Abstract
A novel double deletion in cardiac troponin T (cTnT) of two highly conserved amino acids (Asn-100 and Glu-101) was found in a restrictive cardiomyopathic (RCM) pediatric patient. Clinical evaluation revealed the presence of left atrial enlargement and marked left ventricle diastolic dysfunction. The explanted heart examined by electron microscopy revealed myofibrillar disarray and mild fibrosis. Pedigree analysis established that this mutation arose de novo. The patient tested negative for six other sarcomeric genes. The single and double recombinant cTnT mutants were generated, and their functional consequences were analyzed in porcine skinned cardiac muscle. In the adult Tn environment (cTnT3 + cardiac troponin I), the single cTnT3-ΔN100 and cTnT3-ΔE101 mutations had opposing effects on the Ca2+ sensitivity of force development compared with WT, whereas the double deletion cTnT3-ΔN100/ΔE101 increased the Ca2+ sensitivity + 0.19 pCa units. In addition, cTnT3-ΔN100/ΔE101 decreased the cooperativity of force development, suggesting alterations in intrafilament protein-protein interactions. In the fetal Tn environment, (cTnT1 + slow skeletal troponin I), the single (cTnT1-ΔN110) and double (cTnT1-ΔN110/ΔE111) deletions did not change the Ca2+ sensitivity compared with control. To recreate the patient's heterozygous genotype, we performed a reconstituted ATPase activity assay. Thin filaments containing 50:50 cTnT3-ΔN100/ΔE101:cTnT3-WT also increased the myofilament Ca2+ sensitivity compared with WT. Co-sedimentation of thin filament proteins indicated that no significant changes occurred in the binding of Tn containing the RCM cTnT mutation to actin-Tm. This report reveals the protective role of Tn fetal isoforms as they rescue the increased Ca2+ sensitivity produced by a cTnT-RCM mutation and may account for the lack of lethality during gestation.
Keywords: ATPases, Calcium-binding Proteins, Cardiac Muscle, Cardiac Hypertrophy, Mutant, Protein-Protein Interactions, Restrictive Cardiomyopathy, Skinned Fibers, Troponin
Introduction
Depending on structural and functional characteristics, primary cardiomyopathies are classified into the three most common forms of dilated, hypertrophic, or restrictive cardiomyopathy (DCM,8 HCM, and RCM). RCM is the rarest of these cardiomyopathies and is characterized by a stiffened ventricular myocardium. This manifestation leads to diastolic dysfunction with decreased ventricular filling and subsequent dilation of the atria (1). RCM is much less common than either DCM and HCM, only accounting for 3–5% of all pediatric cardiomyopathies; however, it carries the poorest prognosis with an average 2-year survival rate of less than 50% after diagnosis and often requires heart transplantation (2–5). An interesting feature of cardiomyopathies is the overlap of phenotypes that can exist between them. Both DCM and RCM can have phenotypic overlap with HCM (6–8). To date, less than 20 mutations have been reported in the genes of cardiac desmin, α-actin, β-myosin heavy chain, cardiac troponin T (cTnT), and cardiac troponin I (cTnI) to be linked to RCM (9). In contrast, more than 900 mutations have been associated with HCM (8–11). In light of the catastrophic outcome of RCM, further definition of genotype-phenotype correlations remains an important task (9).
Mutations in the cardiac troponin complex (cTn) have been found to be a significant cause of genetic based cardiomyopathies (8). Cardiac Tn has an important role in regulating cardiac contractility. The cTn complex is constituted by three subunits: troponin C (cTnC), the subunit that confers Ca2+ sensitivity to striated muscle; cTnI, the subunit that prevents myosin interactions with actin; cTnT, that participates in the activation of muscle contraction and links the cTnI-cTnC complex to tropomyosin (Tm) in the thin filament (12). Previously, it was thought that the primary role of cTnT was to anchor cTn to the thin filament. However, it has been established by studying cardiomyopathic mutations that cTnT has multiple roles in muscle contraction such as modulating actomyosin ATPase activity, Ca2+ sensitivity of contraction, and maximal force (8, 13).
Recently, several studies have uncovered important details on the role of troponin T in development and disease. In mice, targeted disruption of TNNT2 causes severe defects of myofibrillogenesis, which leads to early embryonic demise (14, 15). In zebrafish, the mutant silent heart (sih) has been shown to be caused by TNNT2 mutations (16). In both models disassembly of myofibrils and dysregulation in thin filament expression replicate the hallmarks of sarcomere loss and myocyte disarray caused by human TNNT2 mutations (14, 15). A previous in vitro study indicated that the deletion of glutamic acid 96 in cTnT linked to RCM had a large effect on Ca2+ sensitivity of force development (17). In addition to these findings, a recent clinical report indicates the presence of an additional mutation, E136K, in the TNNT2 gene that was identified in a patient diagnosed with RCM (7). Cardiac tissue from the explanted heart showed that vacuolation was present, although it lacked the cardiomyocyte disarray normally associated with cTnT-based cardiomyopathies. Functional studies may further strengthen the association of this mutation with RCM.
The cTnT gene can express four different isoforms due to alternative splicing of exons four and five (18–20). The variability found between isoform 1 (predominant in fetal hearts) and isoform 3 (predominant in adult hearts) lies in the absence of exon 5 from the N-terminal domain of the adult isoform 3 (18, 19). The different cTnT isoforms are temporally expressed during cardiac muscle development and show distinctly altered abilities to modulate Ca2+ sensitivity of the myofilament (20, 21). It has been established that TnI switches isoforms during heart development. The slow skeletal troponin I (SSTnI) isoform is the predominant TnI isoform throughout fetal development (22). In the neonatal heart ssTnI is down-regulated, and the cTnI gene is expressed (23). Upon structural and physiological analysis, it was determined that these TnI isoforms confer distinct contractile properties on the myocardium (24, 25). Studying the physiological effects of the Tn isoforms is important in understanding the development of disease.
