Abstract
The SLC38 family of transporters has in total 11 members in humans and they encode amino acid transporters called sodium-coupled amino acid transporters (SNAT). To date, five SNATs have been characterized and functionally subdivided into systems A (SLC38A1, SLC38A2, and SLC38A4) and N (SLC38A3 and SLC38A5) showing the highest transport for glutamine and alanine. Here we present identification of a novel glutamine transporter encoded by the Slc38a7 gene, which we propose should be named SNAT7. This transporter has l-glutamine as the preferred substrate but also transports other amino acids with polar side chains, as well as l-histidine and l-alanine. The expression pattern and substrate profile for SLC38A7 shows highest similarity to the known system N transporters. Therefore, we propose that SLC38A7 is a novel member of this system. We used in situ hybridization and immunohistochemistry with a custom-made antibody to show that SLC38A7 is expressed in all neurons, but not in astrocytes, in the mouse brain. SLC38A7 is unique in being the first system N transporter expressed in GABAergic and also other neurons. The preferred substrate and axonal localization of SLC38A7 close to the synaptic cleft indicates that SLC38A7 could have an important function for the reuptake and recycling of glutamate.
Keywords: Amino Acid Transport, Glutamate, Glutamine, Neurobiology, Xenopus, SLC38, SNAT, SNAT7, System A, System N
Introduction
Solute carriers (SLCs),3 are the largest group of transporters in the human genome (1, 2). Many of the SLCs are coupled transporters with important functions in cellular processes, including roles as transporters for amino acids and neurotransmitter cycling, whereas others are passive transporters or exchangers (2–4). It has been suggested that there are close to 100 SLC transporters for amino acids in the human genome, of these around 60 have been characterized regarding substrate and the rest are postulated as probable amino acid transporters (5). This large number indicates the importance of having a regulated membrane transport of amino acids over the cell membrane as well as over mitochondrial and vesicular membranes. The SLCs have been categorized into at least 46 different families with varied biochemical properties (5) and recently two more families have been identified (6). In mammals, phylogenetic classification of SLCs show that the SLC proteins form four major phylogenetic groups, termed α, β, γ, and δ, with proteins in each group having a common evolutionary origin. The β-group includes the SLC32, SLC36, and SLC38 families and is the second largest phylogenetic cluster of amino acid transporters. Some members of the β-group are known to be expressed in the brain and most are located in the plasma membrane, whereas SLC32A1 is found in neuronal vesicles. Yet, others are still orphans and very little is known about their tissue distribution, functional characteristics, or substrate specificity (5, 7).
The SLC38 family consists of 11 members. The orphans SLC38A7–11 were only recently identified and have thus far not been characterized regarding their substrate of transport (7), whereas the other members, SLC38A1–6, are functionally classified as sodium-coupled neutral amino acid transporters, also known as sodium-coupled amino acid transporters (SNATs). The SNAT1 (SLC38A1 (8–9)), SNAT2 (SLC38A2 (10–13)), and SNAT4 (SLC38A4 (14, 15)) have been further classified into system A, whereas SNAT3 (SLC38A3) and SNAT5 (SLC38A5) belong to system N transporters (16–18). The system N/A classification depends on the functional properties and patterns of substrate recognition. The SNAT6 (SLC38A6) (19) and other more recently discovered members of the family have not yet been classified according to the N/A system. All the SNAT genes, except SNAT4 (14), are expressed in the brain, with SNAT1 (8, 20) and SNAT2 (21) expressed both in astrocytes and neurons, whereas SNAT3 (16) and SNAT5 (22) are located in glial cells. No exact cellular localization of the protein, or substrate profile, has been reported for the orphan members of the SLC38 family. l-Glutamine is a favored substrate for most members throughout the SLC38 family, and it is suggested that the transporters are part of the glutamine-glutamate cycle in the CNS (16, 18, 23, 24).
The non-essential amino acid glutamate serves as the major excitatory neurotransmitter in the brain. The glutamine-glutamate cycle involves synthesis, release, and recycling of the synaptic transmitter glutamate in both neurons and glial cells to sustain the glutamate neurotransmitter pool and another key function is to circumvent the neurotoxic properties of glutamate (25–27). A number of both known and unknown glutamate and glutamine transporters are involved in this metabolic cycle. However, the exact contribution of the SLC38 proteins in the glutamine-glutamate cycle is still unclear. Several lines of more recent evidence, showing mainly a lack of expression in the nerve terminals, suggests that none of the known SNATs are involved in the glutamate-glutamine cycle in neurons (28, 29).
In this study we report the functional characterization of the SLC38A7 transporter, one of the previously orphan members in the SLC38 family, which we suggest be named SNAT7. The SLC38A7 was found to be expressed in the majority of neurons, both in GABAergic and other neurons, in brain and spinal cord. Functional characterization, by overexpression in Xenopus laevis oocytes, showed that SLC38A7 preferably mediates transport of l-glutamine and l-histidine, among other substrates. We showed that SLC38A7 has a profile resembling the known system N transporters, based on substrate profile, their sodium-dependent transport, and the tissue expression pattern. We also show that SLC38A7 is expressed on axons of neurons, which together with the substrate profile, suggest that SLC38A7 may play a role in sustaining the glutamate neurotransmitter pool in the brain through the glutamine-glutamate cycle.
EXPERIMENTAL PROCEDURES
Tissue Collection and Sectioning
Animal care procedures for C57Bl6/J adult male mice were approved by the local ethical committee in Uppsala and followed the guidelines of European Communities Council Directive (86/609/EEC). Adult male C57Bl6/J mice (Taconic M&B, Denmark) were intraperitoneally injected with a 1:1 mixture of Dormitor (Medetomidine hydrochloride, 70 μg/g body weight, Orion Pharma, Finland) and Ketalar (ketamine hydrochloride, 7 μg/g body weight, Pfizer). Transcardial perfusion was then performed through the left ventricle with phosphate-buffered saline (PBS) followed by 4% formaldehyde (HistoLab, Sweden). The brain was excised and stored in 4% formaldehyde overnight. For free floating tissue sections, the brain was washed in PBS, embedded in 4% agarose and sectioned (70 μm) on a Leica VT1000S vibratome (Leica Microsystems, Germany). Sections were then dehydrated through a series of methanol washes and stored in 100% methanol at −20 °C until further processing. For paraffin-embedded tissue sections the brain was fixed in zinc-formalin (Richard-Allan Scientific) for 18–24 h at 40 °C before dehydration and paraffin infusion (Tissue-Tek vacuum infiltration processor; Miles Scientific). The sections were cut (7 μm) using a Microm 355S STS cool cut microtome, attached on Superfrost Plus slides (Menzel-Gläser, Germany), dried overnight at 37 °C, and stored at 4 °C until use.
Design and Synthesis of RNA Probes
Antisense probe was synthesized from the mouse Slc38a7 EST clone ID 4009949 (Invitrogen). The Slc38a7 gene was cloned into a pcDNA3.1/FLAG vector using standard procedures. The coding sequence was amplified briefly by using forward (5′-GATCGAATTCGGAGCCCAGGTCAGCATCAA-3′) and reverse (5′-GATCCTCGAGTCAGGCCAAGAGATCCACAAAAA-3′) primers with Platinum Pfx proofreading DNA polymerase (Invitrogen), digested using EcoRI and XhoI (Fermentas, Canada), and cloned into the pcDNA3.1/FLAG vector (30) using T4 ligase (Invitrogen). This gave the construct pcDNA3.1/FLAG/Slc38a7. Plasmid cDNA was prepared with the JETstar 2.0 Plasmid Purification Midi Kit/50 (Genomed, Germany), the concentration was measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies), and the clone was sequenced (Eurofins MWG Operon, Germany) and verified correct. The plasmid was cut with SmaI (Fermentas, Canada), and the probe was synthesized using 1 μg of cleaved vector as template with SP6 RNA polymerase in the presence of digoxigenin-11-UTP (Roche Diagnostics). The digoxigenin-labeled mouse Slc38a7 (742 bp) probe was then quantified and controlled for integrity using the Experion RNA StdSens Analysis kit on an Experion automated electrophoresis system (Bio-Rad) and stored at −80 °C.
