Abstract
Porcine surfactant protein D (pSP-D) displays distinctively strong, broad-range inhibitory activity against influenza A virus (IAV). N-Linked glycosylation of the carbohydrate recognition domain (CRD) of pSP-D contributes to the high affinity of this collectin for IAV. To investigate the role of the N-linked glycan further, HEK293E protein expression was used to produce recombinant pSP-D (RpSP-D) that has similar structural and antiviral properties as NpSP-D. We introduced an additional N-linked glycan in the CRD of RpSP-D but this modification did not alter the antiviral activity. Human SP-D is unglycosylated in its CRD and less active against IAV compared with pSP-D. In an attempt to modify its antiviral properties, several recombinant human SP-D (RhSP-D) mutants were constructed with N-linked glycans introduced at various locations within its CRD. To retain lectin activity, necessary for the primary interactions between SP-D and IAV, N-linked glycosylation of RhSP-D was shown to be restricted to the corresponding position in the CRD of either pSP-D or surfactant protein A (SP-A). These N-glycosylated RhSP-D mutants, however, did not show increased neutralization activity against IAV. By developing RhSP-D mutants that also have the pSP-D-specific Ser-Gly-Ala loop inserted in the CRD, we could demonstrate that the N-linked glycan-mediated interactions between pSP-D and IAV involves additional structural prerequisites of the pSP-D CRD. Ultimately, these studies will help to develop highly effective SP-D-based therapeutic and prophylactic drugs against IAV.
Keywords: Antiviral Agents, Glycoprotein, Glycosylation, Innate Immunity, Site-directed Mutagenesis, C-type Lectin, Influenza A Virus, Lung, Sialic Acid
Introduction
Influenza is a highly contagious respiratory disease of humans and animals and new epidemics and pandemics are likely to occur in the future (1). The degree of infection, transmission, and maintenance of influenza A viruses (IAVs)2 in host organisms is mainly determined by the effectiveness of the immunological responses that are crucial for early containment of infection as well as ultimate viral clearance. The first layer of protection against IAV infection in organisms depends on the activation of the innate immune response. This includes cellular defense mechanisms (e.g. neutrophils, macrophages) and soluble factors that can inhibit infection directly, but that also can enhance neutralization and clearance of microorganisms by interacting with host immune cells (2–4). An important class of mammalian innate immune proteins known as “collectins” is involved in this early immune response to limit infection and spread of pathogens at mucosal surfaces of the naive host (5, 6). Well characterized members of these collagenous C-type lectins are the two lung collectins, surfactant proteins A (SP-A) and D (SP-D, reviewed in Ref. 6–8), and the mannan binding lectin (9), which are expressed in the lung (SP-A and SP-D) (10), in extra-pulmonary mucosal tissues (SP-D) (10), and in the liver (mannan binding lectin).
Previously it was shown that SP-A, SP-D, and the scavenger receptor cysteine-rich glycoprotein 340, help to protect lung epithelial cells against infection by IAV (11–13). By now, it is well established that SP-D is particularly important because it exhibits the strongest anti-IAV activity when compared with other collectins as indicated by various in vitro studies (12, 14). Its protective role is also underlined by studies with SP-D knock-out mice (15, 16). SP-D-mediated protection is primarily established by reducing the number of infectious particles via collectin-mediated aggregation of viral particles. This helps to prevent attachment of virus to the host respiratory epithelium, but also induces phagocytic responses resulting in enhanced viral clearance (opsonization). In addition, SP-D is involved in control of pulmonary inflammation at early stages of IAV infection (15–17), prevents deactivation of neutrophils (18), and can also bridge the innate and adaptive immune responses by modulating the function of dendritic cells and T cells (19).
Pigs are known to be highly susceptible to infection by various subtypes of IAVs and may support influenza viral reassortment (20) and interspecies transmission into humans, as illustrated by the 2009 flu pandemic caused by swine origin IAV subtype H1N1 (21). The respiratory epithelial cells in pigs are decorated with ligands for both avian and human IAVs and this at least partially explains why these animals can be considered “mixing vessels” for IAV. To learn more about innate defense mechanisms against IAV in this animal species, we initiated studies into the structure and function of porcine lung collectins, in particular porcine SP-D (pSP-D) (22). Our investigations revealed that asparagine-linked complex-type glycans present in the CRD of lung collectins can result in substantially enhanced IAV neutralization activity in vitro (23). Although Asn-linked glycosylation of the lectin domain is conserved in SP-A from all mammalian species characterized to date, pSP-D is unique in that it is the only SP-D species known to be equipped with a complex sugar in its lectin domain (24). In vitro neutralization studies with IAV revealed that the presence of this sugar moiety in the “pathogen binding domain” of the SP-D molecule not only enhances its antiviral activity, but also broadens the range of viral strains that can be inhibited by pSP-D (23). These studies demonstrated that although the CRD of SP-D plays a key role by binding sugars present on the surface of IAV, an additional mode of interaction can be facilitated by the terminal sialic acid (SA) residues present on this N-glycan that are believed to interact with the SA receptor present on the hemagglutinin (HA) of IAV. Moreover, the type of linkage between the SA residues and the penultimate galactose residues (either α(2,3)-linked or α(2,6)-linked) within this Asn-linked sugar have been shown to be important for neutralization of strains that have a corresponding SA-linkage receptor specificity of their HA receptor (25). The SA linkage specificity of the viral HA is a major determinant of IAV because it restricts infection to certain host species (e.g. human or avian species), due to specified linkage patterns of SAs present on the host pulmonary epithelial cell membrane. Based upon these studies we hypothesized that glycan modifications of collectins, e.g. changing the number, location, and terminal sialylation of asparagine-linked oligosaccharide moieties in the CRD, may substantially enhance recognition and neutralization of IAV and, most probably, other pathogens.
In this paper we describe the mammalian expression of recombinant versions of wild-type pSP-D (RpSP-D) and wild-type hSP-D (RhSP-D) and several derivatives that have N-linked oligosaccharides introduced at different locations in the CRD. The expression, purification, and biochemical characterization of these recombinant SP-Ds (RSP-Ds) was followed by in vitro studies with several IAV strains and showed that RpSP-D, like natural pSP-D (NpSP-D), exhibits distinctively strong inhibitory activity compared with RhSP-D. In an attempt to enhance the moderate antiviral activity of RhSP-D, we developed several derivatives having N-linked glycans introduced in the CRD by site-directed mutagenesis, and investigated their inhibitory activity against IAV. It was shown that the contributory effect of N-linked glycans to the antiviral activity of SP-D against IAV is complex and requires additional intrinsic structural features present in the CRD of pSP-D.
EXPERIMENTAL PROCEDURES
Reagents
iScript cDNA synthesis kit was purchased from Bio-Rad, Taq DNA polymerase supplied by Fermentas (St. Leon-Roth, Germany), TOPO T/A cloning kit by Invitrogen, and the QuikChange II site-directed mutagenesis kit by Stratagene (Bio-connect, Huissen, The Netherlands). Qiagen spin miniprep and midiprep kits, gel extraction kit, and proofstart DNA Polymerase were all provided by Qiagen Benelux BV (Venlo, The Netherlands). All restriction enzymes were supplied by New England Biolabs (van Westburg, Leusden, The Netherlands). The expression vector pUPE101-01 was provided by U-Protein Express (Utrecht, The Netherlands). Dulbecco's modified Eagle's medium (DMEM high glucose), mannan-Sepharose, collagenase (type VII from Clostridium histolyticum), endotoxin-free plasmid mega kit, and BS3 cross-linker (suberic acid bis(3-sulfo-N-hydroxysuccinimide ester, sodium salt) were purchased from Sigma. N-Glycanase and the DIG Glycan Differentiation Kit were both supplied by Roche Diagnostics. The ToxinSensorTM Chromogenic LAL Endotoxin Assay Kit was purchased from GenScript (Piscataway, NJ). Porcine bronchoalveolar lavage was obtained from adult porcine lungs (The Central Laboratory Animal Research Facility Utrecht, Utrecht, The Netherlands).
Construction of Full-length pSP-D cDNA
Total RNA, extracted from adult porcine lung tissue, was used in the iScript cDNA synthesis reaction following the manufacturer's recommendations to obtain porcine lung cDNA. This was used as a template in a standard 50-μl PCR using proofstart DNA polymerase to amplify the full coding sequence of pSP-D including the signal peptide sequence, starting 8 nucleotides downstream the start of transcription (numbers according to Ref. 22), ending 142 nucleotides downstream from the stop codon (total length: 1315 bp). For subcloning purposes, the forward primer included a BsmBI/BamHI restriction site (PSPDFOR, gcgtctcggatcc GCC TGG AGA TTC TGA GCT CTA G. All DNA sequences described in this study are 5′ to 3′ with gene-specific sequences in capital letters) and the reverse primer contained a NotI sequence (PSPDREV, 5′-gcggccgc TGA GGG AGG CGT TCC ATA GGC-3′). The amplified DNA fragment was gel purified and 3′ A-overhangs were added by incubating 10 min at 72 °C in the presence of 1 unit of Taq DNA polymerase and 0.4 mm dATP. The gel purified product was cloned into pCR4-TOPO sequencing vector using a TOPO T/A cloning kit (Invitrogen). Insert-positive clones were isolated and sequenced to rule out any errors. The full-length pSP-D construct was digested with BsmBI/NotI, gel extracted, and ligated into pUPE-101-01 expression vector, a modified version of the pTT3 expression vector (26). No tags were used. Subclones containing the cDNA were identified by BamHI/NotI restriction analysis.