Here we report the first de novo double deletion of asparagine and glutamic acid at residues 100 and 101, respectively, in the TNNT2 gene in an 11-year-old RCM patient. Genetic analysis indicated that the proband was genotype-negative for mutations in six additional RCM susceptibility genes. In addition, pedigree analysis revealed that the mutation arose de novo as it was absent in both parents and two siblings. The clinical manifestation evaluated by a set of different techniques confirmed the presence of atrial enlargement and left ventricular diastolic dysfunction. Histopathological studies of the explanted heart demonstrated myocyte disarray and mild interstitial fibrosis. Functional studies strengthened the association of this deletion with the disease, as it resulted in phenotypic outcomes characteristic of other RCM causing mutations. Surprisingly, when the cTnT-RCM mutation was in the fetal Tn environment, the phenotypic outcome (increase in Ca2+ sensitivity) was rescued. The conclusions are as follows. 1) The functional phenotype can be directly correlated with the diastolic dysfunction seen in the patient. 2) We present a full clinical and genetic characterization coupled with functional studies that assist in linking these de novo mutations with their associated disease. 3) The functional phenotype was recapitulated in an assay containing Tn in a 50:50 mutant to WT ratio, which demonstrates the autosomal dominant effect of the mutation. 4) This report reveals the protective role of the Tn fetal isoforms, as they rescue the increased Ca2+ sensitivity produced by this cTnT-RCM deletion mutation and may account for the lack of lethality during gestation.
EXPERIMENTAL PROCEDURES
Patient and Clinical Evaluation
The index case and her family were recruited at Sainte Justine Hospital into an ongoing research protocol approved by the institution's ethics committee. All participants gave informed consent and were evaluated by family history, physical exam, electrocardiogram, and echocardiogram to measure wall thickness as well as systolic and diastolic function. Tissue Doppler imaging was performed as previously described (26). In addition to routine laboratory tests, the index case underwent stress testing using a modified Bruce protocol as well as right and left heart cardiac catheterization. Magnetic resonance imaging was performed on a 1.5 T system (Symphony, Siemens, Dorval, Canada) to yield optimal visualization of wall thicknesses in four-chamber, short axis, and long axis views. For control purposes, we used an in-house biobank of ethnically matched individuals.
Histological Evaluation
Heart tissue retrieved at catheterization and heart transplantation was used for pathological studies. Briefly, representative fresh heart tissue was formalin-fixed, paraffin-embedded, and stained with hematoxylin phloxine saffron. Moreover, representative adjacent fresh heart tissue was fixed in 3% glutaraldehyde, postfixed in 4% osmium tetroxide, and embedded in epoxy resin using standard procedures. Thick sections were cut and stained with p-phenylenediamine from four embedded tissue blocks suitable for ultra structural examination. Thin sections were stained with uranyl acetate followed by lead citrate and examined with a Philips EM208 electron microscope. Magnifications for light microscopy were 400× and for electron microscopy, 1800x.
DNA/RNA Extraction and Sequencing
Genomic DNA was extracted from peripheral blood samples as well as left and right ventricular tissue obtained upon transplantation using a PUREGENE DNA purification kit (Gentra Systems, Qiagen, Missisauga, ON, Canada) according to the manufacturer's instructions. Total RNA from left and right ventricular tissue was extracted using TRIzol according to standard protocols. cDNA was synthesized from 2 μg of total RNA using oligo-dT primers and an Omniscript reverse transcription kit (Qiagen). The entire open reading frames (ORF) of TNNI3, TNNT2, TPM1, and ACTC were amplified from left ventricular cDNA (primer sequences available upon request). Sequence changes identified in those ORF sequences were further verified by bidirectional sequencing of genomic DNA obtained from both ventricular tissues and lymphocytes. In addition, all coding exons and adjacent splice sites of MYH7, MYBPC3, and TNNC1 were bidirectionally sequenced from genomic DNA. As a control, sequence variants were then tested in the family as well as 100 unaffected healthy control DNAs.
Cardiac Skinned Fiber Studies
Fiber Preparation
Porcine papillary muscle was collected and prepared according to methods previously outlined (17, 27). Strips of left ventricle papillary muscle were extracted and incubated overnight in a pCa 8.0 solution containing 50% glycerol and 1% Triton X-100 at −20 °C; thereafter, they were transferred to a similar replacement solution without Triton X-100 and stored up to 2 weeks at −20 °C.
HcTnT Preparation for Fiber Studies
Adult human cardiac wild type troponin T (HcTnT3-WT) and fetal human cardiac wild type troponin T (HcTnT1-WT) were previously cloned in our laboratory (21) and used as a template for overlapping PCR with primers designed to delete those amino acids affected in the RCM troponin T mutant proteins (HcTnT3-ΔN100, HcTnT3-ΔE101, HcTnT3-ΔN100/ΔE101, HcTnT1-ΔN110, HcTnT1-ΔE111, and HcTnT1-ΔN110/ΔE111). Recombinant human cardiac troponin T, cTnI, ssTnI, and troponin C proteins were prepared as previously reported (17).
Tn-displaced Skinned Cardiac Fibers
To determine the effects of the HcTnT RCM mutants on the various Ca2+-dependent parameters of muscle contraction, the endogenous porcine cTn was displaced with the HcTnT to be studied. Previously, it was shown that human papillary fibers exchanged with human cTn developed similar changes in Ca2+ sensitivity as porcine papillary fibers exchanged with human cTn (28, 29). Therefore, porcine fibers serve as a suitable model to study human disease. Once the fiber was mounted and tested for calcium responsiveness, the fibers were relaxed and submerged into HcTnT solution (3 × 1 h incubation at room temperature with fresh HcTnT). Before incubating the fiber with HcTnT, the protein was diluted by ∼40% using the same buffer as described above without KCl, thus reducing the protein and salt concentration. This procedure results in the loss of Ca2+-activated force, and the fibers begin to demonstrate unregulated force irrespective of the Ca2+ concentration. To restore Ca2+-regulated force development after TnT incubation, fibers were incubated with a preformed cTnI-cTnC or cTnI-cTnC binary complex (30 μm, in relaxing solution) for ∼1 h. Upon reconstitution with the binary complex, the fibers underwent gradual relaxation. Once fully relaxed, the calcium-regulated force was restored (for further details, see Ref. 17).
