Abstract
Biogenesis of the ribbon-like membrane network of the mammalian Golgi requires membrane tethering by the conserved GRASP domain in GRASP65 and GRASP55, yet the tethering mechanism is not fully understood. Here, we report the crystal structure of the GRASP55 GRASP domain, which revealed an unusual arrangement of two tandem PDZ folds that more closely resemble prokaryotic PDZ domains. Biochemical and functional data indicated that the interaction between the ligand-binding pocket of PDZ1 and an internal ligand on PDZ2 mediates the GRASP self-interaction, and structural analyses suggest that this occurs via a unique mode of internal PDZ ligand recognition. Our data uncover the structural basis for ligand specificity and provide insight into the mechanism of GRASP-dependent membrane tethering of analogous Golgi cisternae.
Keywords: Golgi, Membrane Biogenesis, Membrane Fusion, Membrane Proteins, Membrane Trafficking, Protein Structure
Introduction
Golgi biogenesis involves membrane tethering by two multifunctional proteins, GRASP65 and GRASP55, which are differentially localized to cis and medial Golgi cisternae, respectively. GRASP65 is associated with cis Golgi cisternae via both binding to the cis-localized golgin GM130 and insertion of its myristoylated N terminus (1, 2). GRASP55 is primarily on medial Golgi cisternae and binds medial-localized golgin-45 and other proteins and is myristoylated and palmitoylated (3, 4). Each protein contains a conserved N-terminal GRASP domain that mediates self-association, which results in the formation of homotypic tethering complexes that link analogous cisternae in adjacent ministacks (5–8).
The GRASP region is predicted to contain two PDZ-like domains (4, 9). PDZ domains are ubiquitous globular protein-protein interaction modules featuring a hydrophobic binding groove, which interacts with the C terminus of its target ligand, although recognition of internal sequences has also been observed (10, 11). Although recent work supports the presence of PDZ domains within the GRASP module (8), secondary structure predictions indicate significant mismatches to the typical organization of β-strands and α-helices found in eukaryotic PDZs.
Toward elucidating the structural mechanism of GRASP-mediated tethering, we solved the crystal structure of the GRASP domain of GRASP55. Although the GRASP domain was indeed composed of two PDZ domains, the domains were circularly permuted, resulting in overall folds that were structurally more similar to prokaryotic PDZs. This unusual arrangement of a metazoan PDZ revealed that the key β2 strands of the binding grooves lay outside of the previously predicted PDZ-like regions. Significantly, an internal ligand sequence mapped in GRASP65 that binds to its first PDZ domain formed a surface projection that appeared to fit inside a deep depression within the PDZ1-binding pocket. Taken together, these data suggest a unique internal PDZ ligand interaction on opposite sides of the molecule and imply a multimeric tethering mechanism that mediates Golgi biogenesis.
MATERIALS AND METHODS
Constructs
GFP-ActA (8) was cloned into GRASP55 pCS-2 (12) using XbaI. To generate His-tagged GRASP55, GRASP55 was inserted into pRSETB (Invitrogen) using the EcoRI site. Point mutations were introduced using the QuikChange protocol (Stratagene). Two sequential rounds of mutagenesis were used for double point mutations. For His-TEV-GRASP55 1–208, the TEV2 recognition sequence ENLYFQG was inserted upstream of the start codon in HisGRASP55, and a stop codon was introduced after residue 208.
Cell Culture and Immunofluorescence
Transient transfection of HeLa cells was performed with jetPEI (Genesee Scientific, San Diego, CA) according to the manufacturer's specifications, and cells were fixed 24 h after transfection with 3% paraformaldehyde at room temperature or methanol at −20 °C. Immunofluorescence, image capture, and analysis were performed as described previously (8).