Site-directed Mutagenesis
Site-directed mutagenesis was performed on the full-length pSP-D cDNA clone in pCR4-TOPO vector by overlap extension PCR using the QuikChange II site-directed mutagenesis kit. RhSP-D and derivatives were produced using the full-length hSP-D cDNA clone provided by Dr. E. C. Crouch (Washington University, St. Louis, MO) (27). This clone was used as a template for PCR cloning into pCR4-TOPO as described for RpSP-D in the previous section, to introduce BsmBI/BamHI (5′-end) and NotI (3′-end) restriction sites to accommodate subcloning in pUPE-101.01. The following mutagenesis primers were used to generate the N303Q mutation to delete the N-glycosylation site in the CRD of pSP-D resulting in the RpSP-DdNG mutant: PSPDKOFOR, GAC TGA CAT CAA GAC GGA GGG CCA GTT CAC CTA CCC CAC GGG GGA G, with PSPDKOREV, CTC CCC CGT GGG GTA GGT GAA CTG GCC CTC CGT CTT GAT GTC AGT. To generate a Q282N mutation to produce the RpSP-D2N mutant having an additional N-glycan in the CRD (P2N, Fig. 1), the following primers were used: PSPDKIFOR, GAG AAC GAG GCC TTG AGC AAC CTG ACC ACA GCT CAG AAT AAG GC; PSPDKIREV, GCC TTA TTC TGA GCT GTG GTC AGG TTG CTC AAG GCC TCG TTC TC. The full-length human SP-D clone in PCR4-TOPO was used as a template to introduce N-glycosylation sequence motifs at one of six different locations in the CRD of hSP-D (N1, N2, N3, N4, N5, and N6, Fig. 1). Mutagenesis primer pairs used were as follows: HUD1KIFOR, CAA GAC AGC AGG CTT TGT AAA AAA CTT TAC GGA GGC ACA GCT GCT GTG with HUD1KIREV, GCA CAG CAG CTG TGC CTC CGT AAA GTT TTT TAC AAA GCC TGC TGT CTT G (P253N, mutant RhSP-DN1); HUD2KIFOR, CTC CAC GCT CTG CCG CTG AGA ATG CCA CCT TGC AAC AGC TGG TCG TAG C with HUD2KIREV, GCT ACG ACC AGC TGT TGC AAG GTG GCA TTC TCA GCG GCA GAG CGT GGA G (A279T, mutant RhSP-DN2); HUD3KIFOR, GCT GGT CGT AGC TAA GAA CGA GAC TGC TTT CCT GAG CAT GAC TGA TTC C with HUD3KIREV, GGA ATC AGT CAT GCT CAG GAA AGC AGT CTC GTT CTT AGC TAC GAC CAG C (A290T, mutant RhSP-DN3); HUD4KIFOR, GAC TGA TTC CAA GAC AGA GGG CAA CTT CAC CTA CCC CAC AGG AGA G with HUD4KIREV, CTC TCC TGT GGG GTA GGT GAA GTT GCC CTC TGT CTT GGA ATC AGT C (K303N, mutant RhSP-DN4); HUD5KIFOR, CAG GAG AGT CCC TGG TCA ACT ATA CCA ACT GGG CCC CAG GGG AG with HUD5KIREV, CTC CCC TGG GGC CCA GTT GGT ATA GTT GAC CAG GGA CTC TCC TG (S315T and insertion of an Asn codon between Val313 and Tyr314, only performed on mutant RhSP-DN4 as template, yielding RhSP-DN4,5); and finally, HUD6KIFOR, CGG GCA GAG GAC TGT GTG GAG AAC TTC ACC AAT GGC AAG TGG AAT GAC with HUD6KIREV, GTC ATT CCA CTT GCC ATT GGT GAA GTT CTC CAC ACA GTC CTC TGC CCG (I334N, mutant RhSP-DN6). Two RhSP-D double glycan mutants were produced by performing an additional round of site-directed mutagenesis on mutant RhSP-DN4 with primers HUD5KIFOR/HUD5KIREV (RhSP-DN4,5, see above) or HUD6KIFOR/HUD6KIREV (RhSP-DN4,6). In addition, RhSP-D and RhSP-DN4 were used as a template to insert the pSP-D-specific Ser329-Gly330-Ala331 sequence (SGA loop) between residues Ser328 and Glu329 of the CRD of hSP-D, using the following set of mutagenesis primers: HSPDSGAFOR, GCC CAA CGA TGA TGG CGG GTC AAG CGG AGC AGA GGA CTG TGT GGA GAT C, and HSPDSGAREV, GAT CTC CAC ACA GTC CTC TGC TCC GCT TGA CCC GCC ATC ATC GTT GGG C. The two resulting clones were annotated: RhSP-D-SGA and RhSP-DN4-SGA, respectively. After sequence verification, the pCR4-TOPO inserts were digested with BsmBI/NotI, gel purified, and transferred to pUPE101.01 as described for RpSP-D.
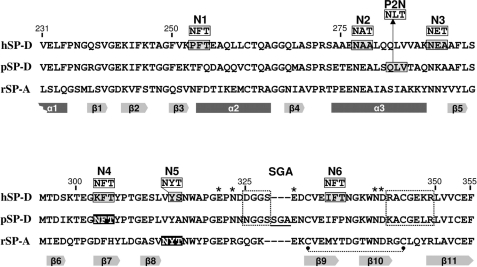
FIGURE 1.
Locations of N-linked glycans in the CRD sequence of pSP-D and hSP-D derivatives. Amino acid sequence alignment of the CRDs of human SP-D (hSP-D, GenBank accession code X65018), porcine SP-D (pSP-D, AF132496), and rat SP-A (rSP-A, U43092) with the numbering according to the human SP-D sequence and dashes to indicate spaces that are inserted to maximize the identity across the alignment. Positions of N-glycosylations introduced via site-directed mutagenesis into the sequence of pSP-D (P2N) and hSP-D (N1–N6) are indicated; sequences subjected to mutation are boxed in gray with the resulting sequences indicated above. N-Glycosylation sequence motifs as present in wild-type pSP-D and the conserved N-glycosylation site in SP-A are shaded black, the SGA insert present in pSP-D is underlined. Secondary structural regions are indicated as gray arrows (β-sheets) and dark grey bars (α-helices) below the sequence, deduced from the crystal structure of rat SP-A (45). SP-D groove regions (boxed with dotted lines), cysteine bridge (dotted line), and key residues necessary for coordination of Ca2+ ions and hydroxyl groups of oligosaccharides (asterisks) are indicated.
Recombinant Expression of Porcine and Human SP-Ds
Recombinant collectins were produced using HEK293 cells stably expressing Epstein-Barr virus nuclear antigen 1. HEK293E cells were transiently transfected as described previously (28). The SP-D sequence-containing pUPE101.01 expression plasmids were purified using an endotoxin-free plasmid maxiprep kit. Expression media containing the secreted proteins of interest were harvested by centrifugation 6 days post-transfection. Large-scale expression media were concentrated 10-fold and diafiltrated into 5 mm Hepes, 0.9% NaCl, 5 mm EDTA, pH 7.4, using the Quixstand Hollow Fiber System (GE Healthcare) using a cartridge with a 10-kDa cutoff.
Purification of Porcine and Human SP-Ds
NpSP-D was isolated from porcine bronchoalveolar lavage fluid as described previously (24) with the exception of Tris-HCl, which was replaced by Hepes (5 mm, pH 7.4) in all buffers. This protocol was also used for the purification of RSP-Ds from the HEK293E cell-free diafiltrated medium but instead of a single overnight incubation at 4 °C, the medium was subjected to batchwise overnight incubation in the presence of 5 mm calcium chloride and mannan-agarose (5-ml bed volume/liter of medium). This procedure was repeated twice to increase yield. Purification of RSP-Ds was performed in the absence of Tween 80 throughout the procedure. Mannan-bound SP-D was eluted with 5 mm Hepes, 0.9% NaCl, 5 mm EDTA, pH 7.4, filtrated through a 5-μm filter and concentrated to ∼5 ml via ultrafiltration with Amicon ultracel centrifugal filters (50-kDa molecular mass cut-off; Millipore). Gel filtration chromatography was performed using an ÄKTA purifier10 system, which was equipped with a Hiload 16/60 Superdex 200 PREP GRADE column. Fractions were analyzed by SDS-PAGE analysis and Western blot analysis. Pooled fractions were further concentrated by ultracel centrifugation and analyzed for protein content. Endotoxin levels were determined with the Toxinsensor LAL assay kit (ranging between 10 and 100 pg/μg of SP-D).
Biochemical Characterization of SP-Ds
SDS-PAGE analysis, Western blot analysis, and enzymatic digestions with collagenase to produce a collagenase-resistant fragment (CRF) of SP-D, and N-glycanase to remove N-linked oligosaccharides from SP-D were performed as described in detail for the characterization of NpSP-D (24). Chemical cross-linking of native SP-D was achieved with BS3 cross-linker (suberic acid bis(3-sulfo-N-hydroxysuccinimide ester): 5 μg of SP-D was dissolved in 5 mm Hepes, 100 mm NaCl, 5 mm EDTA, pH 7.4, BS3 was added to a final concentration of 5 mm and the mixture was incubated for 30 min at RT. The reaction was stopped by adding Laemmli sample buffer and subsequent boiling for 2 min after which the sample was analyzed by gradient (4–15%) SDS-PAGE. The presence of α(2,3)-linked and α(2,6)-linked terminal SA residues was assessed by Western blotting, followed by detection with digoxigenin-labeled lectins according to the instructions supplied by the manufacturer (DIG Glycan Differentiation Kit, Roche Diagnostics). Desialylation of SP-D was carried out with SA-linkage-specific recombinant sialidase as described (25).
Identification of Glycosylated Asn Residues in the CRD of SP-D by Mass Spectrometry
The mannan-purified CRFs of RpSP-D and RpSP-D2N were subjected to N-glycanase treatment to enzymatically remove all the N-linked glycans. As a result, the Asn residues from which the glycans were removed were deaminated to Asp residues, which resulted in an increase in the peptide mass with 1 mass unit, a mass difference resolvable by MS (29). After N-glycan removal, the RpSP-D and RpSP-D2N-derived protein fragments were separated by SDS-PAGE (15%) and the Coomassie-stained bands of interest were excised from the gel and sequentially digested with trypsin and chymotrypsin as described (30). Peptides were analyzed as described by Vogels et al. (31) and results were searched against a custom data base containing 4 proteins (RpSP-D, RpSP-D2N, RpSP-D-N337D, and RpSP-D2N-N316D/N337D) using an in house Mascot server (variable modification: oxidation on Met, His, and Trp; fixed modification: carbamidomethlyl on Ser; 2 missed cleavages allowed, 50 ppm mass accuracy for peptides, 0.8 mass unit for fragments).
Modeling the N-Linked Oligosaccharides in the CRD of hSP-D
The crystal structure of the trimeric neck CRD of human surfactant protein D in complex with α1,2-dimannose (PDB code 3G83 (32)) was used as a template to visualize the approximate locations of the N-linked oligosaccharide as present in the CRD of wild-type pSP-D (Asn303), and the N-linked glycans artificially introduced via site-directed mutagenesis resulting in RpSP-D2N (Asn282 and Asn303, Fig. 2). The N-linked glycosylation motifs that were generated in the different RhSP-D mutants, resulting in potential Asn-linked glycans at positions 253, 277, 288, 303, and 334 and at the Asn residue inserted between Val313 and Tyr314 (all human SP-D numbering), were modeled accordingly using model-building tools for molecular graphics, distributed as “Coot” (33). The atoms of the pentasaccharide core of each N-linked oligosaccharide attached to these Asn residues were rendered in space filling representation using the GlyProt server.
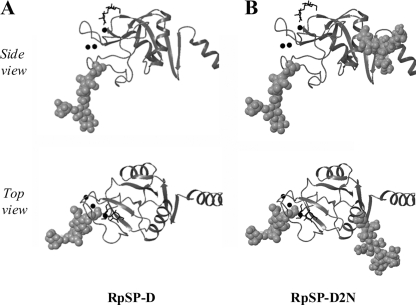
FIGURE 2.