In Situ Hybridization on Free Floating Sections
Non-radioactive in situ hybridization was performed on free floating mouse brain sections by stepwise rehydration from 100% methanol to 0.1% PBT (PBS with 0.1% Tween 20 (Sigma)) and bleached with 6% H2O2 in PBT. The sections were permeabilized with 0.5% Triton X-100 (Sigma), digested in 20 μg/ml of proteinase K (Invitrogen), and post-fixed in 4% formaldehyde (HistoLab, Sweden) with PBT washes between all steps. Sections were then pre-hybridized at 55 °C in hybridization buffer (50% formamide, 5 × SSC, pH 4.5, 1% SDS, 50 μg/ml of tRNA (Sigma), 50 μg/ml of heparin (Sigma), and 0.1% diethyl pyrocarbonate-treated water). The digoxigenin-labeled Slc38a7 probe (1 μg/ml) diluted in hybridization buffer was heat denatured at 80 °C, cooled on ice, and added to the sections for hybridization overnight at 55 °C. The sections were repeatedly washed in the first buffer (50% formamide, 2 × SSC, pH 4.5, 0.1% Tween 20, and 0.1% diethyl pyrocarbonate-treated water) and then the second buffer (50% formamide, 0.2 × SSC, pH 4.5, 0.1% Tween 20, and 0.1% diethyl pyrocarbonate-treated water) at 55 °C. TBST (0.1% Tween 20 in Tris-buffered saline) was used for additional washing prior to a 2-h incubation in blocking solution (1% blocking reagent (Roche Diagnostics) in TBST) followed by incubation in the anti-digoxigenin antibody conjugated to alkaline phosphatase (Roche Diagnostics) diluted 1:5000 in blocking solution overnight at 4 °C. The sections were repeatedly washed in TBST with 2 mm levamisole (Thermo Fisher Scientific) followed by 10 min in NTMT with 2 mm levamisole (100 mm NaCl, 10 mm Tris-HCl, pH 9.5, 50 mm MgCl2, and 0.1% Tween 20) before color development with BM-Purple AP enzyme substrate (Roche Diagnostics) at 37 °C. After mounting in DTG media with antifade (diazabicyclo(2.2.2)octane in glycerol and Tris) the sections were photographed with a Leica MZ16F microscope connected to a Leica DFC300 FX camera (Leica Microsystems, Germany) with the Leica FireCam software.
Fluorescent Immunohistochemistry on Paraffin Sections
Sections were deparaffinized in X-tra Solv (Medite Histotechnik, Germany) and rehydrated through a series of ethanol solutions (100, 95, 75, 50, and 25%) dissolved in H2O, followed by PBS washes. Antigen retrieval was performed by heating the sections to 100 °C in 0.01 m citric acid, pH 6.0, (Sigma) for 10 min. The sections were then washed in PBS, placed in a humidified chamber, and incubated with primary antibody rabbit anti-SLC38A7 (1:200, custom-made polyclonal antibody by Innovagen), mouse anti-NeuN (1:400, Millipore, Sweden), chicken anti-GFAP (1:400, Abcam, United Kingdom), mouse anti-MAP2 (1:500, Sigma), and mouse anti-synaptophysin (1:200, BD Transduction Laboratories) diluted in supermix (Tris-buffered saline, 0.25% gelatin, 0.5% Triton X-100) overnight at 4 °C. After washes in PBS, sections were incubated in 1:200 diluted secondary antibodies (donkey anti-rabbit-594, donkey anti-rabbit-488, goat anti-mouse-594, goat anti-mouse-488, and goat anti-chicken-488 (Invitrogen)) in supermix for 2 h. The sections were further washed in PBS, and then stained with DAPI (1:2500) for 5 min prior to mounting using Mowiol antifade media (33.3% glycerol, 16.7% Mowiol 4–88, 0.02% Thimerosal, and 2% n-propyl gallate in PBS (all from Sigma)). Sections were photographed using a fluorescent microscope (Zeiss Axioplan2 imaging) connected to a camera (AxioCam HRm) with the Carl Zeiss AxioVision version 4.7 software.
Combined in Situ Hybridization/Immunohistochemistry
In situ hybridization was combined with fluorescent immunohistochemistry on paraffin sections. The sections were, as previously described, deparaffinized, rehydrated, post-fixed, and digested in proteinase K. After refixation and washes in PBS, slides were acetylated (1.3% trietholamine (Sigma), 0.06% HCl (Sigma), and 2% acetic anhydride (Fluka, Switzerland) diluted in water). Sections were then permeabilized in 1% Triton X-100 and subsequent rinsed in PBS prior pre-hybridized in hybridization buffer (50% formamide (Sigma), 5× SSC, pH 4.5, 5 × Denhardt's, 250 μg/ml of yeast transfer RNA (Sigma), 500 μg/ml of sheared salmon sperm DNA (Ambion) in 0.1% diethyl pyrocarbonate-treated water) for 2 h at 55 °C. The denatured Slc38a7 probe (0.7 μg/0.5 ml) was added for hybridization overnight at 55 °C. The following day, sections were washed in warm 5× SSC, incubated in 0.2× SSC for 1 h at 55 °C, washed in 0.2× SSC, and pre-blocked in blocking solution (0.1 m Tris-HCl, pH 7.5, 0.15 m NaCl, and 10% albumin bovine serum (Sigma)) for 1 h before incubation overnight in alkaline phosphatase-conjugated anti-digoxigenin Fab fragments (Roche Diagnostics) diluted 1:2500 in blocking solution. The next day, sections were washed in TBST with 2 mm levamisole and NTMT with 2 mm levamisole, as previously described, and color was developed in Fast Red solution (Roche Diagnostics). After color development, the sections were treated as described in the fluorescent immunohistochemistry on paraffin sections protocol, starting after the antigen retrieval step. Primary antibody dilutions were: 1:400 goat anti-GAD67 (Santa Cruz Biotechnology), 1:400 chicken anti-GFAP, 1:400 rabbit anti-NeuN (Chemicon/Millipore), and 1:2500 DAPI (Sigma). Secondary antibodies (Invitrogen) were diluted: 1:200 donkey anti-goat-488, 1:200 goat anti-chicken-488, and 1:200 goat anti-rabbit-488. Sections were photographed and analyzed as previously described.
Cloning into pβ/RN3P Expression Vector
A full-length clone for the Slc38a7 gene was synthesized from the mouse Slc38a7 clone (Invitrogen number 4009949) and inserted in the pcDNA3.1/FLAG vector, as described under “Design and Synthesis of RNA Probes”. The coding sequence was amplified using forward (5′-GATCGAATTCACAGCCACCGGAATGGCCCAGGTCAGCATCAA-3′) and reverse (5′-GATCGCGGCCGCTCAGGCCAAGAGATCCACAAAAA-3′) primers with Platinum Pfx proofreading DNA polymerase (Invitrogen), digested using EcoRI and NotI (Fermentas, Canada), and cloned into pβGFP/RN3P vector (31, 32) by excising the original GFP using EcoRI and NotI and inserting the coding sequence of Slc38a7 using T4 ligase (Invitrogen) into the same sites. Plasmids were extracted and sequenced as above. This gave the construct pβ/RN3P/Slc38a7, which was prepared and sequenced (Eurofins MWG Operon, Germany) to assure integrity.
mRNA Synthesis for Cytosol Injections
The pβ/RN3P/Slc38a7 expression vector was digested with SfiI (Fermentas, Canada) and the concentration was determined using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies). A mMessage Machine T3 kit and the Megaclear kit (Ambion) was used for capped RNA transcription and purification according to the manufacturer's instructions. The concentration was quantified and controlled with the Experion RNA StdSens Analysis kit on an Experion automated electrophoresis system (Bio-Rad) prior to storage of samples at −80 °C.
Oocyte Preparation and Microinjection
Adult unfertilized female African clawed frogs, X. laevis (Nasco), were housed in a controlled environment (21 °C, 12:12 light/dark cycle) with access to salmon pellets (Aller Aqua, Denmark). All animal experiments were approved by the North Stockholm Ethics Committee for Animal Experiments at the Karolinska Institute. Physical anesthesia through hypothermia was used to anesthetize the frog during surgery for removal of ∼3 ml of ovarian tissue. The ovarian tissue was rinsed in OR2 buffer (82 mm NaCl, 2.5 mm KCl, 1 mm CaCl2, 1 mm MgCl2, 1 mm Na2HPO4, and 5 mm HEPES, pH 7.8) and cut into smaller pieces. The oocytes were then rinsed prior to defolliculation by treatment with 7 units of research grade LiberaseTM (Roche Diagnostics) diluted in ∼12.5 ml of OR2 buffer for 1 h and 50 min during slow rotation at 19 °C. After additional rinsing the stage V–VI oocytes were sorted and incubated overnight in OR2 buffer with 10 μg/ml of gentamicin (Sigma) at 19 °C. The oocytes were microinjected into the cytoplasm with ∼36.8 nl of in vitro transcribed Slc38a7 mRNA (∼25 ng) with a micromanipulator (Leica Microsystems, Germany) connected to an microinjection pipette Nanoject II autonanoliter injector (Drummond Scientific Co.) using a Leica MZ75 microscope with a table movable in two dimensions. The injected and the non-injected oocytes, used as controls, were incubated in OR2 buffer for 48 h at 19 °C for overexpression of the SLC38A7 protein by translation.
