Abstract
The Agrobacterium tumefaciens BlcR is a member of the emerging isocitrate lyase transcription regulators that negatively regulates metabolism of γ-butyrolactone, and its repressing function is relieved by succinate semialdehyde (SSA). Our crystal structure showed that BlcR folded into the DNA- and SSA-binding domains and dimerized via the DNA-binding domains. Mutational analysis identified residues, including Phe147, that are important for SSA association; BlcRF147A existed as tetramer. Two BlcR dimers bound to target DNA and in a cooperative manner, and the distance between the two BlcR-binding sequences in DNA was critical for BlcR-DNA association. Tetrameric BlcRF147A retained DNA binding activity, and importantly, this activity was not affected by the distance separating the BlcR-binding sequences in DNA. SSA did not dissociate tetrameric BlcRF147A or BlcRF147A-DNA. As well as in the SSA-binding site, Phe147 is located in a structurally flexible loop that may be involved in BlcR oligomerization. We propose that SSA regulates BlcR DNA-binding function via oligomerization.
Keywords: Allosteric Regulation, Calorimetry, Crystal Structure, DNA-binding Protein, Protein Structure, Transcription Factors, Transcription Regulation
Introduction
Bacteria in the environment are constantly exposed to non-optimal growth conditions, and they often respond to these variations by regulating specific gene expression through the activity of transcription factors. A newly classified transcriptional regulator family, termed the isocitrate lyase transcription regulator-type family (1–4), controls a wide range of cellular activities in response to environmental or growth variations. In responding to small effector ligands, which often serve as conditional cues, IclR2-type proteins alter their DNA binding capacity, leading to differential gene regulation. So far, roughly 500 different IclR-type proteins have been identified from bacteria and archaea and regulate diverse biological processes, including metabolic pathways (4), multidrug resistance (5), aromatic compound degradation (6, 7), pathogenicity (8), sporulation (9–11), amino acid biosynthesis (12), and quorum-sensing signal degradation (13). Members of the IclR family are usually in the range of 240–280 amino acid residues and are composed of two functional domains. The N-terminal domains contain a helix-turn-helix structural feature, which is a prevalent DNA binding motif observed for prokaryotic transcription factors that recognize specific target DNA sequences (14–16). The C-terminal domains include the effector ligand-binding sites, which are predicted to fold into a GAF (cGMP-regulated cyclic nucleotide phosphodiesterases, certain adenyl cyclases, the bacterial transcription factor FhlA) structural motif.
Except for a few systems, the functions of most IclR members are poorly understood. Even in the more intensively studied systems, information on the regulatory DNA sequences, the small effector ligands, and the mode of regulation is rudimentary. The founding member, IclR of Escherichia coli, regulates the aceBAK operon that encodes essential enzymes (isocitrate lyase, malate synthase, and isocitrate dehydrogenase kinase/phosphorylase) for E. coli to utilize acetate or fatty acids as a sole carbon source via the glyoxylate pathway (2, 17). The aceBAK operon is repressed by IclR if the preferred carbon source (glucose) is present and becomes derepressed when acetate or fatty acids are the only carbon source (18). Sequences of the IclR-binding sites in the aceBAK promoter are palindromic (19, 20), and different mechanisms have been proposed for IclR to function as a repressor (21). The cognate effector ligand for IclR is still not clear, although glyoxylate and pyruvate have been shown to bind to IclR; interestingly, these two molecules have the opposite influence on IclR-DNA interaction (22). So far, there is only one structure of a full-length IclR-type protein (from Thermotoga maritima TM0065, termed here as TM-IclR) (23). However, for TM-IclR, information on the regulated operon, the target DNA sequence, and the cognate effector ligand is not known.
BlcR (formerly AttJ) of the plant pathogen Agrobacterium tumefaciens is an IclR-type regulator, negatively controlling the blcABC operon that is responsible for the catabolism of γ-butyrolactone (24–26). The three enzymes (BlcA, a semialdehyde dehydrogenase; BlcB, an alcohol dehydrogenase; and BlcC, a lactonase) convert γ-butyrolactone sequentially to hydroxybutyrate, succinate semialdehyde (SSA), and succinic acid, the latter being integrated into the TCA cycle. Thus, the γ-butyrolactone pathway allows A. tumefaciens to metabolize γ-butyrolactone, abundant in plant exudates, as a carbon source. The lactonase BlcC can also efficiently degrade acylated homoserine lactones, that function as quorum-sensing signals in A. tumefaciens and other diverse Gram-negative bacteria (13). The BlcR-binding site has been mapped to near the blcABC promoter, and SSA has been identified as the cognate effector ligand that affects BlcR DNA binding (26). In addition, an in vitro assay of BlcR function via lactonase activity (BlcC) has been established (27). Thus, BlcR represents an experimentally amenable IclR-type protein for biochemical and structural characterizations, to reveal how the IclR-type proteins recognize their inducing ligands and target DNA sequences, how they interact with DNA, and how the effector ligands modulate this activity. As a first step toward mechanistic understanding of the BlcR regulatory function, we have solved the three-dimensional structure of BlcR by crystallography and examined the SSA and DNA binding activities of the wild type protein and several mutant derivatives. From these structural studies we propose a new model for ligand-dependent control of BlcR activity.
EXPERIMENTAL PROCEDURES
Molecular Cloning and Protein Purification
The blcR coding sequence was PCR-amplified from A. tumefaciens A6 and cloned into the expression vectors pQE9 (Qiagen) with BamHI and PstI sites and pGEX-2T (GE Healthcare) with BamHI and SmaI sites to generate pQE-BlcR and p2T-BlcR, respectively. Generation of site-specific point mutations was performed using the QuikChange kit (Stratagene) on pQE-BlcR. For those mutations (Y133A, T158A, and D210A) that led to insoluble BlcR proteins upon overexpression, genes were subcloned into pTB146 (with SapI and XhoI sites) to generate pTB-BlcR; the vector improved the soluble fraction of BlcR proteins, probably due to the fused SUMO moiety. All mutations were confirmed by DNA sequencing. pQE-BlcR expressed BlcR with a His6 tag at the N terminus. The p2T-BlcR construct expressed BlcR with a thrombin-cleavable GST tag at the N terminus, which yields the native form of BlcR protein after proteolytic removal of the GST tag. The pTB-BlcR plasmids expressed BlcR with a His6-SUMO tag at the N terminus, which can be removed by SUMO protease to give native BlcR. BlcR from pQE-BlcR (and pTB-BlcR) gave a higher yield, but that from p2T-BlcR was used to produce BlcR crystals.