Locations of the N-linked glycans of RpSP-D (A) and RpSP-D2N (B). Ribbon diagrams of the CRD from hSP-D as published (SP-D·dimannose complex, PDB 3G83 (32)) in which the approximate locations of the N-glycosylation site as present in the CRD of RpSP-D (and NpSP-D, panels A), and the extra N-glycan introduced via site-directed mutagenesis (RpSP-D2N) are highlighted. The atoms of the Asn side chain and the pentasaccharide core of the N-linked glycan are shown in space-filling representation. Shown are views from the side of a single monomer in the trimer, and a top view looking down the trimer axis into the lectin binding pocket; part of the neck domain is shown on the right side of each structure. Ca2+-ions are represented by black spheres and the location of the dimannose molecule (black structure) indicates the lectin binding site of the CRD. Modeling details are described under “Experimental Procedures.”
Hemagglutination Inhibition (HAI) Assay
The IAV strains used in this study were grown in chorioallantoic fluid of 10-day-old chicken eggs and purified on a discontinuous sucrose gradient as described previously (34) and after dialysis against PBS, viral stocks were aliquoted and stored at −70 °C. The A/Phillipines/82(H3N2) (Phil) and the bovine serum β-inhibitor-resistant variant, PhilBS were provided by Dr. E. M. Anders (University of Melbourne, Victoria, Australia). The A/Puerto Rico/8/34(H1N1) (PR-8) strain was a kind gift of Dr. J. Abramson (Department of Pediatrics, Bowman Gray School of Medicine, Winston-Salem, NC). The hemagglutination titer of each virus preparation was determined by titration of virus samples in PBS++ (PBS with 1 mm calcium chloride and 0.5 mm magnesium chloride) with thoroughly washed human type O, Rh(−) red blood cells as described (35). HAI was measured by serially diluting collectin preparations (25 μl) in round-bottom 96-well plates using PBS++ as a diluent. After adding 25 μl of IAV (4 hemagglutination units) in each well, the IAV/protein mixture was incubated for 10 min at room temperature. Thereafter, 50 μl of a type O human erythrocyte suspension (in PBS++) was added to each well and plates were incubated for 2 h at RT. The minimal concentration of SP-D required to fully inhibit the hemagglutination activity of the viral suspension was detected by the formation of erythrocyte pellets in wells having the lowest amounts of SP-D. The highest concentrations tested were 2 μg/ml for the pSP-D preparations and 50 μg/ml for the RhSP-D.
Viral Aggregation
Aggregation of IAV particles (Phil82) was assessed for RpSP-D and RpSP-D2N and was carried out as previously described (36). For both collectin preparations, 3.2 μg/ml was mixed with a suspension of virus (Phil) in a final volume of 1 ml. During stirring, the light transmission was monitored for 15 min using an SLM/Aminco 8000C spectrofluorimeter (excitation and emission wavelengths were 350 nm). Viral aggregation resulted in a decline in light transmission and results were expressed as a percentage of control light transmission (virus without RpSP-Ds).
Neutralization of Infectivity Assay Using MDCK Cells
Madin-Darby canine kidney confluent monolayers were prepared in 96-well plates and infected for 45 min at 37 °C with IAV (Phil), which was preincubated for 30 min at 37 °C in the absence (control) or presence of increasing amounts of collectins. After washing with serum-free and glucose-free DMEM, the monolayers were incubated for 7 h at 37 °C, washed again, fixed, and fluorescein isothiocyanate-labeled for IAV nucleoprotein as described (37). Fluorescent foci, which appeared to be single infected cells in general, were counted. Results were expressed as a percentage of control foci present after infection with collectin-treated virus as compared with untreated virus.
Measurement of Binding of Collectins to IAV
Binding of collectins to IAV strains was measured by ELISA in which suspensions of virus (1 μg/ml) were dried in 96-well plates overnight, washed, and incubated with PBS containing 2.5% fatty acid-free BSA for blocking. Subsequently, various collectins (2 μg/ml) diluted in PBS were added and incubated for 30 min at 37 °C. After washing to remove unbound collectins, captured collectins were detected with monoclonal anti-human antibodies (30 min at 37 °C), followed by incubation with horseradish peroxidase-conjugated donkey anti-mouse F(ab′)2 fragments (Jackson Immunochemicals) for 30 min at 37 °C. Bound secondary antibodies were detected with 3,3′,5,5′-tetramethylbenzidine peroxidase substrate (Bio-Rad). The reaction was stopped with 0.5 m H2SO4 and absorbance was measured with an ELISA plate reader at 450 nm. All data points were performed in duplicate.
Neutrophil Uptake
IAV (Phil) was fluorescein isothiocyanate-labeled as described previously (36) and preincubated with various concentrations of RSP-D or with control buffer for 30 min at 37 °C. The mixtures were added to freshly isolated human neutrophils and incubated for 30 min at 37 °C. Trypan blue (0.2 mg/ml) was added to quench extracellular fluorescence. After washing, cells were fixed with paraformaldehyde and the mean neutrophil-associated fluorescence (>1000 cells counted/sample) was measured by flow cytometry. Results were expressed as a percentage of IAV uptake in the absence of collectins. Statistical comparisons for all IAV assays were made using Student's paired, two-tailed t test.
RESULTS
Biochemical Characterization of RpSP-D
The purification of NpSP-D from porcine bronchoalveolar lavage fluid was performed as described previously using mannan affinity chromatography followed by gel filtration chromatography (24). The same approach was used for purification of RpSP-D from HEK293E cell medium transiently transfected with the full-length pSP-D sequence-containing pUPE101.01 expression vector. RpSP-D, like NpSP-D, bound to mannan-agarose in a Ca2+-dependent manner and was analyzed by reducing SDS-PAGE. The RpSP-D preparation showed a major band of ∼50 kDa, comparable with the monomeric size of NpSP-D preparations (Fig. 3A). Western blot analysis of the HEK293E cell-derived medium, using a previously characterized rabbit anti-porcine SP-D IgG antibody (24), showed that after two rounds of mannan-agarose extraction, a RpSP-D fraction was still detectable in the medium that could not be purified with mannan-agarose. The minor bands running at higher molecular weight represent a small fraction of unreducable SP-D as shown by Western blot analysis (Fig. 3A). Digestion of the purified protein with N-glycanase results in a mobility shift identical for NpSP-D and RpSP-D, indicating that the degree of N-glycosylation is similar for both SP-Ds. Analysis of the mannan-purified SP-D fraction by non-reducing SDS-PAGE reveals differences in assembly between NpSP-D and RpSP-D, the latter showing bands that correspond to the size of monomeric, dimeric, and trimeric SP-D. In contrast, NpSP-D migrates as trimers and higher-order multimers (Fig. 3B). Chemical cross-linking of SP-D by amidation of primary amine groups and subsequent analysis by gradient gel electrophoresis shows a band of high molecular weight in both fractions (>500 kDa) and an additional band corresponding to ∼150 kDa in the RpSP-D preparations. Size exclusion chromatography on both pSP-D preparations using a Superdex 200 column also indicated the presence of a second peak running at lower molecular weight present in RpSP-D, in addition to the main peak present in the void volume (>600 kDa) of the column (results not shown). The average yield of RpSP-D purified from 1 liter of HEK293E cell-derived medium ranged from 2 to 2.5 mg.
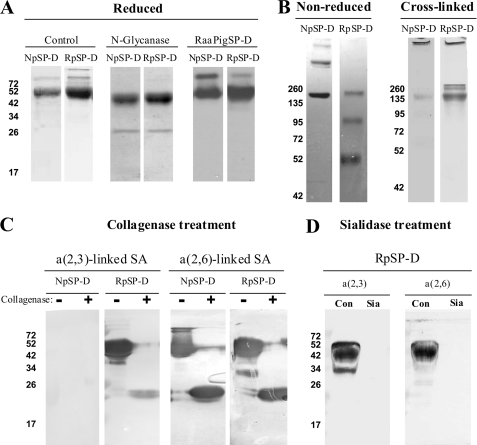
FIGURE 3.
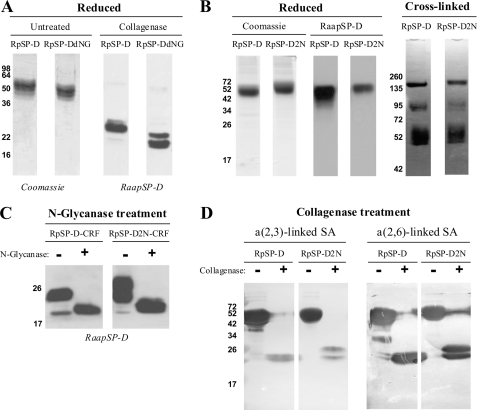
Biochemical characterization of NpSP-D and RpSP-D. A, reducing SDS-PAGE (12%) and Coomassie staining on purified natural porcine SP-D (NpSP-D) and recombinant porcine SP-D (RpSP-D) before (control) and after removal of N-linked oligosaccharide (N-glycanase). Western blot analysis of purified NpSP-D and RpSP-D followed by immunodetection with rabbit anti-porcine SP-D antibodies (RaαPigSP-D). B, gradient (4–15%) SDS-PAGE and Coomassie staining on non-reduced NpSP-D and RpSP-D and the same preparations after treatment with the chemical cross-linker BS3. C, collagenase treatment (+) of NpSP-D and RpSP-D and untreated controls (−) were analyzed by SDS-PAGE (12%) and transferred to NC membranes, followed by detection for the presence of terminal α(2,3)-linked and α(2,6)-linked SA residues using digoxigenin-conjugated lectins M. amurensis agglutinin and S. nigra agglutinin, respectively. D, reducing (12%) SDS-PAGE on RpSP-D after treatment in the absence (Con) or presence of linkage-aspecific sialidase (Sia), followed by Western blot transfer to NC membrane and staining for terminal α(2,3)-linked and α(2,6)-linked SA residues as described above.
To investigate the presence and nature of the N-linked oligosaccharide in the carbohydrate recognition domain (CRD) of RpSP-D, we analyzed a collagenase-treated preparation of RpSP-D by lectin blot analysis and compared the outcome with NpSP-D (Fig. 3C). As expected, collagenase treatment of the pSP-Ds results in a fragment of ∼25 kDa after analysis by SDS-PAGE. As previously described, the oligosaccharides present on NpSP-D are exclusively sialylated with α(2,6)-linked SAs (25), in contrast to RpSP-D, which in addition, also contains α(2,3)-linked SA residues on the oligosaccharide present in its CRD (Fig. 3C). As a control, removal of all SAs via treatment with a linkage-aspecific sialidase results in loss of staining mediated by either Maackia amurensis agglutinin (specific for α(2,3)-linked SAs) or Sambucus nigra agglutinin (specific for α(2,6)-linked SAs; Fig. 3D).
