Abstract
Corin is a cardiac serine protease that activates natriuretic peptides. It consists of an N-terminal cytoplasmic tail, a transmembrane domain, and an extracellular region with a C-terminal trypsin-like protease domain. The transmembrane domain anchors corin on the surface of cardiomyocytes. To date, the function of the corin cytoplasmic tail remains unknown. By examining the difference between human and mouse corin cytoplasmic tails, analyzing their gene sequences, and verifying mRNA expression in hearts, we show that both human and mouse corin genes have alternative exons encoding different cytoplasmic tails. Human corin isoforms E1 and E1a have 45 and 15 amino acids, respectively, in their cytoplasmic tails. In transfected HEK 293 cells and HL-1 cardiomyocytes, corin isoforms E1 and E1a were expressed at similar levels. Compared with isoform E1a, however, isoform E1 was more active in processing natriuretic peptides. By cell surface labeling, glycosidase digestion, Western blotting, and flow cytometry, we found that corin isoform E1 was activated more readily as a result of more efficient cell surface targeting. By mutagenesis, we identified a DDNN motif in the cytoplasmic tail of isoform E1 (which is absent in isoform E1a) that promotes corin surface targeting in both HEK 293 and HL-1 cells. Our data indicate that the sequence in the cytoplasmic tail plays an important role in corin cell surface targeting and zymogen activation.
Keywords: Membrane Enzymes, Peptide Hormones, Protease, Proteolytic Enzymes, Serine Protease, Corin, Cytoplasmic Tail
Introduction
Corin is a cardiac membrane serine protease important in regulating blood pressure and cardiac function (1, 2). In cardiomyocytes, corin cleaves pro-atrial natriuretic peptide (pro-ANP),3 converting it to ANP, a cardiac hormone that promotes natriuresis, diuresis, and vasodilation. Corin deficiency causes spontaneous hypertension and cardiac hypertrophy in mice (3, 4). Recent studies suggest that corin deficiency may contribute to hypertension and heart failure in patients (5–10).
Corin is a multiple-domain protein, consisting of an N-terminal cytoplasmic tail, a single-span transmembrane domain, and an extracellular region that includes two Frizzled-like domains, eight LDL receptor-like domains, one scavenger receptor-like domain, and one C-terminal protease domain (11, 12). Studies indicate that the transmembrane domain is not required for corin catalytic activity but may serve as a mechanism to localize corin on the surface of cardiomyocytes, allowing efficient processing of natriuretic peptides upon their release from the cells (13). In contrast, Frizzled- and LDL receptor-like domains are important for corin to cleave pro-ANP (14). In population genetic studies, gene variants altering amino acids in corin Frizzled domains are associated with hypertension and cardiac hypertrophy in African Americans (6, 8–11).
Natriuretic peptides exist in all vertebrates, reflecting their fundamental importance in controlling salt-water balance (15, 16). Similarly, the corin gene has been found in a variety of species from fruit fly to man (11, 17). Among mammals, corin proteins are well conserved, sharing ∼85% sequence similarities. One notable exception is that the cytoplasmic tail varies markedly among different species. For example, rat and mouse corin cytoplasmic tails differ from the human corin cytoplasmic tail in both length and sequence (12, 18). Such a major difference was striking, but the biological significance was unclear.
In this study, we used bioinformatics to analyze the genomic sequences of the human and mouse corin genes. We identified alternative exons that may encode different cytoplasmic tails of corin. In functional studies, we determined that alternatively spliced corin mRNAs from these exons exist in human and mouse hearts. More importantly, we found that amino acid sequences in the cytoplasmic tail play a critical role in targeting corin to the cell surface.
EXPERIMENTAL PROCEDURES
Cell Culture
HEK 293 cells were cultured in DMEM with 10% FBS. Murine atrial HL-1 myocytes were provided by William Claycomb (Louisiana State University Medical Center) and cultured in Claycomb medium (JRH Biosciences) with 10% FBS and 4 mmol/liter l-glutamine (19). All cells were cultured at 37 °C in humidified incubators with 5% CO2 and 95% air.
RT-PCR
mRNAs from mouse hearts were used to make cDNAs with the SuperScript III first-strand synthesis system (Invitrogen). Human heart cDNAs were from Clontech. PCR to amplify human and mouse corin mRNAs from 5′-exons was carried out with primers F1 (5′-TGA AAC AGT CTC CTG CCC TC-3′), R1 (5′-CAT AGG AAA GCA GGA TCA CCA-3′), F3 (5′-CTA GTG GAC TTG GCT GCA CA-3′), R3 (5′-ACC AGC AAG AGA ACG AGA GC-3′), f1 (5′-CTT GCT CCG GAG GAG TAC A-3′), r1 (5′-CCC TTT TTA ATG TTC CCA CAA A-3′), f2 (5′-ACC TCT CCT GCA GAG TCC CT-3′), and r2 (5′-ATG TTC CCA CAA AGG ACA GC-3′). Amplified PCR fragments were analyzed on agarose gels and confirmed by DNA sequencing.