Steady State and Calcium Dependence of Force Development
Fibers were mounted on to tweezer clips connected to a force transducer on one side and submerged in a 1.3-ml cuvette containing pCa 8.0 solution (10−8 m Ca2+, 1 mm Mg2+, 7 mm EGTA, 2.5 mm MgATP2−, 20 mm MOPS, pH 7.0, 20 mm creatinine phosphate, and 15 units/ml creatine phosphokinase, ionic strength 150 mm). To ensure complete removal of the sarcoplasmic reticulum, the fibers were incubated in pCa 8.0 solution containing 1% Triton X-100 for 30 min before the experiment. After extensive washing with pCa 8.0 solution, the fibers were tested for steady state force development in a pCa 4.0 solution (the same as pCa 8.0 solution, except the Ca2 was 10−4 m) followed by relaxation in the pCa 8.0 solution. Once the fibers were relaxed, they were exposed to solutions of increasing Ca2+ concentration (from pCa 8.0 to 4.0). The various pCa solutions were calculated using the computer program (pCa calculator) developed in our laboratory (30). Data were analyzed using the equation % change in force = 100 × [Ca2+]n/([Ca2+]n + [Ca502+] n), where [Ca502+] is the free Ca2+ concentration that produces 50% force, and n is the Hill coefficient.
Actomyosin-Tm-Tn ATPase Activity
The actomyosin-Tm-Tn ATPase activity assays were performed and plotted as a function of pCa according to Pinto et al. (31). Briefly, porcine cardiac myosin, rabbit skeletal F-actin, and porcine cardiac Tm were prepared as previously described (21). The individual troponin subunits were cloned, expressed, and purified according to Gomes et al. (21) and Pinto et al. (17). The troponin subunits were first dialyzed against 3 m urea, 1 m KCl, 10 mm MOPS, 1 mm DTT, 0.1 mm phenylmethanesulfonyl fluoride, and then twice against the same buffer excluding urea. The protein concentration of the individual subunits was determined using the Coomassie Plus kit (Pierce), and then the subunits were combined in a 1.5:1.3:1 TnT:TnI:TnC molar ratio. After 1 h, the complexes were successively dialyzed against decreasing concentrations of KCl (0.7, 0.5, 0.3, 0.1, 0.05, and 0.025 m). The excess precipitated TnT and TnI that formed during complex formation was removed by centrifugation. Proper stoichiometry was verified by SDS-PAGE before storage of troponin complexes at −80 °C. The ATPase reaction was initiated with the addition of ATP and quenched after 20 min using trichloroacetic acid to a final concentration of 35%. The precipitated assay proteins were removed by centrifugation, and the inorganic phosphate concentration in the supernatant released by ATP hydrolysis was determined. pCa curves were performed under the following 1× conditions (15 mm MOPS, 2 mm EGTA, 1 mm nitrilotriacetic acid, 1 mm free Mg2+, 2.5 mm MgATP2−, ionic strength (μ) = 75 mm (pH 7.0 at 25 °C)) changed slightly in varied pCa solutions (pCa 7.0 (0.457 mm CaCl2), pCa 6.5 (0.968 mm CaCl2), pCa 6.2 (1.305 mm CaCl2), pCa 6.0 (1.500 mm CaCl2), pCa 5.9 (1.583 mm CaCl2), pCa 5.7 (1.720 mm CaCl2), pCa 5.6 (1.778 mm CaCl2), pCa 5.4 (1.865 mm CaCl2), pCa 5.2 (1.932 mm CaCl2), and pCa 4.5 (2.140 mm CaCl2)), calculated according to methods established in our laboratory (30).
Co-sedimentation
Thin filament proteins were combined at the following concentrations: F-actin at 20 μm and Tn/Tm at 2.86 μm to obtain a 7:1:1 molar ratio. The proteins were co-sedimented using an air-driven ultracentrifuge (Beckman Airfuge; rotor A100) for 30 min at 28 p.s.i. (∼120,000 × g). The buffer conditions were either pCa 8.0 or pCa 4.5 as described above.
Statistical Analysis
Ca2+ sensitivity of force development curves were normalized and reported as a percentage of maximal Ca2+-activated force in pCa 4.0. The maximal force recovery was normalized to the initial maximal force of the fiber. The experimental results were analyzed for significance using the unpaired Student's t test at p < 0.05.
RESULTS
Clinical Characteristics of the Index Case and Her Family
The index case was a young girl who first presented at the age of 11 with chest pain and dyspnea upon exertion. She had normal growth parameters and vital signs with a regular heart of 92 beats/min and a blood pressure of 106/52 mm Hg. Physical examination was unremarkable with a normal S1 and S2, no S3, no S4, and no heart murmur. Peripheral pulses and her abdomen were normal.