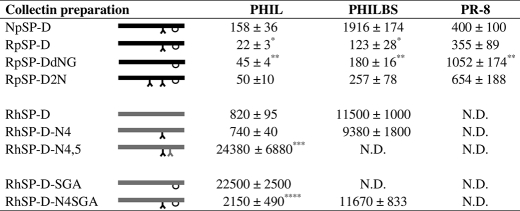
To assess and compare the inhibitory activity of NpSP-D and RpSP-D, HAI was performed on three different strains (Table 1). The susceptibility to inhibition by RpSP-D was: Phil(H3N2)>PhilBS(H3N2)>PR-8(H1N1), in agreement with what generally is found in SP-D-mediated HAI studies on these IAV strains. However, RpSP-D was significantly more active than NpSP-D against Phil and PhilBS.
TABLE 1.
HA inhibition of IAV by porcine and human SP-Ds
Inhibition of HA activity of A/Phillippines/82(H3N2, PHIL), its β-inhibitor-resistant variant (PHILBS) and A/Puerto Rico/34 (H1N1, PR-8) was measured by serially diluting collectin preparations (25 μl/well) in round-bottom 96-well plates using PBS++ as diluent. After adding 25 μl of IAV solution (final concentration: 40 HA units/ml), the IAV/collectin mixture was preincubated for 15 min, followed by addition of 5 μl of human type O erythrocyte suspension in PBS++. The minimal concentration of a collectin, required to fully inhibit HA caused by IAV, was indicated by the formation of a pellet of red blood cells after incubation for 2 h. The entire procedure was carried out at room temperature. All measurements were Ca2+-dependent because addition of EDTA reduced all HA inhibitory activity to 0. SP-Ds analyzed were: natural porcine SP-D (NpSP-D), recombinant porcine SP-D (RpSP-D), RpSP-D with deleted N-glycan (RpSP-DdNG), double-glycosylated RpSP-D (RpSP-D2N), recombinant human SP-D(RhSP-D), N-glycosylated RhSP-D as in pSP-D (RhSP-DN4), double glycosylated RhSP-D (RhSP-DN4,5), RhSP-D with SGA-loop inserted (RhSP-D-SGA); RhSP-DN4 with SGA loop inserted (RhSP-DN4-SGA). Graphical representations are given for all these preparations; black bars, pSP-Ds; grey bars, hSP-Ds; λ = N-linked glycosylation; ∪ = SGA loop region. Values (ng/ml) are mean ± S.E. of at least three experiments; *, P < 0.05 compared to NpSP-D; **, p < 0.05 compared to RpSP-D; ***, p < 0.02 compared to RhSP-D; ****, p < 0.001 compared to RhSP-D-SGA. Statistical comparisons by Student's paired, two-tailed t test.
N.D., not detectable (no inhibition with highest collection concentration tested: 50 μg/ml).
Deletion of the N-Glycosylation Site in the CRD of RpSP-D
As a control, the N-glycan deletion mutant RpSP-DdNG was produced to assess the contribution of the “wild-type” N-linked glycan in the CRD of RpSP-D against IAV. SDS-PAGE analysis (12%) showed a slightly lower molecular mass of the RpSP-DdNG monomer compared with RpSP-D (Fig. 4A). This difference in size was also observed for their respective CRFs, and the size of the CRF from RpSP-DdNG (main band of ∼20 kDa) corresponds well with the size of the N-deglycosylated CRF derived from RpSP-D. Taken together, these results indicate that the N-glycan moiety in the CRD is absent in the RpSP-DdNG variant.
FIGURE 4.
Biochemical analysis of RpSP-D, RpSP-DdNG, and RpSP-D2N. A, reducing 12% SDS-PAGE analysis of RpSP-D and RpSP-DdNG followed by Coomassie staining (left two lanes); both preparations were also treated with collagenase and subjected to 15% SDS-PAGE followed by Western blot analysis and immunostaining with rabbit anti-porcine SP-D antibodies and goat anti-rabbit HRP conjugate (right two lanes). B, comparison of RpSP-D and RpSP-D2N monomeric size after reducing 12% SDS-PAGE analysis, followed by Coomassie staining (left two lanes) or by Western blot analysis and subsequent anti-pSP-D immunostaining as described above (center two lanes); both preparations were also analyzed by 4–15% gradient gel electrophoresis after chemical cross-linking with BS3 (right two lanes). C, effect of N-deglycosylation on the CRF of RpSP-D and RpSP-D2N, analyzed by 12% SDS-PAGE followed by Western blotting and stained with rabbit anti-porcine SP-D antibodies. D, collagenase-treated (+) and untreated controls (−) of RpSP-D and RpSP-D2N were subjected to analysis by SDS-PAGE (12%) and transferred to NC membranes, followed by detection for the presence of terminal α(2,3)-linked and α(2,6)-linked SA residues by staining with digoxigenin-conjugated lectins M. amurensis agglutinin and S. nigra agglutinin, respectively, and digoxigenin detection.
The HAI activity of RpSP-DdNG was measured against three strains of IAV and compared with that of RpSP-D (Table 1). It was found for all strains that the deletion mutant was significantly less active. The drop in activity was most pronounced for PR-8, the IAV strain that is known to be poorly glycosylated.
Introduction of a Second N-Glycosylation Site in the CRD of RpSP-D
The role of the N-linked, SA-rich oligosaccharide in the CRD of pSP-D appears important because it contributes to the anti-IAV activity of this SP-D species. In an attempt to enhance the N-linked glycan-mediated activity of pSP-D we generated a mutant, RpSP-D2N, that has an extra N-linked oligosaccharide inserted in the CRD (“P2N,” Fig. 1), located on helix α3, which forms part of one side of the CRD flanking the core β-sheets, as shown in Fig. 2B. After transfection, RpSP-D2N was expressed by HEK293E cells at levels comparable with RpSP-D and could be purified by mannan-affinity chromatography in the presence of 5 mm calcium chloride. Biochemical characterization of this mutant by SDS-PAGE analysis included analysis of the reduced protein and showed that RpSP-D2N monomers migrate at a slightly higher molecular mass compared with RpSP-D, as illustrated by Coomassie-stained gels and Western blot analysis followed by detection with rabbit anti-porcine SP-D (Fig. 4B). Chemical cross-linking and gradient (4–15%) SDS-PAGE analysis showed identical patterns of multimers for both RpSP-Ds, although all bands visualized in the RpSP-D2N lane migrate slightly slower compared with the bands derived from RpSP-D (Fig. 4B, right two lanes). The higher molecular mass observed for RpSP-D2N suggests that RpSP-D2N indeed contains an additional N-linked glycan. Additional proof was acquired by comparative analysis of the CRFs derived from RpSP-D and RpSP-D2N, before and after treatment with N-glycanase (Fig. 4C). In the case of RpSP-D, collagenase treatment results in a major digestion product of ∼22–24 kDa which, after subsequent treatment with N-glycanase, shifts toward 20 kDa. This result shows that, as expected, an N-linked glycosylation is present in the CRD of RpSP-D. In the case of RpSP-D2N, however, collagenase digestion results in two major, partly overlapping CRF-bands: one with a mass similar to the band present in RpSP-D-CRF, and one that migrates at a higher molecular mass of ∼25–27 kDa. N-Glycanase treatment results in a shift of both bands toward a single molecular mass as present in N-glycanase-treated RpSP-D-CRF. These results show that RpSP-D2N is heterogeneously glycosylated, having either one or two N-linked glycans present on its CRD.
LC-MS/MS was performed on the N-deglycosylated tryptic peptides derived from the CRF of RpSP-D and RpSP-D2N to assess whether the potential N-glycosylation sites were indeed glycosylated. This is reflected by a difference of 1 mass unit due to the conversion of an N-glycosylated Asn residue to an Asp residue after enzymatic removal of the glycan moiety. MS/MS spectral data (m/z values) were produced on four deglycosylated peptides for which the amino acid sequences were confirmed by data base searching against a custom built data base: 300TEGNFTYPTGEPLVY314 (present in both RpSP-D and RpSP-D2N), 273SETENEALSQLVTAQNK289 (present in RpSP-D), and 273SETENEALSNLTTAQNK289 (present in RpSP-D2N). The underlined Asn residues in the sequences were all showing a 1 mass unit increase in peptide mass (data not shown), which proved that the (wild-type) N-glycosylation motif Asn303-Phe301-Thr302 present in RpSP-D and RpSP-D2N indeed contained an N-linked oligosaccharide at residue Asn303. Importantly, the Asn282-Leu283-Thr284 sequence, which was generated into RpSP-D2N by site-directed mutagenesis, was shown to be successfully N-glycosylated at residue Asn282.
To investigate the presence of SAs and distribution of SA-linkages, RpSP-D and RpSP-D2N were, after treatment in the absence (−) or presence (+) of collagenase, subjected to reducing SDS-PAGE analysis and after transfer to nitrocellulose, stained for the presence of α(2,3)-linked SAs or α(2,6)-linked SAs (Fig. 4D). The obtained results show similar patterns for RpSP-D and RpSP-D2N, as can be seen for staining of the intact monomers as well as for the CRFs. Again, doublets are present in the RpSP-D2N-CRF lanes, suggesting that both N-glycosylation sites are not fully glycosylated.
Antiviral Activity of RpSP-D2N Versus RpSP-D against IAV
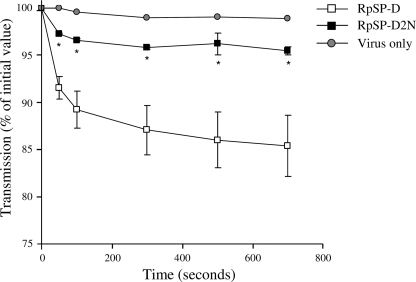
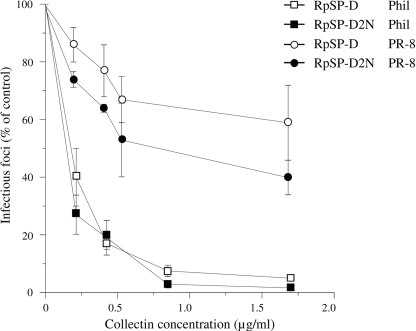
The HAI activity of both RpSP-Ds was measured against IAV strains Phil, PhilBS, and PR-8 (Table 1). No statistically significant differences in HAI activity could be measured between RpSP-D2N and RpSP-D against all three strains. Viral aggregation activity induced by RpSP-D or RpSP-D2N was assessed using the Phil strain. As shown in Fig. 5, RpSP-D (3.2 μg/ml) caused aggregation of IAV particles, whereas the extent of IAV aggregation caused by RpSP-D2N was significantly less, reaching values comparable with those of the control (virus only). In addition, we measured and compared the inhibitory activity of the two RpSP-Ds against two IAV strains, Phil and PR-8, in a fluorescent focus assay using MDCK cells (Fig. 6). Neutralization of infectivity mediated by RpSP-D and RpSP-D2N showed identical profiles against the Phil strain. The PR-8 strain proved to be slightly more susceptible for inhibition by RpSP-D2N compared with RpSP-D, although statistical analysis did not reveal any significant differences at the various collectin concentrations tested.