Plasmids of pQE-BlcR, p2T-BlcR, and pTB-BlcR were transformed into BL21DE3 cells for overexpression. Cells were grown at 37 °C in LB medium to A600 of 0.4–0.5, induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside and allowed to continue growth at room temperature for 4–5 h. To purify BlcR (or mutants) from pQE-BlcR, cells were suspended in 50 mm Tris-Cl, pH 7.5 (room temperature), 5 mm imidazole, and 300 mm NaCl and lysed using a microfluidizer (Microfluidics). The clear lysate was loaded onto an Ni2+ affinity column (GE Healthcare), and a linear imidazole gradient (5–500 mm) was applied. The eluted His6-BlcR was dialyzed into a solution containing 300 mm NaCl and 50 mm Tris-Cl, pH 7.5, at 4 °C overnight and loaded onto a heparin column (GE Healthcare). Most of the protein did not bind to the resin. The unbound fraction was concentrated and further purified using a gel filtration column (Superdex200, GE Healthcare) in 50 mm Tris-Cl, pH 7.5, 300 mm NaCl, and 0.5 mm EDTA. Purification of BlcR from pTB-BlcR was similar except for an additional procedure between the Ni2+ affinity and heparin chromatographic steps. The eluted His6-SUMO-BlcR from the Ni2+ column was digested with SUMO protease at 4 °C overnight, dialyzed to 50 mm Tris-Cl, pH 7.5, 5 mm imidazole, and 300 mm NaCl, and reloaded onto an Ni2+ column. The unbound fraction was concentrated and subjected to the heparin purification followed by gel filtration step. To purify native BlcR from p2T-BlcR, cells were lysed in 140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.3, and the clear lysate was incubated with glutathione-Sepharose 4B resin (GE Healthcare) at 4 °C overnight. After extensive washing, thrombin was added to the resin containing solution, and the cleavage was carried out for 24 h. The unbound fraction containing BlcR was dialyzed into 50 mm Tris-Cl, pH 7.5, 150 mm NaCl, and 0.5 mm EDTA, concentrated, and further purified via a Superdex200 column.
Crystallization and Structural Determination
Purified BlcR (from p2T-BlcR construct) was concentrated to ∼20 mg/ml in 50 mm Tris-Cl, pH 7.5, 150 mm NaCl, and 0.5 mm EDTA and subjected to crystallization by hanging drop vapor diffusion at room temperature. 2 μl of BlcR was mixed with 2 μl of the reservoir solution. Crystals were obtained under three conditions: PEG (9% PEG 400, 30 mm MgCl2, 30 mm MES, pH 5.6), ethanol (18% ethanol, 45 mm NaCl, 30 mm MES, pH 5.6), and isopropyl alcohol solutions (9% isopropyl alcohol, 15 mm MgCl2, 30 mm MES, pH 5.6). The cryo conditions for the PEG crystals were 30% PEG 400, 50 mm MES, pH 5.6, and 50 mm MgCl2; for the ethanol crystals, conditions were 30% glycerol, 20% ethanol, 50 mm MES, pH 5.6, and 25 mm MgCl2; and for the isopropyl alcohol crystals, conditions were 30% glycerol, 9% isopropyl alcohol, 50 mm MES, pH 5.6, and 25 mm MgCl2. The isopropyl alcohol crystals were highly twinned, and diffraction data (2.0 Å) could not be processed. Both the PEG and the ethanol crystals belonged to the same space group (p212121) with the same cell dimensions (51.1 × 74.6 × 141.6 Å), but the PEG crystals diffracted to higher resolution (1.79 Å) than those formed in ethanol (2.5 Å). Our structural studies were thus carried out using the PEG crystals. To obtain phase information, the PEG-BlcR crystals were soaked with various heavy atom compounds (platinum, mercury, and gold). The structure was solved using the diffraction data (2.2 Å) from the gold-soaked crystals by a combination of single isomorphous replacement and anomalous scattering using Phenix (28). The structure was refined using the native data set, using Phenix (28) and CNS (29). The Rw and Rf are 18.71 and 21.83%, respectively.
Isothermal Titration Calorimetry (ITC)
To generate the DNA duplex, a 51-nt single-stranded oligonucleotide containing both IR1 and IR2 (5′-CCATAGTTCACTCTAATGATTCAAGTTCAATTAGTTGAACTCTAATGCGGG-3′) and its complementary oligonucleotide (Integrated DNA Technologies) were incubated at 98 °C, and the temperature was slowly decreased to allow optimal annealing. The sequence of the 50-bp DNA is the same as that of the 51-bp DNA except that nucleotide 29 (A, underlined) is omitted, and the 52-bp DNA has a G base inserted after nucleotide 29. ITC experiments were carried out at 25 °C in a VP-ITC titration calorimeter system (MicroCal, Northampton, MA). To study SSA binding to BlcR, 30 aliquots of 10-μl samples of SSA (∼500 μm) were injected into 1.4 ml of a BlcR solution (∼50 μm in monomer) at 900-s intervals. To study DNA binding to BlcR, 30 aliquots of 10-μl samples of ∼200 μm BlcR were injected into the DNA solution (∼5 μm) at 420-s intervals. To study DNA binding to F147A BlcR in the presence of SSA, the protein was incubated with excess SSA (monomeric F147A BlcR/SSA = 1:6), and 30 aliquots of 10-μl samples of BlcR-SSA (28.85 μm, tetramer) were injected into 1.4 ml of DNA solution (3.09 μm) at 420-s intervals. Proteins and DNA were in 50 mm Tris-Cl, pH 7.5, 300 mm NaCl, and 0.5 mm EDTA. Data were processed with Origin (OriginLab, Northampton, MA), and the base line-corrected binding isotherm was used to derive thermodynamic parameters of the binding process.
Differential Scanning Fluorimetry (DSF)
25 μl of BlcR proteins (50 μm, monomeric concentration) without SSA or with various ratios (1:2, 1:5; 1:10; monomeric BlcR/SSA) in 50 mm Tris-Cl, pH 7.5, 300 mm NaCl, and 0.5 mm EDTA was mixed with SYPRO orange dye. Thermal unfolding, in a 25–85 °C range with 0.5 °C intervals, was monitored at 610 nm (λex = 492 nm) using Mx3005P QPCR systems (Agilent Technologies).
Electromobility Shift Assay (EMSA)
The infrared dye-labeled blcABC promoter (214 bp) of A. tumefaciens A6 was generated using two primers: 5′-ATGCACTTTCCTTGACACACTTGG-3′ (IRD700-labeled; MWG Biotech) and 5′-CGATGATCATCGAGTTGG-3′ (Integrated DNA Technologies). This fragment extends across the 55-bp intergenic region between the divergent blcR and blcABC operons, from 38 bp downstream of the blcR start codon to 121 bp downstream of the blcA start codon. The PCR products were purified using a PCR purification kit (Qiagen). BlcR was incubated with 27.2 nm infrared dye-labeled blcABC promoter DNA in reaction buffer (2 mm Tris-Cl, pH 8.0, 0.2 mm EDTA, 0.2 mm DTT, 12 mm potassium glutamate, 4 μg/ml BSA, 2% glycerol) at various concentrations. In studies using SSA, stock SSA solution (pH 7.6) was added into the protein/DNA (with final concentrations of 27.2 nm DNA, 300 nm BlcR monomer) solutions to the desired concentrations. The reactions were subjected to a 4% non-denaturing polyacrylamide gel in 20 mm Tris acetate, pH 8.0, and 0.5 mm EDTA. The gel was scanned, and band intensities were digitized using the Odyssey infrared image system (LI-COR).
RESULTS
Structure of BlcR
Overall Structure of BlcR
BlcR was a dimer in solution and remained so when crystallized. For both monomers, the N-terminal ∼20 residues and the C-terminal ∼3 residues are not resolved in the structure (see Table 1 for crystallographic statistics). Two other regions display structural flexibility that may bear functional significance. A region close to the SSA ligand-binding site is disordered in both monomers (residues 139–144 for monomer A and 143–145 for B). A loop connecting α2 and α3, including potential DNA-interacting residues, is not well structured in monomer A with no electron density for residues Lys59 and Ser60.