Expression Plasmids
A human corin cDNA fragment containing the full-length E1a isoform was amplified by PCR from human heart libraries (Clontech) using Phusion polymerase with sense primer 5′-ATG GGC AGG GTT TCC TTC AG-3′ and antisense primer 5′-GTT TAG GAG AAA GGT CTG GAT GTA AAT C-3′. Plasmids expressing human corin mutants D1–D4, D4a–D4g, AANN, DDAA, and AAAA were made by site-directed mutagenesis using pcDNAhCorin plasmid as a template. To facilitate protein detection, a C-terminal V5 tag was added to recombinant proteins. Plasmids expressing human wild-type corin and cleavage site mutant R801A were described previously (20).
Transfection, Immunoprecipitation, and Western Blotting
HEK 293 and HL-1 cells were transfected with plasmids using FuGENE reagents (Roche Diagnostics) or Lipofectamine 2000 (Invitrogen) according to the manufacturers' instructions. Cells were cultured for 24–48 h and lysed in buffer containing 50 mmol/liter Tris-HCl (pH 8.0), 150 mmol/liter NaCl, 1% (v/v) Triton X-100, and a protease inhibitor mixture (1:100 dilution; Sigma). Immunoprecipitation and Western blotting were performed as described previously (20). Membranes were developed using ECL reagents (Denville Scientific) and exposed to x-ray films. The absorbance of bands representing corin zymogen and the protease domain was measured by densitometry, and the percentage of the activated fragments was calculated using computer software (Bio-Rad). To ensure that the x-ray films were not overexposed, each Western blot had at least two x-ray films exposed for shorter and longer time periods, and calculated ratios from them had to be consistent if the data were used. As we did not include an internal standard for the linearity of ECL signals in each Western blot, values from the densitometric analysis were taken as an estimate.
Natriuretic Peptide Processing
Conditioned medium containing human pro-ANP was prepared from a stable HEK 293 cell line, added to HEK 293 cells expressing corin, and incubated at 37 °C for 30 min. Pro-ANP and ANP in the medium were analyzed by immunoprecipitation and Western blotting. The percentage of pro-ANP to ANP conversion was calculated as described previously (21).
Cell Surface Protein Labeling
HEK 293 cells expressing corin were labeled with 1 mmol/liter sulfo-NHS-biotin (Pierce) in PBS (pH 8.0) at 4 °C for 5 min. The reaction was quenched with 100 mmol/liter glycine in PBS. The cells were lysed in buffer containing 50 mmol/liter Tris-HCl (pH 8.0), 150 mmol/liter NaCl, 1% (v/v) Triton X-100, and a protease inhibitor mixture (1:100 dilution). Streptavidin-Sepharose beads (30 μl) were added to the cell lysate (150 μl), and the mixture was rotated at 4 °C for 2 h. After washing, the beads were boiled in sample buffer. Proteins were analyzed by SDS-PAGE and Western blotting using an anti-V5 antibody.
Glycosidase Digestion
Corin proteins were expressed in HEK 293 cells. The cell lysate or membrane proteins (10 μg) were added to 10 μl of glycoprotein denaturing buffer (New England Biolabs) and heated at 100 °C for 10 min. Reaction buffer containing peptide N-glycosidase F (PNGase F; 1 unit; New England Biolabs) or endoglycosidase H (Endo H; 1 unit; New England Biolabs) was added to the mixture and incubated at 37 °C for 2 h. Proteins were analyzed by SDS-PAGE and Western blotting.
Flow Cytometry
Transiently transfected HEK 293 cells expressing the corin E1 and E1a isoforms on the cell surface were analyzed by flow cytometry using an anti-V5 antibody, followed by a FITC-conjugated secondary antibody. Life gating was performed using pyridinium iodide (Sigma). All data were acquired on a Cytomics FC500 flow cytometer and analyzed using CXP2.0 software (Beckman Coulter, Inc.).
Statistical Analysis
All data are presented as means ± S.D. Statistical analysis was done with Student's t test. A p value of <0.05 was considered to be statistically significant.