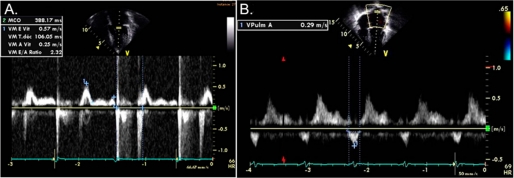
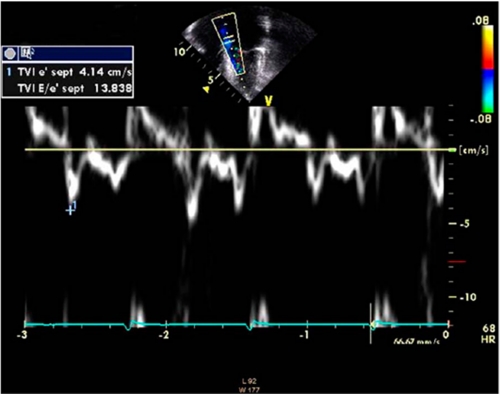
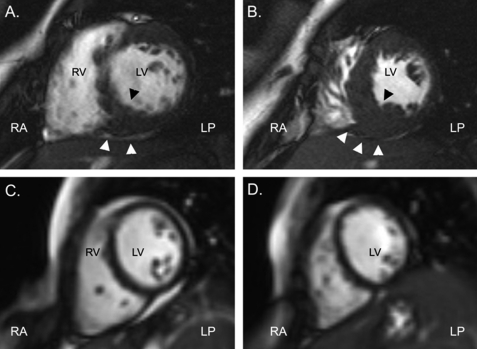
An electrocardiogram revealed sinus rhythm, left atrial enlargement, biventricular hypertrophy and diffuse T-wave changes (data not shown). The echocardiogram showed that the heart was structurally normal but with massive left atrial enlargement. Wall thicknesses measured by M-mode were in the normal range. Left ventricular dimensions and systolic function were also normal; however, the midseptum was mildly hypertrophic (maximal thickness = 15 mm) but demonstrated no regional wall motion abnormalities or left ventricular outflow tract obstruction. Marked left ventricular diastolic dysfunction was evidenced by an abnormal left ventricular inflow pattern with decreased atrial filling velocity (0.25 m/s, normal >0.5 m/s), a decreased e-wave deceleration time (106 ms, normal >150 ms), an abnormal e/a wave ratio of 2.32 (normal <2), and a decreased isovolumic relaxation time (25 ms, normal >70 ms) (Fig. 1A). Furthermore, we observed reversal of the pulmonary venous a Doppler wave, indicative of elevated end-diastolic pressures in the left ventricle (Fig. 1B). We also measured the early diastolic mitral annulus velocity e′ by tissue Doppler, which reflects the velocity of the mitral annulus in the longitudinal direction. With an e′ value of 4.14 cm/s, we found this velocity to be significantly lower than normal (normal, 13.8–15.2 cm/s) (26). With a mitral E wave velocity of 57 cm/s, this yields a severely depressed septal E/e′ ratio of 13.83 (normal, 6.3–7.0) (26) (Fig. 2 and supplemental Fig. 1). Taken together, these findings were consistent with a restrictive left ventricular filling pattern and elevated left-ventricular end-diastolic pressure (32). Magnetic resonance imaging demonstrated a structurally normal heart with left atrial enlargement, normal ventricular size, and with no evidence of pericardial abnormality. Focal thickening of the myocardium was seen at several levels, in particular at the mid-ventricular and apical inferior segments, with a maximal thickness measured at 17 mm (Fig. 3). No such thickening was observed in the anteroseptal, anterolateral, and inferolateral segments or in the right ventricle. Late enhancement was not seen. A modified Bruce exercise test induced marked ST-segment depression and chest pain, indicative of ischemia (data not shown). The test was stopped at the beginning of step 3 at a heart rate of 166 beats/min, corresponding to a decreased exercise capacity of 8.3 metabolic equivalents at 79% that of the predicted maximal heart rate. Repeated Holter EKG recordings did not show any evidence of ventricular arrhythmia (data not shown).
FIGURE 1.
Doppler imaging. A, shown is a pulsed wave Doppler image of left ventricular inflow at the level of the mitral valve, demonstrating restrictive physiology. B, shown is a pulsed wave Doppler image of pulmonary venous inflow into the left atrium. Note clearly positive s and d waves followed by a negative a wave.
FIGURE 2.
Tissue Doppler sampling at the septal position in an apical 4 chamber view; note the severely altered E/e′ ratio. The mitral E-wave velocity is 57 cm/s (Fig. 1), and the early mitral annulus e′ velocity is 4.34 cm/s, giving a severely altered E/ e′ ratio of 13.8.
FIGURE 3.
Cardiac magnetic resonance findings. All frames were taken at end-diastole. A, short-axis cut at mid-ventricular level is shown. B, short-axis cut at a more apical level is shown. C and D, short axis cuts at comparable levels to A and B in a sex- and age-matched subject with normal cardiac anatomy and function are shown. Note the fine structure of the left ventricular myocardium. RA, right anterior; LP, left posterior; RV, right ventricle; LV, left ventricle. Arrowheads denote focal thickening of an inferior septal segment.
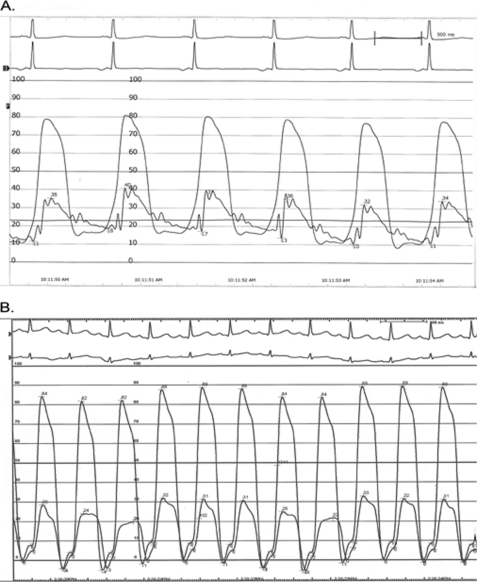
Etiologic investigations revealed a normal karyotype, normal plasma amino acids, and urine organic acids. Results of the major laboratory tests were within normal range except for a markedly elevated plasma level of pro-brain natriuretic peptide at 3000 ng/liter (upper limit of normal, 115 ng/liter) (33). Cardiac catheterization indicated a normal cardiac index and normal systemic and pulmonary vascular resistance. We observed a rapid decline in left ventricular pressure at the onset of diastole, with a rapid rise to plateau in early diastole, reminiscent of the square root sign; the systolic right ventricular pressure was elevated with marked elevation of right ventricular diastolic pressure (Fig. 4A and Table 1). The tracing of a pediatric patient without significant heart disease is shown in Fig. 4B. We interpreted this pattern as indicative of a severely decreased compliance of the left ventricle and subsequent increase of right ventricular pressure. Left ventricular angiography showed the typical “ballerina foot” configuration seen in septal hypertrophy (data not shown). Endomyocardial biopsy revealed moderate myocyte hypertrophy with mild interstitial fibrosis but no evidence of inflammatory infiltrate, glycogen deposits, fibroelastosis, amyloidosis, or hemochromatosis. In light of these results, the patient was diagnosed with RCM with mid- and inferoseptal hypertrophy.