TABLE 1.
Crystallographic data collection and refinement statistics
| Parameters | Values |
|
|---|---|---|
| BlcR (native) | BlcR (gold derivative) | |
| X-ray diffraction data | ||
| Space group | P212121 | P212121 |
| Unit cell dimensions | a = 51.155 Å | a = 51.074 Å |
| b = 74.773 Å | b = 74.596 Å | |
| c = 141.808 Å | c = 141.607 Å | |
| α, β, γ = 90° | α, β, γ = 90° | |
| Resolution (Å) | 50-1.79 | 50-2.2 |
| Unique reflections | 48,827 | 28,271 |
| Redundancya | 5.3 (2.0) | 7.1 (6.5) |
| Completeness (%)a | 94.0 (71.6) | 99.9 (99.8) |
| Rsym (%)a,b | 8.1 (38.9) | 10.9 (62.1) |
| I/σa | 25.1 (2.3) | 25.5 (1.0) |
| Refinement statistics | ||
| Reflections in working set | 46,106 | |
| Reflections in test set | 1892 | |
| Rwork (%)c | 18.71 | |
| Rfree (%)d | 21.83 | |
| Non-hydrogen atoms | 3732 | |
| Waters | 399 | |
| Root mean square deviation from ideal values | ||
| Bond length (Å) | 0.007 | |
| Bond angles (degrees) | 1.037 | |
a The numbers in parenthesis refer to the last shell (1.90-1.79 Å for the wild type and 2.24–2.20 Å for the gold-soaked crystal).
b Rsym = ΣhklΣj|Ij(hkl) − 〈I(hkl)〉|/ΣhklΣjIj(hkl), where Σhkl denotes the sum over all reflections and Σj is the sum over all equivalent and symmetry-related reflections.
c Rwork = Σhkl‖Fo(hkl)| − |Fc(hkl)‖/Σhkl|Fo(hkl) is the crystallographic R-factor.
d Rfree = Σhkl‖Fo(hkl)| − |Fc(hkl)‖/Σhkl|Fo(hkl)| is calculated from reflections (∼5% of the total reflections) belonging to a test set of randomly selected data.
Each monomer is organized into two domains, linked by a helix (α4) (Fig. 1A). The N-terminal domain (NTD) contains the helix(α2)-turn-helix(α3) DNA binding motif. The C-terminal domain (CTD) bears the GAF structural fold and presumably contains the binding site for the SSA ligand. Two BlcR monomers associate via their α1, α3, and α4 helices of the NTD, and the CTD does not contribute to dimer formation. Along the dimeric interface, side chains of hydrophobic residues from one molecule (Val26 and residues Val33 and Leu36 of α1) intercalate with those from the other molecule (Ile22 of α1 and Leu65, Val68, Leu72, and Leu74 of α3), forming a highly intertwined hydrophobic tower (Fig. 1B). Polar interactions are found at the bottom of the tower; Asp37 of one monomer is hydrogen-bonded to both Trp92 and His88 of the other monomer. Thus, nonpolar interactions prominently contribute to dimer formation, although hydrogen bonds further secure the association and seal the hydrophobic interface. A total surface of 3877 Å2 is buried as a result of dimerization. Both the folding and the organization of the two domains as well as the dimeric interaction are consistent with those of TM-IclR (23), which parallel with sequence analysis showing a sequence identity of 25.0%, or a sequence similarity of 44.9%, between BlcR and TM-IclR.
FIGURE 1.
Structure of apo-BlcR. A, the crystal structure of the dimeric BlcR. One monomer is colored in orange and the other in green. α3 is the putative DNA-binding helix. Regions (mainly loops of β3/β4 and β4/β5) that interact with the neighboring BlcR dimers in crystal are colored in magenta. Part of the β4/β5 loop is structurally unresolved and is presented as a dotted line. B, the hydrophobic dimeric interface. Residues involved in dimeric interaction are shown with side chains. Asp37 forms two intermolecular hydrogen bonds with His88 and Trp92 of the other monomer. One monomer is colored in green and the other one in gray. C, the putative SSA-binding site with key residues highlighted with their side chains displayed. D, the structural role of Phe147. Phe147 is located at a structurally flexible loop and is part of a hydrophobic path leading to the SSA-binding site. Side chains of the residues lining the hydrophobic path are shown, and the structurally unresolved β4/β5 loop is presented as a dotted line. The two bound water molecules occupying the SSA-binding site are shown as spheres.
The dimeric BlcR in the crystal is structurally symmetric; the two monomers of BlcR are related by a rotational dyad and are superimposable with a root mean square deviation of 1.75 Å. This is in direct contrast with TM-IclR (23). The NTDs of TM-IclR are symmetric like BlcR, but the linker helix (α4) of TM-IclR adopts two different conformations that lead to two different orientations of the CTDs with respect to their corresponding NTDs. This asymmetric structure, resulting from crystal packing, allows the two CTDs of dimeric TM-IclR to interact extensively with those from the neighboring TM-IclR molecule in the crystal to form an apparent TM-IclR tetramer. For BlcR, no such tetramer is found in crystal. Instead, the BlcR dimers are arranged according to the crystallographic symmetry to form infinite filaments; both BlcR monomers interact with one neighboring BlcR dimer. Unlike the extensive intermolecular contact in TM-IclR crystals, the BlcR dimers within the filaments interact mainly via two loops, the β3/β4 and β4/β5 loops, in the CTDs (Fig. 1A). Three charged-charged interacting pairs, Glu127, Arg152, and Arg148 of one BlcR dimer aligned in the respective order with Arg148, Arg152, and Glu127 of the interacting BlcR dimer, dominate the intermolecular interface. Glu127, Arg148, and Arg152 are located in the β3/β4 loop, the β4/β5 loop, and β5, respectively. It is noted that the BlcR β4/β5 loop is structurally flexible, and structural flexibility may be of functional importance (see below).
Structure of the DNA-binding Domain
The N-terminal domain folds into a canonical winged helix-turn-helix DNA binding structure, preceded by a third helix (α1). The symmetrical arrangement of the two α3 helices (the DNA recognition helix) is consistent with the palindromic properties of the target DNA sequence. The distance between the two α3 helices is rather short (∼31 Å between the Cα atoms of Pro58 of monomer A and Lys59 of monomer B; Lys59 of monomer A is disordered), consistent with small gaps within the two half-sites of the proposed BlcR-binding sequences (26) (for details, see below). Overall, the structure of the dimeric N-terminal domains of BlcR is superimposable with that of TM-IclR with a root mean square deviation of 1.75 Å.
In the NTD, the conserved residues among the IclR family (supplemental Fig. S1) are associated with protein folding and structural stability. They are either within the hydrophobic dimeric interface (e.g. Val26, Leu29, Val33, Leu36, Asp37, Leu74, and Ile85) or involved in folding of the BlcR NTD (e.g. Ile35, Leu36, Leu38, Val39, Leu46, Leu51, Leu55, Leu57, Leu65, Leu66, and Met69). Residue 59 of α3 is largely conserved as Lys, Arg, or Gln, which may promote BlcR binding to DNA. Lys59 (as well as Ser60) of monomer A is not resolved in the structure, suggesting structural adaptability of this residue upon interacting with DNA.
