeLa cells and in fixed cells immunostained for synuclein (E). α- and β-synuclein produce mitochondrial fragmentation (p < 0.0001), whereas γ-synuclein does not (D). n = 39–50 cells per group from three independent transfections. F, cells were cotransfected with synuclein and either siRNA to human synuclein (Silencer Select s13204 (1), s13205 (2), and s13206 (3)) or control siRNA (Silencer Select negative control 1 (con)). siRNA2 and -3 (but not siRNA1 or control siRNA) effectively reduce synuclein expression (supplemental Fig. S3) and block mitochondrial fragmentation (n = 18–27 cells per group from two independent transfections, p < 0.0001).
Expression of the soluble cytosolic CFP has no effect on mitochondrial morphology (Fig. 1, A and B). In addition, cotransfection of effective (but not ineffective) siRNA to human α-synuclein (supplemental Fig. S2) completely blocks fragmentation produced by the human protein (Fig. 1F), supporting the specificity of the effect. Similar to wild type human α-synuclein, the A53T and E46K mutants associated with familial PD also fragment mitochondria, but the A30P mutant, which does not bind membranes in vivo, has no discernible effect on the mitochondrial morphology of transfected cells (Fig. 1, A and B) despite equivalent expression by Western analysis (supplemental Fig. S2), providing an additional control for the effect of the wild type protein. To assess the dose dependence of effects on mitochondrial morphology, we also immunostained the fixed cells for α-synuclein. Fig. 1C shows that only the mitochondria in cells with no or low levels of wild type α-synuclein remain unfragmented, indicating that moderate expression suffices to produce fragmentation. We also find that β-synuclein causes less fragmentation than α-synuclein, and γ-synuclein has very little if any effect on mitochondrial morphology (Fig. 1, D and E). Because β- and γ-synuclein have not been found to cause PD, the relative specificity for α-synuclein supports the relevance of these observations for degeneration.
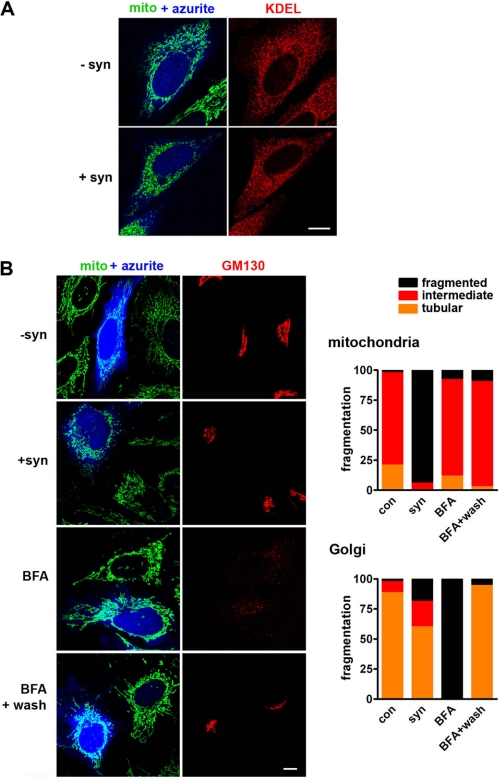
Work in yeast has shown that wild type α-synuclein can disrupt multiple membrane trafficking pathways, including transport from the endoplasmic reticulum to the Golgi complex (12, 46). The A53T mutant also disrupts the morphology of both Golgi complex and mitochondria in vivo (22). However, Fig. 2 shows that in mammalian cells exhibiting clear mitochondrial fragmentation, α-synuclein has no effect on the morphology of the endoplasmic reticulum. α-Synuclein does cause a small increase in fragmentation of the Golgi complex, but the Golgi remains normal in the vast majority of cells with fragmented mitochondria (Fig. 2B). This effect is also much smaller than that produced by the fungal metabolite brefeldin A, which causes reversible reabsorption of the Golgi complex into the endoplasmic reticulum (47) without any effect on mitochondrial morphology (Fig. 2B). We also failed to observe any effect of synuclein on the morphology of lysosomes or on the microtubule cytoskeleton (supplemental Fig. S3).
FIGURE 2.
α-Synuclein does not substantially affect endoplasmic reticulum or Golgi morphology. A, HeLa cells were cotransfected with cDNAs encoding azurite, mitoGFP, and either vector control (−syn) or α-synuclein (+syn). 48 h after transfection, cells were fixed, and the endoplasmic reticulum was identified by staining for the KDEL receptor. α-Synuclein has no effect on the morphology of the endoplasmic reticulum. Scale bar indicates 10 μm. B, HeLa cells stably expressing mitoGFP were cotransfected with cDNAs encoding azurite and either vector control (−syn) or α-synuclein (+syn). Control transfected cells were also treated with 5 mg/ml brefeldin A (BFA) for 60 min before imaging and then imaged again 4 h after wash out. 48 h after transfection, cells were fixed and stained for the Golgi matrix protein GM130. Random cells were classified blind to transfection in terms of mitochondrial and Golgi morphology. α-Synuclein produces mitochondrial fragmentation (p < 0.0001), whereas control, BFA, and BFA following wash do not. In contrast, BFA causes dramatic fragmentation of the Golgi complex far greater than that by synuclein (p < 0.0001), and this does not affect mitochondrial morphology. n = 24–111 cells per group from four independent transfections.
Peroxisomes use some of the same machinery for fission as mitochondria, including the dynamin-related protein Drp1 (48), raising the possibility that α-synuclein might also affect peroxisomes. However, α-synuclein does not affect the area or morphology of peroxisomes (supplemental Fig. S4). Because the punctate morphology of peroxisomes might make it difficult to detect an increase in fragmentation, we investigated this further using a dominant negative mutant of Drp1 (K38A) that increases the tubulation of both mitochondria and peroxisomes (48). α-Synuclein again fails to alter the morphology of these more tubular peroxisomes although it does fragment the more tubular mitochondria (supplemental Fig. S4). The effect of α-synuclein thus appears remarkably specific for mitochondria.
Although we have not detected any synuclein-immunoreactive aggregates after transfection (data not shown), α-synuclein accumulates in the Lewy bodies and dystrophic neurites of PD (8, 49) and can also accumulate in cultured cells under certain circumstances (50, 51), raising the possibility that aggregation may trigger the change in mitochondrial morphology. We thus compared the effects of synuclein with those of a mutant form of huntingtin (exon 1 with 72 CAG repeats) that is prone to aggregation (52). In contrast to cells expressing α-synuclein that show mitochondrial fragmentation, cells with either diffuse huntingtin or focal aggregates do not differ from control in mitochondrial morphology (p > 0.5 for both diffuse and aggregated huntingtin versus control) (supplemental Fig. S5). Thus, the effect of α-synuclein on mitochondrial morphology cannot simply represent a nonspecific effect of protein aggregation.
Mitochondrial Fragmentation Precedes a Decline in Oxidative Phosphorylation
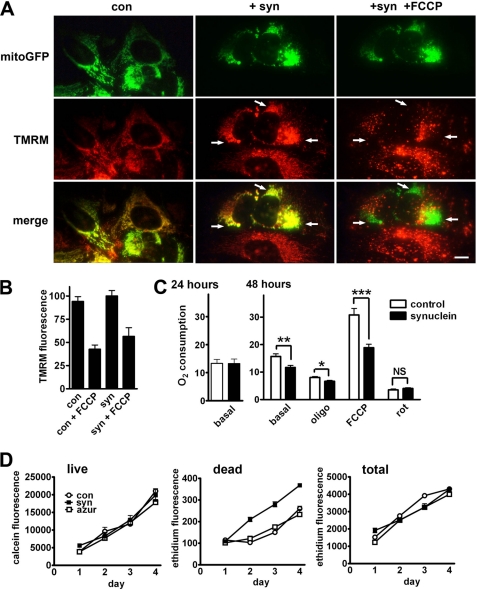
Cell injury and mitochondrial dysfunction in particular can produce mitochondrial fragmentation (53), and synuclein has recently been reported to impair complex I activity in cultured cells and in vivo (24, 54), raising the possibility that α-synuclein affects mitochondrial morphology indirectly, simply by producing toxicity. We thus examined the effect of α-synuclein on mitochondrial function and cell viability. To assess mitochondrial membrane potential, we used the dye TMRM (55). In cells cotransfected with mitoGFP, we find that despite the mitochondrial fragmentation in synuclein-expressing cells, TMRM stains mitoGFP-labeled compartments to the same extent as controls (Fig. 3, A and B). Treatment with the proton ionophore FCCP redistributes TMRM away from GFP+ mitochondria (Fig. 3A), indicating that we could have detected a decline in membrane potential induced by synuclein if this had occurred. To address the possibility that high levels of synuclein may impair mitochondrial membrane potential, we also labeled cells expressing α-synuclein and GFP with TMRM, then sorted for both GFP fluorescence (as a surrogate for synuclein) and TMRM. The supplemental Fig. 6A shows that membrane potential does not decline even at the highest levels of GFP and hence synuclein expression. Thus, even the most fragmented mitochondria in cells with the highest level of synuclein expression maintain their membrane potential.