RESULTS
Alternative Exons in Human and Mouse Corin Genes
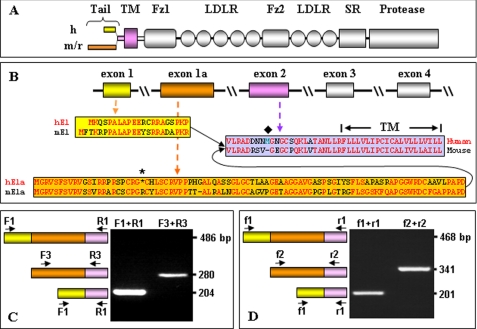
The cytoplasmic tail in mouse and rat corin differs markedly from that in human corin in length and amino acids (Fig. 1A). To examine whether the difference is due to the use of alternative exons, we analyzed genomic sequences of the human and mouse corin genes by BLAST using protein sequences from mouse and human corin cytoplasmic tails. The results showed that both human and mouse corin genes contain exon 1 and an alternative exon, designated exon 1a, in their 5′-regions (Fig. 1B). The amino acid sequences encoded by exon 1 or exon 1a and exon 2 are shown in Fig. 1B. Apparently, the published human corin N-terminal sequence was from exon 1, whereas that of mouse corin was from exon 1a (Fig. 1B). The results suggest that human and mouse corin may have two isoforms that differ in their cytoplasmic tails.
FIGURE 1.
Alternative exons in the corin gene. A, protein domain structure and different cytoplasmic tails of human (h), mouse (m), and rat (r) corin. Tail, cytoplasmic tail; TM, transmembrane; Fz, Frizzled; LDLR, LDL receptor; SR, scavenger receptor. B, alternative exons encoding N-terminal sequences of human and mouse corin. Protein sequences from different exons are color-coded. The asterisk indicates a stop codon. The diamond indicates Met-30. C and D, RT-PCR analysis of alternatively spliced corin mRNA in human (C) and mouse (D) hearts. Oligonucleotide primers based on sequences of different exons are indicated. Predicted PCR fragments from different exon combinations are shown on the left. Negative controls without mRNA templates and positive controls of GAPDH mRNA are not shown.
Alternatively Spliced Corin mRNAs in Hearts
We performed RT-PCR using primers based on sequences from exons 1, 1a, and 2 of the human and mouse corin genes (Fig. 1, C and D). From human and mouse hearts, PCR fragments corresponding to corin mRNAs spliced from exons 1 and 2 and exons 1a and 2 were amplified (Fig. 1, C and D) and confirmed by DNA sequencing. In human hearts, the PCR fragment from exons 1 and 2 was more abundant, whereas in mouse hearts, the fragments from exons 1 and 2 and exons 1a and 2 were at similar levels (Fig. 1, C and D). The data were confirmed by real-time quantitative PCR (data not shown). In contrast, no fragments corresponding to mRNAs from exons 1, 1a, and 2 were detected in either human or mouse hearts. The result shows the presence of alternatively spliced corin mRNAs in the heart, encoding isoforms with different cytoplasmic tails.
Expression and Functional Analysis of Human Corin Isoforms
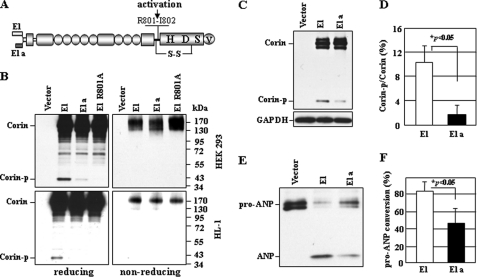
We made plasmids expressing human corin isoforms with cytoplasmic tails encoded by exon 1 (isoform E1) or exon 1a (isoform E1a). Because human corin exon 1a has a stop codon (Fig. 1B, asterisk), isoform E1a is predicted to start at Met-30 (Fig. 1B, diamond) with a shorter cytoplasmic tail, whereas isoform E1 is predicted to start at Met-1 with a longer cytoplasmic tail (Figs. 1B and 2A). In transfected HEK 293 cells, isoforms E1 and E1a were expressed at similar levels, as shown by Western blotting (Fig. 2B, upper panels). An ∼40-kDa band (Corin-p) was more abundant in samples from isoform E1 than in those from isoform E1a. This band represents the activated corin protease fragment (Fig. 2A), which is absent in activation cleavage site mutant R801A or when SDS-PAGE was done under nonreducing conditions (Fig. 2B, upper right panel), which allowed the fragment to be connected to the propeptide by a disulfide bond (Fig. 2A). In Western blots with shorter exposure times, corin zymogen bands (Corin) from isoforms E1 and E1a were similar (Fig. 2C), indicating that the difference in the cytoplasmic tail in these isoforms was too small to be detected by Western blotting. By densitometric analysis, the activated corin protease fragment represented 10.3 ± 2.7 and 1.6 ± 1.5%, respectively, of the total E1 and E1a proteins (Fig. 2D).