Protein Expression, Purification, and Crystallization
Purified GRASP55 residues 1–208 were obtained by expression of His-TEV-GRASP55 1–208 in BL21(pLys) Escherichia coli induced with 0.5 mm isopropyl-β-d-thiogalactopyranoside. Clarified cell lysate after lysis (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 10% glycerol, 10 mm imidazole, 1 mm PMSF, 7 mm 2-mercaptoethanol), sonication, and centrifugation was incubated with nickel-agarose (Invitrogen) for 2 h at room temperature and washed with a series of salt concentrations from 300 to 50 mm NaCl. The protein was eluted with 250 mm imidazole and incubated with TEV protease overnight while dialyzing against 25 mm Tris, pH 8.0, 50 mm NaCl, 5% glycerol, 5 mm 2-mercaptoethanol. The resulting solution was passed over a nickel-agarose column, collected and concentrated by ultrafiltration, and separated by size exclusion chromatography on a Superdex G75 gel filtration column (Amersham Biosciences) equilibrated in 10 mm Tris, pH 8.0, 100 mm NaCl, 7 mm 2-mercaptoethanol, 10 mm tris(2-carboxyethyl)phosphine. Peak fractions were concentrated to 10 mg/ml. The purified protein was crystallized by vapor diffusion using the sitting drop method. The sample was mixed with an equal volume of mother liquor containing 0.2 m ammonium acetate, 0.1 m imidazole, 25% polyethylene glycol 400, 10% polyethylene glycol 20,000. Crystals appeared in about 2–3 days and reached full size in 1–2 weeks. The crystals were frozen in liquid nitrogen before mounting in a nitrogen stream at 100 K.
Structure Determination
Initial crystals of selenium-methionine substituted GRASP55 1–208 were screened at the University of Pittsburgh School of Medicine Crystallography Facility using a Saturn 944 CCD detector (Rigaku). High resolution single anomalous dispersion data were collected at the National Synchrotron Light Source beamline X25 and processed with HKL2000 (13). Two of the three selenium positions were located using HKL2MAP (14), and an initial model was built into an experimental electron density map using RESOLVE as implemented with the PHENIX crystallographic software package (15). Subsequent model building was performed using COOT, and refinement was performed using REFMAC5 as implemented with the CCP4i suite of crystallographic software (16–18). Crystallographic and refinement statistics are given in supplemental Table 1.
RESULTS
The GRASP Domain Is a Tandem Array of Circularly Permuted PDZ Domains
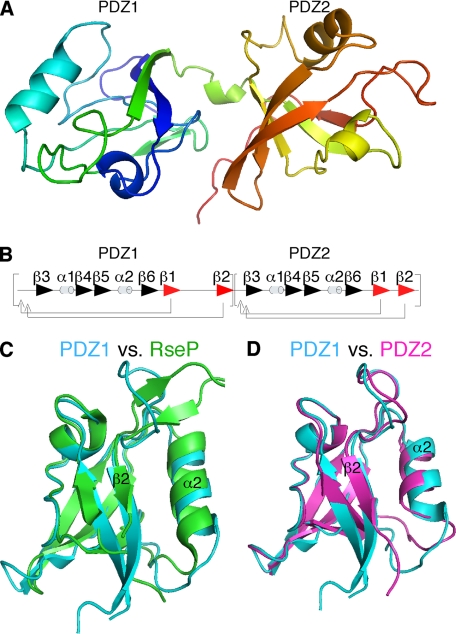
The GRASP domain lacks the typical βββαββαβ secondary structure organization of eukaryotic PDZ domains. Nevertheless, our structure determination of the GRASP55 GRASP domain at 1.65 Å resolution revealed two juxtaposed globular domains, each containing a combination of β-sheets and α-helices reminiscent of a partially opened β-sandwich typical of PDZ domains (Fig. 1A). As with other PDZ domains, each PDZ of GRASP55 contained a recognizable peptide-binding groove that was formed between an α-helix and a β-strand. Remarkably, unlike other eukaryotic PDZ domains, including those used to model GRASP55, each binding groove was formed by the final, rather than the second, β-strand within the fold. This divergence revealed a circular permutation from a typical eukaryotic PDZ domain (Fig. 1B) and indicated that the binding grooves were partially excluded from the previously predicted PDZ-like domains (4).
FIGURE 1.
Structure of the GRASP domain and comparison of its PDZ domains. A, graphic structure of GRASP55 1–208 showing globular domains of PDZ1 (left) and PDZ2 (right) separated by a short α-helix. B, ordering of secondary structural elements within GRASP55 PDZ1 and PDZ2 indicates a circular permutation of a typical eukaryotic PDZ domain. C, graphic representation of GRAS55 PDZ1 (blue) and RseP PDZ2 (green) demonstrates high structural overlap. D, comparison of G55 PDZ1 (blue) and G55PDZ2 (purple) shows high overall similarity with one another.