FIGURE 3.
Mitochondrial fragmentation precedes any disturbance in mitochondrial function or toxicity. A and B, HeLa cells were transfected with mitoGFP and either empty vector control (con) or α-synuclein (+syn) and then loaded with the membrane potentially sensitive dye TMRM (1 nm) for 1 h before imaging live. Selected on the basis of GFP, TMRM fluorescence was quantified in individual cells. Treatment with the proton ionophore FCCP (2.5 μm) for 4 min before re-imaging causes a marked redistribution of TMRM fluorescence away from mitochondria (arrows). Scale bar indicates 10 μm. Quantitation of TMRM fluorescence (B) shows no effect of synuclein. The values indicate mean ± S.E. n = 12–49 cells per condition from four independent transfections. C, COS cells were transfected with either vector control or α-synuclein. 1–2 days later, oxygen consumption was measured in the basal state (10 mm glutamate and 2.5 mm malate) and, after addition of the ATP synthase inhibitor oligomycin (oligo) (1 μg/ml), the proton ionophore FCCP (1 μm) and the complex I inhibitor rotenone (rot) (500 nm). Synuclein impairs base-line respiration at 48 h but not 24 h, and the effect is greater in mitochondria uncoupled with FCCP. *, p < 0.01; **, p < 0.005; ***, p < 0.001; NS, not significant by one-way analysis of variance and Newman-Keuls post hoc test. n = 4–9 experiments per group. D, COS cells were transfected with vector control, α-synuclein, or azurite (azur), and after 16 h, replated onto 96-well plates. At 24, 48, 72, and 96 h after transfection, the cells were treated with either calcein green-AM (1 μm) to count live cells, ethidium bromide (5 μm) to count dead cells, or after 70% methanol followed by ethidium bromide to count total cells. α-Synuclein also has no effect on cell survival at 24 h, when fragmentation has already occurred in essentially all cells (supplemental Fig. S8B). By 48 h, however, there is a small increase in the number of dying cells that persists for the duration of the experiment (p < 0.01 versus control and azurite at 48, 72, and 96 h by one-way analysis of variance and Newman-Keuls post hoc test). Data show mean ± S.E., n = 6 wells per group.
Because mitochondria generate reactive oxygen species, and increased reactive oxygen species such as superoxide can accompany mitochondrial fragmentation under certain conditions (56), we also measured superoxide levels. The supplemental Fig. 6B shows that despite marked mitochondrial fragmentation, α-synuclein does not increase superoxide levels. Indicating the potential to detect a change in superoxide, expression of K38A DRP1 reduces the levels, as shown previously (56).
To assess the effects of α-synuclein on oxidative phosphorylation, we used transfection into COS cells, which confers the more efficient expression (in ∼70% cells) required for biochemical studies. By 24 h after transfection, essentially all cells expressing synuclein show fragmented mitochondria (supplemental Fig. S7A) but no change in the basal rate of oxygen consumption (Fig. 3C, left). The effect of α-synuclein on mitochondrial morphology thus precedes any change in mitochondrial membrane potential, reactive oxygen species, or respiration. Indeed, the lack of effect on mitochondrial function seemed remarkable considering the dramatic change in mitochondrial morphology, and previous work has shown that a primary disturbance of mitochondrial dynamics can eventually impair function (57). Consistent with this, we find that at 48 h after transfection into COS cells, α-synuclein reduces both basal and maximal (assessed in the presence of FCCP) rates of respiration (Fig. 3C). In addition, α-synuclein has no effect on cell survival 1 day after transfection, but by 48 h, there is a significant increase in the number of dying cells (Fig. 3D). However, the increase is small relative to the total number of cells, most of which survive (at 96 h, ∼9% of cells treated with synuclein are dead versus 6% in control groups). Although the effect of synuclein on mitochondrial morphology appears primary, it can thus eventually result in toxicity.
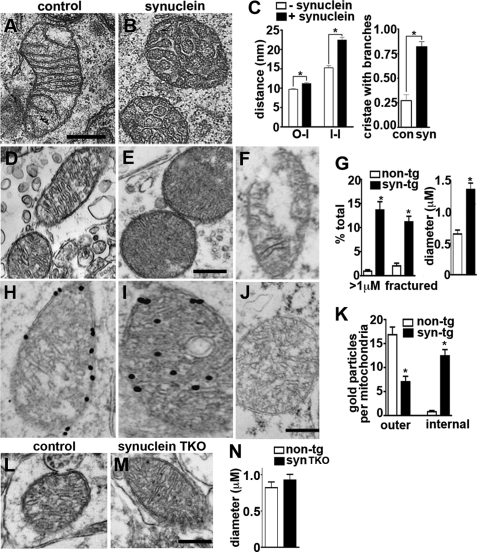
We also used electron microscopy to characterize the effect of α-synuclein on mitochondrial ultrastructure. Using flow cytometry to identify transfected cells, we found that α-synuclein converts the elongated, thin mitochondria observed in controls to shorter structures with increased cross-sectional diameter (Fig. 4, A and B), as early as 24 h after transfection, before any changes in mitochondrial membrane potential or respiration (Fig. 3). In particular, the mitochondria show disordered cristae, with enlarged and irregular intermembrane and intercristal spaces (Fig. 4, A and B).
FIGURE 4.
Synuclein disrupts mitochondrial ultrastructure. A–C, COS cells were transfected with mitoGFP and either vector control or α-synuclein and sorted at 18 h for GFP fluorescence, and cells falling into the top quartile of expression were plated onto aclar discs. Cultured for an additional 6 h, the cells were fixed in 2.5% glutaraldehyde and examined by electron microscopy. A and B, α-synuclein disrupts mitochondrial ultrastructure. Scale bar indicates 500 nm. C, random collection of mitochondria were analyzed for the distances between the outer and inner mitochondrial membranes (O-I) and between adjacent inner membranes (I-I), and the data presented as mean ± S.E., n = 138–176 for O-I, and 222–335 for I-I. C, synuclein also increases the proportion of cristae that branch at least once while traversing the diameter of the mitochondrion. Bars indicate mean ± S.E., n = 55–56 cristae per group from two independent transfections. *, p < 0.01 by two one-way analysis of variance and Newman-Keuls post hoc test. con, control. D–G, ultrastructural analysis shows mitochondria with well organized cristae in the midbrain neurons of 6-month-old nontransgenic mice (D), but disorganized cristae in the mitochondria of α-synuclein transgenic (tg) mice (E). α-Synuclein transgenic mitochondria also show an increase in cross-section diameter (D, E, and G) and discontinuous (fractured) outer membranes (F and G). *, p < 0.01 by unpaired two-tailed Student's t test (n = 4 per group). The size bar indicates 0.25 μm. H–J, immunoelectron microscopy for α-synuclein in the midbrain neurons of 6-month nontransgenic (H) and α-synuclein transgenic (I) mice shows 10 nm gold particles in association with the circumferential membranes of non-transgenic mitochondria, with no labeling by IgG control (J). In synuclein transgenic mice, however, most of the gold particles localize to internal membranes. The bar graph (K) indicates the distribution of gold particles over outer and internal mitochondrial membranes. *, p < 0.01 by unpaired two-tailed Student's t test (n = 4 per group). The size bar indicates 0.5 μm. L–N, ultrastructural analysis shows no difference in the morphology of mitochondria in midbrain neurons from control versus synuclein TKO mice. The size bar indicates 0.5 μm.