FIGURE 2.
Corin isoform expression and activation in HEK 293 and HL-1 cells. A, corin domain structure with alternative cytoplasmic tails. Active residues His (H), Asp (D), and Ser (S) in the protease domain and a C-terminal V5 tag (V) are indicated. The arrow indicates the activation cleavage site at Arg-801. A disulfide bond (S-S) connecting the propeptide and the protease domain is shown. B, corin isoform expression and activation analyzed by Western blotting under reducing and nonreducing conditions with recombinant proteins from transfected HEK 293 and HL-1 cells. Vector-transfected cells and activation cleavage site mutant R801A were used as controls. Corin-p, activated corin protease fragment. C, isoform E1 and E1a expression in HEK 293 cells. D, percentage of the activated protease versus zymogen fragments estimated by densitometry. Data are means ± S.D. from three independent experiments. E, Western analysis of pro-ANP processing by isoforms E1 and E1a. F, estimation of pro-ANP processing by densitometric analysis of Western blots. Data are means ± S.D. from three independent experiments.
We verified this result in physiologically relevant HL-1 cardiomyocytes, which showed similar expression levels of isoforms E1 and E1a but markedly reduced levels of the ∼40-kDa fragment in isoform E1a (Fig. 2B, lower panels). In a pro-ANP processing assay, HEK 293 cells expressing isoform E1 had significantly higher activity compared with cells expressing isoform E1a (Fig. 2, E and F). The data indicate that the corin E1 isoform is more readily activated than the E1a isoform in transfected HEK 293 and HL-1 cells.
Cell Surface Targeting
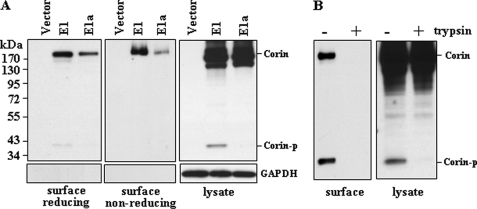
Corin is a transmembrane protein, and its activation depends on cell surface expression (21, 23). We examined the membrane expression of corin isoforms by biotin labeling of cell surface proteins and by Western analysis. Cell surface expression of isoform E1a was much less abundant than that of isoform E1 as indicated in reducing and nonreducing gels (Fig. 3A, left and middle panels). In contrast, the levels of isoforms E1 and E1a were comparable in cell lysates (Fig. 3A, right panel). Used as a control, GAPDH was detected in cell lysates, but not in labeled surface proteins. As another control, the biotin-labeled membrane corin proteins, but not those in cell lysates, were removed when the cells were treated with trypsin before being lysed for Western analysis (Fig. 3B).
FIGURE 3.
Cell surface expression of corin isoforms. A, Western analysis of biotin-labeled cell surface proteins in transfected HEK 293 cells expressing corin isoforms E1 and E1a under reducing (left panel) and nonreducing (middle panel) conditions. As a control, total corin proteins in cell lysates were verified (right panel). GAPDH protein was used as another control for cell surface protein labeling (lower panels). B, Western analysis of biotin-labeled cell surface proteins and total lysates from cells with or without trypsin treatment before lysis. Corin-p, activated corin protease fragment.
Glycosidase Digestion and Flow Cytometry
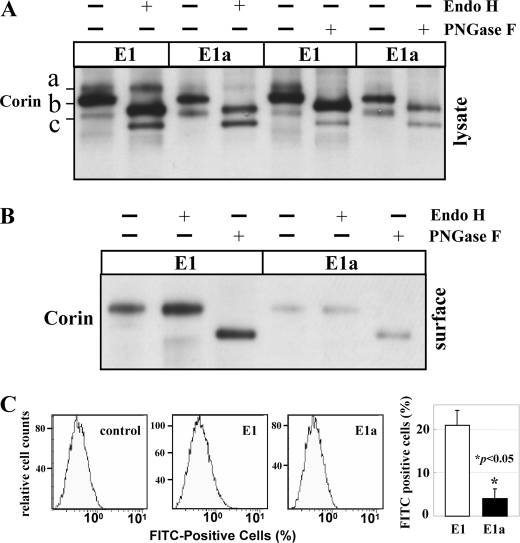
We further studied corin trafficking by glycosidase digestion. On Western blots, corin proteins from transfected cells migrated as three distinct bands (a, b, and c) under high resolution (Fig. 4A). Band a was Endo H-resistant but PNGase F-sensitive, indicating that the protein fragment contained N-glycans, traveled through late Golgi compartments, and possibly reached the cell surface. In contrast, bands b and c were sensitive to both Endo H and PNGase F, suggesting that these protein species remained in the endoplasmic reticulum or early Golgi compartments. Band a was significantly less abundant in isoform E1a than in isoform E1, indicating that fewer isoform E1a molecules were membrane-bound. Consistently, biotin-labeled cell surface corin proteins, which corresponded to band a and were Endo H-resistant but PNGase F-sensitive, were more abundant in samples from isoform E1 compared with isoform E1a (Fig. 4B). Densitometric analysis showed that the difference was significant (p < 0.01, n = 3). By flow cytometry, we confirmed more corin molecules on the cell surface in HEK 293 cells expressing isoform E1 compared with isoform E1a (Fig. 4C).