Surprisingly, the GRASP55 domains exhibited highest structural similarity to prokaryotic PDZ domains. For example, GRASP55 PDZ1 matched the second PDZ domain of RseP (Fig. 1C), which is a bacterial protein that also contains two circularly permuted tandem PDZ domains (19). Interestingly, superimposing the structures of the two PDZ domains of GRASP55 revealed remarkable overlap, despite differences in the secondary structure assignments using the PyMOL software (Fig. 1D). These results demonstrate that the GRASP domain is composed of two structurally similar PDZ domains distinct from typical eukaryotic PDZ domains and suggest that new sequence analysis criteria may reveal a host of unidentified eukaryotic PDZ domains.
Sequence Comparison of GRASP55 and GRASP65 and Organelle Tethering
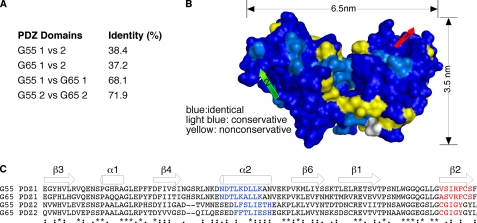
Despite the structural similarity of the two GRASP55 PDZ domains, the sequence identity is only 38% (Fig. 2A). Interestingly, the percentage of identity between GRASP55 and GRASP65 within the PDZ domains is ∼70% (Fig. 2A), raising the possibility that the functions of the corresponding PDZ domains of GRASP55 and GRASP65 are conserved. Indeed, when we modeled the structure of GRASP65 using GRASP55 as a template, non-conserved residues between the two proteins were excluded largely from the area of the putative binding grooves (Fig. 2, B and C).
FIGURE 2.
Comparison of GRASP55 and GRASP65 PDZ domains. A, the percentage of identical residues between individual PDZ domains of G55 and G65 is shown. B, G65 modeled from the G55 structure showing residues that are identical (dark blue), conserved (light blue), and non-conserved (yellow) indicates highest similarity within the binding grooves of PDZ1 (red arrow) and PDZ2 (green arrow). C, sequence alignment of the four individual PDZ domains of G55 and G65 indicates highest similarity within ligand-binding domains formed by the second α-helix (shown in blue) and second β-strand (shown in red). Asterisks indicate sequence identity across domains, two dots indicate highly conservative changes, one dot denotes moderately conservative changes, and zero dots indicate little or no similarity between residues.
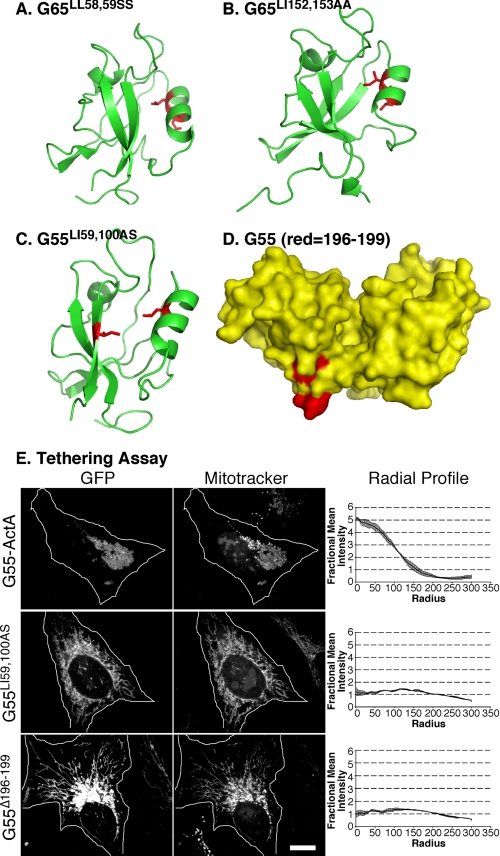
Accordingly, we sought to determine whether our previous disrupting mutations in the putative PDZ1- and PDZ2-binding grooves of GRASP65 (8, 20) actually mapped to the binding grooves of GRASP65 modeled using the GRASP55 structure as a template. Strikingly, the PDZ1 pocket mutant G65LL58,59 that blocked membrane tethering mapped to residues facing the groove of PDZ1 (Fig. 3A), and the G65LI152,153AA that disrupted GRASP65 Golgi localization mapped to residues facing the groove of PDZ2 (Fig. 3B). Thus, these GRASP65 activities likely result from bona fide and functionally distinct PDZ interactions.