Immunohistochemistry on Injected Oocytes
The SLC38A7 overexpressing and control oocytes were embedded in Killik (Bio-Optica, Italy) and frozen to −80 °C prior to sectioning in 12-μm thin sections in a cryostat (HM 560, Microm International, Germany). The immunohistochemistry was performed by first fixating the oocytes in 4% paraformaldehyde for 15 min. The sections were then rinsed 3 times with PBS and pre-blocked in supermix for 1 h. The oocytes were incubated with our custom-made primary SLC38A7 antibody diluted 1:200 in supermix overnight at 4 °C. The next day the oocytes were washed in PBS and incubated in secondary donkey anti-rabbit-488 (Invitrogen) diluted 1:400 in supermix for 2 h in RT. The oocytes were mounted and photographed as described for fluorescent immunohistochemistry on paraffin sections.
Transport Assay
The SLC38A7 overexpressing and control oocytes were transferred onto a 96-well plate with one oocyte/well and incubated in KRH buffer (118 mm NaCl, 4.8 mm KCl, 1.2 mm MgSO4, 2.5 mm CaCl2, 10 mm HEPES, adjusted to pH 7.8) for 15 min. The l-glutamine concentration for maximum transport analysis was performed with total concentrations of 0.2, 0.4, 1.2, 5.2, 25.2, and 125.2 μm l-glutamine of which 0.2 μm was l-[3H]glutamine (PerkinElmer Life Sciences) at a fixed concentration and the remaining was cold l-glutamine. The screen with 3H-labeled l-amino acids l-glutamine, l-arginine, l-alanine, l-serine, l-lysine, and l-aspartic acid, was performed by incubating the oocytes for 60 min in a total concentration of 1.2 μm amino acid (0.2 μm 3H-labeled amino acid with 1 μm un-labeled amino acid). During the time course the oocytes were incubated in 0.2 μm l-[3H]glutamine (PerkinElmer Life Sciences) diluted in KRH buffer for 2, 5, 10, 20, 30, 40, 60, and 120 min. The sodium ion dependence and the lithium ion tolerance of the l-glutamine transport was investigated by adding 0.2 μm l-[3H]glutamine diluted in three different buffers to the oocytes: KRH buffer and KRH with 118 mm NaCl replaced by either equimolar LiCl (Sigma) or choline chloride (BDH, United Kingdom). Substrate preference analysis was measured in a competition study where 0.2 μm l-[3H]glutamine was used as a tracer together with 1 μm unlabeled l-glutamine in the presence of a number of 0.12 mm competing unlabeled l-amino acids, α-(methyl)aminoisobutyric acid (MeAIB), or neurotransmitters (pH adjusted to 7.8, all substrates were from Sigma or Fluka, Switzerland). During all tests the oocytes were incubated in the 3H-labeled substrate for 1 h, except the time course, and the transport was stopped with ice-cold KRH buffer washes. The cells were lysed in 10% SDS and transferred into scintillation tubes with scintillation solution (Zinsser Analytic, Germany) for counting the radioactive decay in a Packard 1900CA Tri-carb liquid scintillation analyzer (Beckman). All functional analyses were performed in groups of 7–12 oocytes per assay at 29 °C, with the exception of substrate preference analysis where 12–52 oocytes were used in each group. All experiments were repeated at least 3 times. The p values for the collected data were presented as unpaired t tests with a 95% confidence interval of mean ± S.D. between uptake into SLC38A7 overexpressing oocytes and endogenous uptake into control oocytes (non-injected). To visualize the substrate preference for SLC38A7 as specific transport, normalized values were calculated by dividing the 1.2 μm radiolabeled l-glutamine controls without competing substrate, with the actual value for the competing substrate per oocyte.
Western Blot
We performed Western blot analysis of SLC38A7 in brain tissue from adult, male C57Bl6/J mice (Taconic M&B, Denmark). Briefly, tissue was homogenized in homogenization buffer (50 mm Tris, 150 mm NaCl, 4 mm MgCl, 0.5 mm EDTA, 2% Triton X-100, and 1 mm protease inhibitor PMSF (Sigma) diluted in isopropyl alcohol). Protein concentrations were determined by the protein assay DC (Bio-Rad) according to the manufacturer's instructions. Equal amounts of protein (200 μg or 13 μg/μl) were separated, together with PageRuler prestained protein ladder (Fermentas, Canada), on a Mini-Protean TGX gel (4–10%, Bio-Rad) in running buffer (0.1% SDS, 0.025 Tris base, and 0.192 m glycine) by gel electrophoresis. The proteins were transferred to a Immobilon-P PVDF membrane (Millipore) in transfer buffer (0.025 Tris base, 0.192 m glycine, and 20% methanol) and pre-blocked for 1 h in blocking buffer (5% nonfat dry milk (Bio-Rad) diluted in 1.5 m NaCl, 0.1 m Tris, 0.05% Tween 20, pH 8.0). The membrane was cut in the middle, giving two membranes with equally loaded protein samples. One-half of the membrane was hybridized with the primary antibody against SLC38A7 (diluted 1:200, rabbit anti-SLC38A7, Innovagen, Sweden). The other half of the membrane was hybridized with SLC38A7 primary antibody pre-blocked with an excess of the same synthetic peptide (sequence (NH2-)CVMSKEPDGASGSPW(-CONH2)) that was used to generate the antibody. The hybridization was then performed overnight at 4 °C. After washes in water, the membranes were incubated for 1 h with horseradish peroxidase-conjugated secondary antibody (diluted 1:10000, goat anti-rabbit, Invitrogen) followed by detection with the enhanced chemiluminescent (ECL) method. The membranes were incubated for 3 min in a 1:1 mixture of luminol/enhancer and peroxidase buffer solutions (Immun-Star HRP, Bio-Rad) and developed on high performance chemiluminescence film (GE Healthcare).
Localization of FLAG-SLC38A7 in Transfected Cells
Two cell culture dishes of 50% confluent HEK 293 cells were transiently transfected with 5.3 μg of the pcDNA3.1/FLAG/Slc38a7 vector (described under “Experimental Procedures”) and with a mock vector as control, using fast forward transfection with the Attractene transfection reagent according to the manufacturer's instructions (Qiagen). The successful transfection was verified with immunohistochemistry and Western blot 48 h post-transfection. Poly-l-lysine- (Sigma) coated (10 μg/ml) slides were added to the bottom of the cell culture dishes prior transfection, and immunohistochemistry was performed by first rinsing the slides in PBS. The cells were then fixated in 4% paraformaldehyde, pre-blocked in supermix (Tris-buffered saline, 0.25% gelatin, 0.5% Triton X-100), and incubated in primary antibody rabbit anti-SLC38A7 (1:200, Innovagen, Sweden) and mouse anti-FLAG M2 (1:200, Sigma) diluted in supermix overnight at 4 °C. The next day, cells were repeatedly rinsed in PBS and incubated in secondary antibody goat anti-rabbit-594 and goat anti-mouse-488 (Invitrogen) for 2 h. After washing in PBS, cells were stained with DAPI (1:2500) and mounted in DTG DABCO/Tris/Glycerol (pH 8.6) or 1,4-diazabicyclo[2.2.2] octane/Tris/Glycerol (pH 8.6). Images were captured on a Zeiss LSM 510 Meta confocal microscope (Zeiss, Germany) and analyzed with ImageJ (33). The Western blot was performed as described under “Western Blot” under “Experimental Procedures” with the following changes. Cellular lysates were performed by washing the transfected cells in PBS prior to incubation in 500 μl of lysis buffer (50 mm Tris, pH 8.0, 1% Triton X-100, 150 mm NaCl, and 1 mm PMSF (Sigma) diluted in isopropyl alcohol) for 10 min at 4 °C. The cells were scraped from the dishes, transferred to tubes, and disrupted by pipetting. The disrupted cells were then centrifuged at 13,000 × g for 15 min at 4 °C. Both the supernatant and pellet were collected, concentration was determined and used in the Western blot. Briefly, crude extraction of proteins (50 μg) were separated on a gel and transferred to a membrane. The membrane was cut in the middle, giving two membranes with equally loaded protein samples. One-half of the membrane was hybridized with the primary antibody rabbit anti-SLC38A7 (1:200, Innovagen), whereas the other half of the membrane was hybridized with the mouse anti-FLAG (M2, 1:1500, Sigma). The membranes were incubated in the secondary antibody goat anti-rabbit-HP or the goat anti-mouse-HP (1:10000, Invitrogen) followed by detection with the enhanced chemiluminescent (ECL) method.