Structure of the SSA-binding Domain
BlcR CTD contains the binding site for ligand SSA. The domain folds into a three-level structure: a curved six-stranded β-sheet (β3–β8) is situated between a two-helical layer (α5 and α9) and a three-helical layer (α6, α7, and α8). The region following β4, the β4/β5 loop (residues 137–149), is partially disordered in the crystal, with residues 139–144 in monomer A and 143–145 in monomer B unresolved structurally, and, as mentioned earlier, is involved in intermolecular contact in crystallized BlcR dimers (Fig. 1A). Moreover, the loop is part of the SSA-binding site, because two putative SSA-binding residues, Tyr133 and Phe147 (see below), are related to this region. A dual role of the β4/β5 loop in mediating intermolecular contact and forming an SSA-binding site may be functionally significant because it offers a mechanism for SSA regulation on DNA binding activity of BlcR.
The surface underneath the concave β-sheet has been shown to bind small ligands in the structural studies of the ligand-binding domains of the IclR family members (22, 23, 30). For BlcR, the residues lining the putative SSA-binding surface include Thr122, Tyr133, Phe147, Thr158, Thr160, Asp210, and Cys220 (Fig. 1C), all except Phe147 being polar. The surface polarity is consistent with the polar/charged nature of SSA. Two water molecules (separated by 3.65 Å) are found within the cavity formed by these residues; one H2O is hydrogen-bonded to both Cys220 (2.82 Å) and Asp210 (2.62 Å), and the other is hydrogen-bonded to both Tyr133 (3.18 Å) and Thr160 (3.08 Å) (Fig. 1D). When bound, SSA (molecular mass of 102 Da) may replace the two ordered H2O molecules to occupy the cavity. To confirm that this cavity is the SSA-binding site and to assess the contribution of individual residues to SSA binding, we have mutated several putative residues (Y133A, F147A, T158A, D210A, and C220A) and characterized various biochemical properties for these mutant proteins, including associations of SSA and DNA.
Mutational Effects on Protein Stability
DSF (31) was used to study thermal unfolding of various BlcR proteins. Based on DSF, the melting temperature (Tm) of the wild type BlcR was found to be 54.5 ± 0.5 °C. Mutations in the SSA-binding site generally lowered the Tm of BlcR mutants by 5–10 °C (Table 2). The lowered Tm values suggest that mutations destabilize the overall structure of BlcR and that all of the mutants are less stable than the wild type.
TABLE 2.
Effect of mutations on binding of BlcR with SSA
| Proteins |
Tma |
ITC Kd | |||
|---|---|---|---|---|---|
| 1:0b | 1:1 | 1:5 | 1:10 | ||
| °C | μm | ||||
| BlcR | 54.5 | 57 | 58.4 | 60 | 0.7 ± 0.1 |
| BlcRY133A | 44.0 | 44.5 | 49.5 | 49.5 | 19.3 ± 3.1 |
| BlcRF147A | 45.5 | 46.5 | 49 | 50 | 3.9 ± 0.3 |
| BlcRT158A | 49.5 | 53.5 | 56 | 56.5 | 1.6 ± 0.3 |
| BlcRD210A | 48.0 | 48.0 | 48.0 | 48.0 | Weakc |
| BlcRC220A | 47.0 | 47.0 | 47.0 | 47.0 | NDd |
a Melting temperatures are from DSF studies.
b BlcR monomer/SSA ratio.
c Heat exchange was too low for accurate ITC analysis.
d ND, not detected.
Association of SSA with BlcR
The presence of SSA generally increased Tm of BlcR proteins by 5–7 °C (Table 2); the change in Tm suggests specific binding of SSA to the BlcR proteins. Notably, the SSA effect on Tm was not observed for both BlcRD210A and BlcRC220A mutants even in the presence of excess SSA (1:10 molar ratio), suggesting that both mutants had weak affinity for SSA or did not bind SSA at all.
Binding of SSA to BlcR proteins was further analyzed using ITC. SSA bound to the wild type BlcR with a dissociation constant (Kd) of 0.7 μm (25 °C), which is comparable with our earlier reported value (Kd of 0.36 μm, 28 °C) (25). It is noted that the binding molar ratio (N) was significantly low (dimeric BlcR/SSA = 1:1.10; supplemental Fig. S2A), suggesting that either one of the two SSA-binding sites in dimeric BlcR or 50% of the purified BlcR was defective in binding SSA. Deficiency in SSA binding may be due to structural modification at the binding site during protein purification or to preoccupation of endogenous SSA in the binding site. To remove residual SSA that might be incorporated during cell growth, purified BlcR was dialyzed extensively before being subjected to ITC studies; however, the N ratio remained low. Thus, the low SSA occupancy is not due to contamination but probably to structural perturbations around the binding site during protein expression and purification. For example, the β4/β5 loop, which is related to the SSA-binding site, is structurally more flexible in monomer A than that in monomer B, and such conformational differences may account for different SSA binding behaviors between the two monomers, leading to a 50% decrease in SSA binding capacity.
Consistent with the lack of SSA effect on thermal unfolding of BlcRC220A in the DSF experiments, no heat exchange was observed in the titration of SSA to BlcRC220A (supplemental Fig. S2B), indicating the central role of Cys220 in SSA association. Similarly, the very small heat exchange in the ITC experiments of SSA titration to BlcRD210A (data not shown) suggested a critical role for Asp210 in SSA binding. Mutations of Y133A, T158A, and F147A resulted in various degrees of decrease in the binding affinity for SSA, with Kd values of 19.3, 1.6, and 3.7 μm (Table 2), respectively, compared with 0.7 μm for wild type BlcR. Together, these findings indicate that the C-terminal cavity is the binding site for SSA and that Cys220 and Asp210 are the critical residues, whereas Tyr133, Phe147, and Thr158 are involved in SSA binding.
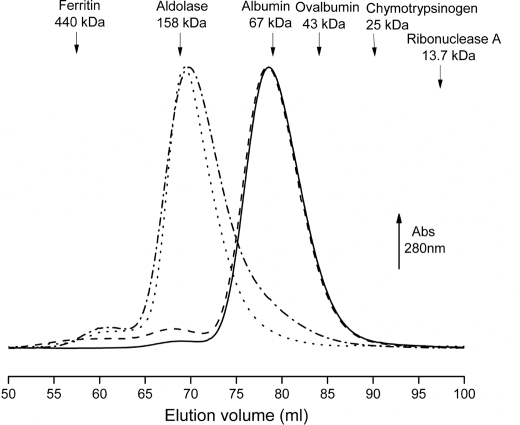
Oligomeric States of BlcR
In solution, BlcR migrated in gel filtration with a molecular species corresponding to a size of ∼76.5 kDa (Fig. 2), suggesting that in the absence of SSA BlcR existed as dimer in solution (molecular mass for monomeric BlcR is 29.8 kDa). This is in contrast with E. coli IclR; it exists as tetramer, dimer, and monomer or in an oligomeric equilibrium of these three states (19, 32). To examine whether SSA influenced BlcR oligomerization, purified apo-BlcR was incubated over a range of SSA concentrations (with molar ratios of monomeric BlcR/SSA = 1:1, 1:5, and 1:10), and subjected to gel filtration analysis. With the presence of 150 μm SSA in the mobile phase, the elution profiles of SSA-BlcR remained the same as that without SSA (Fig. 2), indicating that SSA did not affect the dimeric form of BlcR. No ligand-induced effect on protein oligomeric state is also observed in Pseudomonas putida TtgV, also an IclR-type protein (5).