FIGURE 3.
Binding groove and surface mutations that block PDZ function. A and B, representations of GRASP65 modeled after the crystal structure of GRASP55 showing binding groove residues (red) that blocked tethering (A) and Golgi localization (B). C, representation of the pocket residues (shown in red) introduced into GRASP55 PDZ1. D, a surface representation of GRASP55 highlighting the internal ligand residues on PDZ2 that we targeted for deletion (shown in red). E, HeLa cells expressing the wild-type construct (G55-GFP-ActA), the pocket mutant construct (G55LI59,100AS), or the deleted internal ligand construct (G55Δ196–199) were analyzed after a 24-h transfection. Cells were imaged using GFP fluorescence to identify transfected proteins and MitoTrackerTM to identify mitochondrial membranes. Quantification was by radial profile analysis (2, 7, 8), which reports averages corresponding to the fraction of total fluorescence present in each concentric circle drawn from the centroid.
To investigate whether GRASP55 mediates organelle tethering using a mechanism similar to that of GRASP65, we made targeted mutations using the crystal structure as a guide and assayed the functional consequences. First, one residue each within α2 and β2 of the PDZ1-binding groove was mutated, and for this purpose, we chose two prominently exposed hydrophobic residues facing the interior of the groove (Fig. 3C). Next, we targeted what we believed would be the ligand that binds the PDZ1 groove for tethering. Recent work identified a 20-residue peptide at the end of the GRASP domain of GRASP65, Cys192–Lys212, that binds the GRASP65 PDZ1-binding groove, and mutations within the stretch 194IGYGYL199 blocked binding, tethering activity, and Golgi ribbon formation (21). The core sequence of this internal PDZ ligand and most of its surrounding residues are conserved in GRASP55; thus, this sequence (Fig. 3D) was deleted.
To measure the organelle-tethering activity of the mutated groove or ligand constructs, we used a previously described assay in which the protein is targeted to the outer membrane of mitochondria by expressing a fusion protein containing the mitochondrial targeting sequence of ActA (8). When expressed in HeLa cells, the wild-type GRASP55 construct induced mitochondrial clustering (Fig. 3E, G55-ActA), indicating that, like GRASP65, GRASP55 tethered membranes. In contrast, expression of the PDZ1 pocket mutant failed to cluster mitochondria (Fig. 3E, G55LI59,100AS). Thus, similar to GRASP65, the membrane-tethering activity of GRASP55 was dependent on the binding groove of PDZ1, implying a PDZ interaction. Further, the ligand-deleted construct also failed to cluster mitochondria (Fig. 3E, G55Δ196–199). Thus, the internal ligand mapped in GRASP65, and conserved in GRASP55, is also required for GRASP55-tethering activity, indicating the presence of an internal PDZ ligand in PDZ2 that mediates self-interaction by binding the PDZ1-binding groove.
The GRASP Self-interaction Suggests a Novel Mode of Internal PDZ Ligand Binding
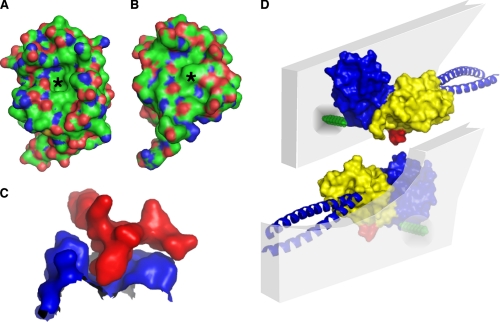
To gain insight into the binding mechanism of the internal PDZ ligand of the GRASP domain, we analyzed the structural features of the PDZ1-binding groove as well as the ligand. Interestingly, the interface between the second α-helix and second β-strand of the PDZ1-binding domain contains a deep depression and resembles more of a pocket than a groove (Fig. 4A). By contrast, analysis of the PDZ2 groove indicates a phenylalanine at position 150 that occludes the pocket (Fig. 4B) and thus likely provides a mechanism of ligand binding specificity between the two PDZ domains. Moreover, the core sequence of the internal ligand (196YGYL199) forms a conspicuous surface protrusion near the C terminus of PDZ2 that appears well designed to fit into the pocket of PDZ1 (Fig. 4C). Taken together, these observations suggest a novel mode of internal PDZ ligand binding of the GRASP domain that bypasses the requirement for a free C terminus.