RESULTS
Slc38a7 Has Abundant mRNA Expression in the Brain
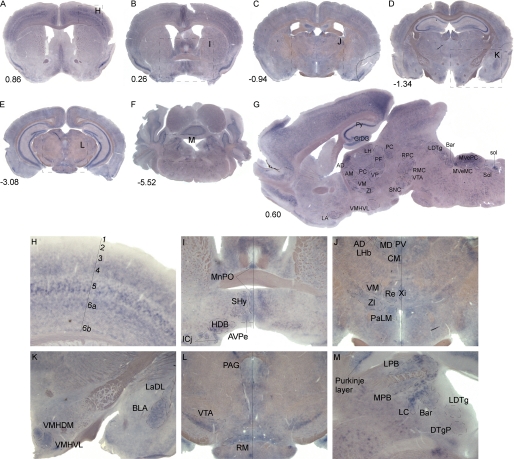
A detailed in situ hybridization tissue mapping with the probe against the Slc38a7 mRNA was performed in mouse brain. The probe design and quality control of the mRNA probe synthesis from the cDNA3.1/FLAG/Slc38a7 vector are shown under “Experimental Procedures,” and supplemental Fig. 1, A and B, respectively. The digoxigenin-labeled antisense Slc38a7 mRNA probe hybridized sequence specific to 742 bp, corresponding to 5 exons of the gene. The mRNA probe was controlled, showing a single peak of undergraded mRNA at ∼750 bp. The in situ hybridization method confirmed high levels of Slc38a7 expression in gray but not white matter. The screen showed that the overall expression pattern was high in the hippocampus, especially in the granular layer of dentate gyrus cells and the pyramidal cell layer of the hippocampus (Py), amygdala, thalamus, hypothalamus, in the layer of Purkinje cells in the cerebellum and the layers of cortex. Particular strong expression was found in neurons of the ventromedial hypothalamus, basolateral amygdala, ventral tegmental area, and locus coeruleus, see Fig. 1, A–M.
FIGURE 1.
Slc38a7 mRNA expression in mouse brain. Floating in situ hybridization using 1 μg/ml of digoxigenin-labeled mouse Slc38a7 probe on coronal sections with bregma numbers (A–F), close up pictures (H–M), and the sagittal section with interaural lateral number (G). Abbreviations and described brain regions are collected from Franklin and Paxinos (59). High Slc38a7 expression is found in G; dentate gyrus cells (GrDG), pyramidal cell layer of hippocampus (Py), thalamic nucleus (AD), lateral hypothalamic area (LH), parafascicular thalamic nucleus (PF), paracentral thalamic nucleus (PC), ventral pallidum (VP), ventromedial thalamic nucleus (VM), anteromedial thalamic nucleus (AM), zona incerta (ZI), ventromedial hypothalamic nucleus (VMH), ventrolateral part (VMHVL), lateroanterior hypothalamic nucleus (LA), substantia nigra pars compacta (SNC), ventral tegmental area (VTA), red nucleus, magnocellular part (RMC), red nucleus, parvicellular part (RPC), laterodorsal tegmental nucleus (LDTg), Barrington's nucleus (Bar), medial vestibular nucleus, magnocellular part (MVeMC), medial vestibular nucleus, parvicellular part (MVePC), nucleus of the solitary tract (Sol), and solitary tract (sol) are indicated. H, cortical layers 3–5, 6a, and 6b. I, median preoptic nucleus (MnPO), septohypothalamic nucleus (SHy), nucleus of the horizontal limb of the diagonal band (HDB), islands of Calleja (Icj,) and anteroventral periventricular nucleus (AVPe). J, anterodorsal thalamic nucleus (AD), lateral habenular nucleus (LHb), mediodorsal thalamic nucleus (MD), paraventricular thalamic nucleus (PV), central medial thalamic nucleus (CM), ventromedial thalamic nucleus (VM), zona incerta (ZI), reuniens thalamic nucleus (Re), xiphoid thalamic nucleus (Xi), and lateral magnocellular part (PaLM). K, ventromedial hypothalamic nucleus, central part (VMHDM), ventromedial hypothalamic nucleus, ventrolateral part (VMHVL), basolateral amygdaloid nucleus, anterior part (BLA), and lateral amygdaloid nucleus, dorsolateral part (LaDL). L, periaqueductal gray (PAG), ventral tegmental area (VTA), and retromammillary nucleus (RM). M, Purkinje layer, lateral parabrachial nucleus (LPB), medial parabrachial nucleus (MPB), locus coeruleus (LC), Barrington's nucleus (Bar), dorsal tegmental nucleus, pericentral part (DTgP), and laterodorsal tegmental nucleus (LDTg).
Expression of SLC38A7 mRNA and Protein in Excitatory and Inhibitory Neurons
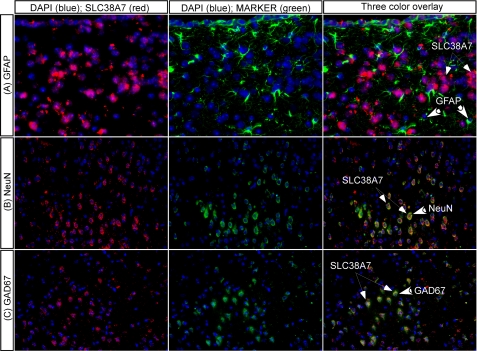
In situ hybridization combined with immunohistochemistry showed co-localization of Slc38a7 mRNA with neuronal markers, indicating expression of Slc38a7 in both excitatory and inhibitory neurons in the mouse brain. Co-staining with antibody against glial fibrillary acidic protein (GFAP), a marker predominantly found in glial cells (34), did not show overlap with the Slc38a7 mRNA expression (Fig. 2A). The co-localization of Slc38a7 mRNA and neuronal marker NeuN, a marker of neuron-specific nuclear proteins of most neuronal cell types (35), showed on the other hand, highly overlapping expression (Fig. 2B). All cells stained with NeuN were co-localized with Slc38a7, but not all cells expressing the Slc38a7 mRNA were stained using the neuronal marker. We used GAD67, a marker for inhibitory GABAergic neurons (36), combined with Slc38a7 mRNA expression to confirm neuronal expression (Fig. 2C). The Slc38a7 expression combined with GAD67 showed highly overlapping expression.
FIGURE 2.
Neuronal localization of Slc38a7. In situ hybridization combined with immunohistochemistry on 7-μm paraffin mouse brain sections for co-localization of Slc38a7 mRNA and known cellular markers. Co-expression with the glial cell marker GFAP are shown on the first row (A), the neuronal protein markers NeuN are shown on the second row (B), and GAD67 on the third row (C). The cell nuclei marker DAPI was stained in blue, the Slc38a7 mRNA in red, whereas other markers were labeled in green. The mRNA expression of Slc38a7 only shows overlap with the excitatory and inhibitory neuronal marker (×20 objective), and not with the glial cell marker (×40 objective). The thick arrows indicate cells expressing the marker and thin arrows Slc38a7 expressing cells.
We generated a rabbit polyclonal antibody and used Western blot analysis for investigating the specificity, see supplemental Fig. S2A. Additional specificity studies were performed by immunohistochemistry and Western blot on transfected cells expressing a FLAG-SLC38A7 fusion protein, see supplemental Fig. S2, B and C.