FIGURE 5.
Aggregation of IAV (Phil) by RpSP-D and RpSP-D2N. IAV particles, suspended in buffer, were mixed at t = 0 with 3.2 μg/ml of RpSP-D or RpSP-D2N (control: virus only) and the continuously stirred mixture was monitored for light transmission and expressed as % of light transmission at t = 0. Measurements were performed in duplicate and values are mean ± S.E. Statistical analysis was carried out by the Student's two-tailed t test. *, p < 0.05.
FIGURE 6.
Inhibition of IAV infectivity of MDCK cells mediated by RpSP-D and RpSP-D2N. Fluorescent focus assays were performed to assess the inhibitory activity of RpSP-D and RpSP-D2N against the Phil (squares) and PR-8 (circles) strains of IAV on infectivity of MDCK cells. A fixed amount of virus was preincubated with increasing concentrations of collectin for 30 min at 37 °C, added to monolayers of MDCK cells, and incubated for 45 min at 37 °C. After washing and fixation, the monolayers were labeled with monoclonal Ab to IAV nucleoprotein and fluorescein isothiocyanate-labeled goat anti-mouse IgG. The numbers of infectious foci were counted and expressed as a percentage of the number of infectious foci formed in the control incubation (IAV added to monolayer in absence of collectin). Values are mean ± S.E. of three experiments.
Recombinant Expression of CRD-glycosylated RhSP-D Derivatives
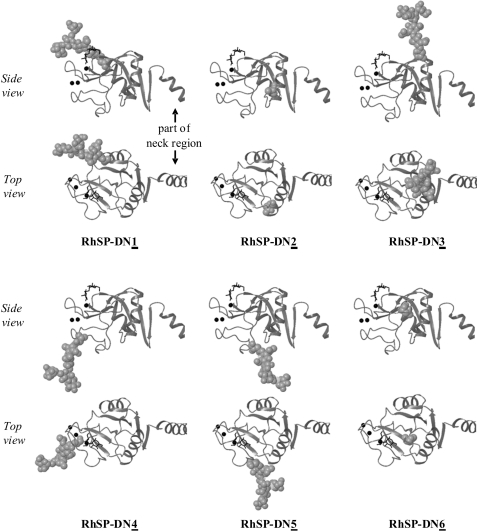
To investigate the feasibility of enhancing the anti-IAV properties of RhSP-D by introducing SA-mediated neutralization of IAV in addition to the lectin-mediated antiviral activity of hSP-D, several N-glycosylated RhSP-D mutants were produced by site-directed mutagenesis. These mutants differ in the location of the introduced oligosaccharide (Fig. 7, N1 to N6), as well as in the number of oligosaccharides that were generated into the CRD of RhSP-D (single and double N-glycosylated mutants). Two of these mutants were N-glycosylated at locations in the CRD of RhSP-D, which correspond to “naturally” occurring N-glycosylation sites in the CRD of C-type lectins known to date: RhSP-DN4, which is N-glycosylated at the site as uniquely present in pSP-D (Fig. 1, Asn303), and RhSP-DN4,5, which has in addition the oligosaccharide inserted at the location of the conserved N-glycosylation site as present in the CRD of SP-A (Fig. 1, Asn313). Based upon the crystal structures of hSP-D and rSP-A and the overall homology of the C-type lectin domains, these N-linked oligosaccharides appear to be located at adjacent surface loop regions, projecting outward, distal from the ligand binding site, and well exposed to enable interactions with the environment (e.g. pathogens, Fig. 7). Unfortunately we were unable to produce RhSP-DN5 and therefore this derivative is not included in this study. The N-linked oligosaccharides of two other mutants, RhSP-DN1 and RhSP-DN3, are positioned in line with the symmetry axis of the trimeric neck region on top of the CRD, more proximal to the ligand binding site and therefore possibly interfering with the accessibility for glycans to approach the carbohydrate binding site of the CRD. The N-glycosylation sites chosen for the remaining two mutants (RhSP-DN2 and RhSP-DN6) are expected to be buried inside the CRD structure and are likely to hamper interactions with glyco-modifying enzymes during post-translational processing of the protein.
FIGURE 7.
Structural model of N-linked glycan positions in the CRD of the RhSP-D mutants. Ribbon diagrams as detailed in the legend to Fig. 2, showing six different locations of the N-linked oligosaccharides introduced by site-directed mutagenesis into the CRD of RhSP-D, producing mutants RhSP-DN1, RhSP-DN2, RhSP-DN3, RhSP-DN4, RhSP-DN5, and RhSP-DN6. The glycosylation sites of mutants RhSP-DN2 and RhSP-DN5 are buried and therefore only the Asn side chains are depicted for these two mutants. Two double N-glycosylated mutants were also generated, RhSP-DN4,5 and RhSP-DN4,6, which had the two N-glycosylation sequons introduced as indicated by both numbers. Shown are views from the side of a single monomer in the trimer, and a top view looking down the trimer axis into the lectin binding pocket; part of the neck domain is shown on the right side of each structure.
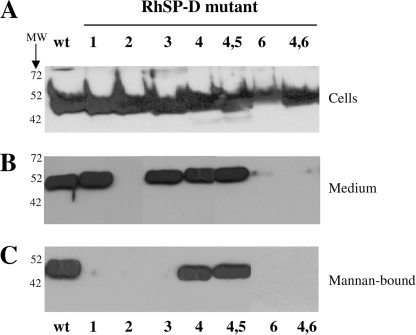
Small scale (4 ml) expressions in HEK293E cells of wild-type RhSP-D and all N-glycosylated derivatives were analyzed by reducing SDS-PAGE (12%) and Western blotting, followed by immunostaining using rabbit anti-human SP-D antibodies (Fig. 8). As illustrated in panel A, each RhSP-D-derived construct, which was transfected into HEK293E cells resulted in synthesis of RhSP-D because SP-D-specific immunostaining of an ∼50-kDa band was observed in samples derived from lysed HEK293E cells. Analysis of the expression medium derived from these transfected cells showed the absence of anti-hSP-D-reactive protein for RhSP-DN2, RhSP-DN6, and RhSP-DN4,6 (panel B). Mannan-affinity purification of all medium revealed that only (wild-type) RhSP-D, RhSP-DN4, and RhSP-DN4,5 could be purified with mannan-agarose in the presence of calcium chloride.
FIGURE 8.
Western blot analysis of N-glycosylated RhSP-D derivatives. Reducing SDS-PAGE (12%) analysis of RhSP-D(wt) and mutant versions (1, 2, 3, 4, 5, and 6 and double glycosylated mutants 4,5 and 4,6; indicated with bold characters) with N-glycosylation sites introduced by site-directed mutagenesis in the CRD of hSP-D as illustrated in Fig. 7. After transfer to nitrocellulose, proteins were immunostained with rabbit anti-human SP-D followed by goat anti-rabbit HRP. A, HEK293E cells that were transiently transfected with RhSP-D wild-type or RhSP-D mutant and expressing RhSP-Ds for 4 days, were collected by centrifugation. Cell pellets were redissolved in β-mercaptoethanol containing Laemmli buffer and equal amounts of these lysed cells were loaded onto the gel; B, analysis of medium (20 μl) derived from HEK293E cells as described for panel A; C, expression medium collected from the different RhSP-D-expressing HEK293E cells were treated with mannan-agarose in the presence of 5 mm calcium chloride, incubated overnight at 4 °C, and after several washes, incubated with 5 mm EDTA containing buffer to release RhSP-Ds, which were bound Ca2+ dependently to mannan; 5 vol % of each EDTA-eluted fraction was applied in each lane.
Characterization and Antiviral Activity of RhSP-DN4, the N-Glycosylated Analog of pSP-D
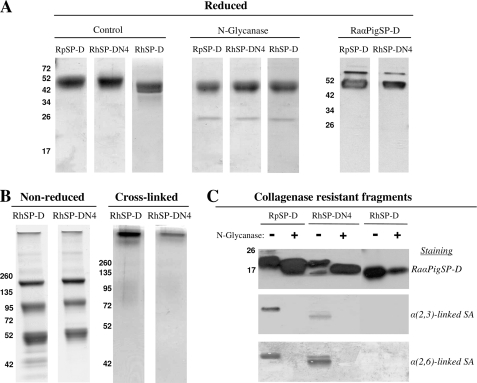
Because RhSP-DN4 was the first N-glycosylated RhSP-D derivative shown to retain Ca2+-dependent lectin activity, we performed a large scale expression of this variant in HEK293E cells to compare the activity of this construct with wild-type RhSP-D and wild-type RpSP-D. This allowed us to study the contributory effect of an N-linked oligosaccharide in the CRD of RhSP-D. These purified recombinant SP-Ds were first characterized and compared for monomeric size, N-glycanase sensitivity, and immunoreactivity (Fig. 9, panel A). Like RpSP-D, the RhSP-DN4 monomer migrates slower than RhSP-D, whereas N-glycanase treatment reduces the molecular mass of RpSP-D and RhSP-DN4 to that of RhSP-D. Immunostaining was similar for RpSP-D and RhSP-DN4 when using rabbit anti-porcine SP-D antibodies. Next, analysis by nonreducing SDS-PAGE as well as chemical cross-linking of RhSP-D and RhSP-DN4 revealed no major differences in assembly between RhSP-D and RhSP-DN4 (Fig. 9, panel B). Finally, the presence of N-linked glycans in the CRD was determined by analyzing the CRFs from RpSP-D, RhSP-D, and RhSP-DN4 before (−) and after (+) treatment with N-glycanase, followed by staining with antibodies against SP-D or with lectins specifically recognizing SA linkages (Fig. 9, panel C). N-Glycanase treatment reduced the molecular mass of the CRFs derived from RpSP-D and RhSP-DN4 but did not affect that of RhSP-D. Furthermore, the CRFs from RpSP-D and RhSP-DN4, which differ slightly in molecular mass, were both positive for α(2,3)-linked SAs or α(2,6)-linked SAs and all staining was lost after treatment with N-glycanase. The CRD of RhSP-D is not N-glycosylated and therefore the RhSP-D-derived CRF does not show any staining for SAs. Of note, the relative amount of α(2,3)-linked SAs appears higher for RpSP-D in comparison with RhSP-DN4.