FIGURE 4.
Glycosidase digestion and flow cytometric analysis of corin isoform proteins. Recombinant corin isoform E1 and E1a proteins were expressed in HEK 293 cells. Cell lysates (A) and biotin-labeled surface proteins (B) were digested with Endo H or PNGase F. Glycosidase-digested corin proteins were analyzed by Western blotting using an anti-V5 antibody. Corin isoforms on the surface of transiently transfected cells were examined by flow cytometry with an anti-V5 antibody as described under “Experimental Procedures.” C, after background subtraction (control cells), the percentage of cells that were FITC-positive on the surface was calculated and is presented in a bar graph (right). The values are representative of data from three independent experiments.
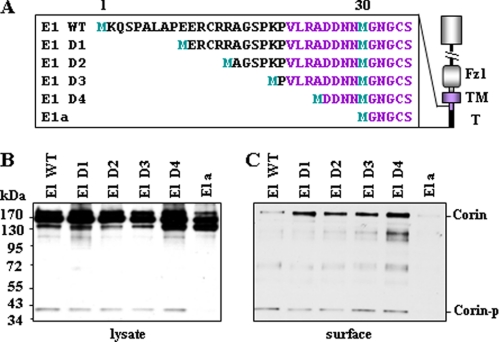
Cytoplasmic Tail Sequences Important for Membrane Targeting
The difference in membrane targeting efficiency between isoforms E1 and E1a suggested that amino acids between Met-1 and Met-30 play a role in corin trafficking (Fig. 5A). We constructed a series of plasmids (E1 D1–D4) for corin mutants with shorter cytoplasmic tails (Fig. 5A) and examined their expression and cell surface targeting in transfected HEK 293 cells. Deleting amino acids up to Ala-25 did not significantly alter corin activation cleavage and cell surface targeting (Fig. 5, B and C), suggesting that the residues between Asp-26 and Asn-29 may be important.
FIGURE 5.
Analysis of human corin mutants with shortened cytoplasmic tails. A, illustration of N-terminal sequences of corin deletion mutants. Plasmids expressing these mutants were transfected in HEK 293 cells. Fz, Frizzled; TM, transmembrane. T, tail. Corin proteins in total cell lysates (B) and biotin-labeled cell surface proteins (C) were analyzed by Western blotting as described under “Experimental Procedures.” Corin-p, activated corin protease fragment.
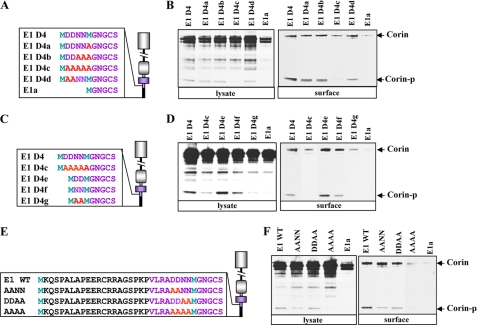
We made another set of mutant plasmids by replacing residues between Asp-26 and Met-30 with Ala (E1 D4a–D4d) (Fig. 6A). Western analysis of mutant proteins from transfected cells showed that corin cell surface expression and activation cleavage were reduced in E1 D4c, but not in E1 D4a, D4b, and D4d (Fig. 6B), indicating that specific amino acids, but not the length of four amino acids in the cytoplasmic tail, were important for membrane targeting. We made the additional corin mutants E1 D4e–D4g (Fig. 6C) and found that activation cleavage and membrane targeting were reduced in E1 D4g, but not in E1 D4e and D4f (Fig. 6D), indicating that the Asp-Asp or Asn-Asn motif was important. We verified the result by replacing these residues in the context of the full-length cytoplasmic tail (Fig. 6E). The results showed that the Asp-Asp or Asn-Asn motif was sufficient, although slightly less efficient than Asp-Asp-Asn-Asn, for targeting corin to the cell surface (Fig. 6F).
FIGURE 6.