Synuclein Fragments Mitochondria in Midbrain Neurons and Promotes Neural Degeneration
To determine whether α-synuclein also produces mitochondrial fragmentation in neurons in vivo, we used transgenic mice expressing the human protein (41). Importantly, transgenic animals expressing the wild type protein show no neural degeneration and modest behavioral deficits (58). Furthermore, we examined them at a relatively young age (6 months). Relative to controls, the mitochondria of midbrain neurons from α-synuclein transgenic mice have a larger diameter, with disorganized cristae (Fig. 4, D, E and G), very similar to the results by light and electron microscopy in non-neural cell lines (Fig. 4, A–C). In addition, a number of mitochondria from transgenic neurons appear fractured, with a loss of continuity at the outer membrane (Fig. 4, F and G). To assess a role for endogenous synuclein in base-line mitochondrial morphology, we also examined synuclein TKO mice lacking α-, β-, and γ-synuclein isoforms. As described previously (59), young synuclein null mice are viable, fertile, and grossly normal. In contrast to the transgenic mice, we observed no difference in the morphology of ventral midbrain mitochondria between synuclein TKO mice and controls at 5 months of age (Fig. 4, L–N).
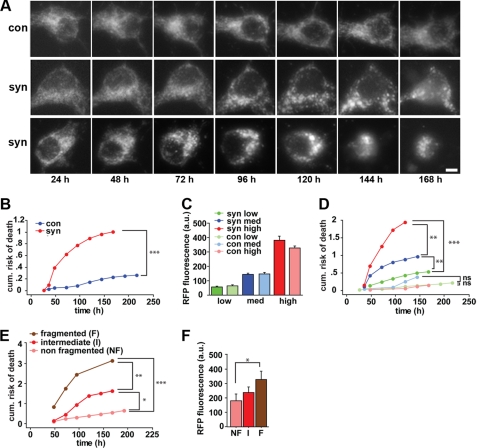
To determine whether the mitochondrial fragmentation produced by synuclein might predispose to neural degeneration, we examined the survival of live cells as a function of synuclein expression and mitochondrial morphology. Cotransfecting hippocampal neurons with mitoGFP to label mitochondria, and mRFP to track the survival of individual neurons over time (supplemental Fig. S8) (29, 34), we find that the mitochondrial morphology of control neurons does not change significantly over the 10 days after transfection (Fig. 5A, top panels). In contrast, cells transfected with human α-synuclein show mitochondrial fragmentation that can occur either late (Fig. 5A, middle panels) or early (Fig. 5A, bottom panels). We also find that cells transfected with human α-synuclein show a dose-dependent increase in the risk of death (Fig. 5, B–D). When stratified prospectively in terms of mitochondrial morphology, increasing fragmentation also predicts an increased risk of death (Fig. 5, E and F). Thus, wild type human α-synuclein exerts a dose-dependent toxicity on cultured neurons, and mitochondrial fragmentation precedes cell death by several days.
FIGURE 5.
Mitochondrial fragmentation by synuclein precedes neuronal death. Rat hippocampal neurons were transfected 5 days after plating with mRFP and either α-synuclein (syn) or vector control (con) (A–F), without or with mitoGFP (A, E, and F). MitoGFP (A) or mRFP (not shown) were then imaged on a daily basis using an automated microscope. Scale bar is 10 μm. B, cumulative risk of death curves indicate that primary neurons expressing synuclein have a significantly greater risk of death than control cells (***, p < 0.0001, log-rank test). n = 112–115 neurons per group. The experiment was repeated three times with similar results. C, mRFP fluorescence of individual neurons was measured 24 h after transfection and used as a surrogate for synuclein expression. Based on mRFP fluorescence, neurons were stratified into three expression groups (low, medium, and high). a.u., arbitrary units of fluorescence. Within each of these groups, synuclein (dark) and control (light) transfected neurons showed no significant difference in mRFP fluorescence. D, cumulative risk of death curves indicates that neurons expressing higher levels of synuclein (red and blue) have a dose-dependent greater risk of death than controls expressing equivalent mRFP. **, p < 0.005; ***, p < 0.0001; ns, not significant, n = 22–46 neurons per group. E, cumulative risk of death curves show that neurons with fragmented mitochondria at 48 h have a progressively greater risk of death than neurons displaying intermediate or nonfragmented mitochondria (*, p < 0.05; **, p < 0.005; ***, p < 0.001). E and F, neurons with fragmented mitochondria express progressively higher levels of synuclein (*, p < 0.05).
Synuclein Promotes Mitochondrial Fission through a Drp1-independent Pathway
How does α-synuclein affect mitochondrial morphology? Synuclein could produce mitochondrial fragmentation by decreasing the rate of mitochondrial fusion or by increasing fission through either a Drp1-dependent or -independent pathway. Live imaging of transfected cells shows that fragmentation occurs gradually over hours, with the fragmentation state reflecting a balance between the rate of fusion and fission (supplemental Movies S1–S3). Similarly, overexpression of fusion proteins fails to inhibit fragmentation by synuclein (supplemental Fig. S9A), consistent with a role for synuclein in fission rather than fusion.
We further tested the role of α-synuclein in a direct assay for mitochondrial fusion (supplemental Fig. S7B). Using polyethylene glycol to fuse cells overexpressing α-synuclein and either mitoGFP or mitoDsRed (60), we began to detect fusion by 4 h in controls, with complete fusion by 6.5 h. Cells expressing α-synuclein fuse as well but with a longer latency. However, mitochondrial fragmentation increases the number of individual mitochondria, requiring more events to achieve fusion of all the fragments. Indeed, expression of the fission protein Drp1 slows fusion to the same or perhaps a greater extent than synuclein, strongly suggesting that synuclein does not cause an intrinsic defect in mitochondrial fusion. We infer from this result that synuclein acts by promoting fission.
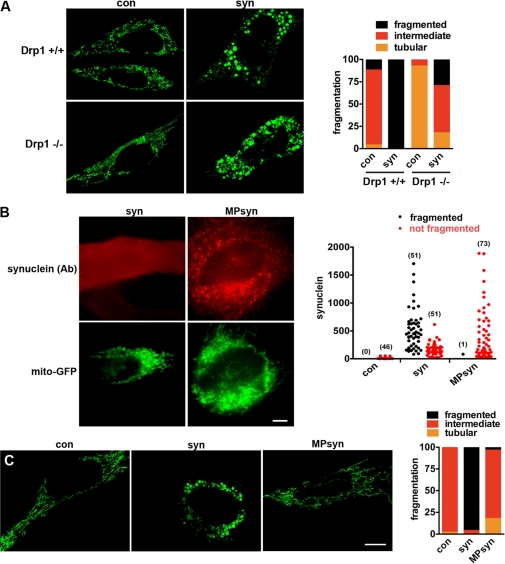
To determine whether synuclein affects mitochondrial morphology through the known fission machinery, we both transfected cells overexpressing the dominant negative mutant of Drp1 (K38A) and used cells entirely lacking Drp1. As anticipated, the loss of Drp1 causes a shift in base-line mitochondrial morphology toward increased tubulation. Despite the complete absence of Drp1, however, synuclein still increases mitochondrial fragmentation (Fig. 6A and supplemental Fig. 9B). Consistent with this and the specificity of the effect for mitochondria, synuclein does not affect the morphology of peroxisomes (supplemental Fig. S4), which also rely on Drp1 for fission. Mitochondrial depolarization also triggers fission independent of Drp1 (53) but produces characteristic circular disks during fragmentation that we did not observe with α-synuclein (supplemental Fig. S1B).
FIGURE 6.
Synuclein fragments mitochondria through a direct interaction independent of Drp1. A, immortalized Drp1 KO and control mouse embryonic fibroblasts were cotransfected with cDNAs encoding azurite (to identify transfected cells), mitoGFP (to identify mitochondria), and either empty vector control (con) or wild type α-synuclein (syn). Forty eight hours after transfection, cells were selected on the basis of azurite fluorescence and imaged live, and the images were randomized, and mitochondrial morphology was classified as fragmented, tubular, or intermediate blind to the genotype of transfection. Scale bar indicates 10 μm. The bar graph shows the percentage of cells in each group. At base line, cells lacking Drp1 have more tubulated mitochondria (p < 0.0001). However, synuclein produces robust mitochondrial fragmentation even in the absence of Drp1 (p < 0.0001). n = 13–34 cells per group from two independent transfections. B and C, HeLa cells were cotransfected with azurite (to identify transfected cells), mitoGFP (to identify mitochondria), and either untagged α-synuclein (syn) or α-synuclein targeted to the secretory pathway by fusion to the N-terminal myristoylation/palmitoylation signal of Lyn kinase (MPsyn). B, 48 h after transfection, cells were fixed and immunostained for α-synuclein, and those with a range of synuclein levels (selected blind to mitochondrial morphology) imaged and classified in terms of mitochondrial morphology. The number of cells in each group is indicated in parentheses. In contrast to syn, MPsyn fails to fragment mitochondria even when expressed at high levels. C, cells were selected on the basis of azurite fluorescence and imaged live, and the images were randomized and mitochondrial morphology was classified as fragmented, tubular, or intermediate blind to the genotype of transfection. Scale bar indicates 10 μm. The bar graph shows the percentage of cells in each group. syn fragments mitochondria (p < 0.0001), whereas control and MPsyn do not. n = 28–50 cells per group from two independent transfections.