FIGURE 4.
Simultaneous right and left ventricular pressure recordings. x axis, time; y axis, pressure in mm Hg. A, note the elevated diastolic pressures in the left ventricle as well as the elevated right ventricular systolic and diastolic pressures, as reported in Table 1. B, comparative tracing of a pediatric patient without hemodynamically significant heart disease. Note that end-diastolic pressures in both ventricles are lower by ∼10 mm Hg.
TABLE 1.
Summary of pressures encountered at catheterization during initial diagnostic workup
| Systolic/end-diastolic | Mean | Upper limit of normal | |
|---|---|---|---|
| Right atrium | 7 | 5 | |
| Right ventricle | 36/12 | 15–25/0–8 | |
| Pulmonary artery | 36/13 | 15–25/8–15 | |
| Pulmonary capillary wedge | 23 | 6–12 | |
| Left ventricle | 79/17 | 120/8 |
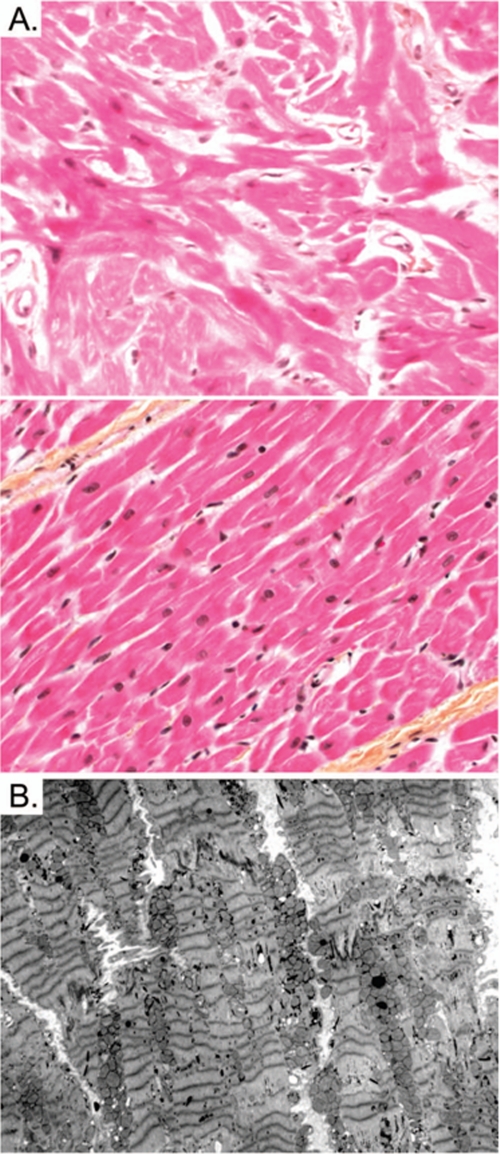
During the following year she was treated with metoprolol (2 mg/kg/day) and aspirin (80 mg three times a day). Her clinical condition deteriorated rapidly with frequent chest pain and increasing dyspnea (New York Heart Association functional class III). The patient ultimately underwent cardiac transplantation 15 months after her initial presentation. Gross anatomic examination of the explanted heart revealed mild hypertrophy, which was marked only at the septal level, consistent with pre-transplantation imaging. Histology revealed mild hypertrophy, mild interstitial fibrosis, and varying degrees of disarray of the myocytes (Fig. 5A). Transmission electron microscopy showed misalignment of Z-bands and unequal Z-Z band distances (“streaming”) (Fig. 5B). In the normal human cardiomyocyte, Z-bands are well aligned and have equal Z-Z band distances (34). After cardiac transplantation, the patient performed well with normal growth and development. Endomyocardial biopsy revealed no evidence of rejection.
FIGURE 5.
Histopathological studies. A, extensive cardiomyocyte disarray is seen in light microscopy using hematoxylin phloxine saffron stain (top) in comparison with normal histology (bottom) at 400×-fold magnification. B, transmission electron microscopy shows extensive Z-disc streaming but normal mitochondrial morphology.
There was no family history of cardiac or neuromuscular disease. Clinical investigations including electrocardiogram and echocardiogram in both of her parents and two brothers were normal.
Genetic Investigations
A heterozygous in-frame deletion in exon 9 of TNNT2 (c.297–302 AATGAGdeletion) was found in the index case that resulted in the deletion of asparagine and glutamic acid at amino acid positions 100 and 101, respectively (p.Asn100_Glu101del) (Fig. 6). This mutation was confirmed in her genomic DNA from both ventricular tissue and lymphocytes. Its absence in both parents and her two brothers confirmed the de novo occurrence of this genetic alteration. This deletion mutation was not present in 192 analyzed healthy controls and has not yet been reported. Sequence and structural analysis shows that this mutation is located in a highly conserved position across many species. Three reported TNNT2 mutations associated with RCM (E136K, I79N, and ΔE96) and most of the HCM, causing mutations in TNNT2, are located in this region (Fig. 6). No mutations involving the genomic sequence of TNNI3, ACTC, MYH7, TPM1, MYBPC3, and TNNC1 were detected in the index case except for the known polymorphisms given in Table 2.
FIGURE 6.
Mutational analysis. A, pherogram was obtained upon sequencing of exon 9. The deleted sequence is highlighted in gray. The identity of the sequence change was further verified by subcloning (data not shown). B, the mutation is located in a highly conserved domain of TNNT2, as illustrated across multiple species including human, mouse, rat, chicken, and zebrafish. Note that this sequence reflects the adult isoform cTnT3. However, if the mutations are present in the fetal isoform cTnT1, the numbering would be shifted by 10 amino acids due to the presence of exon 5. AA, amino acid position. Mutations associated with HCM are marked in black (top), and mutations linked to RCM are shown in blue (bottom).
TABLE 2.