Analysis of additional human corin mutants with replaced amino acids. Shown are corin mutants E1 D4a–D4d (A and B), E1 D4e–D4g (C and D), and AANN, DDAA, and AAAA (E and F) and their expression in HEK 293 cells. Corin proteins in cell lysates (left panels in B, D, and F) and biotin-labeled cell surface proteins (right panels in B, D, and F) were analyzed by Western blotting. Corin-p, activated corin protease fragment.
DISCUSSION
Alternative splicing is a basic mechanism regulating gene expression and creating genomic diversity (24, 25). More than half of human genes are estimated to have alternatively spliced mRNAs (26–28), which produce proteins with different stability, transport efficiency, and extracellular localization. Corin belongs to the type II transmembrane serine protease family, defined by the presence of a transmembrane domain near the N terminus and an extracellular protease domain at the C terminus (29, 30). The function of type II transmembrane serine proteases is predicted to be carried out on the cell surface. All type II transmembrane serine proteases contain a cytoplasmic tail of various lengths, but its functional significance was poorly understood (29, 30).
Like trypsin, corin is made as a one-chain zymogen that is converted proteolytically to a two-chain active enzyme (13). The enzyme(s) responsible for corin activation is unknown. Previously, we (21) and others (22, 23) found that rat and mouse corin proteins are more readily activated than human corin in transfected HEK 293 cells and cardiomyocytes, but the reason for such a difference was unclear. Mouse and rat corin proteins are homologous to human corin, and their sequences around the zymogen activation cleavage site are identical to that of human corin, suggesting that other elements in the corin protein may influence its activation.
The apparent difference in cytoplasmic tail length and sequence between mouse/rat and human corin suggested that corin isoforms with different N termini may exist. This hypothesis was supported by identifying an alternative exon 1 in the 5′-regions of the mouse and human corin genes, which was confirmed by RT-PCR with mRNAs from hearts (Fig. 1). In principle, corin mRNAs may be spliced in a continuous fashion from exons 1, 1a, and 2. However, such mRNAs would contain in-frame stop codons, making them nonfunctional. In our study, we detected only mRNAs from exons 1 and 2 or exons 1a and 2, indicating that mRNAs from a combination of exons 1, 1a, and 2 may not exist.
We studied human corin isoforms E1 and E1a, which contain 45 and 15 amino acids, respectively, in their cytoplasmic tails. In both HEK 293 and HL-1 cells, human corin isoform E1 was more readily activated than isoform E1a. Cell surface labeling, glycosidase digestion, and flow cytometry showed that the difference was due mainly to their efficiencies in cell surface targeting. The data are consistent with previous findings indicating that corin activation depends on cell surface expression (21–23). By testing a series of mutants, we found that it is specific amino acids (but not the length) in the cytoplasmic tail that are critical for corin cell surface targeting.
Short motifs in cytoplasmic tails are known to act as signals to sort proteins to the cell surface or different subcellular compartments (31–34). For example, diacidic motifs are known to serve as endoplasmic reticulum export signals in human ion channel proteins (35). By site-directed mutagenesis, we identified a DDNN motif at residues 26–29 to be critical for the cell surface targeting of human corin. Within this sequence, apparently either the DD or NN motif is sufficient. Interestingly, similar DD or NN motifs are found in the cytoplasmic tails of other type II transmembrane serine proteases. For example, matriptase and matriptase-3 have the NN motif and spinesin has the DD motif in their cytoplasmic tails (36–39). It remains to be tested if such motifs play a role in the cell surface targeting of these proteases. Further studies also are needed to understand the potential role of such motifs in membrane insertion, endoplasmic reticulum export, and membrane protein recycling. Low levels of human corin isoform E1a, which lacks the DDNN motif, were detected on the cell surface. It therefore appears that this motif may promote, but is not absolutely required for, corin cell surface targeting. Consistent with this notion, the DD/NN motif is not present in the cytoplasmic tail of rat and mouse corin, suggesting that other sequences may also regulate corin cell surface targeting in these species.
As shown by RT-PCR, both corin isoforms E1 and E1a exist in human hearts (Fig. 1). The expression of isoform E1 appears to be higher than that of isoform E1a. If human corin isoform E1 is more active than isoform E1a, it will be interesting to determine whether mechanisms exist to regulate corin isoform expression. It is possible that the expression of these corin isoforms is altered selectively under physiological or pathological conditions, resulting in enhanced or reduced corin activities. In animal models of and patients with heart failure, corin mRNA and protein expression appears to be increased, but corin activity did not increase proportionally (40–42). Recently, soluble corin was detected in human blood (43–45), and its levels were found to be lower in patients with heart failure (5, 7, 46). It remains to be determined whether alternative expression of corin isoforms contributes to different levels of corin protein and/or activity in these patients, which may be part of the pathological mechanisms underlying their heart disease.