Requirement for a Direct Interaction with Mitochondrial Membrane
How does α-synuclein promote mitochondrial fission? Synuclein preferentially binds to mitochondria in vitro (27), suggesting that the effect on mitochondrial fragmentation may result from a direct interaction. To test this possibility, we introduced a dual myristoylation/palmitoylation sequence from Lyn kinase (61) that targets synuclein to the secretory pathway. The lipid anchor completely blocks fragmentation at even the highest levels of synuclein expressed (Fig. 6, B and C), supporting the requirement for a direct interaction with mitochondria. The fragmentation is also blocked by either the A30P mutation or the replacement of multiple N-terminal threonines by lysine, both of which disrupt the association of α-synuclein with membranes (Fig. 1B) (13, 23, 62, 63). Furthermore, deletion of the C-terminal 20, 30, and 41 residues all fail to block the mitochondrial fragmentation produced by α-synuclein (supplemental Fig. S10), indicating that the C terminus is not required. However, a larger truncation of the final 60 residues, which extend into the membrane-binding domain, does disrupt fragmentation. The N-terminal membrane-binding domain of synuclein is thus both necessary and sufficient for the effects on mitochondrial morphology.
To assess further the direct interaction of synuclein with mitochondrial membranes in vivo, we used immunoelectron microscopy. In the midbrain of nontransgenic mice immunostained for α-synuclein, gold particles over mitochondria localize primarily to membranes at the circumference (Fig. 4, H and K). Only occasional gold particles labeled internal structures. Interestingly, α-synuclein transgenic mice show much more intense labeling of internal membranes (Fig. 4, I and K). CA3 hippocampal neurons showed similar labeling patterns (data not shown).
α-Synuclein Fragments and Clusters Artificial Membranes Containing Cardiolipin
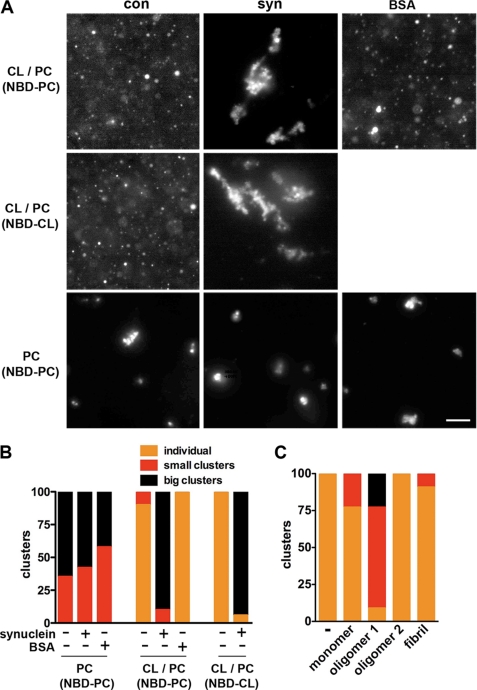
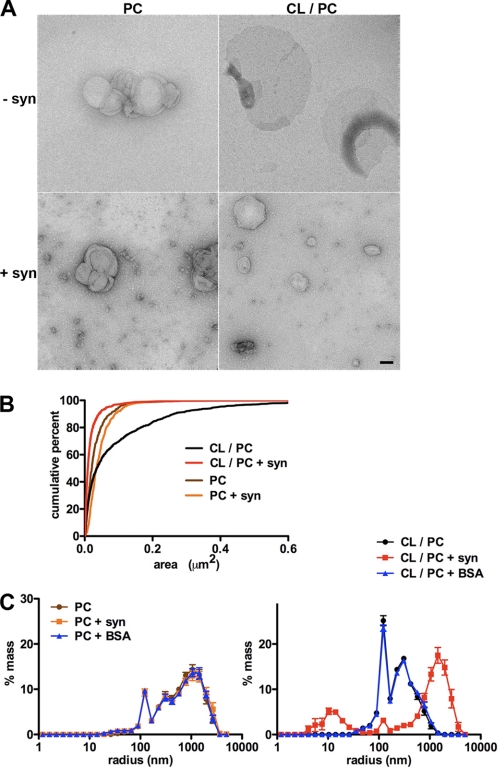
Independence from Drp1 (Fig. 6 and supplemental Figs. S4 and S9B) together with requirements for the membrane-binding N terminus of synuclein (Fig. 1 and supplemental Fig. S10) and a direct interaction with mitochondria (Figs. 4, H–J, and 6, B and C) (27) suggest that synuclein may fragment mitochondria by directly binding and disrupting mitochondrial membranes. To test this possibility in a system free of other proteins, we incubated recombinant human α-synuclein with artificial membranes containing the acidic phospholipid cardiolipin (CL) that is enriched in mitochondria and, like other acidic phospholipids, promotes the interaction of synuclein with artificial membranes (27). Using membranes labeled with trace amounts of the fluorophore nitrobenzoxadiazole (NBD), fluorescence microscopy shows that in the absence of synuclein, liposomes containing CL and phosphatidylcholine (1:1 or 1:3) stay dispersed and, in many cases, flatten onto the substrate (Fig. 7). In the presence of 20 μm synuclein, however, large clusters quickly form (Fig. 7, A and B), and electron microscopy shows that the remaining individual liposomes are much smaller with synuclein than those without (Fig. 8). Furthermore, all the effects of synuclein are specific for membranes that contain CL, and liposomes containing PC alone form small, stable clusters that are not affected by synuclein. Bovine serum album (BSA) and aggregated mutant huntingtin also have no effect on the morphology of fluorescent liposomes containing CL (Fig. 7, A and B, and supplemental Fig. S11). To quantify the changes in vesicle size, we used light scattering and found that synuclein reduces the size of membranes containing CL but not control membranes lacking this phospholipid (Fig. 8C). In addition, light scattering shows that synuclein can generate larger membranous structures (Fig. 8C), presumably due to aggregation (Fig. 7, A and B).
FIGURE 7.
Recombinant, oligomeric α-synuclein clusters artificial membranes. A, artificial liposomes containing either CL and PC (1:1) or PC alone were incubated for 5 min without protein, with a combination of monomeric and trimeric synuclein (0.3 mg/ml), or with BSA (0.6 mg/ml), and the fluorescence of trace NBD conjugated to the N terminus of either 1,2-dioleoyl-sn-glycero-3-phosphocholine or heart cardiolipin (NBD-CL) visualized by light microscopy. Scale bar indicates 10 μm. B, quantitative analysis of liposome clustering. Individual indicates that essentially all liposomes in a field are isolated; small clusters indicate 2–10 liposomes per cluster, and big clusters indicates more than 10 liposomes per cluster. Liposomes containing CL and PC alone or with BSA remain isolated and in many cases flatten onto the substrate. In the presence of synuclein, however, large clusters form rapidly (p < 0.0001 versus control for CL/PC liposomes). Synuclein has no effect on liposomes containing PC alone. n = 21–22 fields per group. C, CL liposomes were incubated with 80 μm synuclein monomer, small oligomer (oligomer 1), mature oligomer (oligomer 2), or fibrils, and the extent of clustering was quantified. Only oligomer 1 results in clustering (p < 0.0001). 17–20 fields per group.
FIGURE 8.
Recombinant synuclein drives the fission of artificial membranes. A, liposomes were incubated for 5 min at room temperature in the presence or absence of synuclein (20 μm, monomeric and trimeric) and visualized by EM. Scale bar indicates 100 nm. B, random fields of isolated liposomes were photographed, and their areas were quantified. Cumulative frequency distribution shows that synuclein reduces the mean area for those containing CL/PC but not those with PC alone. n = 263–1768 liposomes per group. C, analysis by light scattering shows that synuclein (0.3 mg/ml) but not BSA (0.3 mg/ml) causes the emergence of two liposome populations, one smaller than the original, and the other larger. The graphs indicate mean ± S.E., n = 11–12 wells per group from three independent experiments.