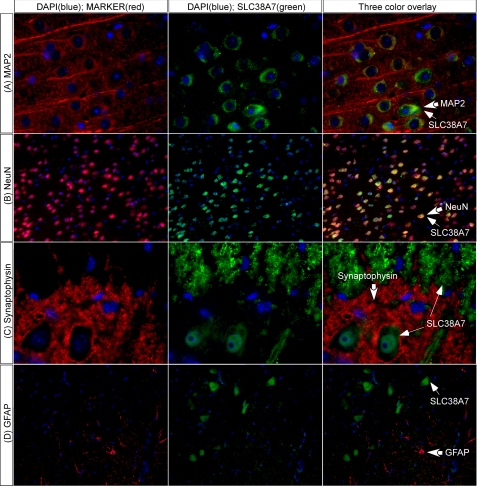
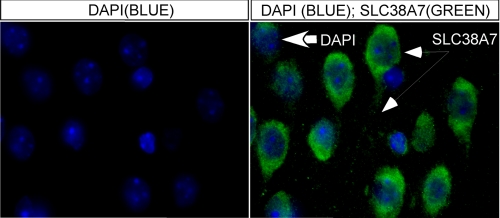
We performed double immunohistochemistry on mouse brain and spinal cord for localization of the SLC38A7 protein in the CNS (see Fig. 3). The protein co-localization was performed with SLC38A7 antibody and markers for neurons, NeuN, and the neuron-specific cytoskeletal protein marker microtubule-associated protein 2 (MAP2), astrocytes, the GFAP marker, and vesicles, the marker of axonal nerve terminals and synapses synaptophysin (37, 38). The SLC38A7 protein showed neuronal expression in brain, and co-localized with both MAP2 and NeuN (Fig. 3, A and B). The lack of co-localization with synaptophysin in spinal cord suggests that SLC38A7 does not overlap with nerve terminals in the spinal cord but the SLC38A7 protein is found in neuronal axons (Fig. 3C). SLC38A7 expression did not overlap with cells positive for GFAP, neither in brain (data not shown) nor spinal cord (Fig. 3D). Fig. 4 shows cellular localization of the SLC38A7 protein, with high expression in the soma and axon.
FIGURE 3.
SLC38A7 protein expression in neurons. Immunohistochemistry on adult mouse brain (row A and row B at bregma −1.82) and spinal cord (row C and row D at lumbar area L1) sections using SLC38A7 antibody stained in green and blue cell nucleus staining with 4′,6-diamidino-2-phenylindole (DAPI) (A–D). The antibody markers are stained in red; A, microtubule-associated protein 2 (MAP2); B, neuronal nuclei protein (NeuN); C, synaptophysin; and D, GFAP. A, the first row shows co-localization of SLC38A7 and MAP2 in brain cortex illustrated neuronal staining (×63 objective). B, the second row visualize that SLC38A7 is predominantly expressed in neurons. NeuN was used to stain most neuronal cell types, which is shown as highly overlapping staining (×20 objective). C, the third row demonstrates the presynaptic vesicle protein synaptophysin, found in all nerve terminals. The SLC38A7-labeled cells were found in motor neurons and neuronal axons but not in the nerve terminals in the spinal cord (×63 objective). D, in the last row, GFAP was used to stain glial cells, which show a line around the walls of the ventricle and the walls of the whole spinal cord. The image illustrating the SLC38A7 transporter and GFAP showed no co-localization (×20 objective). Thick arrows indicate cells labeled with the marker and thin arrows indicate the SLC38A7 protein expressing cells.
FIGURE 4.
Cellular localization of SLC38A7. The cellular localization of SLC38A7 was shown by using immunohistochemistry with the primary SLC38A7 antibody, shown in green on the right panel (thin arrows) and with blue DAPI staining on both the left and right panels (thick arrows), performed on 12-μm paraffin-embedded mouse brain sections at bregma −1.82. The high magnification (×100 objective) fluorescent microscope image illustrates SLC38A7 protein expression in the cortex in the neuronal membrane of the cell body, the axon, and the cytoplasm around the DAPI-stained nuclei.
Overexpression of SLC38A7 in X. laevis Oocytes
Functional characterization was performed by overexpressing SLC38A7 in oocytes to identify coupled transport properties and the preferable substrates of transport. The cloned pβ/RN3P/Slc38a7 vector, containing a β1-globin ribosome binding site for higher protein expression efficiency, was used for synthesis of mRNA (supplemental Fig. S1C) and quality controlled to verify that the mRNA was pure and not degraded (supplemental Fig. S1D). The mRNA was microinjected in oocytes and translated to the SLC38A7 protein. Immunohistochemistry on Slc38a7-injected and non-injected oocytes, with the SLC38A7 antibody, showed the overexpressed transporter to be localized close to the cell membrane of the injected oocytes, indicating SLC38A7 expression in the membrane (supplemental Fig. S1E).
l-Glutamine Uptake by SLC38A7 Increases with Time
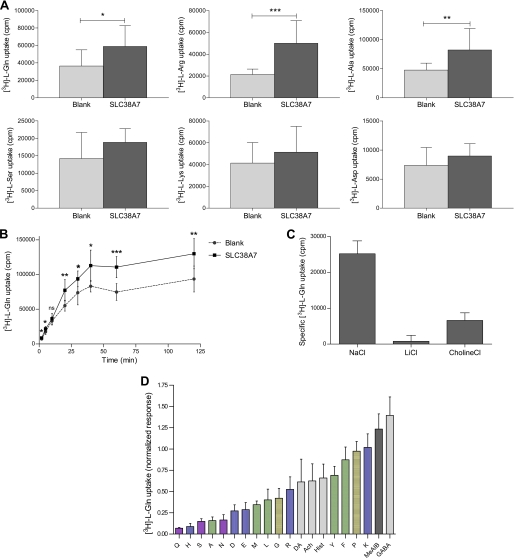
We investigated the characteristics of SLC38A7 by measuring radiolabeled substrate uptake in SLC38A7 expressing oocytes and control oocytes. A screen with 3H-labeled l-amino acids (l-glutamine, l-arginine, l-alanine, l-serine, l-lysine, and l-aspartic acid) revealed that the uptake for SLC38A7 was specific for l-glutamine, l-arginine, and l-alanine (Fig. 5A). The amino acid l-glutamine was chosen as a radioactive substrate for the subsequent experiments. A time course showed increasing amounts of l-glutamine uptake into SLC38A7 expressing oocytes over time (Fig. 5B). At least 2 min of l-glutamine uptake was needed to see a significant difference (p < 0.05) between uptake into SLC38A7 expressing oocytes and endogenous uptake into control oocytes. We found that the specific l-glutamine uptake into SLC38A7 expressing cells was increased with longer incubation times, but no drastic increase was seen between 60 and 120 min of incubation. The specific l-glutamine uptake at 2 min was only 4% of the specific uptake at 60 min. To have a large dynamic range when measuring specific uptake (by subtracting the counts for the control oocytes from the Slc38a7-injected oocytes), 60 min of incubation was chosen for all uptake assays. This indicated that SLC38A7 is a relatively low capacity transporter.
FIGURE 5.
Time course, sodium dependence, and substrate profile of SLC38A7. A, screen of 3H-labeled l-amino acid uptake into 38A7 expressing and control oocytes (mean ± S.D., n = 9–11 oocytes). The first row shows l-glutamine, l-arginine, and l-alanine, whereas the second row shows l-serine, l-lysine, and l-aspartic acid. Unpaired t tests with a 95% confidence interval of mean values showed that l-glutamine was preferred as the most stable substrate in the SLC38A7 uptake assays (n = 3) and chosen as substrate in the following experiments (*, p < 0.05; **, p < 0.01; ***, p < 0.001). B, uptake of 0.2 μm l-[3H]glutamine into 38A7 expressing oocytes. Different times of incubation were used; 2, 5, 10, 20, 30, 40, 60, and 120 min (mean ± S.D., n = 6–8 oocytes). SLC38A7 showed increasing and saturable transport over time. An unpaired t test with a 95% confidence interval of mean values, when comparing uptake into SLC38A7 expressing oocytes and endogenous uptake into control oocytes (non-injected), showed that 60 min of l-glutamine uptake have the highest significant difference (p = 0.0011). C, the uptake of l-[3H]glutamine (0.2 μm) was measured in Slc38a7-injected oocytes and non-injected control oocytes (mean ± S.D., n = 10–22 oocytes) in buffers containing sodium chloride (NaCl), lithium chloride (LiCl), or choline chloride (choline Cl). Mean counts were compared between Slc38a7-injected oocytes and control oocytes and an unpaired t test showed that specific uptake of SLC38A7 is sodium dependent (p < 0.0001) and can partially be replaced with choline (p = 0.0067) but not lithium. D, in the last graph l-[3H]glutamine (1.2 μm) was used in a substrate preference analysis with a range of 100-fold higher (1.2 mm) competing amino acids, neurotransmitters, and MeAIB. The SLC38A7 protein showed the most preferable transport, shown as the most competition, of the amino acid l-glutamine of all tested substrates (mean ± S.D., n = 8–52 oocytes). Single letter amino acid abbreviations are indicated for l-glutamine, l-histidine, l-serine, l-alanine, l-aspargine, l-aspartic acid, l-glutamic acid, l-methionine, l-leucine, l-glycine, arginine, l-tyrosine, l-phenylalanine, l-proline, and l-lysine, dopamine (DA), acetylcholine (ACh), and γ-aminobutyric acid (GABA). The color codes on amino acid side chains are polar uncharged (purple), electrically charged (blue), hydrophobic (green), special cases (yellow), and the neurotransmitters are shown in light gray and MeAIB in dark gray. The specific l-glutamine transport of SLC38A7 are presented as counts per minute in A–C, and as the normalized response of l-glutamine transport by dividing the actual transport with the positive control (no competing substrate) and thereafter calculating the specific transport in D.