FIGURE 2.
Gel filtration elution profiles of wild type BlcR and BlcRF147A in the absence and presence of SSA. The protein was incubated with SSA in a molar ratio of 1:10 (monomeric BlcR/SSA), and 150 μm SSA was included in the mobile phase (50 mm Tris-Cl, pH 7.5, 300 mm NaCl, and 0.5 mm EDTA) of a Superdex200 column. Solid line, BlcR without SSA; dashed line, BlcR with SSA; dashed and dotted line, BlcRF147A without SSA; dotted line, BlcRF147A with SSA. Proteins used for molecular weight calibration are shown at their elution volumes.
Mutations of Y133A, T158A, D210A, and C220A did not alter the dimeric state of BlcR. Remarkably, the F147A mutation led to a tetrameric form of BlcR with a molecular size of ∼150 kDa based on gel filtration (Fig. 2). Moreover, the tetrameric form of BlcRF147A was not affected by the presence of SSA (Fig. 2).
Association of DNA with BlcR
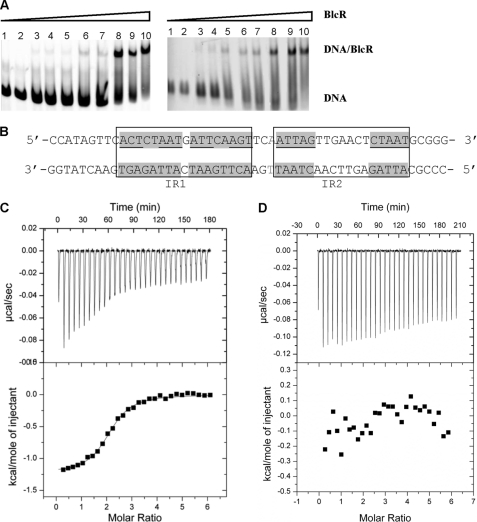
Using EMSA to investigate the DNA-BlcR association, we found that BlcR formed a single complex with the 214-bp blcABC promoter DNA over the protein concentrations used (Fig. 3A), consistent with our earlier studies (13, 25). Multiple BlcR-DNA complexes have also been reported (26). In that study, the blcABC promoter sequence is from a different species of A. tumefaciens (R10 versus A6 in our studies). Importantly, our titration studies showed that binding of BlcR to DNA was highly cooperative, as shown in Fig. 3A, suggesting that more than one BlcR dimer bound to DNA and that their binding to DNA was highly correlated. The estimated apparent Kd (Kd,app), the concentration of dimeric BlcR at which 50% of the full-length 214-bp DNA was BlcR-bound, is ∼120 nm. To localize the BlcR-binding site on the blcABC promoter, we used a 51-bp DNA duplex (Fig. 3B) containing two pairs of inverted repeats (IRs) that were mapped previously as the BlcR-binding site (26). EMSA studies using this 51-bp DNA fragment also demonstrated the high binding cooperativity of BlcR to DNA and yielded a Kd,app of 120 nm (Fig. 3A, right). The presence of similar binding behaviors, particularly the same Kd,app, indicates that the BlcR-binding site on blcABC promoter is contained within the 51-bp fragment and that binding of BlcR to the promoter is reasonably approximated by that to the 51-bp DNA.
FIGURE 3.
Binding of BlcR with DNA. A, EMSA studies of BlcR binding to the 214-bp blcABC promoter DNA (left) and to the 51-bp DNA (right). Concentrations of dimeric BlcR in lanes 1–10 were as follows: 0, 5, 65, 80, 100, 105, 107.5, 125, 150, and 200 nm, respectively. The DNA was maintained at 27.2 nm. B, sequence of the 51-bp DNA near the blc promoter. Bases that are related in the imperfect inverse repeats (IR1 and IR2) are in boxes, and those with perfect palindromic relations are shown with a gray background. The base highlighted in gray was omitted in the 50-bp DNA. C and D, ITC studies of titration of the 51- and 50-bp DNA to BlcR. Top panels, heat exchange during each injection. A negative peak indicates that the reaction is exothermic. Bottom panels, integrated area within each injection peak.
To examine the thermodynamic properties of BlcR-DNA interaction, we used ITC studies to show that BlcR bound to this 51-bp DNA with a Kd of 490 nm (Fig. 3C). The higher affinity, or the lower binding constant, of BlcR-DNA derived from EMSA than from ITC may be correlated with the lower salt concentration used in EMSA than in ITC studies (see “Experimental Procedures”); low ionic strength promotes protein-DNA interaction. ITC results also indicated that two BlcR dimers were bound with one DNA molecule (Table 3). Given the high binding cooperativity of BlcR to DNA shown by EMSA (Fig. 3A), binding of one BlcR dimer to DNA must facilitate a second BlcR dimer to associate with DNA. Together, our EMSA and ITC studies suggest that BlcR tetramerizes on DNA. Members of the IclR family, including E. coli IclR and P. putida TtgV, are also found to bind their promoter DNA in tetrameric form (5, 32, 33).
TABLE 3.
Effect of mutations on binding of BlcR to DNA
ITC-derived thermodynamic parameters of BlcR-DNA interaction are shown. Unless indicated, DNA is the 51-bp duplex; sequence of the 50 bp is the same as that of the 51 bp except that one base pair (highlighted in gray in Fig. 3B) is omitted; sequence of the 52 bp has one GC pair inserted after the highlighted base pair.
| Samples | Kda | ΔH | ΔS | Stoichiometry (N)b |
|---|---|---|---|---|
| kcal/mol | cal/mol | |||
| BlcR | 490 ± 53 | −1.23 ± 0.1 | 24.8 | 2.1 ± 0.1 |
| BlcRY133A | 260 ± 23 | −10.8 ± 0.1 | −6.0 | 2.3 ± 0.1 |
| BlcRF147A | 54 ± 6 | −19.9 ± 0.2 | −33.4 | 1.0 ± 0.1 |
| BlcRT158A | 370 ± 45 | −11.5 ± 0.1 | −9.0 | 2.3 ± 0.1 |
| BlcRC220A | 340 ± 26 | −8.7 ± 0.1 | 0.5 | 2.1 ± 0.1 |
| BlcRF147A with SSAb | 32 ± 2 | −15.5 ± 0.2 | −17.6 | 1.2 ± 0.1 |
| BlcRF147A with 50 bp | 68 ± 7 | −18.0 ± 0.2 | −27.4 | 0.8 ± 0.1 |
| BlcRF147A with 52 bp | 93 ± 8 | −17.9 ± 0.2 | −27.7 | 0.8 ± 0.1 |
a Protein concentration expressed in dimeric form except for BlcRF147A, which is in tetrameric form.
b Molar ratio of monomeric F147A/SSA = 1:6.