Synuclein occupies a number of different states, from natively unfolded monomers in solution to an elongated α-helix on membrane binding (27, 64, 65) and β-sheet structure in fibrils (66). To determine which of these forms might be responsible for the effect of synuclein on membranes, we used dynamic light scattering and size-exclusion chromatography to analyze the preparation of recombinant synuclein used for the experiments in vitro. The analysis reveals a mixture composed predominantly of trimers and/or tetramers but also some monomers (supplemental Fig. S12, A and B). Because dynamic light scattering and size-exclusion chromatography both depend on conformation in addition to size, we also used equilibrium analytical ultracentrifugation to determine mass independent of conformation. The results indicate a mean molecular mass of 34 kDa, consistent with a mixture of monomeric and trimeric α-synuclein (supplemental Fig. S12C).
To assess further the role of different conformations in the fragmentation of mitochondria by α-synuclein, we isolated monomeric, oligomeric (intermediate and mature), and fibrillar forms of the recombinant human protein, and we characterized them by light scattering, size-exclusion chromatography, electron microscopy, and FTIR (supplemental Fig. S12). Samples were also analyzed by thioflavin fluorescence, which stained only the fibril fraction (data not shown). A heterogeneous mixture of intermediate oligomers (oligomer 1) clustered the artificial membranes, whereas monomer, mature oligomer, and fibrils all failed to produce any discernible effect (Fig. 7C). Smaller oligomeric forms thus appear responsible for the effect of synuclein on membrane morphology and, by inference, on mitochondria.
DISCUSSION
Synuclein Has a Primary Effect on Mitochondrial Morphology
Previous work has shown effects of synuclein on a variety of cell membranes. In yeast, expression of human synuclein causes the aggregation of intracellular vesicles involved in endoplasmic reticulum-Golgi transport and secretion (13, 46). In mammalian cells, however, we now find that α-synuclein has an early, specific effect on the morphology of mitochondria. We observe no change in the morphology of endoplasmic reticulum, lysosomes, peroxisomes, or microtubule cytoskeleton. We detect some changes in the Golgi complex, but these appear modest and occur in only a small fraction of the cells with fragmented mitochondria. We also observe mitochondrial fragmentation in mammalian neurons as well as non-neural cells, very similar to a recent report using neuroblastoma cells and Caenorhabditis elegans (67). Although similar biochemical mechanisms may account for the effect on membranes in yeast and metazoan cells, the specific membranes targeted thus differ.
The changes in mitochondrial morphology occur in the absence of overt toxicity or even mitochondrial dysfunction. Previous work has indeed shown that a normal mitochondrial membrane potential is compatible with fragmentation (68, 69). Recent studies have also begun to suggest that synuclein interacts specifically with mitochondria. Although synuclein generally behaves like a soluble protein in mammalian cells, it has been reported to colocalize with mitochondria, either constitutively or after cell injury (21, 23–26, 70). A sensitive FRET-based reporter for membrane binding also indicates a specific interaction with mitochondria (27). However, the functional consequences of this interaction have remained unclear. Synuclein has been reported to influence mitochondrial complex I activity (24, 54), but we find that at a time when fragmentation has already occurred, mitochondrial membrane potential and respiration remain normal.
Effect of Synuclein on Mitochondrial Morphology Involves a Direct Interaction
How does α-synuclein fragment mitochondria? A30P and T6K mutations block both mitochondrial fragmentation produced by synuclein and the interaction of synuclein with membranes in vivo (13, 23, 62, 63), and deletion of the C terminus has no effect. Mitochondrial fragmentation thus requires the N-terminal membrane-binding domain of synuclein. Consistent with a direct interaction, targeting to the plasma membrane blocks the effect of synuclein on mitochondrial morphology. In addition, our data suggest that the effect of synuclein on mitochondrial morphology involves an increase in fission rather than a block in fusion as recently suggested (67). More numerous, fragmented mitochondria presumably take longer to fuse than large, tubular mitochondria. but when controlled for size, mitochondria fragmented by overexpression of synuclein do not differ in the rate of fusion from those fragmented by fission protein Drp1, which should not affect fusion. Although by exclusion synuclein thus appears to drive mitochondrial fission, it does not apparently require Drp1, suggesting a novel mechanism. Analysis of the Drp1 knock-out indeed demonstrates the existence of Drp1-independent mechanisms for fission, with their relative importance varying in different neuronal populations (31). Consistent with a Drp1-independent mechanism, the ability of synuclein to fragment artificial membranes in vitro shows that synuclein can act alone to fragment membranes, and another report has recently described similar fragmentation of large liposomes by synuclein (71). Synuclein thus belongs to a growing number of proteins that directly affect membrane dynamics.
Peripheral membrane proteins like synuclein may influence membrane curvature through several distinct mechanisms. First, synuclein forms an amphipathic helix on membrane binding (72), and the insertion of an amphipathic α-helix directly into the lipid bilayer can cause membrane buckling (73). Second, the clustering of lipids with a particular headgroup can influence the size of acyl chain relative to headgroup and hence membrane curvature. Indeed, synuclein has been found to cluster the acidic phospholipid phosphatidylglycerol (74), and presumably has a similar effect on cardiolipin. Interestingly, the protein tBid is also hypothesized to regulate the curvature of mitochondrial membranes by binding to cardiolipin (75). Alternatively, changes in the conformation of synuclein may drive membrane deformation; synuclein adopts a broken helix on small membranes such as detergent micelles (76) but an extended α-helical conformation on binding to large liposomes and mitochondria (27, 65).
The polymerization of membrane-associated coat proteins such as clathrin, COP1, and COP2 may also drive membrane deformation (77, 78), and synuclein has been suggested to polymerize on membranes (79). We now find that the effect of synuclein on artificial membranes specifically involves a small oligomer. Monomer, large oligomer, and fibril are not active, but preparations containing a small oligomer (trimer or tetramer) are responsible for the activity observed in vitro. A small oligomer may therefore also cause the fragmentation of mitochondria in cells.
What accounts for the selective effect of α-synuclein on mitochondrial morphology? Synuclein has an affinity for the widely distributed lipid phosphatidic acid that is equivalent to that for the mitochondrion-specific cardiolipin (27). However, the ability of synuclein to interact with artificial membranes in vitro depends on a high lipid:protein ratio, and the content of acidic phospholipid (mostly cardiolipin) in mitochondrial membranes may exceed that of other organelles (80–83). The specific effects of synuclein on mitochondrial morphology may thus simply reflect the unusually high concentration of acidic phospholipid. Indeed, the inner mitochondrial membrane contains more cardiolipin than the outer, and we now find that the disruption of mitochondrial morphology is accompanied by a redistribution of synuclein from the outer surface of mitochondria to internal structures, suggesting that access to the inner membrane contributes to changes in mitochondrial morphology.
Normal Versus Pathologic Function
Does the effect on mitochondrial morphology reflect a normal function of α-synuclein? We have not detected a change in the morphology of midbrain mitochondria from synuclein TKO mice, but we also cannot exclude a subtle effect, and RNAi for synuclein was recently found to make mitochondria more tubular in a neural cell line (67). In addition, we cannot exclude compensatory effects by other proteins that promote fission such as Drp1 (84, 85). Endogenous synuclein may also influence the delivery, accumulation, or function of mitochondria at presynaptic boutons, affecting transmitter release (59, 86, 87).
However, the effects of overexpressed α-synuclein on mitochondrial morphology may reflect a strictly pathologic role. The amount of synuclein bound to mitochondria increases dramatically in PD, specifically in the brain regions that undergo degeneration (24). Consistent with this, previous work has shown that a disturbance in the machinery controlling mitochondrial dynamics can result in neural degeneration. A defect in fusion leads eventually to mitochondrial dysfunction in a cell line (57), and mutations in the fusion proteins mitofusin 2 and Opa1 cause, respectively, Charcot-Marie-Tooth neuropathy and optic atrophy (69, 88, 89). Fragmentation produced by the high levels of α-synuclein that occur in essentially all forms of classical PD might thus be expected to impair mitochondrial function and result in cell death. Consistent with this possibility, respiration deteriorates 2 days after transfection of synuclein into COS cells. We also find that overexpression of synuclein results in the death of neurons. By stratifying transfected neurons prospectively in terms of synuclein expression, we observe that highly expressing cells have a much higher likelihood of death. Furthermore, cotransfection of a mitochondrial reporter shows that mitochondrial fragmentation predicts cell death. The effects on mitochondrial morphology may thus contribute to the role of synuclein in neural degeneration.