FIGURE 4.
Model of GRASP-mediated tethering at the Golgi membrane. A, G55 PDZ1 contains a deep depression (asterisk) within its ligand-binding domain. B, G55 PDZ2 has a phenylalanine at position 150 (asterisk) making the binding domain shallower. C, surface view of residues 192–204 of PDZ2 (shown in red) docked into the binding pocket of PDZ1 (shown in blue), demonstrating the fit of the two surfaces. D, schematic rendition of GRASP domains in two apposing membranes with myristoylation (green segment) and golgin partner (blue coiled-coils) hypothetically placed.
DISCUSSION
The GRASP domain structure reveals two circularly permuted eukaryotic PDZ domains that show overall structural similarity yet confer distinct biological functions. For GRASP65, PDZ2 targets the molecule to cis Golgi membranes by binding a PDZ ligand present at the C terminus of cis-localized GM130, whereas PDZ1 likely binds a similarly targeted GRASP65 molecule on an adjacent membrane, thereby forming a trans complex that bridges the two membranes. A parallel reaction may take place on medial Golgi cisternae involving GRASP55. Strikingly, the tethering interface of each self-interaction involves a deep binding pocket in PDZ1 and a surface protuberance on PDZ2 comprising an internal PDZ ligand.
The GRASP domain internal ligand is unique in both its conformation and its position. PDZ domain proteins are widespread in eukaryotes and eubacteria and typically bind to C-terminal peptide ligands. Relatively few cases of binding to internal ligands are documented, but at least two different mechanisms have emerged that permit the internal ligand to bypass the requirement for a free C terminus. The internal ligand in neuronal NOS makes a sharp β-turn to avoid the carboxylate-binding loop in the groove of its binding partner syntrophin (10), and the binding of Pals-1 to the groove of Par-6 deforms the carboxylate loop to alleviate the steric hindrance (11).
Two prominent features of the GRASP internal ligand are its sharply curved backbone and the outward projection of its tyrosine and leucine side chains, which suggests a novel type of internal PDZ ligand interaction for two reasons. First, the carboxylate-binding loop is avoided by a surface projection inherently formed via the side chains of two critical residues rather than a previously observed β-turn topology. Second, the completion of the β-sheet does not occur following ligand binding as the sharply curved backbone of the ligand is incompatible with β-strand formation. Instead, the ligand-binding domain of PDZ1 forms a deep, pocket-like depression that appears designed to accommodate the outward projection of the internal ligand in PDZ2.
The PDZ1 and PDZ2 domains of GRASP65 and GRASP55, in total, present four similar binding grooves, yet experimental evidence indicates the functional importance of specificity in their interactions. The homologous PDZ domains in the two GRASP paralogs are more similar to each other than PDZ1 is to PDZ2, as would be expected because the two isoforms are believed to have evolved from a common ancestor that already contained tandem PDZ domains. This divergence is likely the basis of specificity, for example, of GRASP65 PDZ1 mediating self-interaction while PDZ2 binds GM130. What is less clear is what prevents GRASP65 from forming stable complexes with GRASP55 (22). Although there are minor differences in the PDZ1 grooves of the two GRASPs that could contribute to specificity in interactions with their respective internal ligands, the internal ligands themselves are nearly identical. One possibility is that non-conserved residues present outside the core of the ligand sequence also contribute to binding. Another possibility is that the differential localization of GRASP65 and GRASP55 in the Golgi restricts their ability to make heterotypic contacts. Interestingly, heterotypic interaction would provide a satisfying explanation for the contribution of the GRASP proteins to stacking of Golgi cisternae (1, 22).