SLC38A7 Mediates Sodium-dependent Amino Acid Transport
Sodium ion dependence and tolerance for lithium or choline ion substitution was explored to characterize the coupled transport properties of SLC38A7 (Fig. 5C). In the presence of sodium ions the l-glutamine uptake into SLC38A7 overexpressing oocytes was significantly higher than uptake into control oocytes (p < 0.0001). Replacement of sodium ions with lithium ions showed that lithium is not able to drive the coupled transport of l-glutamine by SLC38A7. The replacement of sodium ions with choline ions showed that choline is able, to a lower extent (23% of sodium driven transport), to drive transport of l-glutamine with an uptake significantly greater for SLC38A7 compared with control oocytes (p = 0.0067).
SLC38A7 Transports Polar Amino Acids and Alanine
A competition analysis was performed with amino acids, neurotransmitters, and MeAIB, the specific inhibitor of system A, to examine the substrate profile for SLC38A7. First, to find the total concentration that gives the highest specific uptake of l-glutamine into oocytes and to perform saturation kinetics, we conducted an experiment with different total concentrations of l-glutamine. We did not reach saturation of the transporter at the available concentrations of l-[3H]glutamine (supplemental Fig. S3) and attempts to obtain Vmax and Km from this data showed that we were still in the linear range at 5 μm. This suggests that the Km is significantly higher than 5 μm and possibly in the same range as measured for other SNATs, 200–1000 μm (24, 39, 40). However, the test showed that a total concentration of 1.2 μm l-glutamine, containing 0.2 μm l-[3H]glutamine together with 1.0 μm non-labeled l-glutamine, gave the highest specific uptake (p = 0.0003) into cells expressing the SLC38A7 transporter. Therefore, a fixed total concentration of 1.2 μm l-glutamine (with 0.2 μm l-[3H]glutamine) was used in the competition analysis with a range of 100-fold higher concentration of competing substrates (Fig. 5D). The competition experiment showed that SLC38A7 transport of l-[3H]glutamine can be inhibited by amino acids with polar uncharged side chains, like l-glutamine, l-serine, and l-aspargine, the positively charged l-histidine, as well as the negatively charged l-aspartic acid and l-glutamic acid. Amino acids with hydrophobic side chains, like l-alanine, competed to a lower extent. Neurotransmitters like dopamine, acetylcholine, and histamine did to some low extent inhibit the l-[3H]glutamine uptake by SLC38A7, but MeAIB, the analog for system A, or GABA did not inhibit the uptake of l-glutamine. The substrate profile of SLC38A7, by inhibiting the l-[3H]glutamine uptake, was ranked: l-glutamine > l-histidine > l-serine > l-alanine > l-aspargine > l-aspartic acid > l-glutamic acid > l-methionine > l-leucine > l-glycine.
DISCUSSION
We have previously identified Slc38a7 as a gene that appears to be encoding a novel protein similar to the SNATs (7). In this study, we clarify the detailed CNS expression pattern for SLC38A7 and identify this protein as an l-glutamine transporter. We also characterize the substrate profile. We show that SLC38A7 is a sodium-coupled amino acid transporter of system N type, and we suggest this protein should be named SNAT7, to adhere with the SNAT nomenclature.
We used in situ hybridization to scan Slc38a7 mRNA expression in the mouse brain to identify localization in specific cell populations. We identified strong expression of Slc38a7 in ventromedial hypothalamus, basolateral amygdala, ventral tegmental area, and locus coeruleus, areas known to be involved in feeding (41, 42). Within the cortex, the expression of Slc38a7 appears in layers, and in the hippocampus the gene was expressed in the granular layer of dentate gyrus cells and pyramidal cell layer of hippocampus, areas that all support selective expression by neurons. The Slc38a7 gene also showed expression in the layer of Purkinje cells in cerebellum, known to be GABAergic (43). This expression pattern suggests that Slc38a7 has a role in pathways involved in both excitatory and inhibitory neurotransmission.
The localization of the previously characterized SNATs to either neurons or astrocytes (16, 22), or in some cases both (8, 20, 21), made us interested in investigating the nature of the cell types that express SLC38A7. We used mRNA expression of Slc38a7 in combination with NeuN, a marker of neuron-specific proteins (35), and the glial cell marker GFAP (34). All cells stained with NeuN were co-localized with Slc38a7 but not all cells expressing Slc38a7 were stained with the neuronal marker, indicating expression of Slc38a7 in GABAergic and glutamatergic neurons and in additional cells that could be neuronal precursor cells. Consequently, Slc38a7 co-localized to cells stained with NeuN and was not expressed in astrocytes. The GABAergic expression was additionally confirmed by co-localization with GAD67, a marker of inhibitory GABAergic neurons (36).
We also used a custom-made antibody for detection of the SLC38A7 protein. The mRNA expression pattern of Slc38a7 was confirmed with the similar protein expression pattern of SLC38A7 indicating epitope specificity of the polyclonal SLC38A7 antibody. This was confirmed with Western blot studies (supplemental Fig. S2A). The ∼70 kDa protein detected with Western blot was a larger protein than the expected 49.91 kDa. A possible explanation for this could be post-translational modifications at palmitoylation or glycosylation sites. Additional proof for specificity of the custom-made SLC38A7 antibody was performed with immunohistochemistry and Western blot studies on cells expressing a FLAG-SLC38A7 fusion protein (supplemental Fig. S2, B and C). The labeling of the SLC38A7 protein and the FLAG tag co-localized in the transfected cells and detected the same band on the Western blot, indicating epitope specificity of the antibody.
Immunohistological double labeling with the nerve cell marker (NeuN) confirmed SLC38A7 localization in neurons, and the total overlap indicated expression in all neurons in the CNS, both glutamatergic and GABAergic. The cellular distribution of SLC38A7 showed expression in soma and on the cellular axon. To investigate whether SLC38A7 was expressed on neuronal terminals, we conducted double labeling studies using SLC38A7 and synaptophysin, a marker for presynaptic vesicle protein (38). The absence of co-localization for the marker and SLC38A7 indicated that the transporter is not expressed in neuronal terminals, whereas the spinal cord expression showed clear expression in motor neurons and neuronal axons.
The known SNATs are classified to belong to either systems N or A. The classical system A transporters are known to be expressed ubiquitously in mammalian tissues. SNAT1, which belong to system A, is expressed in a number of peripheral tissues and in the CNS, and is highly concentrated to neurons (8, 9). However, the system N transporter SNAT3 is strongly expressed in brain, liver, and kidney and located on astrocytes in the brain (16). The classical system N transporters have a more restricted expression pattern than system A transporters, often with high expression in liver. We previously showed that Slc38a7 is highly expressed in liver (7) using quantitative real time PCR and the results in this study showed neuronal SLC38A7 expression in both glutamatergic and GABAergic neurons in the brain.
It is known that the characterized members of the SNAT family use sodium for the coupled transport of amino acids. A characteristic of the system N transporters is that they can partially use lithium as a substitute to sodium, whereas the classic system A transporters were first suggested to not tolerate lithium substitution (44, 45). However, all of the known SNATs belonging to system A were later shown to tolerate lithium substitution (46, 47) and sensitivity to lithium substitution is therefore not a good diagnostic criteria for systems A or N. Here we investigated if the SLC38A7 transporter mediated coupled transport of l-glutamine and sodium and/or lithium ions into cells. The results showed that this sodium-dependent transport cannot be substituted with lithium. If anything, this could be an indication of a system A transporter but considering the conflicting data, we suggest that lithium sensitivity should not be considered as a criteria for the classification. These findings encourage further detailed studies to investigate if the transport is pH-dependent and to clarify if the l-glutamine SLC38A7 transport stimulates sodium-hydrogen exchange and to show that SLC38A7 belongs to the system N/A classification and not the ASC system.