If the effects of synuclein on mitochondrial morphology contribute to the pathogenesis of PD, why does the A30P mutation, which causes familial PD, apparently fail to influence mitochondrial morphology? We have previously found that the A30P mutant does not show any enrichment at the synapse (90) and may exhibit significantly increased levels in the cell body (91). These higher levels may indeed offset the reduced intrinsic ability of A30P α-synuclein to influence mitochondrial morphology. At the same time, the penetrance of this point mutant is lower than the others that cause PD (92).
The recent identification of a role for recessive PD genes PARK2 (parkin) and PINK1 in mitochondrial autophagy suggests interesting parallels to the effect of synuclein on mitochondrial morphology (84, 85, 93, 94). Indeed, fragmentation may be required for efficient mitochondrial autophagy. However, it remains unclear whether parkin and PINK1 have direct effects on mitochondrial morphology or simply operate downstream in the clearance mechanism (84, 85, 93–97, 99). In contrast, α-synuclein appears to promote mitochondrial fragmentation through a direct effect on the membrane. Interestingly, the loss of both fusion proteins Mfn1 and -2 has already been shown to result in the depolarization of mitochondria and the recruitment of parkin (4, 57), suggesting that parkin and PINK1 may be required to protect against the effects of synuclein overexpression. However, the autosomal recessive forms of PD produced by loss of parkin and PINK1 differ clinically and pathologically from idiopathic PD. In particular, most cases of recessive PD do not appear to involve the accumulation of synuclein or indeed Lewy bodies (100, 101).
The requirement for oligomeric synuclein to fragment artificial membranes in vitro further suggests that oligomeric forms account for the effects on mitochondrial morphology in cells. Originally considered a monomeric protein with no native structure (102), synuclein has been shown to form oligomers in cells, in transgenic animals, and in PD (54, 98), and some of these forms may lie on the pathway to fibrillization. Although prominent in the pathology of PD, synuclein aggregates are also very difficult to detect after transfection into a wide variety of cells. The robust effect on mitochondrial morphology now suggests an important role for oligomeric synuclein even in the absence of aggregates. Because the monomer clearly binds to membranes, the effect of oligomeric but not monomeric synuclein on membrane morphology further suggests that binding alone does not suffice to produce fragmentation.
Supplementary Material
Acknowledgments
We thank Kurt Thorn for help with optical imaging; Marty Bigos and The Gladstone Institutes Flow Cytometry Core for cell sorting; Marc Diamond for use of a fluorescent plate reader; Aarvin Dar for assistance with light scattering; Gregor Lotz and Paul Muchowski for mutant huntingtin; Scott Hansen and Dyche Mullins for help with analytical ultracentrifugation; Edward Rockenstein for the α-synuclein transgenic mice; and Reena Zalpuri and Margarita Trejo for the electron microscopy. We also thank Bipasha Mukherjee for technical assistance, Akanksha Bapna for assistance with size-exclusion chromatography, and Rebecca Seal for comments on the manuscript.
This work was supported, in whole or in part, by Grant GM089853 from the National Institutes of Health (to H. S.), Grant 2R01NS39074 from NINDS and Grants 1KO8NS062954-01A1 (to K. N.), and NS057096 from NINDS (to E. M.), Grants P01AG022074, AG18440, and AG022074 from NIA, and Grant DA10154 from NIDA. This work was also supported by a postdoctoral fellowship from the Hillblom Foundation, an Alzheimer's Disease Research Center pilot grant, and a Burroughs-Wellcome Medical Scientist Fund Career Award (to K. N.), the University of California San Francisco Medical Scientist Training Program and a predoctoral fellowship from the Hillblom Foundation (to V. M. N.), postdoctoral fellowships from the Hillblom Foundation (to G. S.) and the Taube-Koret Center (to S. F.), and a Brain Disorders Award from the McKnight Foundation (to R. H. E.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S12 and Movies S1–S3.
- PD
- Parkinson disease
- MPTP
- 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- CL
- cardiolipin
- PC
- phosphocholine
- NBD
- nitrobenzoxadiazole
- TMRM
- tetramethylrhodamine methyl ester
- TKO
- triple KO
- CFP
- cyan fluorescent protein.
REFERENCES
- 1. Schapira A. H., Mann V. M., Cooper J. M., Dexter D., Daniel S. E., Jenner P., Clark J. B., Marsden C. D. (1990) J. Neurochem. 55, 2142–2145 [DOI] [PubMed] [Google Scholar]
- 2. Bender A., Krishnan K. J., Morris C. M., Taylor G. A., Reeve A. K., Perry R. H., Jaros E., Hersheson J. S., Betts J., Klopstock T., Taylor R. W., Turnbull D. M. (2006) Nat. Genet. 38, 515–517 [DOI] [PubMed] [Google Scholar]
- 3. Valente E. M., Abou-Sleiman P. M., Caputo V., Muqit M. M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A. R., Healy D. G., Albanese A., Nussbaum R., González-Maldonado R., Deller T., Salvi S., Cortelli P., Gilks W. P., Latchman D. S., Harvey R. J., Dallapiccola B., Auburger G., Wood N. W. (2004) Science 304, 1158–1160 [DOI] [PubMed] [Google Scholar]
- 4. Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Polymeropoulos M. H., Lavedan C., Leroy E., Ide S. E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., Stenroos E. S., Chandrasekharappa S., Athanassiadou A., Papapetropoulos T., Johnson W. G., Lazzarini A. M., Duvoisin R. C., Di Iorio G., Golbe L. I., Nussbaum R. L. (1997) Science 276, 2045–2047 [DOI] [PubMed] [Google Scholar]
- 6. Krüger R., Kuhn W., Müller T., Woitalla D., Graeber M., Kösel S., Przuntek H., Epplen J. T., Schöls L., Riess O. (1998) Nat. Genet. 18, 106–108 [DOI] [PubMed] [Google Scholar]
- 7. Zarranz J. J., Alegre J., Gómez-Esteban J. C., Lezcano E., Ros R., Ampuero I., Vidal L., Hoenicka J., Rodriguez O., Atarés B., Llorens V., Gomez Tortosa E., del Ser T., Muñoz D. G., de Yebenes J. G. (2004) Ann. Neurol. 55, 164–173 [DOI] [PubMed] [Google Scholar]
- 8. Spillantini M. G., Schmidt M. L., Lee V. M., Trojanowski J. Q., Jakes R., Goedert M. (1997) Nature 388, 839–840 [DOI] [PubMed] [Google Scholar]
- 9. Singleton A. B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., Lincoln S., Crawley A., Hanson M., Maraganore D., Adler C., Cookson M. R., Muenter M., Baptista M., Miller D., Blancato J., Hardy J., Gwinn-Hardy K. (2003) Science 302, 841. [DOI] [PubMed] [Google Scholar]
- 10. Ibáñez P., Bonnet A. M., Débarges B., Lohmann E., Tison F., Pollak P., Agid Y., Dürr A., Brice A. (2004) Lancet 364, 1169–1171 [DOI] [PubMed] [Google Scholar]
- 11. Feany M. B., Bender W. W. (2000) Nature 404, 394–398 [DOI] [PubMed] [Google Scholar]
- 12. Cooper A. A., Gitler A. D., Cashikar A., Haynes C. M., Hill K. J., Bhullar B., Liu K., Xu K., Strathearn K. E., Liu F., Cao S., Caldwell K. A., Caldwell G. A., Marsischky G., Kolodner R. D., Labaer J., Rochet J. C., Bonini N. M., Lindquist S. (2006) Science 313, 324–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Outeiro T. F., Lindquist S. (2003) Science 302, 1772–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Willingham S., Outeiro T. F., DeVit M. J., Lindquist S. L., Muchowski P. J. (2003) Science 302, 1769–1772 [DOI] [PubMed] [Google Scholar]
- 15. Zhou W., Hurlbert M. S., Schaack J., Prasad K. N., Freed C. R. (2000) Brain Res. 866, 33–43 [DOI] [PubMed] [Google Scholar]