Because BlcR existed as a dimer in solution, we next examined whether DNA played a role in orchestrating BlcR tetramerization. The 51-bp DNA contains two IR pairs (IR1 and IR2 in Fig. 3B), and a straightforward model is that each IR pair is associated with one BlcR dimer. In this side-by-side tetramerization model, the two DNA-bound BlcR dimers are probably involved in direct protein-protein interaction due to the short gap (3 bp) separating IR1 and IR2. Protein oligomerization via direct protein-protein contact may account for the high cooperative binding of BlcR to DNA observed in the EMSA studies. However, for a productive protein-protein interaction between the two DNA-bound BlcR dimers for tetramerization, the arrangement of IR1 and IR2 along DNA is critical. To test this hypothesis, we altered the inter-IR gap by either deleting or adding one base pair and found that BlcR did not interact with the 50- or 52-bp DNA variants as indicated by the lack of heat exchange in ITC experiments (e.g. see Fig. 3D). Together, our DNA-binding studies indicate that tetrameric BlcR is the DNA-binding active form and suggest that DNA promotes BlcR tetramerization via optimal positioning of the two IR sequences that comprise the BlcR-binding site.
Like the wild type BlcR, BlcRY133A, BlcRT158A, and BlcRC220A bound to the 51-bp DNA with two dimers, consistent with the notion of the tetrameric state as the DNA-binding form and with similar affinity (Table 3). ITC analysis of BlcRD210A interaction with DNA was inconclusive because the reaction isotherm could not be fit into a valid biological model.
Enhanced DNA Binding by BlcRF147A
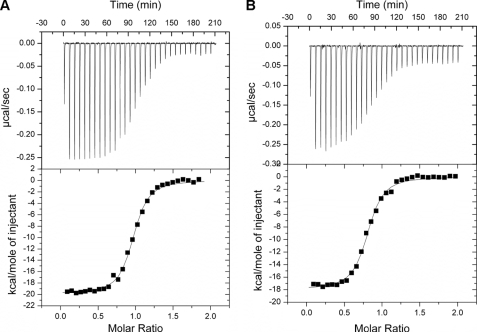
Strikingly, the tetrameric BlcRF147A bound to DNA in a 1:1 molar ratio (tetrameric BlcR/DNA = 0.94:1) and with a 10-fold stronger binding affinity (Kd of 54 nm) than the wild type (Kd of 490 nm; Fig. 4A and Table 3). The fact that a preassembled tetramer, BlcRF147A, bound DNA with an enhanced affinity was consistent with the model of tetrameric BlcR as the DNA-binding form and suggested that BlcRF147A simulates the DNA-bound BlcR tetramer. Supporting the similarity between BlcRF147A and the DNA-bound tetrameric wild type BlcR, EMSA studies show that the BlcRF147A-DNA complex migrated at the same electrophoretic rate as the BlcR-DNA complex (Fig. 5A, lanes 2 and 8). Moreover, 1-bp alterations of the inter-IR distance in DNA did not affect binding of BlcRF147A to the 50- and 51-bp DNA variants because BlcRF147A was able to bind both the 50- and 52-bp DNA fragments with similar affinity (Kd of 68 and 93 nm; see also Table 3 and Fig. 4B) as the 51-bp DNA. It is noteworthy that binding of BlcRF147A to these DNA variants was higher than that of the wild type BlcR with the canonical 51-bp DNA. These results further support the model of DNA-induced BlcR tetramerization and the importance of BlcR tetramerization in binding to DNA.
FIGURE 4.
DNA binding activity of tetrameric BlcRF147A. Shown are isotherms of the 51-bp DNA with BlcRF147A (A) and the 50-bp DNA with BlcRF147A (B). A and B are as described in the legend to Fig. 3, C and D.
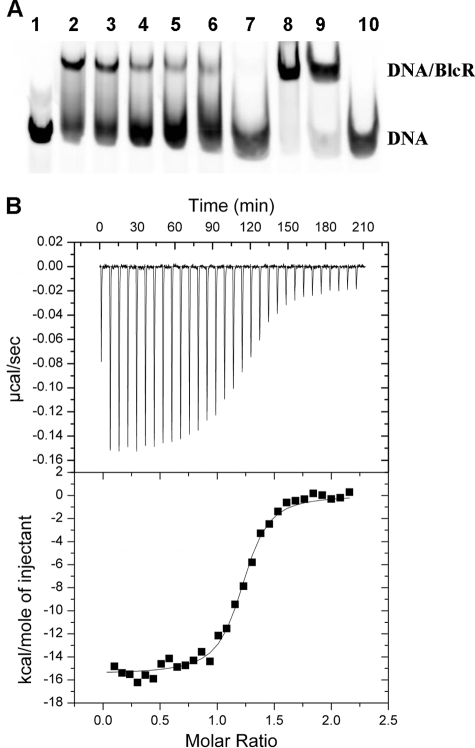
FIGURE 5.
Effect of SSA on DNA binding activity of BlcR and BlcRF147A. A, EMSA studies. Lane 1, 214-bp DNA. Lanes 2–7, SSA effect on BlcR-DNA; SSA concentrations were as follows: 0, 0.01, 0.1, 1, 10, and 100 mm, respectively. Lanes 8 and 9, SSA effect on BlcRF147A-DNA; SSA concentrations were 0 and 100 mm, respectively. Lane 10, DNA with 100 mm SSA. The DNA concentration remained 27.2 nm. B, ITC studies of titrating BlcRF147A into the 51-bp DNA in the presence of excess SSA (BlcR monomer/SSA = 1:6). A and B are as described in the legend to Fig. 3, C and D.
Effect of SSA on DNA Binding of BlcR and BlcRF147A
The EMSA results in Fig. 5A show that SSA reduced DNA binding activity of BlcR, as reported previously (25, 26). The similar binding affinity of BlcR for SSA (Kd of 0.7 μm) and for DNA (Kd of 490 nm) may account for the large amount of SSA required to effectively dissociate BlcR-DNA complex. Strikingly, even at high SSA concentration, the BlcRF147A-DNA complex remained stable (Fig. 5A, lane 9). Consistently, we found from ITC studies that BlcRF147A retained high affinity for DNA in the presence of excess SSA with a Kd of 32 nm (Fig. 5B). Given that SSA did not dissociate tetrameric BlcRF147A (Fig. 2), these results reinforce the model in which BlcR binds to DNA in a tetrameric form and DNA plays an important role in promoting BlcR tetramerization.
DISCUSSION
Proteins of the IclR family are newly classified bacterial transcriptional factors that regulate a wide range of cellular activities in response to environmental conditions. Despite their prevalence, information is scarce on the genetic elements they control, the signals they respond to, and the mechanisms by which they function. BlcR is among the limited number of IclR-type proteins for which activity has been characterized at the molecular level. Here we determined the crystal structure of dimeric BlcR and confirmed the SSA-binding site by mutational studies. Our biochemical studies demonstrated that tetrameric BlcR is the DNA-bound form and suggested that DNA may serve as a scaffold to facilitate BlcR tetramerization. The BlcRF147A mutant, locked in a tetrameric state, mimicked the DNA-bound BlcR tetrameric form in that it bound DNA with higher than the wild type affinity. We further demonstrated that elements in DNA (inter-IR distance), important for DNA-induced BlcR tetramerization in the wild type BlcR, did not affect DNA binding of this preassembled tetramer. SSA decreased binding affinity of BlcR for DNA; however, it did not reduce the BlcRF147A-DNA interaction, probably because SSA did not dissociate the BlcRF147A tetramer. Finally, our structural analysis revealed a dual role of Phe147 in SSA binding and tetramerization. Together, our biochemical and structural analyses offer a mechanism for SSA regulation on the DNA binding activity of BlcR. Results from our studies here provide a basic molecular model for the largely uncharacterized yet functionally important IclR transcription factors.