The system A transporters show substrate selectivity of amino acid analog MeAIB across the cell membrane (48), as well as a broad range of amino acids such as l-glycine, l-alanine, l-cysteine, and l-glutamine (11, 49), whereas system N has l-glutamine, l-histidine, and l-aspargine as first choice substrates (44, 46, 50). We performed a competition study in which the ability of various nonradiolabeled substances to influence the specific transport of l-[3H]glutamine was assessed. The competition assay showed that the transport of l-[3H]glutamine was inhibited to the greatest extent by l-histidine and l-alanine but also by amino acids with polar side chains like l-glutamine, l-serine, and l-aspargine. Moreover, when competing with MeAIB, the hallmark of system A (51), and neurotransmitters, such as dopamine, acetylcholine, histamine and GABA, we found that these are not preferred substrates for SLC38A7. This shows that SLC38A7 bears the hallmarks of system N, with preference for l-glutamine, l-histidine, and l-aspargine, and is insensitive to MeAIB, but the preferred substrate profile of SLC38A7 is unusually broad compared with other known system N transporters. We suggest SLC38A7 to be a new system N amino acid transporter on the basis of the expression profile, the high expression in liver, and substrate selectivity. The substrate profile for SLC38A7 most closely resembles those of SNAT3 (16, 18) and SNAT5 (22), both transporters of l-glutamine classified into the system N subfamily. However, we found that SLC38A7 is expressed in both glutamatergic and GABAergic neurons in the brain, like SNAT1 (8, 9) and SNAT2 (10, 12), whereas SNAT3 and SNAT5 expression is restricted to glial cells in brain and to liver and kidney. Accumulatively, this suggests SLC38A7 to be the first system N transporter found in all neurons in brain. On the other hand, the fact that the SLC38A7 does not tolerate lithium for sodium substitution and the unusually broad substrate profile makes us believe that the SLC38A7 transporter shares some of the physiological features of both the classical N and A systems.
Glutamine functions as a precursor for nucleic acids and nucleotides and is used in protein synthesis and also as a precursor for the neurotransmitter glutamate in neurons, being an intermediate in the glutamate-glutamine cycle (23, 52). Glutamine is present in large amounts in the cerebrospinal fluid and blood plasma and plays not only an essential role in the brain, but also it is essential for ammonia detoxification in the liver (4, 53). Glutamate synthesized from glutamine in astrocytes has been shown to mainly be released from glutamatergic neurons into the synapse (54). In addition, glutamate can be further converted into GABA via enzymes GAD67 or GAD65, which are expressed by GABAergic neurons. Uptake of glutamine released from astrocytes is essential for the neuronal supply of glutamine (55), therefore, glutamine transporters are clearly key proteins for all neurons. System A transporters have been shown to be important in mediating the uptake of glutamine by neurons (8, 46), whereas system N transporters release glutamine by astrocytes or glial cells (16, 18). Activities classified as system N-like have also been studied in neurons (56). Most of the system A/N glutamine transporters in the SLC38 family, because they prefer l-glutamine as substrate, have been suggested to be part of the synthesis of the neurotransmitter glutamate, and are therefore likely involved in altering both GABA and glutamate levels as well as the cycling of glutamate (4, 8, 18, 46). Sundberg et al. (7) previously performed detailed quantitative real time PCR analysis in a range of rat brain and peripheral tissues to establish the mRNA expression profile for all members of the β-group. Most of the SLC38 family members have relatively broad expression, with Slc38a7 detected at low or intermediate levels in almost every tissue investigated, including the CNS, except with higher levels in the liver (7).
The CNS and the liver are tissues where it is known that glutamine and glutamate metabolism is of profound importance for normal cellular functions (25, 52, 57). However, the exact contribution of the SLC38 proteins in the glutamine-glutamate cycle has yet to be clarified (28, 29). We showed neuronal localization of the SLC38A7 transporter with high expression in the brain at the membrane of the cell body and in axons, and the favored substrate was shown to be l-glutamine. The neuronal localization of other SNAT protein members has only been found between the soma and dendritic branches (somatodendritic) in glutamatergic and GABAergic neurons for SNAT1 (20, 24) and both somatodendritic and in axons for SNAT2 (21). However, the SNAT1 transporter was previously shown to be located at the synapse (58). The preferred substrate and axonal localization of SLC38A7 close to the synaptic cleft, where transmitter synthesis and vesicular packaging of transmitters take place, indicates that SLC38A7 could have an important function in the cycle of glutamate. An important related question is whether the characterized SNATs are responsible for sustaining the glutamine and GABA neurotransmitter pools, independent of the indirect localization in the synaptic cleft, or if SLC38A7 and other orphan transporters play a more direct role in the cycle.
In conclusion, these findings suggest that a function of SLC38A7, a novel member of the SLC38 family, is a sodium-coupled amino acid transport in glutamatergic and GABAergic neurons in the brain. The preferable substrates of transport, l-Glu > l-His > l-Ser > l-Ala > l-Asn > l-Asp, the tissue expression pattern, and the sodium-dependent transport indicates that SLC38A7 is a novel member of the system N amino acid transporter system. It is likely that this transporter plays a role in the glutamine-glutamate cycle. Characterization of novel amino acid transporters, such as SLC38A7, may provide insight into key events in neurobiology, including neuronal metabolism and neurotransmitter cycling.
Supplementary Material
Acknowledgments
We thank Sahar Roshanbin for assistance with the transport assay, Victoria Gridlund for assistance with double in situ hybridization, and Mari-Anne Carlsson for assistance with paraffin embedding of brain tissue.
This work was supported in part by Swedish Research Council Grant VR-NT 2008-3228 (to R. F.), Åhlens Foundation, The Novo Nordisk Foundation, The Göran Gustafsson Foundation, Engkvist Foundation, and the Magnus Bergvall Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Data S1–S3.
- SLC
- solute carrier
- MeAIB
- α-(methyl)amino isobutyric acid
- GFAP
- glial fibrillary acidic protein
- SNAT
- sodium-coupled amino acid transporters.