Acknowledgments
We thank Dr. William Claycomb for providing HL-1 cells and Dr. Ningzheng Dong for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants HL089298, HL089298-S1, and HD064634. This work was also supported by grants from the Ralph Wilson Medical Foundation, the Bakken Heart-Brain Institute, and the Priority Academic Program Development of Jiangsu Higher Education Institutions and by National Natural Science Foundation of China Grants 31070716 and 30811130467.
- ANP
- atrial natriuretic peptide
- PNGase F
- peptide N-glycosidase F
- Endo H
- endoglycosidase H.
REFERENCES
- 1. Wu Q. (2007) Front. Biosci. 12, 4179–4190 [DOI] [PubMed] [Google Scholar]
- 2. Wu Q., Xu-Cai Y. O., Chen S., Wang W. (2009) Kidney Int. 75, 142–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chan J. C., Knudson O., Wu F., Morser J., Dole W. P., Wu Q. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nigrovic P. A., Gray D. H., Jones T., Hallgren J., Kuo F. C., Chaletzky B., Gurish M., Mathis D., Benoist C., Lee D. M. (2008) Am. J. Pathol. 173, 1693–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dong N., Chen S., Yang J., He L., Liu P., Zheng D., Li L., Zhou Y., Ruan C., Plow E., Wu Q. (2010) Circ. Heart Fail. 3, 207–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dries D. L., Victor R. G., Rame J. E., Cooper R. S., Wu X., Zhu X., Leonard D., Ho S. I., Wu Q., Post W., Drazner M. H. (2005) Circulation 112, 2403–2410 [DOI] [PubMed] [Google Scholar]
- 7. Ibebuogu U. N., Gladysheva I. P., Houng A. K., Reed G. L. (2011) Circ. Heart Fail. 4, 114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rame J. E., Drazner M. H., Post W., Peshock R., Lima J., Cooper R. S., Dries D. L. (2007) Hypertension 49, 857–864 [DOI] [PubMed] [Google Scholar]
- 9. Rame J. E., Tam S. W., McNamara D., Worcel M., Sabolinski M. L., Wu A. H., Dries D. L. (2009) Circ. Heart Fail. 2, 541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang W., Liao X., Fukuda K., Knappe S., Wu F., Dries D. L., Qin J., Wu Q. (2008) Circ. Res. 103, 502–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pan J., Hinzmann B., Yan W., Wu F., Morser J., Wu Q. (2002) J. Biol. Chem. 277, 38390–38398 [DOI] [PubMed] [Google Scholar]
- 12. Yan W., Sheng N., Seto M., Morser J., Wu Q. (1999) J. Biol. Chem. 274, 14926–14935 [DOI] [PubMed] [Google Scholar]
- 13. Knappe S., Wu F., Masikat M. R., Morser J., Wu Q. (2003) J. Biol. Chem. 278, 52363–52370 [DOI] [PubMed] [Google Scholar]
- 14. Knappe S., Wu F., Madlansacay M. R., Wu Q. (2004) J. Biol. Chem. 279, 34464–34471 [DOI] [PubMed] [Google Scholar]
- 15. McCormick S. D., Bradshaw D. (2006) Gen. Comp. Endocrinol. 147, 3–8 [DOI] [PubMed] [Google Scholar]
- 16. Tsukada T., Takei Y. (2006) Gen. Comp. Endocrinol. 147, 31–38 [DOI] [PubMed] [Google Scholar]
- 17. Irving P., Troxler L., Heuer T. S., Belvin M., Kopczynski C., Reichhart J. M., Hoffmann J. A., Hetru C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 15119–15124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Langenickel T. H., Pagel I., Buttgereit J., Tenner K., Lindner M., Dietz R., Willenbrock R., Bader M. (2004) Am. J. Physiol. Heart Circ. Physiol. 287, H1516–H1521 [DOI] [PubMed] [Google Scholar]
- 19. Claycomb W. C., Lanson N. A., Jr., Stallworth B. S., Egeland D. B., Delcarpio J. B., Bahinski A., Izzo N. J., Jr. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2979–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang J., Wu S., Wang W., Chen S., Peng J., Zhang X., Wu Q. (2011) J. Biol. Chem. 286, 10066–10072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liao X., Wang W., Chen S., Wu Q. (2007) J. Biol. Chem. 282, 27728–27735 [DOI] [PubMed] [Google Scholar]