FIGURE 9.
SDS-PAGE analysis of RhSP-D and RhSP-DN4 as compared with RpSP-D. A, purified RhSP-D and RhSP-DN4 were analyzed by reducing SDS-PAGE (12%, Coomassie stained) and compared with RpSP-D before (Control) and after removal of N-linked oligosaccharides (N-Glycanase). RpSP-D and RhSP-DN4 were also analyzed by Western blotting followed by immunodetection with RaαPigSP-D and RaαHuSP-D antibodies, respectively; B, gradient (4–15%) SDS-PAGE and Coomassie staining on non-reduced RhSP-D and RhSP-DN4 (left two panels) and the same preparations after treatment with the chemical cross-linker BS3 (right two panels); C, collagenase-treated RpSP-D, RhSP-D, and RhSP-DN4 were subjected to digestion by N-glycanase (+) and applied to 12% SDS-PAGE analysis alongside untreated controls (−), followed by Western blotting. Subsequently, these membranes were either immunostained with RaαPigSP-D (followed by detection with goat anti-rabbit HRP), or stained for the presence of terminal α(2,3)-linked or α(2,6)-linked SA residues using DIG-conjugated lectins M. amurensis agglutinin and S. nigra agglutinin, respectively.
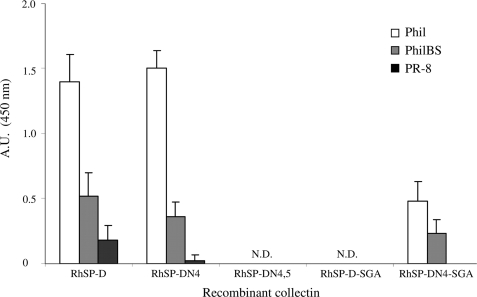
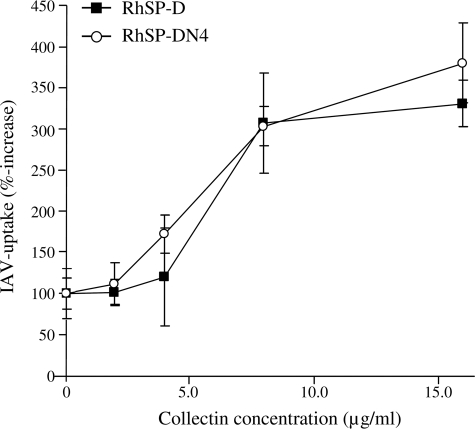
The inhibitory activity of RhSP-D against IAV was compared with that of RhSP-DN4 by the use of four different functional assays (i-iv). (i) Measurement of the antiviral activity by HAI showed no differences between RhSP-D and RhSP-DN4 for IAV strains Phil, PhilBS, and PR-8 and both RhSP-D and RhSP-DN4 were less active compared with RpSP-D (Table 1). (ii) Binding affinity for these three strains, as determined by ELISA measurements, also showed no differences between RhSP-D and RhSP-DN4 (Fig. 10). (iii) SP-D-mediated protection of MDCK cells against infectivity by IAV (Phil strain) again did not reveal any differences between the wild-type and the N-glycosylated recombinant variant (Fig. 11). (iv) Finally, we determined whether introduction of the N-glycan in the CRD of hSP-D might have an effect on the SP-D-mediated interactions between neutrophils and IAV, but the increased uptake of IAV caused by RhSP-DN4 was almost identical to that of RhSP-D (Fig. 12). “Superglycosylation” of RhSP-D via introduction of two N-linked glycans in its CRD, generating the RhSP-DN4,5 mutant, resulted in substantial loss of HAI activity (Table 1), decreased binding affinity to IAV (Fig. 10), and reduction in neutralizing activity against IAV as measured by the fluorescent focus assay using MDCK cells (Fig. 11).
FIGURE 10.
Binding of RhSP-Ds to IAV as measured by ELISA. Suspensions of three IAV strains (Phil, PhilBS, and PR-8) were allowed to dry in 96-well plates and after washings, several RhSP-Ds were diluted in PBS++ (2 μg/ml) and added to the wells. After incubation for 30 min at 37 °C, plates were washed and bound collectins were detected with monoclonal antibodies raised against human SP-D, followed by donkey anti-mouse HRP-labeled secondary antibodies and staining with tetramethylbenzidine substrate. The reaction was stopped with 0.5 m H2SO4 and absorbance was measured at 450 nm. Measurements were performed in quadruplicate; values are mean ± S.E. N.D., not detectable.
FIGURE 11.
Neutralization of IAV infectivity by RhSP-D and derivatives. Infectivity of IAV for MDCK cells was assessed using the fluorescent focus forming assay (details under “Experimental Procedures” and Fig. 6, legend). The inhibitory activity of increasing concentrations of RhP-D and four derivatives was assessed against the Phil strain, to study the impact of N-glycosylation and insertion of the SGA loop in the CRD of hSP-D. The numbers of infectious foci were counted and expressed as a percentage of the number of infectious foci formed in the control incubation (IAV added to monolayer in absence of collectin). Values are mean ± S.E. of five experiments.
FIGURE 12.
Effect of RhSP-D and RhSP-DN4 on IAV-induced neutrophil activity. Fluorescein isothiocyanate-labeled Phil virus was incubated in the absence or presence of increasing amounts of collectins for 30 min at 37 °C and this mixture was subsequently added to human neutrophils. After incubation for 45 min at 37 °C, trypan blue was added to quench any extracellular fluorescence, and after fixation with paraformaldehyde, the neutrophil-associated fluorescence was measured by flow cytometry. Uptake in the absence of collectins (control) was set to 100%. Results are mean ± S.E. for two experiments.
Functional Analysis of SGA-loop Containing RhSP-D Mutants
To investigate other porcine-specific structural features present in the CRD of pSP-D that might be involved in the functionality of the N-linked sugar present in this domain, two additional RhSP-D derivatives were produced containing a small loop region of 3 amino acids (Ser329-Gly330-Ala331, Fig. 1) that, when aligned with the sequence of pSP-D, was inserted at the corresponding location in the CRD of hSP-D. These collectins were produced based upon the wild-type hSP-D (yielding RhSP-D-SGA) and the N4-glycosylated variant (RhSP-DN4-SGA) and were tested for their inhibitory activity against IAV. Insertion of the SGA loop into hSP-D almost completely abrogated the activity of this collectin as illustrated by the data obtained with RhSP-D-SGA. This was observed for all IAV assays used (HAI (Table 1), IAV binding via ELISA (Fig. 10), and inhibition of infectivity using MDCK cells (Fig. 11)). Interestingly, N4-glycosylation of this inactive mutant restored the inhibitory activity of the obtained RhSP-DN4-SGA mutant toward levels as obtained for wild-type RhSP-D.
DISCUSSION
HEK293E Cells Produce RpSP-D Structurally and Functionally Similar to NpSP-D
Previously it was shown that the anti-IAV activity of pSP-D is much stronger compared with human SP-D (23) and it was demonstrated that this was partly due to the unique SA-rich N-linked glycan present in the CRD of pSP-D that interacts with the SA-recognizing IAV-receptor. To study this in more detail, a HEK293E cell expression system was successfully applied to produce recombinant equivalents of pSP-D, hSP-D, and several glyco-modified derivatives. Because N-linked protein glycosylation was of key importance for this study, HEK293 type E cells were used to ensure modification of our RSP-Ds with mature, wild-type sialoglycan structures. Transient expression was preferred to provide flexibility, efficiency, and high yield production of the various RSP-Ds used in this study (1–5 mg/liter of expression medium). For all constructs, the native signal sequence of SP-D was used to target the protein to the secretory pathway of the HEK293E cells. We also tested the strong artificial cystatin signal peptide (38) for expression of RpSP-D but this did not result in differences with regard to expression levels and assembly (results not shown). This is the first study on the production and characterization of RpSP-D and therefore, several important structural and functional properties of this protein were analyzed and compared with the porcine lung lavage-derived NpSP-D. Like NpSP-D, RpSP-D could be isolated from the calcified medium by sugar affinity chromatography using mannan-agarose. RpSP-D displayed a combination of structural features that are known to be characteristic for collectins: C-type lectin activity, collagenase sensitivity, trimerization, and oligomerization of trimeric subunits. Furthermore, RpSP-D was immunoreactive for a polyclonal antibody that was raised against NpSP-D in rabbits. Like NpSP-D, RpSP-D was N-glycosylated and results obtained with the RpSP-D-derived CRF showed the presence of an SA-rich oligosaccharide in the CRD. N-Glycosylation of the CRD of RpSP-D was also confirmed by mass spectrometry analysis. Interestingly, in the case of RpSP-D this oligosaccharide was equipped with both α(2,3)-linked and α(2,6)-linked terminal SA residues, in contrast to NpSP-D that exclusively contained α(2,6)-linked terminal SA residues (Fig. 3C). This is probably caused by differences in activity and specificity of sialyltransferases between NpSP-D-producing porcine pulmonary epithelial cells and RpSP-D-producing HEK293E cells. In terms of anti-IAV potency, heterogeneity of SA linkages of SAs on RpSP-D is considered favorable because it will enable RpSP-D to interact with HA receptors recognizing α(2,3)-linked SAs (preferentially recognized by avian strains) as well as α(2,6)-linked SAs (preferentially recognized by human strains). Consequently, RpSP-D could have broader-range anti-IAV activity compared with NpSP-D although this requires more extensive studies that cover a wider range of IAV strains. In line with this notion was the observation that the antiviral activity of RpSP-D was significantly higher than that of NpSP-D (Table 1). The HAI activity of RpSP-DdNG, a mutant that lacks the N-linked glycan as found in the CRD of pSP-D, was significantly less active than the wild-type RpSP-D for all three strains measured (Table 1). This observation underlined the previously found contributory effect of the N-linked glycan on NpSP-D to its anti-IAV activity (23). Contribution of the N-linked glycan on RpSP-D appeared to be most pronounced against the PR-8 strain. This strain is poorly glycosylated, making it less susceptible for lectin-mediated interactions. Therefore, SA-mediated interactions between RpSP-D and PR-8 play a more prominent role compared with RpSP-D-mediated interactions with IAV strains.