IR1 and IR2 Comprise a Single Binding Site for BlcR
Our findings suggest that IR1 and IR2 comprise a single binding site for BlcR. Because these sequences are the only ones identified thus far for BlcR, and our understanding of the DNA sequence requirements for BlcR binding is limited, it is conceivable that additional binding sites that we do not yet recognize are distantly located on DNA. In the E. coli lac operon (34), three operator sites, the primary O1 and the auxiliary O2 and O3, are located distantly at +11, −82, and +412 bp sites on DNA, and binding of both O1 and at least one of either O2 or O3 by the repressor LacI is required for effective repression. LacI exists as a tetramer, and the distal O2 and O3 can be brought to O1 via a DNA looping mechanism (35) in which one dimer within the LacI tetramer may bind to O1 while the other dimer associates with either O2 or O3 (36). The high stability of the LacI tetramer offsets the unfavorable energetics of forming a DNA loop structure. However, this protein tetramerization-driven DNA looping is not applicable to BlcR, because wild type BlcR is stable in solution as a dimer and apparently only forms a tetramer when binding to its target DNA. No additional distal sites are required for this tetramerization as it occurs on the 51-bp fragment. Thus, we consider that all of the DNA sequence elements required for tetrameric binding by BlcR are located near the promoter, as observed in the blcABC promoter, rather than widely separated as for the lac operon. However, it remains a formal possibility that currently unrecognized distal DNA-binding sites might alter BlcR that is primarily bound at the blcABC site.
Our findings that modifications of the inter-IR distance abolished BlcR-DNA interaction support the model that the two IRs are composed of one single BlcR-binding site. Based on the enhanced DNA binding of the preassembled BlcRF147A tetramer, we interpret that the importance of proper spatial arrangement of IR1 and IR2 is to engage the two BlcR dimers in direct protein-protein contact to form tetramer on DNA. Binding-induced protein oligomerization is an effective way to increase cooperativity in protein-DNA interaction and therefore to improve DNA binding specificity of transcription regulators. These findings on the specific positioning of IR1 and IR2 in the BlcR-binding site may be generally applicable to members of the IclR family and may aid in the identification of promoter sequences for the IclR-type regulators.
The β4/β5 Loop in the SSA-binding Domain Is a Potential Protein Interface for Forming the DNA-binding Tetramer
As mentioned, the crystallized TM-IclR forms a fortuitous asymmetric tetramer via the ligand-binding CTD, and regions corresponding to the β3/β4 and β4/β5 loops of BlcR are included in the large tetrameric interface of TM-IclR. Similarly, we found that the BlcR CTD (encompassing the SSA-binding domain) was involved in intermolecular contact between BlcR dimers in the crystal. The limited intermolecular interface is formed by the β3/β4 and β4/β5 loops (highlighted in Fig. 1A) mainly via electrostatic interactions: Glu127 (of β3/β4 loop), Arg148 (of β4/β5 loop), and Arg152 (of β5) of one BlcR dimer pair with Arg148, Glu127, and Arg152 of the other BlcR dimer, respectively. Although the observed intermolecular interactions of both TM-IclR and BlcR result from crystal packing, they suggest that regions in the ligand-binding CTD are capable of engaging in specific protein-protein interactions. The total buried intermolecular interface between BlcR dimers in the crystal structure is limited (596 Å2); however, the weak intermolecular interaction can be reinforced by external factors. For example, geometric constraints imposed by appropriately positioning the BlcR-binding sites on DNA can bring the two DNA-bound BlcR dimers close in proximity, optimal for productive protein-protein interaction.
Intriguingly, within the BlcR β4/β5 loop, residues 137–149 are structurally flexible (unresolved residues: 139–144 of monomer A and 143–145 of monomer B) because part of the loop is disordered in the crystal. Phe147, although located immediately next to the structurally unresolved region, is well ordered; its side chain projects into a hydrophobic path formed by Tyr133, Met151, Leu153, Ile214, Cys220, and Met218 (Fig. 1D). The docking of Phe147 to the hydrophobic path may serve to anchor both the C terminus of the β4/β5 loop and β5 to the core β-sheet structure of CTD. Substitution of Phe147 to a small residue, such as alanine, may compromise the anchoring function at position 147, resulting in further increase in conformational flexibility of the β4/β5 loop. The additional structural flexibility may allow the β4/β5 loop to adopt the conformation required for productive intermolecular contact in the tetramer. Thus, in addition to crystal packing analysis, our structural analysis accounts for BlcRF147A tetramerization, lending further support to the prediction that the β4/β5 loop is involved in tetramer formation.
SSA Allostery on DNA Binding Activity of BlcR
The hydrophobic path within which the BlcR Phe147 residue resides is part of the SSA-binding site, and the F147A mutation led to a ∼5-fold decrease in affinity for SSA. This functional role for Phe147 in SSA binding, combined with its structural importance via the β4/β5 loop in the intermolecular interaction (above), provides a mechanism by which SSA may modulate the DNA binding activity of BlcR. SSA occupancy at the SSA-binding site may relay via its interaction with Phe147 (and perhaps other residues within this binding region) to restrict the structural flexibility of the β4/β5 loop, undermining the already weak intermolecular interaction between BlcR dimers and interfering with formation of the DNA-bound, active tetramer. Thus, association with SSA allosterically prevents BlcR from associating with DNA. Interestingly, the tetrameric interaction in BlcRF147A appears to be strong enough to offset the disruptive effect from SSA association because the presence of SSA did not dissociate BlcRF147A tetramer and did not inhibit BlcR binding to DNA.
Associations with small ligands can allosterically control the DNA binding activity of transcription regulators by modifying conformations of the DNA-binding domains. In the trp repressor, binding of the cognate ligand l-tryptophan aligns the two DNA recognition helices (helix E) of the dimeric trp repressor at an optimal distance and orientates them in a conformation that facilitates their interaction with DNA (37, 38). The two DNA-binding domains of the trp repressor dimer are structurally independent and do not contribute to the dimer interface. In contrast, BlcR dimerizes via the DNA-binding domains, and their engagement in extensive interactions (Fig. 1B) prevents structural rearrangements of the domain, including α3, the putative DNA recognition helix. Thus, the stable dimeric interaction precludes the mechanism that SSA affects the conformation of α3, thereby regulating the DNA function of BlcR. Alternatively, many transcriptional regulators bind DNA as oligomers, and ligand association alters the oligomeric states, thereby affecting their DNA binding activity, in turn regulating transcription of target genes. ToxT, the virulence transcription regulator of Vibrio cholerae, binds to DNA most likely as dimer to activate its target genes (39). Virstatin has been found to inhibit ToxT function by stabilizing ToxT in monomeric form (39). In addition, cis-palmitoleic acid was found to occupy the putative hydrophobic dimeric interface to occlude ToxT from dimerization, accounting for its inhibition of DNA binding and a corresponding decrease in V. cholerae virulence (40). In this study, we show that, although BlcR exists as a dimer in solution, the tetrameric BlcR is the DNA-bound form and that BlcR tetramerization is mediated via DNA. Because the putative tetramerization interface overlaps with the SSA-binding site, we propose that SSA association distorts the tetrameric interface, destabilizing the already compromised metastable tetramer. As a result, SSA binding dissociates the DNA-mediated BlcR tetramer into the dimeric form, disrupts the cooperative DNA binding from the two BlcR dimers, and causes dissociation of BlcR-DNA. Our model is supported by results from tetrameric BlcRF147A. As a highly stable tetramer, BlcRF147A appears to overcome the disruptive effect of SSA binding on tetramerization because it remains as a tetramer and bound to DNA in the presence of excess SSA.