Compilation of sequencing results
All positions are given according to the coordinates of the assembly NCBI36/hg18. No polymorphisms were found in ACTC, TNNC1, and TPM1. SNP, single-nucleotide polymorphism.
| Gene | Chromosome | Position | rsID | Patient genotype | Type of SNP |
|---|---|---|---|---|---|
| bp | |||||
| MYBPC3 | 11 | 47328174 | rs3729986 | C/T | V158M Known polymorphism |
| MYH7 | 14 | 22951760 | rs12147570 | G/T | 5′-UTR |
| MYH7 | 14 | 22951984 | rs2284651 | C/T | Intronic |
| MYH7 | 14 | 22952026 | rs7149517 | G/T | Intronic |
| MYH7 | 14 | 22953024 | rs3729833 | C/T | Intronic |
| MYH7 | 14 | 22955727 | rs3729829 | A/G | Intronic |
| MYH7 | 14 | 22955837 | rs3729828 | A/G | Intronic |
| MYH7 | 14 | 22956104 | rs3729825 | C/T | Intronic |
| MYH7 | 14 | 22957485 | rs7159367 | C/T | Intronic |
| MYH7 | 14 | 22958505 | rs2277475 | A/T | Intronic |
| MYH7 | 14 | 22968867 | rs735711 | C/T | Synonymous |
| TNNI3 | 19 | 60359459 | rs3729711 | G/T | Synonymous |
Functional Characterization
To ensure that the mutation found in the index case is causative of the disease, we performed functional experiments at the cellular level. We used a well established methodology, porcine skinned cardiac fibers exchanged with exogenous Tn, to evaluate possible changes in the Ca2+ sensitivity of force development as well as maximal force. The RCM mutations were introduced into two different isoforms of cTnT. cTnT isoform 3 found in adult hearts has asparagine and glutamic acid deletions at positions 100 and 101, and in the fetal cTnT isoform 1 the deletions would occur at positions 110 and 111. The altered sequence of these two isoforms is due to exon 5 (which contains 10 amino acids) that is present in isoform 1 but not in isoform 3. As shown in Fig. 7 and Table 3, skinned fibers displaced with the single mutant HCTnT3-ΔN100 and reconstituted with cTnI-cTnC complex displayed a significant increase in the Ca2+ sensitivity of force development (pCa50) along with a significant decrease in thin filament cooperativity (nHill) (pCa50 = 5.96 ± 0.03; nHill = 1.84 ± 0.08) compared with HCTnT3-WT (pCa50 = 5.67 ± 0.01; nHill = 2.83 ± 0.12). However, skinned fibers displaced by the single mutant HCTnT3-ΔE101 and reconstituted with cTnI-cTnC showed a significant decrease in the Ca2+ sensitivity of force development and no changes in the Hill coefficient (pCa50 = 5.39 ± 0.01; nHill = 3.14 ± 0.32) compared with fibers displaced with HCTnT3-WT. The two single mutant proteins when incorporated into the fiber shifted the Ca2+ sensitivity of force development to the same extent but in opposite directions (see ΔpCa50 in Table 3). Surprisingly, fibers incorporated with the double RCM mutant (HCTnT3-ΔN100/ΔE101) and cTnI-cTnC significantly increased the Ca2+ sensitivity of force development and decreased the cooperativity of the thin filament activation, pCa50 = 5.86 ± 0.02 and nHill = 1.94 ± 0.09, respectively, compared with its WT (Fig. 7 and Table 3). Furthermore, the maximal force recovery was also evaluated after incorporation of exogenous TnT and reconstitution with TnI-TnC (see “Experimental Procedures”). Fibers containing the single mutant HcTnT3-ΔN100 or the RCM double mutant HcTnT3-ΔN100/ΔE101 and reconstituted with the cTnI-cTnC complex did not display changes in the recovery of maximal force compared with their control (Table 3). However, fibers incorporated with the single mutant HcTnT3-ΔE101 and reconstituted with cTnI-cTnC complex drastically reduced the maximal force recovery (28.40 ± 1.98%) compared with WT (50.81 ± 3.36%) (Table 3). When the skinned fibers were reconstituted with the fetal troponin isoforms (HcTnT1 and ssTnI), the RCM double mutant (HcTnT1-ΔN110/ΔE111) and the single mutant (HcTnT1-ΔN110) did not significantly change the Ca2+ sensitivity of force development compared with HcTnT1-WT (Fig. 7 and Table 3). Therefore, the single mutant HcTnT1-ΔE111 decreased the Ca2+ sensitivity of force development compared with WT (Fig. 7 and Table 3). The Hill coefficient was not significantly different between the mutants when compared with the WT control (Table 3). The maximal force recovery was increased in fibers displaced with the single mutant HcTnT1-ΔN110 and the double RCM mutants HcTnT1-ΔN110/ΔE111 when reconstituted with ssTnl-cTnC compared with the WT (Table 3). To ensure that the exogenous recombinant TnT was fully incorporated into the thin filament, we measured the Ca2+ unregulated force after TnT incubation/treatment. The amount of exogenous TnT that displaced the whole native Tn complex was measured by the degree that the fiber became Ca2+ unregulated, i.e. generated force in the absence of Ca2+. This occurs because TnI is no longer present to inhibit myosin and actin interactions. The % of Ca2+ unregulated force, as measured by the ratio FpCa8/FpCa4 × 100 after TnT treatment (FpCa8 and FpCa4 are the forces at pCa 8.0 and pCa 4.0 solutions, respectively) can be directly correlated with the displacement of native Tn according to Pinto et al. (17). All the HcTnT3 proteins (adult isoform) that were studied displaced more than 90% of the native Tn complex (Table 3), which demonstrates that they associate normally with the other thin filament proteins. Yet the HcTnT1-ΔN110/Δ111 and the HcTnT1-Δ110 had a significantly reduced ability to displace the native Tn complex compared with the HcTnT1-WT (Table 3).
FIGURE 7.
Measurements of the Ca2+ sensitivity of contraction in porcine skinned cardiac fibers. Upper panels, shown are normalized pCa-force relationship in fibers displaced with HcTnT3 single mutants or RCM double mutant and reconstituted with cTnI-cTnC binary complex compared with their control (HcTnT3-WT) (adult environment). Lower panels, shown are normalized pCa-force relationship in fibers displaced with HcTnT1 single mutants or RCM double mutant and reconstituted with cTnI-cTnC binary complex compared with their control (HcTnT1-WT) (fetal environment). See Table 3 for maximal force recovery, Hill coefficient (nH), and maximal unregulated force values. Data in each experiment are an average of 6–13 experiments, and the mean is shown as ±S.E.