- 16. Tanaka Y., Engelender S., Igarashi S., Rao R. K., Wanner T., Tanzi R. E., Sawa A. L., Dawson V., Dawson T. M., Ross C. A. (2001) Hum. Mol. Genet. 10, 919–926 [DOI] [PubMed] [Google Scholar]
- 17. Giasson B. I., Duda J. E., Quinn S. M., Zhang B., Trojanowski J. Q., Lee V. M. (2002) Neuron 34, 521–533 [DOI] [PubMed] [Google Scholar]
- 18. Lee M. K., Stirling W., Xu Y., Xu X., Qui D., Mandir A. S., Dawson T. M., Copeland N. G., Jenkins N. A., Price D. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8968–8973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dauer W., Kholodilov N., Vila M., Trillat A. C., Goodchild R., Larsen K. E., Staal R., Tieu K., Schmitz Y., Yuan C. A., Rocha M., Jackson-Lewis V., Hersch S., Sulzer D., Przedborski S., Burke R., Hen R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14524–14529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ellis C. E., Murphy E. J., Mitchell D. C., Golovko M. Y., Scaglia F., Barceló-Coblijn G. C., Nussbaum R. L. (2005) Mol. Cell. Biol. 25, 10190–10201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin L. J., Pan Y., Price A. C., Sterling W., Copeland N. G., Jenkins N. A., Price D. L., Lee M. K. (2006) J. Neurosci. 26, 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin X., Parisiadou L., Gu X. L., Wang L., Shim H., Sun L., Xie C., Long C. X., Yang W. J., Ding J., Chen Z. Z., Gallant P. E., Tao-Cheng J. H., Rudow G., Troncoso J. C., Liu Z., Li Z., Cai H. (2009) Neuron 64, 807–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cole N. B., Dieuliis D., Leo P., Mitchell D. C., Nussbaum R. L. (2008) Exp. Cell Res. 314, 2076–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Devi L., Raghavendran V., Prabhu B. M., Avadhani N. G., Anandatheerthavarada H. K. (2008) J. Biol. Chem. 283, 9089–9100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li W. W., Yang R., Guo J. C., Ren H. M., Zha X. L., Cheng J. S., Cai D. F. (2007) Neuroreport 18, 1543–1546 [DOI] [PubMed] [Google Scholar]
- 26. Shavali S., Brown-Borg H. M., Ebadi M., Porter J. (2008) Neurosci. Lett. 439, 125–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakamura K., Nemani V. M., Wallender E. K., Kaehlcke K., Ott M., Edwards R. H. (2008) J. Neurosci. 28, 12305–12317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Voglmaier S. M., Kam K., Yang H., Fortin D. L., Hua Z., Nicoll R. A., Edwards R. H. (2006) Neuron 51, 71–84 [DOI] [PubMed] [Google Scholar]
- 29. Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 30. Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. (2002) Science 296, 913–916 [DOI] [PubMed] [Google Scholar]
- 31. Wakabayashi J., Zhang Z., Wakabayashi N., Tamura Y., Fukaya M., Kensler T. W., Iijima M., Sesaki H. (2009) J. Cell Biol. 186, 805–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakamura K., Bindokas V. P., Kowlessur D., Elas M., Milstien S., Marks J. D., Halpern H. J., Kang U. J. (2001) J. Biol. Chem. 276, 34402–34407 [DOI] [PubMed] [Google Scholar]
- 33. Saudou F., Finkbeiner S., Devys D., Greenberg M. E. (1998) Cell 95, 55–66 [DOI] [PubMed] [Google Scholar]
- 34. Arrasate M., Finkbeiner S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3840–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uversky V. N., Yamin G., Souillac P. O., Goers J., Glaser C. B., Fink A. L. (2002) FEBS Lett. 517, 239–244 [DOI] [PubMed] [Google Scholar]
- 36. Legleiter J., Lotz G. P., Miller J., Ko J., Ng C., Williams G. L., Finkbeiner S., Patterson P. H., Muchowski P. J. (2009) J. Biol. Chem. 284, 21647–21658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Quinlan M. E., Hilgert S., Bedrossian A., Mullins R. D., Kerkhoff E. (2007) J. Cell Biol. 179, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. De Franceschi G., Frare E., Bubacco L., Mammi S., Fontana A., de Laureto P. P. (2009) J. Mol. Biol. 394, 94–107 [DOI] [PubMed] [Google Scholar]
- 39. Chandra S., Fornai F., Kwon H. B., Yazdani U., Atasoy D., Liu X., Hammer R. E., Battaglia G., German D. C., Castillo P. E., Südhof T. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14966–14971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ninkina N., Papachroni K., Robertson D. C., Schmidt O., Delaney L., O'Neill F., Court F., Rosenthal A., Fleetwood-Walker S. M., Davies A. M., Buchman V. L. (2003) Mol. Cell. Biol. 23, 8233–8245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rockenstein E., Mallory M., Hashimoto M., Song D., Shults C. W., Lang I., Masliah E. (2002) J. Neurosci. Res. 68, 568–578 [DOI] [PubMed] [Google Scholar]
- 42. Rockenstein E., Mallory M., Mante M., Sisk A., Masliaha E. (2001) J. Neurosci. Res. 66, 573–582 [DOI] [PubMed] [Google Scholar]
- 43. Marongiu R., Spencer B., Crews L., Adame A., Patrick C., Trejo M., Dallapiccola B., Valente E. M., Masliah E. (2009) J. Neurochem. 108, 1561–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ohi M., Li Y., Cheng Y., Walz T. (2004) Biol. Proced. Online 6, 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zheng Q. S., Braunfeld M. B., Sedat J. W., Agard D. A. (2004) J. Struct. Biol. 147, 91–101 [DOI] [PubMed] [Google Scholar]
- 46. Soper J. H., Roy S., Stieber A., Lee E., Wilson R. B., Trojanowski J. Q., Burd C. G., Lee V. M. (2008) Mol. Biol. Cell. 19, 1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lippincott-Schwartz J., Donaldson J. G., Schweizer A., Berger E. G., Hauri H. P., Yuan L. C., Klausner R. D. (1990) Cell 60, 821–836 [DOI] [PubMed] [Google Scholar]
- 48. Koch A., Thiemann M., Grabenbauer M., Yoon Y., McNiven M. A., Schrader M. (2003) J. Biol. Chem. 278, 8597–8605 [DOI] [PubMed] [Google Scholar]
- 49. Spillantini M. G., Crowther R. A., Jakes R., Hasegawa M., Goedert M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6469–6473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Engelender S., Kaminsky Z., Guo X., Sharp A. H., Amaravi R. K., Kleiderlein J. J., Margolis R. L., Troncoso J. C., Lanahan A. A., Worley P. F., Dawson V. L., Dawson T. M., Ross C. A. (1999) Nat. Genet. 22, 110–114 [DOI] [PubMed] [Google Scholar]
- 51. Ostrerova-Golts N., Petrucelli L., Hardy J., Lee J. M., Farer M., Wolozin B. (2000) J. Neurosci. 20, 6048–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shao J., Welch W. J., Diprospero N. A., Diamond M. I. (2008) Mol. Cell. Biol. 28, 5196–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. De Vos K. J., Allan V. J., Grierson A. J., Sheetz M. P. (2005) Curr. Biol. 15, 678–683 [DOI] [PubMed] [Google Scholar]
- 54. Loeb V., Yakunin E., Saada A., Sharon R. (2010) J. Biol. Chem. 285, 7334–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ward M. W., Rego A. C., Frenguelli B. G., Nicholls D. G. (2000) J. Neurosci. 20, 7208–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu T., Robotham J. L., Yoon Y. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2653–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen H., Chomyn A., Chan D. C. (2005) J. Biol. Chem. 280, 26185–26192 [DOI] [PubMed] [Google Scholar]
- 58. Fleming S. M., Salcedo J., Fernagut P. O., Rockenstein E., Masliah E., Levine M. S., Chesselet M. F. (2004) J. Neurosci. 24, 9434–9440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Greten-Harrison B., Polydoro M., Morimoto-Tomita M., Diao L., Williams A. M., Nie E. H., Makani S., Tian N., Castillo P. E., Buchman V. L., Chandra S. S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 19573–19578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Choi S. Y., Huang P., Jenkins G. M., Chan D. C., Schiller J., Frohman M. A. (2006) Nat. Cell Biol. 8, 1255–1262 [DOI] [PubMed] [Google Scholar]