Introduction of a Second N-Glycosylation Site in RpSP-D Does Not Enhance Its antiviral Activity
Not much is known about the importance of the location of the N-linked glycan in pSP-D and how this determines interactions with (the HA of) IAV. As a first step, we introduced a second N-glycosylation site in the CRD of RpSP-D, located on the α3-helix of the CRD by site-directed mutagenesis (Fig. 1). The trimeric organization of a SP-D subunit would theoretically result in a cluster of oligosaccharides located near the core of the neck trimer, in addition to the more peripherally located wild-type Asn303-linked glycan. The RpSP-D2N mutant has lectin activity and is well assembled. Analysis by mass spectrometry confirmed that the Q282N mutation results in N-glycosylation of Asn282, in addition to the wild-type oligosaccharide present at Asn303, which was also confirmed by mass spectrometry. SDS-PAGE analysis of the CRF showed the presence of 2 bands positive for SA staining, which indicated that superglycosylation was incomplete, most likely due to limitations of the HEK293E cell glycosylation capacity when overexpressing RpSP-D2N. The presence of an additional SA-containing glycan could provide a wider range of interactions with the SA receptor of IAV and, consequently, increase the antiviral activity. However, HAI measurements showed no differences compared with RpSP-D and neutralization of infectivity showed only slightly enhanced activity against the poorly glycosylated PR-8 strain of IAV. To some extent, this could reflect the higher density of SA residues present on RpSP-D2N, resulting in stronger interactions with a strain that is less susceptible to lectin-mediated interactions with SP-D. Remarkably, introduction of the second N-linked glycan resulted in an almost complete loss of the viral aggregating activity, which could be caused by enhanced repulsion between RpSP-D2N molecules due to the high density of negatively charged SA residues.
N-Glycosylation of RhSP-D Is Restricted to Specific Locations within the CRD to Retain C-type Lectin Activity
The antiviral activity of hSP-D relies on interactions between the CRD and viral glycans that are present on the spike protein HA and neuraminidase of IAV (39, 40). Poorly glycosylated IAVs, like the recently circulating pandemic H1N1 strains, show high resistance against neutralization by human collectins including hSP-D (41) and this may result in an enhanced risk of infection of the host lung epithelium by such strains. It was shown by studies with SP-A and pSP-D, in which lung collectins N-glycosylated in their CRD are more effective in binding and neutralizing poorly glycosylated IAVs (23, 42), likely due to interactions between the terminal SA residues present on the N-linked glycan, and the SA receptor of the viral HA. Based upon these observations and with the goal of development of hSP-D-based drugs that exhibit strong broad-range antiviral activity, we pursued the production of RhSP-D variants with artificially introduced N-linked glycans in their CRDs to investigate the impact of these N-linked glycans on the inhibitory activity against IAV. To learn more about the location of the N-linked glycan and how this relates with antiviral activity, we designed and expressed seven different RhSP-D mutants with N-glycosylation sequons introduced at different locations in the CRD via site-directed mutagenesis. Although all RhSP-D derivatives were synthesized by HEK293E cells, as shown by the presence of hSP-D-specific immunostaining in all cell lysate fractions, the mutants with N-linked sugars at positions 2 or 6 were not secreted in the medium despite having a secretory signal sequence similar to the other mutants. Because the positions of these two N-glycosylation sequons appears to be buried inside the structure, this could have lead to protein misfolding. Therefore, it is assumed that these RhSP-D mutants were directed into the ER-associated degradation pathway (e.g. degradation via proteosomes), rather than transported to the Golgi for further N-glycan processing and secretion (43). In contrast, mutants RhSP-DN1, RhSP-DN3, RhSP-DN4, and RhSP-DN4,5 were successfully secreted by HEK293E cells. Molecular modeling showed that the glycan moiety of mutants RhSP-DN1 and RhSP-DN3 were located in relative proximity of the carbohydrate binding pocket of the CRD, which could interfere with accessibility for carbohydrate ligands. Because we were not able to purify these mutants by mannan affinity chromatography, we hypothesize that this interference caused the lack of interaction between RhSP-DN1 and RhSP-DN3 with mannan. However, we did succeed in producing mannan affinity-purified N-glycosylated RhSP-Ds with N-glycans located at positions in the CRD that, upon sequence alignment, matched with the locations of “native” N-linked glycans as can be found in the CRDs of pSP-D (RhSP-DN4) and SP-A (N5, in this study only expressed as part of a double N-glycosylated mutant, RhSP-DN4,5). These findings suggest that N-glycosylation of a C-type lectin-derived CRD is restricted to specific locations where linkage to a complex N-glycan moiety should not interfere with: 1) secondary and tertiary structural conformation; 2) integrity and accessibility of the ligand binding site; and 3) trimerization, hence a peripheral localization of the oligosaccharide, relative to the coiled coil neck domain, is preferred.
Addition of an N-Linked Glycan in the CRD of RhSP-D Similar to pSP-D, Does Not Enhance Its Inhibitory Activity against IAV
In terms of N-glycosylation profiles, the RhSP-DN4 mutant can be regarded as the “porcinized” variant of hSP-D with potentially enhanced antiviral activity. First, the biochemical properties of RhSP-DN4 were thoroughly investigated and compared with those of RpSP-D and RhSP-D (Fig. 9). Like RpSP-D, the RhSP-DN4 monomer was larger than RhSP-D and stained positive for both types of SA-linkages in the CRD, which is lost after N-glycanase treatment, similar to what is observed for RpSP-D. Furthermore, N-glycosylation did not cause any major changes in protein conformation and assembly because immunoreactivity, oligomerization, and C-type lectin activity remained unaltered. The antiviral activity of RhSP-DN4 was assessed by different assays including HAI of IAV, neutralization of IAV infectivity using MDCK cells, and uptake of IAV by neutrophils. Unexpectedly, all antiviral assays used revealed no statistical differences between the IAV neutralizing properties of RhSP-DN4 and RhSP-D and almost identical IAV-binding profiles were found for Phil, PhilBS, and PR-8 as determined by ELISA. Therefore, we concluded that the presence of an SA-rich N-linked oligosaccharide in the CRD of hSP-D at the same location as in the CRD of pSP-D did not enhance the neutralizing activity of hSP-D and remained substantially lower than that of RpSP-D as determined by HAI activity measurements. The addition of a second glycan to the RhSP-DN4 mutant, yielding RhSP-DN4,5, almost completely abrogated the antiviral properties and binding of IAV was no longer detectable (Fig. 10). Although RhSP-DN4,5 was purified by mannan affinity chromatography, indicating that the introduced N-linked glycans did not interfere with the potential of the CRD to bind viral glycans, the addition of the N-linked glycan at the position where SP-A is N-glycosylated (position 5, Fig. 1) blocked binding to IAV completely. We assume that differences in glycan configuration between mannose residues as part of a polysaccharide (mannan), and the sugars as covalently linked on the HA of IAV, may account for this discrepancy. Because we were unable so far to produce a RhSP-D mutant having only glycan N5, it remains unclear whether the complete loss of viral interactions was specifically due to the location of the N5 glycan, or resulted from combined steric hindrance caused by the two N-linked glycans in the CRD of RhSP-DN4,5.
N-Glycan-mediated Activity of SP-D against IAV Requires Additional Structural Features of the CRD
Because, in contrast to pSP-D, the N-linked sialoglycan of RhSP-DN4 did not show any contributory effect to the antiviral activity, we concluded that other pSP-D-specific structural elements in the CRD must be involved. Therefore, we also studied the role of the insertion of 3 amino acids Ser329-Gly330-Ala331, referred to as the “SGA loop.” This unusual loop region within the CRD is absent in SP-D species other than pigs and to investigate its functional importance, two mutants were generated: RhSP-D-SGA and RhSP-DN4-SGA, which both have the SGA residues inserted in the CRD of RhSP-D at the location identical to where this loop is located in the CRD of pSP-D. Remarkably, the various functional assays that were used (ELISA test for viral binding, HAI of IAV, neutralization of IAV infectivity) showed that insertion of the SGA loop region into hSP-D reduced the antiviral activity of the resulting RhSP-D-SGA mutant to near zero levels. This is in contrast to the effects observed in previous studies in which the bovine collectin CL-43-specific “RAK” motif was inserted near the conserved groove region of hSP-D, similar to the SGA modification of hSP-D as performed in this study. Although insertion of RAK into hSP-D substantially increased binding affinity for mannose-containing viral glycoconjugates (44), insertion of the SGA loop apparently induced changes in the groove region of the CRD such that viral carbohydrate recognition by RhSP-D-SGA was impaired. Obviously, it is complicated to predict the exact conformational effects of insertions within loop regions and characterization of the crystal structure of pSP-D complexed with ligand will be required to learn more about the role of the SGA loop region in pSP-D for glycan recognition. Unexpectedly, despite the disruptive effect of the SGA insertion on the antiviral activity of hSP-D, the N-glycosylated variant of the SGA-containing mutant, RhSP-DN4-SGA, showed restored antiviral activity to levels comparable with that of RhSP-D. Somehow, the presence of the N-linked glycan compensated for the loss of lectin-mediated antiviral activity caused by insertion of the SGA loop. Presumably, this restored activity was mainly due to interactions between the sialylated oligosaccharide introduced into the CRD of this mutant, and the SA-recognizing HA receptor of IAV, but this requires further investigation.
The N-Glycan-mediated Neutralization of IAV by Collectins Involves Diverse Mechanisms
Despite the strong homology between the CRDs of the C-type lectins, our findings revealed that simultaneous interactions between the CRD and viral glycans on one hand, and between the SA-rich N-linked glycan and the HA receptor on the other hand, requires not only a finely tuned position of the N-linked glycan but also additional structural prerequisites of the CRD. Apparently, the conformation of pSP-D meets both requirements and our findings with RhSP-DN4 demonstrate that it is relatively complex to mimic both in hSP-D. Interestingly, the N-linked glycan, which is conserved in the CRD of SP-A, interacts in a Ca2+-independent manner with IAV and it was found that removal of this glycan completely abrogates the antiviral activity of SP-A, in contrast to pSP-D where N-glycan-mediated and lectin-mediated interactions with IAV operate synergistically (11, 23). This marked difference in functionality and working mechanism of CRD-linked glycans may result from variations in CRD conformations between the collectins (e.g. SP-A versus SP-D), species-specific differences (e.g. porcine versus human), variations in ligand specificity of the CRDs (e.g. affinity for viral sugars), and last but not least, the density and orientation of available N-linked glycans on the assembled protein. The oligomeric shape and dimensions of SP-A facilitates unidirectional orientation of 18 relatively clustered N-linked glycans that may interact simultaneously with a single virion. In contrast, the molecular structure of dodecameric pSP-D determines that a single trimeric subunit enables only three sialoglycans to interact with the SA receptors of a single IAV particle. This could explain why the lectin-mediated interactions of SP-D play a primary role in binding IAV, enabling additional secondary interactions mediated by a sialoglycan moiety as present in pSP-D to broaden and enhance interactions with IAV.