TABLE 3.
Summary of pCa-force relationship curves in reconstituted porcine skinned fibers at pH 7.0
The pCa50, nH, maximal force, and % Ca2+ unregulated force values are the average of many independent fiber experiments, and the errors are the S.E. values. The Ca2+- unregulated force was calculated by the equation (FpCa8/FpCa4) × 100, where the FpCa8 and FpCa4 are the force at pCa 8.0 and pCa 4.0 solutions, respectively. p values compare the RCM mutant against the TnT3-WT or TnT1-WT. HSSTnI, human SSTnI.
| TnT | TnI | pCa50 | ΔpCa50a | Hill coefficient, nH | % Maximal force | % Ca2+ unregulated force | No. experiments |
|---|---|---|---|---|---|---|---|
| HCTnT3-WT | HCTnI | 5.67 ± 0.01 | 2.83 ± 0.12 | 50.81 ± 3.36 | 90.30 ± 2.75 | 12 | |
| HCTnT3-ΔN100/ΔE101 | HCTnI | 5.86 ± 0.02b | +0.19 | 1.94 ± 0.09b | 51.30 ± 2.90 | 91.79 ± 3.95 | 10 |
| HCTnT3-ΔN100 | HCTnI | 5.96 ± 0.03b | +0.29 | 1.84 ± 0.08b | 54.73 ± 3.09 | 93.77 ± 3.03 | 13 |
| HCTnT3-ΔE101 | HCTnI | 5.39 ± 0.01b | −0.28 | 3.14 ± 0.32 | 28.40 ± 1.98b | 99.33 ± 0.45b | 8 |
| HCTnT1-WT | HSSTnI | 5.78 ± 0.02 | 2.05 ± 0.11 | 44.43 ± 1.56 | 96.46 ± 1.62 | 11 | |
| HCTnT1-ΔN110/ΔE111 | HSSTnI | 5.82 ± 0.02 | +0.04 | 2.11 ± 0.05 | 55.12 ± 3.08b | 87.60 ± 3.76b | 8 |
| HCTnT1-ΔN110 | HSSTnI | 5.82 ± 0.03 | +0.04 | 2.40 ± 0.10 | 56.02 ± 2.09b | 88.42 ± 4.22b | 6 |
| HCTnT1-ΔE111 | HSSTnI | 5.49 ± 0.04b | −0.29 | 2.83 ± 0.33 | 21.83 ± 4.30b | 95.13 ± 2.29 | 6 |
a TnT3-RCM pCa50 − TnT3-WT pCa50 or TnT1-RCM pCa50 −TnT1-WT pCa50.
b p < 0.05.
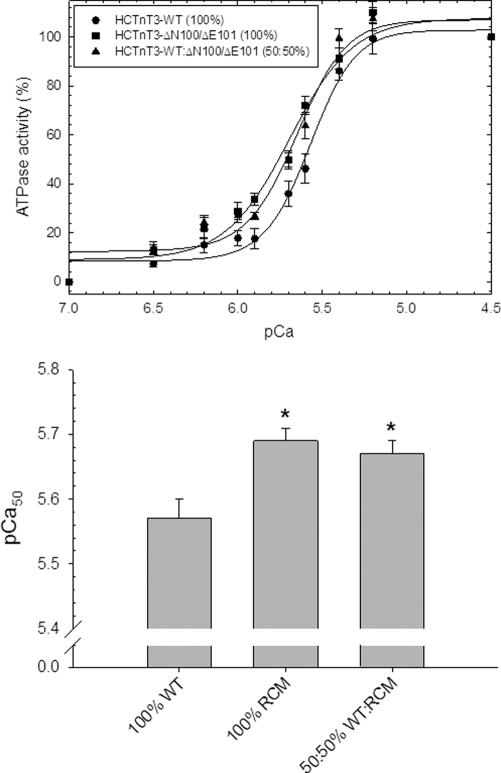
Fig. 8 shows the activation of the myosin ATPase at increasing concentrations of Ca2+ (pCa). It was found that cTn complexes containing 100% of the RCM mutant HcTnT3-ΔN100/ΔE101 significantly increased the Ca2+ sensitivity of the reconstituted filament by +0.12 pCa units compared with 100% HcTnT3-WT. To mimic the expected mutant/WT ratios found in the heterozygous patient, filaments were reconstituted with 50% HcTnT3-WT, 50% HcTnT3-ΔN100/ΔE101 and also significantly increased the Ca2+ sensitivity (+0.10 pCa units) compared with 100% HcTnT3-WT. The cooperativity was unaltered in these in vitro experiments. The pCa50 and nHill values are displayed in Table 3.
FIGURE 8.
Ca2+ dependence of myosin ATPase activity in the reconstituted filaments. The experiments were performed using 0.6 μm cardiac myosin, 3.5 μm skeletal actin, 1.0 μm cardiac Tm, and 1.0 μm cardiac Tn. The buffer conditions were described under “Experimental Procedures.” A, the graph shows relative values (%) of ATPase activity as a function of pCa. The basal ATPase activity at pCa 7.0 was considered 0%, and the intermediate points were normalized to the maximal ATPase activity at pCa 4.5, which was considered 100%. B, the graph shows the pCa50 from each condition. The filaments containing the 100% WT Tn complex showed pCa50 = 5.57 ± 0.03 and nHill = 3.13 ± 0.36. The filaments containing 100% ΔN100/ΔE101 Tn complex yielded pCa50 = 5.69 ± 0.02 and nHill = 2.55 ± 0.17, whereas filaments containing 50% WT and 50% ΔN100/ΔE101 Tn complex yielded pCa50 = 5.67 ± 0.02 and nHill = 2.85 ± 0.35. Each curve represents an average of six to eight experiments, and error is reported as the mean ± S.E.