Supplementary Material
Acknowledgments
We thank Dr. Cheng Kao's laboratory for the DSF experiments and Melissa Illingworth for generating several of the BlcR mutants.
This research was supported in part by National Science Foundation Grant MCB-0416447 (to L. C.) and by National Institutes of Health Grant R01-GM80546 (to C. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (code 3MQ0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- IclR
- isocitrate lyase transcription regulator
- SSA
- succinate semialdehyde
- SUMO
- small ubiquitin-like modifier
- ITC
- isothermal titration calorimetry
- DSF
- differential scanning fluorimetry
- NTD
- N-terminal domain
- CTD
- C-terminal domain
- IR
- inverted repeat.
REFERENCES
- 1. Galinier A., Nègre D., Cortay J. C., Marcandier S., Maloy S. R., Cozzone A. J. (1990) Nucleic Acids Res. 18, 3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sunnarborg A., Klumpp D., Chung T., LaPorte D. C. (1990) J. Bacteriol. 172, 2642–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krell T., Molina-Henares A. J., Ramos J. L. (2006) Protein Sci. 15, 1207–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Molina-Henares A. J., Krell T., Eugenia Guazzaroni M., Segura A., Ramos J. L. (2006) FEMS Microbiol. Rev. 30, 157–186 [DOI] [PubMed] [Google Scholar]
- 5. Guazzaroni M. E., Krell T., Gutiérrez del Arroyo P., Vélez M., Jiménez M., Rivas G., Ramos J. L. (2007) J. Mol. Biol. 369, 927–939 [DOI] [PubMed] [Google Scholar]
- 6. Romero-Steiner S., Parales R. E., Harwood C. S., Houghton J. E. (1994) J. Bacteriol. 176, 5771–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsoi T. V., Plotnikova E. G., Cole J. R., Guerin W. F., Bagdasarian M., Tiedje J. M. (1999) Appl. Environ. Microbiol. 65, 2151–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reverchon S., Nasser W., Robert-Baudouy J. (1991) Mol. Microbiol. 5, 2203–2216 [DOI] [PubMed] [Google Scholar]
- 9. Jiang H., Kendrick K. E. (2000) J. Bacteriol. 182, 5521–5529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yamazaki H., Ohnishi Y., Horinouchi S. (2003) J. Bacteriol. 185, 1273–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Traag B. A., Kelemen G. H., Van Wezel G. P. (2004) Mol. Microbiol. 53, 985–1000 [DOI] [PubMed] [Google Scholar]
- 12. Brune I., Jochmann N., Brinkrolf K., Hüser A. T., Gerstmeir R., Eikmanns B. J., Kalinowski J., Pühler A., Tauch A. (2007) J. Bacteriol. 189, 2720–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang H. B., Wang L. H., Zhang L. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4638–4643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harrison S. C. (1991) Nature 353, 715–719 [DOI] [PubMed] [Google Scholar]
- 15. Pabo C. O., Sauer R. T. (1992) Annu. Rev. Biochem. 61, 1053–1095 [DOI] [PubMed] [Google Scholar]
- 16. Brennan R. G., Matthews B. W. (1989) J. Biol. Chem. 264, 1903–1906 [PubMed] [Google Scholar]
- 17. Maloy S. R., Nunn W. D. (1982) J. Bacteriol. 149, 173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kornberg H. L. (1966) Biochem. J. 99, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nègre D., Cortay J. C., Galinier A., Sauve P., Cozzone A. J. (1992) J. Mol. Biol. 228, 23–29 [DOI] [PubMed] [Google Scholar]
- 20. Pan B., Unnikrishnan I., LaPorte D. C. (1996) J. Bacteriol. 178, 3982–3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yamamoto K., Ishihama A. (2003) Mol. Microbiol. 47, 183–194 [DOI] [PubMed] [Google Scholar]
- 22. Lorca G. L., Ezersky A., Lunin V. V., Walker J. R., Altamentova S., Evdokimova E., Vedadi M., Bochkarev A., Savchenko A. (2007) J. Biol. Chem. 282, 16476–16491 [DOI] [PubMed] [Google Scholar]
- 23. Zhang R. G., Kim Y., Skarina T., Beasley S., Laskowski R., Arrowsmith C., Edwards A., Joachimiak A., Savchenko A. (2002) J. Biol. Chem. 277, 19183–19190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carlier A., Chevrot R., Dessaux Y., Faure D. (2004) Mol. Plant Microbe Interact. 17, 951–957 [DOI] [PubMed] [Google Scholar]
- 25. Wang C., Zhang H. B., Wang L. H., Zhang L. H. (2006) Mol. Microbiol. 62, 45–56 [DOI] [PubMed] [Google Scholar]
- 26. Chai Y., Tsai C. S., Cho H., Winans S. C. (2007) J. Bacteriol. 189, 3674–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang L. H., Weng L. X., Dong Y. H., Zhang L. H. (2004) J. Biol. Chem. 279, 13645–13651 [DOI] [PubMed] [Google Scholar]
- 28. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 29. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 30. Walker J. R., Altamentova S., Ezersky A., Lorca G., Skarina T., Kudritska M., Ball L. J., Bochkarev A., Savchenko A. (2006) J. Mol. Biol. 358, 810–828 [DOI] [PubMed] [Google Scholar]
- 31. Niesen F. H., Berglund H., Vedadi M. (2007) Nat. Protoc. 2, 2212–2221 [DOI] [PubMed] [Google Scholar]
- 32. Donald L. J., Chernushevich I. V., Zhou J., Verentchikov A., Poppe-Schriemer N., Hosfield D. J., Westmore J. B., Ens W., Duckworth H. W., Standing K. G. (1996) Protein Sci. 5, 1613–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Donald L. J., Hosfield D. J., Cuvelier S. L., Ens W., Standing K. G., Duckworth H. W. (2001) Protein Sci. 10, 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oehler S., Eismann E. R., Krämer H., Müller-Hill B. (1990) EMBO J. 9, 973–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schleif R. (1992) Annu. Rev. Biochem. 61, 199–223 [DOI] [PubMed] [Google Scholar]
- 36. Lewis M., Chang G., Horton N. C., Kercher M. A., Pace H. C., Schumacher M. A., Brennan R. G., Lu P. (1996) Science 271, 1247–1254 [DOI] [PubMed] [Google Scholar]
- 37. Zhang R. G., Joachimiak A., Lawson C. L., Schevitz R. W., Otwinowski Z., Sigler P. B. (1987) Nature 327, 591–597 [DOI] [PubMed] [Google Scholar]
- 38. Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. (1988) Nature 335, 321–329 [DOI] [PubMed] [Google Scholar]
- 39. Häse C. C., Mekalanos J. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3183–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lowden M. J., Skorupski K., Pellegrini M., Chiorazzo M. G., Taylor R. K., Kull F. J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2860–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.