- 61. Inoue T., Heo W. D., Grimley J. S., Wandless T. J., Meyer T. (2005) Nat. Methods 2, 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cole N. B., Murphy D. D., Grider T., Rueter S., Brasaemle D., Nussbaum R. L. (2002) J. Biol. Chem. 277, 6344–6352 [DOI] [PubMed] [Google Scholar]
- 63. Fortin D. L., Nemani V. M., Voglmaier S. M., Anthony M. D., Ryan T. A., Edwards R. H. (2005) J. Neurosci. 25, 10913–10921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Perrin R. J., Woods W. S., Clayton D. F., George J. M. (2000) J. Biol. Chem. 275, 34393–34398 [DOI] [PubMed] [Google Scholar]
- 65. Bertoncini C. W., Jung Y. S., Fernandez C. O., Hoyer W., Griesinger C., Jovin T. M., Zweckstetter M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1430–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Uversky V. N., Li J., Fink A. L. (2001) J. Biol. Chem. 276, 10737–10744 [DOI] [PubMed] [Google Scholar]
- 67. Kamp F., Exner N., Lutz A. K., Wender N., Hegermann J., Brunner B., Nuscher B., Bartels T., Giese A., Beyer K., Eimer S., Winklhofer K. F., Haass C. (2010) EMBO J. 29, 3571–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen H., Detmer S. A., Ewald A. J., Griffin E. E., Fraser S. E., Chan D. C. (2003) J. Cell Biol. 160, 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Niemann A., Ruegg M., La Padula V., Schenone A., Suter U. (2005) J. Cell Biol. 170, 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhang L., Zhang C., Zhu Y., Cai Q., Chan P., Uéda K., Yu S., Yang H. (2008) Brain Res. 1244, 40–52 [DOI] [PubMed] [Google Scholar]
- 71. Varkey J., Isas J. M., Mizuno N., Jensen M. B., Bhatia V. K., Jao C. C., Petrlova J., Voss J. C., Stamou D. G., Steven A. C., Langen R. (2010) J. Biol. Chem. 285, 32486–32493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. George J. M., Jin H., Woods W. S., Clayton D. F. (1995) Neuron 15, 361–372 [DOI] [PubMed] [Google Scholar]
- 73. Gallop J. L., Jao C. C., Kent H. M., Butler P. J., Evans P. R., Langen R., McMahon H. T. (2006) EMBO J. 25, 2898–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pandey A. P., Haque F., Rochet J. C., Hovis J. S. (2009) Biophys. J. 96, 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Epand R. F., Martinou J. C., Fornallaz-Mulhauser M., Hughes D. W., Epand R. M. (2002) J. Biol. Chem. 277, 32632–32639 [DOI] [PubMed] [Google Scholar]
- 76. Ulmer T. S., Bax A., Cole N. B., Nussbaum R. L. (2005) J. Biol. Chem. 280, 9595–9603 [DOI] [PubMed] [Google Scholar]
- 77. Bigay J., Gounon P., Robineau S., Antonny B. (2003) Nature 426, 563–566 [DOI] [PubMed] [Google Scholar]
- 78. Zimmerberg J., McLaughlin S. (2004) Curr. Biol. 14, R250–R252 [DOI] [PubMed] [Google Scholar]
- 79. Lee H. J., Choi C., Lee S. J. (2002) J. Biol. Chem. 277, 671–678 [DOI] [PubMed] [Google Scholar]
- 80. Daum G. (1985) Biochim. Biophys. Acta 822, 1–42 [DOI] [PubMed] [Google Scholar]
- 81. Sperka-Gottlieb C. D., Hermetter A., Paltauf F., Daum G. (1988) Biochim. Biophys. Acta 946, 227–234 [DOI] [PubMed] [Google Scholar]
- 82. Liu J., Dai Q., Chen J., Durrant D., Freeman A., Liu T., Grossman D., Lee R. M. (2003) Mol. Cancer Res. 1, 892–902 [PubMed] [Google Scholar]
- 83. Hovius R., Thijssen J., van der Linden P., Nicolay K., de Kruijff B. (1993) FEBS Lett. 330, 71–76 [DOI] [PubMed] [Google Scholar]
- 84. Yang Y., Ouyang Y., Yang L., Beal M. F., McQuibban A., Vogel H., Lu B. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7070–7075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Poole A. C., Thomas R. E., Andrews L. A., McBride H. M., Whitworth A. J., Pallanck L. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1638–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nemani V. M., Lu W., Berge V., Nakamura K., Onoa B., Lee M. K., Chaudhry F. A., Nicoll R. A., Edwards R. H. (2010) Neuron 65, 66–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Burré J., Sharma M., Tsetsenis T., Buchman V., Etherton M. R., Südhof T. C. (2010) Science 329, 1663–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Züchner S., Mersiyanova I. V., Muglia M., Bissar-Tadmouri N., Rochelle J., Dadali E. L., Zappia M., Nelis E., Patitucci A., Senderek J., Parman Y., Evgrafov O., Jonghe P. D., Takahashi Y., Tsuji S., Pericak-Vance M. A., Quattrone A., Battaloglu E., Polyakov A. V., Timmerman V., Schröder J. M., Vance J. M., Battologlu E. (2004) Nat. Genet. 36, 449–451 [DOI] [PubMed] [Google Scholar]
- 89. Alexander C., Votruba M., Pesch U. E., Thiselton D. L., Mayer S., Moore A., Rodriguez M., Kellner U., Leo-Kottler B., Auburger G., Bhattacharya S. S., Wissinger B. (2000) Nat. Genet. 26, 211–215 [DOI] [PubMed] [Google Scholar]
- 90. Fortin D. L., Troyer M. D., Nakamura K., Kubo S., Anthony M. D., Edwards R. H. (2004) J. Neurosci. 24, 6715–6723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Saha A. R., Hill J., Utton M. A., Asuni A. A., Ackerley S., Grierson A. J., Miller C. C., Davies A. M., Buchman V. L., Anderton B. H., Hanger D. P. (2004) J. Cell Sci. 117, 1017–1024 [DOI] [PubMed] [Google Scholar]
- 92. Krüger R., Kuhn W., Leenders K. L., Sprengelmeyer R., Müller T., Woitalla D., Portman A. T., Maguire R. P., Veenma L., Schröder U., Schöls L., Epplen J. T., Riess O., Przuntek H. (2001) Neurology 56, 1355–1362 [DOI] [PubMed] [Google Scholar]
- 93. Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R. L., Kim J., May J., Tocilescu M. A., Liu W., Ko H. S., Magrané J., Moore D. J., Dawson V. L., Grailhe R., Dawson T. M., Li C., Tieu K., Przedborski S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Narendra D., Tanaka A., Suen D. F., Youle R. J. (2009) Autophagy 5, 706–708 [DOI] [PubMed] [Google Scholar]
- 95. Clark I. E., Dodson M. W., Jiang C., Cao J. H., Huh J. R., Seol J. H., Yoo S. J., Hay B. A., Guo M. (2006) Nature 441, 1162–1166 [DOI] [PubMed] [Google Scholar]
- 96. Park J., Lee S. B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J. M., Chung J. (2006) Nature 441, 1157–1161 [DOI] [PubMed] [Google Scholar]
- 97. Exner N., Treske B., Paquet D., Holmström K., Schiesling C., Gispert S., Carballo-Carbajal I., Berg D., Hoepken H. H., Gasser T., Krüger R., Winklhofer K. F., Vogel F., Reichert A. S., Auburger G., Kahle P. J., Schmid B., Haass C. (2007) J. Neurosci. 27, 12413–12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sharon R., Bar-Joseph I., Frosch M. P., Walsh D. M., Hamilton J. A., Selkoe D. J. (2003) Neuron 37, 583–595 [DOI] [PubMed] [Google Scholar]
- 99. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) Nat. Cell. Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 100. Mori H., Kondo T., Yokochi M., Matsumine H., Nakagawa-Hattori Y., Miyake T., Suda K., Mizuno Y. (1998) Neurology 51, 890–892 [DOI] [PubMed] [Google Scholar]
- 101. Gispert S., Ricciardi F., Kurz A., Azizov M., Hoepken H. H., Becker D., Voos W., Leuner K., Müller W. E., Kudin A. P., Kunz W. S., Zimmermann A., Roeper J., Wenzel D., Jendrach M., García-Arencíbia M., Fernández-Ruiz J., Huber L., Rohrer H., Barrera M., Reichert A. S., Rüb U., Chen A., Nussbaum R. L., Auburger G. (2009) PLoS. One 4, e5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Weinreb P. H., Zhen W., Poon A. W., Conway K. A., Lansbury P. T., Jr. (1996) Biochemistry 35, 13709–13715 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.