A significant feature of the GRASP domain structure is that the internal ligand for self-interaction is on the surface opposite that of the PDZ1 groove to which it binds (Fig. 4D). This provides a structural explanation of the previously observed tendency to form multimers (23) and suggests that interdigitation of the molecules may occur during tethering. It also provides a critical restriction in modeling the orientation of the molecule on the membrane because both the ligand and the groove need to be available for interactions in trans. PDZ interactions involve a specific orientation of the groove and ligand; thus, restriction of the orientation of the binding partners will impact binding. Given its dual anchoring on the membrane by myristic acid insertion and golgin binding, it is likely that GRASP domain orientation is restricted. Indeed, removal of either anchor promotes interactions in cis and blocks tethering (20). Thus, identification of the membrane orientation will be a critical step in elucidating the in vivo tethering complex that links Golgi cisternae.
In conclusion, the structure of the GRASP domain reveals unusual metazoan PDZ folds in which one PDZ contains a unique internal peptide ligand that, in an intermolecular reaction bridging adjacent membranes, inserts into a pocket in the binding groove of the other PDZ. Further, the structure provides a platform for future analyses of homotypic membrane tethering and other reactions carried out by the multifunctional GRASP proteins.
Supplementary Material
Acknowledgments
We thank Collin Bachert and Somshuvra Mukhopadhyay for critical reading of the manuscript. Some data for this study were measured at beamline X25 at the National Synchrotron Light Source where financial support comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the United States Department of Energy and from the NCI, National Institutes of Health, Grant P41RR012408.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-56779 (to A. D. L.).
The atomic coordinates and structure factors (code 3RLE) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- TEV
- tobacco etch virus
- G55
- GRASP55
- G65
- GRASP65.
REFERENCES
- 1. Barr F. A., Puype M., Vandekerckhove J., Warren G. (1997) Cell 91, 253–262 [DOI] [PubMed] [Google Scholar]
- 2. Puthenveedu M. A., Bachert C., Puri S., Lanni F., Linstedt A. D. (2006) Nat. Cell Biol. 8, 238–248 [DOI] [PubMed] [Google Scholar]
- 3. Short B., Preisinger C., Körner R., Kopajtich R., Byron O., Barr F. A. (2001) J. Cell Biol. 155, 877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kuo A., Zhong C., Lane W. S., Derynck R. (2000) EMBO J. 19, 6427–6439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Y., Satoh A., Warren G. (2005) J. Biol. Chem. 280, 4921–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xiang Y., Wang Y. (2010) J. Cell Biol. 188, 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feinstein T. N., Linstedt A. D. (2008) Mol. Biol. Cell 19, 2696–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sengupta D., Truschel S., Bachert C., Linstedt A. D. (2009) J. Cell Biol. 186, 41–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barr F. A., Nakamura N., Warren G. (1998) EMBO J. 17, 3258–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hillier B. J., Christopherson K. S., Prehoda K. E., Bredt D. S., Lim W. A. (1999) Science 284, 812–815 [PubMed] [Google Scholar]
- 11. Penkert R. R., DiVittorio H. M., Prehoda K. E. (2004) Nat. Struct. Mol. Biol. 11, 1122–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jesch S. A., Lewis T. S., Ahn N. G., Linstedt A. D. (2001) Mol. Biol. Cell 12, 1811–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 14. Pape T., Schneider T. (2004) J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 15. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 17. Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 18. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 19. Inaba K., Suzuki M., Maegawa K., Akiyama S., Ito K., Akiyama Y. (2008) J. Biol. Chem. 283, 35042–35052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bachert C., Linstedt A. D. (2010) J. Biol. Chem. 285, 16294–16301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sengupta D., Linstedt A. D. (2010) J. Biol. Chem. 285, 39994–40003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shorter J., Watson R., Giannakou M. E., Clarke M., Warren G., Barr F. A. (1999) EMBO J. 18, 4949–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y., Seemann J., Pypaert M., Shorter J., Warren G. (2003) EMBO J. 22, 3279–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.