Taken together, this study provides novel insights into the contribution of N-linked sialoglycans to collectin-mediated neutralizing activity of IAV that was shown to require a complex of structural prerequisites. By producing a structurally well characterized and functionally active RpSP-D, which displays distinctively strong inhibitory activity against IAV, we may learn more about the mechanisms that underlie the antiviral properties of this intriguing SP-D species. Large scale purification of RpSP-D will help us to clarify the three-dimensional structure of the nCRD of pSP-D by crystal structure analysis. Ultimately, these studies will aid in designing a novel class of antivirals based upon a human C-type lectin that exhibits high potential to prevent and treat influenza effectively in humans.
Acknowledgments
We greatly appreciate the gift of the full-length hSP-D cDNA clone from Dr. E. C. Crouch. We also thank Sanne Raadsveld for technical assistance.
This work was supported by European Union FP6 Specific Targeted Research Project Grant LSHM-CT-2004-512093.
- IAV
- influenza A virus
- CRD
- carbohydrate recognition domain
- CRF
- collagenase-resistant fragment
- HAI
- hemagglutination inhibition
- MDCK
- Madin-Darby canine kidney
- NpSP-D
- natural porcine SP-D
- RhSP-D
- recombinant human SP-D
- RpSP-D
- recombinant porcine SP-D
- SA
- sialic acid
- SP-A
- surfactant protein A
- SP-D
- surfactant protein D.
REFERENCES
- 1. Lipatov A. S., Govorkova E. A., Webby R. J., Ozaki H., Peiris M., Guan Y., Poon L., Webster R. G. (2004) J. Virol. 78, 8951–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McGill J., Heusel J. W., Legge K. L. (2009) J. Leukoc. Biol. 86, 803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ehrhardt C., Seyer R., Hrincius E. R., Eierhoff T., Wolff T., Ludwig S. (2010) Microbes Infect. 12, 81–87 [DOI] [PubMed] [Google Scholar]
- 4. White M. R., Doss M., Boland P., Tecle T., Hartshorn K. L. (2008) Expert Rev. Clin. Immunol. 4, 497–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haagsman H. P., Hogenkamp A., van Eijk M., Veldhuizen E. J. (2008) Neonatology 93, 288–294 [DOI] [PubMed] [Google Scholar]
- 6. Kuroki Y., Takahashi M., Nishitani C. (2007) Cell. Microbiol. 9, 1871–1879 [DOI] [PubMed] [Google Scholar]
- 7. Wright J. R. (2005) Nat. Rev. Immunol. 5, 58–68 [DOI] [PubMed] [Google Scholar]
- 8. Kishore U., Greenhough T. J., Waters P., Shrive A. K., Ghai R., Kamran M. F., Bernal A. L., Reid K. B., Madan T., Chakraborty T. (2006) Mol. Immunol. 43, 1293–1315 [DOI] [PubMed] [Google Scholar]
- 9. Dommett R. M., Klein N., Turner M. W. (2006) Tissue Antigens 68, 193–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Herías M. V., Hogenkamp A., van Asten A. J., Tersteeg M. H., van Eijk M., Haagsman H. P. (2007) Mol. Immunol. 44, 3324–3332 [DOI] [PubMed] [Google Scholar]
- 11. Benne C. A., Kraaijeveld C. A., van Strijp J. A., Brouwer E., Harmsen M., Verhoef J., van Golde L. M., van Iwaarden J. F. (1995) J. Infect. Dis. 171, 335–341 [DOI] [PubMed] [Google Scholar]
- 12. Hartshorn K. L., Crouch E. C., White M. R., Eggleton P., Tauber A. I., Chang D., Sastry K. (1994) J. Clin. Invest. 94, 311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hartshorn K. L., Ligtenberg A., White M. R., van Eijk M., Hartshorn M., Pemberton L., Holmskov U., Crouch E. (2006) Biochem. J. 393, 545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hartshorn K. L., White M. R., Shepherd V., Reid K. B. M., Jensenius J. C., Crouch E. C. (1997) Am. J. Physiol. Lung Cell. Mol. Physiol. 273, L1156-L1166 [DOI] [PubMed] [Google Scholar]
- 15. Hawgood S., Brown C., Edmondson J., Stumbaugh A., Allen L., Goerke J., Clark H., Poulain F. (2004) J. Virol. 78, 8565–8572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. LeVine A. M., Whitsett J. A., Hartshorn K. L., Crouch E. C., Korfhagen T. R. (2001) J. Immunol. 167, 5868–5873 [DOI] [PubMed] [Google Scholar]
- 17. White M. R., Crouch E., Vesona J., Tacken P. J., Batenburg J. J., Leth-Larsen R., Holmskov U., Hartshorn K. L. (2005) Am. J. Physiol. Lung Cell. Mol. Physiol. 289, L606-L616 [DOI] [PubMed] [Google Scholar]
- 18. Hartshorn K. L., Reid K. B., White M. R., Jensenius J. C., Morris S. M., Tauber A. I., Crouch E. (1996) Blood 87, 3450–3461 [PubMed] [Google Scholar]
- 19. Pastva A. M., Wright J. R., Williams K. L. (2007) Proc. Am. Thorac. Soc. 4, 252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scholtissek C. (1994) Eur. J. Epidemiol. 10, 455–458 [DOI] [PubMed] [Google Scholar]
- 21. Garten R. J., Davis C. T., Russell C. A., Shu B., Lindstrom S., Balish A., Sessions W. M., Xu X., Skepner E., Deyde V., Okomo-Adhiambo M., Gubareva L., Barnes J., Smith C. B., Emery S. L., Hillman M. J., Rivailler P., Smagala J., de Graaf M., Burke D. F., Fouchier R. A., Pappas C., Alpuche-Aranda C. M., López-Gatell H., Olivera H., López I., Myers C. A., Faix D., Blair P. J., Yu C., Keene K. M., Dotson P. D., Jr., Boxrud D., Sambol A. R., Abid S. H., St. George K., Bannerman T., Moore A. L., Stringer D. J., Blevins P., Demmler-Harrison G. J., Ginsberg M., Kriner P., Waterman S., Smole S., Guevara H. F., Belongia E. A., Clark P. A., Beatrice S. T., Donis R., Katz J., Finelli L., Bridges C. B., Shaw M., Jernigan D. B., Uyeki T. M., Smith D. J., Klimov A. I., Cox N. J. (2009) Science 325, 197–20119465683 [Google Scholar]
- 22. van Eijk M., Haagsman H. P., Skinner T., Archibold A., Reid K. B., Lawson P. R., Archibold A. (2000) J. Immunol. 164, 1442–1450 [DOI] [PubMed] [Google Scholar]
- 23. van Eijk M., White M. R., Crouch E. C., Batenburg J. J., Vaandrager A. B., van Golde L. M., Haagsman H. P., Hartshorn K. L. (2003) J. Immunol. 171, 1431–1440 [DOI] [PubMed] [Google Scholar]
- 24. van Eijk M., van de Lest C. H., Batenburg J. J., Vaandrager A. B., Meschi J., Hartshorn K. L., van Golde L. M., Haagsman H. P. (2002) Am. J. Respir. Cell Mol. Biol. 26, 739–747 [DOI] [PubMed] [Google Scholar]
- 25. van Eijk M., White M. R., Batenburg J. J., Vaandrager A. B., van Golde L. M., Haagsman H. P., Hartshorn K. L. (2004) Am. J. Respir. Cell Mol. Biol. 30, 871–879 [DOI] [PubMed] [Google Scholar]
- 26. Durocher Y., Perret S., Kamen A. (2002) Nucleic Acids Res. 30, E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hartshorn K. L., Chang D., Rust K., White M., Heuser J., Crouch E. C. (1996) Am. J. Physiol. Lung Cell. Mol. Physiol. 271, L753–L762 [DOI] [PubMed] [Google Scholar]
- 28. Morlot C., Hemrika W., Romijn R. A., Gros P., Cusack S., McCarthy A. A. (2007) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 689–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khoshnoodi J., Hill S., Tryggvason K., Hudson B., Friedman D. B. (2007) J. Mass Spectrom. 42, 370–379 [DOI] [PubMed] [Google Scholar]
- 30. Aye T. T., Scholten A., Taouatas N., Varro A., Van Veen T. A., Vos M. A., Heck A. J. (2010) Mol. Biosyst. 6, 1917–1927 [DOI] [PubMed] [Google Scholar]
- 31. Vogels M. W., van Balkom B. W., Kaloyanova D. V., Batenburg J. J., Heck A. J., Helms J. B., Rottier P. J., de Haan C. A. (2011) Proteomics 11, 64–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Crouch E., Hartshorn K., Horlacher T., McDonald B., Smith K., Cafarella T., Seaton B., Seeberger P. H., Head J. (2009) Biochemistry 48, 3335–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Hartshorn K. L., Collamer M., Auerbach M., Myers J. B., Pavlotsky N., Tauber A. I. (1988) J. Immunol. 141, 1295–1301 [PubMed] [Google Scholar]
- 35. Hartshorn K. L., Sastry K., White M. R., Anders E. M., Super M., Ezekowitz R. A., Tauber A. I. (1993) J. Clin. Invest. 91, 1414–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hartshorn K. L., Sastry K., Brown D., White M. R., Okarma T. B., Lee Y. M., Tauber A. I. (1993) J. Immunol. 151, 6265–6273 [PubMed] [Google Scholar]
- 37. Hartshorn K. L., Holmskov U., Hansen S., Zhang P., Meschi J., Mogues T., White M. R., Crouch E. C. (2002) Biochem. J. 366, 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barash S., Wang W., Shi Y. (2002) Biochem. Biophys. Res. Commun. 294, 835–842 [DOI] [PubMed] [Google Scholar]
- 39. Hartshorn K. L., White M. R., Voelker D. R., Coburn J., Zaner K., Crouch E. C. (2000) Biochem. J. 351, 449–458 [PMC free article] [PubMed] [Google Scholar]
- 40. Reading P. C., Tate M. D., Pickett D. L., Brooks A. G. (2007) Adv. Exp. Med. Biol. 598, 279–292 [DOI] [PubMed] [Google Scholar]
- 41. Job E. R., Deng Y. M., Tate M. D., Bottazzi B., Crouch E. C., Dean M. M., Mantovani A., Brooks A. G., Reading P. C. (2010) J. Immunol. 185, 4284–4291 [DOI] [PubMed] [Google Scholar]
- 42. Mikerov A. N., White M., Hartshorn K., Wang G., Floros J. (2008) Med. Microbiol. Immunol. 197, 9–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 44. Crouch E., Tu Y., Briner D., McDonald B., Smith K., Holmskov U., Hartshorn K. (2005) J. Biol. Chem. 280, 17046–17056 [DOI] [PubMed] [Google Scholar]
- 45. Head J. F., Mealy T. R., McCormack F. X., Seaton B. A. (2003) J. Biol. Chem. 278, 43254–43260 [DOI] [PubMed] [Google Scholar]