- 22. Gladysheva I. P., King S. M., Houng A. K. (2008) Biochem. Biophys. Res. Commun. 373, 130–135 [DOI] [PubMed] [Google Scholar]
- 23. Gladysheva I. P., Robinson B. R., Houng A. K., Kováts T., King S. M. (2008) J. Mol. Cell. Cardiol. 44, 131–142 [DOI] [PubMed] [Google Scholar]
- 24. Modrek B., Resch A., Grasso C., Lee C. (2001) Nucleic Acids Res. 29, 2850–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu X., Yang D., Ding J. H., Wang W., Chu P. H., Dalton N. D., Wang H. Y., Bermingham J. R., Jr., Ye Z., Liu F., Rosenfeld M. G., Manley J. L., Ross J., Jr., Chen J., Xiao R. P., Cheng H., Fu X. D. (2005) Cell 120, 59–72 [DOI] [PubMed] [Google Scholar]
- 26. Black D. L. (2003) Annu. Rev. Biochem. 72, 291–336 [DOI] [PubMed] [Google Scholar]
- 27. Johnson J. M., Castle J., Garrett-Engele P., Kan Z., Loerch P. M., Armour C. D., Santos R., Schadt E. E., Stoughton R., Shoemaker D. D. (2003) Science 302, 2141–2144 [DOI] [PubMed] [Google Scholar]
- 28. Sultan M., Schulz M. H., Richard H., Magen A., Klingenhoff A., Scherf M., Seifert M., Borodina T., Soldatov A., Parkhomchuk D., Schmidt D., O'Keeffe S., Haas S., Vingron M., Lehrach H., Yaspo M. L. (2008) Science 321, 956–960 [DOI] [PubMed] [Google Scholar]
- 29. Bugge T. H., Antalis T. M., Wu Q. (2009) J. Biol. Chem. 284, 23177–23181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hooper J. D., Clements J. A., Quigley J. P., Antalis T. M. (2001) J. Biol. Chem. 276, 857–860 [DOI] [PubMed] [Google Scholar]
- 31. Bonifacino J. S., Traub L. M. (2003) Annu. Rev. Biochem. 72, 395–447 [DOI] [PubMed] [Google Scholar]
- 32. Borgese N., Brambillasca S., Colombo S. (2007) Curr. Opin. Cell Biol. 19, 368–375 [DOI] [PubMed] [Google Scholar]
- 33. Brown D., Breton S. (2000) Kidney Int. 57, 816–824 [DOI] [PubMed] [Google Scholar]
- 34. Pandey K. N. (2009) Front. Biosci. 14, 5339–5360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mikosch M., Homann U. (2009) Curr. Opin. Plant Biol. 12, 685–689 [DOI] [PubMed] [Google Scholar]
- 36. Lin C. Y., Anders J., Johnson M., Sang Q. A., Dickson R. B. (1999) J. Biol. Chem. 274, 18231–18236 [DOI] [PubMed] [Google Scholar]
- 37. Szabo R., Netzel-Arnett S., Hobson J. P., Antalis T. M., Bugge T. H. (2005) Biochem. J. 390, 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takeuchi T., Shuman M. A., Craik C. S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11054–11061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yamaguchi N., Okui A., Yamada T., Nakazato H., Mitsui S. (2002) J. Biol. Chem. 277, 6806–6812 [DOI] [PubMed] [Google Scholar]
- 40. Chen S., Sen S., Young D., Wang W., Moravec C. S., Wu Q. (2010) Am. J. Physiol. Heart Circ. Physiol. 299, H1687–H1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jiang W., Cai D. Y., Pan C. S., Qi Y. F., Jiang H. F., Geng B., Tang C. S. (2005) Eur. J. Pharmacol. 507, 153–162 [DOI] [PubMed] [Google Scholar]
- 42. Tran K. L., Lu X., Lei M., Feng Q., Wu Q. (2004) Am. J. Physiol. Heart Circ. Physiol. 287, H1625–H1631 [DOI] [PubMed] [Google Scholar]
- 43. Dong N., Dong J., Liu P., Xu L., Shi S., Wu Q. (2010) Clin. Chim. Acta 411, 1998–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ichiki T., Huntley B. K., Heublein D. M., Sandberg S. M., McKie P. M., Martin F. L., Jougasaki M., Burnett J. C., Jr. (2011) Clin. Chem. 57, 40–47 [DOI] [PubMed] [Google Scholar]
- 45. Peleg A., Jaffe A. S., Hasin Y. (2009) Clin. Chim. Acta 409, 85–89 [DOI] [PubMed] [Google Scholar]
- 46. Shrestha K., Troughton R. W., Borowski A. G., Yandle T. G., Richards A. M., Klein A. L., Tang W. H. (2010) J. Card. Fail. 16, 621–627 [DOI] [PubMed] [Google Scholar]