REFERENCES
- 1. He L., Vasiliou K., Nebert D. W. (2009) Hum. Genomics 3, 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hediger M. A., Romero M. F., Peng J. B., Rolfs A., Takanaga H., Bruford E. A. (2004) Pflugers Arch. 447, 465–468 [DOI] [PubMed] [Google Scholar]
- 3. Hundal H. S., Taylor P. M. (2009) Am. J. Physiol. Endocrinol. Metab. 296, E603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mackenzie B., Erickson J. D. (2004) Pflugers Arch. 447, 784–795 [DOI] [PubMed] [Google Scholar]
- 5. Fredriksson R., Nordström K. J., Stephansson O., Hägglund M. G., Schiöth H. B. (2008) FEBS Lett. 582, 3811–3816 [DOI] [PubMed] [Google Scholar]
- 6. Kusuhara H., Sugiyama Y. (2009) Drug Metab. Pharmacokinet. 24, 37–52 [DOI] [PubMed] [Google Scholar]
- 7. Sundberg B. E., Wååg E., Jacobsson J. A., Stephansson O., Rumaks J., Svirskis S., Alsiö J., Roman E., Ebendal T., Klusa V., Fredriksson R. (2008) J. Mol. Neurosci. 35, 179–193 [DOI] [PubMed] [Google Scholar]
- 8. Varoqui H., Zhu H., Yao D., Ming H., Erickson J. D. (2000) J. Biol. Chem. 275, 4049–4054 [DOI] [PubMed] [Google Scholar]
- 9. Chaudhry F. A., Schmitz D., Reimer R. J., Larsson P., Gray A. T., Nicoll R., Kavanaugh M., Edwards R. H. (2002) J. Neurosci. 22, 62–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yao D., Mackenzie B., Ming H., Varoqui H., Zhu H., Hediger M. A., Erickson J. D. (2000) J. Biol. Chem. 275, 22790–22797 [DOI] [PubMed] [Google Scholar]
- 11. Sugawara M., Nakanishi T., Fei Y. J., Huang W., Ganapathy M. E., Leibach F. H., Ganapathy V. (2000) J. Biol. Chem. 275, 16473–16477 [DOI] [PubMed] [Google Scholar]
- 12. Reimer R. J., Chaudhry F. A., Gray A. T., Edwards R. H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7715–7720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hatanaka T., Huang W., Ling R., Prasad P. D., Sugawara M., Leibach F. H., Ganapathy V. (2001) Biochim. Biophys. Acta 1510, 10–17 [DOI] [PubMed] [Google Scholar]
- 14. Sugawara M., Nakanishi T., Fei Y. J., Martindale R. G., Ganapathy M. E., Leibach F. H., Ganapathy V. (2000) Biochim. Biophys. Acta 1509, 7–13 [DOI] [PubMed] [Google Scholar]
- 15. Desforges M., Lacey H. A., Glazier J. D., Greenwood S. L., Mynett K. J., Speake P. F., Sibley C. P. (2006) Am. J. Physiol. Cell Physiol. 290, C305–312 [DOI] [PubMed] [Google Scholar]
- 16. Chaudhry F. A., Reimer R. J., Krizaj D., Barber D., Storm-Mathisen J., Copenhagen D. R., Edwards R. H. (1999) Cell 99, 769–780 [DOI] [PubMed] [Google Scholar]
- 17. Nakanishi T., Kekuda R., Fei Y. J., Hatanaka T., Sugawara M., Martindale R. G., Leibach F. H., Prasad P. D., Ganapathy V. (2001) Am. J. Physiol. Cell Physiol. 281, C1757–1768 [DOI] [PubMed] [Google Scholar]
- 18. Cubelos B., González-González I. M., Giménez C., Zafra F. (2005) Glia 49, 230–244 [DOI] [PubMed] [Google Scholar]
- 19. Gu S., Roderick H. L., Camacho P., Jiang J. X. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3230–3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Melone M., Quagliano F., Barbaresi P., Varoqui H., Erickson J. D., Conti F. (2004) Cereb. Cortex 14, 562–574 [DOI] [PubMed] [Google Scholar]
- 21. González-González I. M., Cubelos B., Giménez C., Zafra F. (2005) Neuroscience 130, 61–73 [DOI] [PubMed] [Google Scholar]
- 22. Nakanishi T., Sugawara M., Huang W., Martindale R. G., Leibach F. H., Ganapathy M. E., Prasad P. D., Ganapathy V. (2001) Biochem. Biophys. Res. Commun. 281, 1343–1348 [DOI] [PubMed] [Google Scholar]
- 23. Bröer S., Brookes N. (2001) J. Neurochem. 77, 705–719 [DOI] [PubMed] [Google Scholar]
- 24. Mackenzie B., Schäfer M. K., Erickson J. D., Hediger M. A., Weihe E., Varoqui H. (2003) J. Biol. Chem. 278, 23720–23730 [DOI] [PubMed] [Google Scholar]
- 25. Daikhin Y., Yudkoff M. (2000) J. Nutr. 130, 1026S-1031S [DOI] [PubMed] [Google Scholar]
- 26. Danbolt N. C. (2001) Prog. Neurobiol. 65, 1–105 [DOI] [PubMed] [Google Scholar]
- 27. Peng L., Hertz L., Huang R., Sonnewald U., Petersen S. B., Westergaard N., Larsson O., Schousboe A. (1993) Dev. Neurosci. 15, 367–377 [DOI] [PubMed] [Google Scholar]
- 28. Conti F., Melone M. (2006) Neurochem. Int. 48, 459–464 [DOI] [PubMed] [Google Scholar]
- 29. Rae C., Hare N., Bubb W. A., McEwan S. R., Bröer A., McQuillan J. A., Balcar V. J., Conigrave A. D., Bröer S. (2003) J. Neurochem. 85, 503–514 [DOI] [PubMed] [Google Scholar]
- 30. Kowanetz M., Lönn P., Vanlandewijck M., Kowanetz K., Heldin C. H., Moustakas A. (2008) J. Cell Biol. 182, 655–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lemaire P., Garrett N., Gurdon J. B. (1995) Cell 81, 85–94 [DOI] [PubMed] [Google Scholar]
- 32. Gelius B., Wrange O. (2001) Exp. Cell Res. 265, 319–328 [DOI] [PubMed] [Google Scholar]
- 33. Abramoff M. D., Magelhaes P. J., Ram S. J. (2004) Biophotonics Int. 11, 36–42 [Google Scholar]
- 34. Reeves S. A., Helman L. J., Allison A., Israel M. A. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 5178–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mullen R. J., Buck C. R., Smith A. M. (1992) Development 116, 201–211 [DOI] [PubMed] [Google Scholar]
- 36. Kaufman D. L., Houser C. R., Tobin A. J. (1991) J. Neurochem. 56, 720–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huber G., Matus A. (1984) J. Neurosci. 4, 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jahn R., Schiebler W., Ouimet C., Greengard P. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 4137–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Umapathy N. S., Dun Y., Martin P. M., Duplantier J. N., Roon P., Prasad P., Smith S. B., Ganapathy V. (2008) Invest. Ophthalmol. Vis. Sci. 49, 5151–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tanaka K., Yamamoto A., Fujita T. (2005) Neurosci. Lett. 378, 70–75 [DOI] [PubMed] [Google Scholar]
- 41. Abizaid A., Liu Z. W., Andrews Z. B., Shanabrough M., Borok E., Elsworth J. D., Roth R. H., Sleeman M. W., Picciotto M. R., Tschöp M. H., Gao X. B., Horvath T. L. (2006) J. Clin. Invest. 116, 3229–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Williams G., Bing C., Cai X. J., Harrold J. A., King P. J., Liu X. H. (2001) Physiol. Behav. 74, 683–701 [DOI] [PubMed] [Google Scholar]
- 43. McIntire S. L., Reimer R. J., Schuske K., Edwards R. H., Jorgensen E. M. (1997) Nature 389, 870–876 [DOI] [PubMed] [Google Scholar]
- 44. Gu S., Roderick H. L., Camacho P., Jiang J. X. (2001) J. Biol. Chem. 276, 24137–24144 [DOI] [PubMed] [Google Scholar]
- 45. Fei Y. J., Sugawara M., Nakanishi T., Huang W., Wang H., Prasad P. D., Leibach F. H., Ganapathy V. (2000) J. Biol. Chem. 275, 23707–23717 [DOI] [PubMed] [Google Scholar]
- 46. Chaudhry F. A., Reimer R. J., Edwards R. H. (2002) J. Cell Biol. 157, 349–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gu S., Adan-Rice D., Leach R. J., Jiang J. X. (2001) Genomics 74, 262–272 [DOI] [PubMed] [Google Scholar]
- 48. Christensen H. N. (1990) Physiol. Rev. 70, 43–77 [DOI] [PubMed] [Google Scholar]
- 49. Barker G. A., Ellory J. C. (1990) Exp. Physiol. 75, 3–26 [DOI] [PubMed] [Google Scholar]
- 50. Christensen H. N. (1985) J. Membr. Biol. 84, 97–103 [DOI] [PubMed] [Google Scholar]
- 51. Zhang Z., Grewer C. (2007) Biophys. J. 92, 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Newsholme P., Lima M. M., Procopio J., Pithon-Curi T. C., Doi S. Q., Bazotte R. B., Curi R. (2003) Braz. J. Med. Biol. Res. 36, 153–163 [DOI] [PubMed] [Google Scholar]
- 53. Haüssinger D. (1990) Biochem. J. 267, 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sibson N. R., Dhankhar A., Mason G. F., Behar K. L., Rothman D. L., Shulman R. G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2699–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Laake J. H., Slyngstad T. A., Haug F. M., Ottersen O. P. (1995) J. Neurochem. 65, 871–881 [DOI] [PubMed] [Google Scholar]
- 56. Tamarappoo B. K., Raizada M. K., Kilberg M. S. (1997) J. Neurochem. 68, 954–960 [DOI] [PubMed] [Google Scholar]
- 57. Watford M. (2000) J. Nutr. 130, 983S-987S [DOI] [PubMed] [Google Scholar]
- 58. Armano S., Coco S., Bacci A., Pravettoni E., Schenk U., Verderio C., Varoqui H., Erickson J. D., Matteoli M. (2002) J. Biol. Chem. 277, 10467–10473 [DOI] [PubMed] [Google Scholar]
- 59. Franklin K. B. J., Paxinos G. (2007) The Mouse Brain in Stereotaxic Coordinates, Third Edition Ed., Elsevier, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.