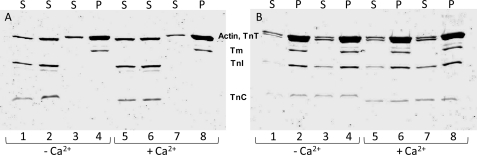
We performed an actin:Tm:Tn co-sedimentation assay at saturating stoichiometric ratio (7 actins, 1 Tm, and 1 Tn) to investigate whether the TnT RCM mutation interfered with the ability of the Tn complex to bind to the thin filament. Fig. 9 shows no discernable alterations in the ability of the Tn complex containing the RCM mutant to bind to actin:Tm compared with the WT Tn complex. In addition, no changes were observed in the protein content of the pellet and supernatant of thin filaments containing WT or mutant HcTnT either in the presence or absence of Ca2+. These data corroborate the skinned fiber data that indicated that the various HcTnTs were able to effectively displace the native Tn complex.
FIGURE 9.
Co-sedimentation of actin, tropomyosin, and troponin complex containing either WT or mutant TnT. The experimental conditions were outlined under “Experimental Procedures.” Samples were separated by 15% SDS-PAGE. S, supernatant; P, pellet. A, lanes 1 and 5, WT troponin complex supernatant (pellet not shown); lanes 2 and 6, ΔN100/ΔE101troponin complex supernatant (pellet not shown); lanes 3 and 7, actin-tropomyosin supernatant; lanes 4 and 8, actin-tropomyosin pellet. B, lanes 1 and 5, actin-tropomyosin-WT troponin supernatant; lanes 2 and 6, actin-tropomyosin-WT troponin pellet; lanes 3 and 7, actin-tropomyosin-ΔN100/ΔE101 troponin supernatant; lanes 4 and 8, actin-tropomyosin-ΔN100/ΔE101 troponin pellet. Lanes 1–4 were performed at a low Ca2+ concentration. Lanes 5–8 were performed at a high Ca2+ concentration.
DISCUSSION
Over the past two decades, molecular genetics analyses have revealed the key role of mutations in sarcomeric protein genes in diseases such as DCM and HCM. Differential effects of mutations on sarcomeric properties such as myofilament sliding, Ca2+ sensitivity, or ATPase activity have emerged as a mechanism underlying those phenotypes (8, 9). Recent studies have shown that RCM is part of the spectrum of sarcomeric gene mutations, including pediatric RCM (7, 35–40). In the largest series of pediatric RCM reported to date, mutation screening of eight sarcomeric genes and desmin suggested a predominant role of mutations in TNNI3, TNNT2, and ACTC in disease pathogenesis (7). Here we identify a de novo deletion in TNNT2 (p.Asn100_Glu101del in exon 9) in an RCM patient and provide a detailed clinical, biochemical, and genetic analysis that validates this TNNT2 mutation as the causative allele. In addition, we present a set of functional data that supports the protective role of fetal troponin isoforms in the developing heart (41).
Interestingly, the morphological findings in this patient indicate the heart was not uniformly affected despite the germ line character of the mutation. The right ventricle was not hypertrophied, and myocyte fiber disarray was more pronounced in biopsied tissue from areas with focal hypertrophy, such as evidenced by echocardiography and magnetic resonance imaging (Figs. 3 and 5). Interestingly, these observations suggest that a certain overlap of the restrictive and hypertrophic phenotypes may have coexisted in this patient. Clearly, imaging studies obtained at the onset of symptoms did not show a degree of hypertrophy proportional to the symptoms manifested by the patient. However, mild localized hypertrophy at the septal level and the absence of obstruction of the left ventricular outflow tract were consistent with an overlapping phenotype of HCM in this patient with predominant features of RCM. Also, the mild hypertrophy and prominent fiber disarray is in line with the histopathological data reported in a previous study of TnT mutations (42). Despite the considerable degree of structural and functional anomalies, we did not observe any arrhythmia in our patient. This does not rule out that such a predisposition may have existed but that the patient was transplanted before such an occurrence. Also, we have learned that changes in the myofilament Ca2+ sensitivity can induce changes in the intracellular Ca2+ cycling as well as development of arrhythmias in mice (43, 44). The amelioration of altered intracellular Ca2+ levels would provide a good therapeutic target to strategically correct cardiomyopathic disease (45). In addition, Peña et al. (46) have shown that increasing the Serca2a gene expression in a tropomyosin HCM animal model improves heart function and postpones the onset of the disease.
Functional studies with deletions of each single residue and also with the RCM double deletion were performed. The Ca2+ sensitivity measured in reconstituted skinned cardiac fibers using the adult troponin background (HcTnT3 and cTnI) proved very interesting, as the effects of the single cTnT deletions (Asn-100 and Glu-101) were opposing, with ΔN100 displaying a dominant phenotype. The single deletion E101 decreased the Ca2+ sensitivity and maximal force, whereas the single deletion Asn-100 increased the Ca2+ sensitivity, which is a hallmark for cTn mutations linked to RCM (8). Interestingly, the combination of the two deletions followed the trend of the functional phenotype manifested by the single deletion of N100. In fact, the deletion of E101 partially rescues a potentially more severe phenotype contributed by ΔN100. The evaluation of the Ca2+ sensitivity of force development in fibers displaced and reconstituted with fetal troponin isoforms revealed their protective role in maintenance of normal physiological parameters. The double RCM mutant displayed the same Ca2+ sensitivity and degree of cooperativity during activation of the thin filament as its control. These findings are in accordance with the known effects of ssTnI, which assists with maintenance of normal cardiac function during extreme conditions such as acidosis (41, 47, 48). The effect of the TnT isoforms on the regulation of cardiac muscle has also been studied (20, 21). However, presently in the literature there is no consensus that TnT isoforms provide a cardioprotective effect. Therefore, we can only speculate that in our system ssTnI may be solely responsible for protecting the heart against deleterious effects of the RCM mutation. The maximal force recovery was increased in fibers displaced with the fetal HcTnT-RCM double mutant and reconstituted with ssTnI in comparison to WT. Previously, we have shown this same functional phenotypic change in a cTnI RCM mouse model (49).
Furthermore, biochemical studies revealed that incorporation of 50% of the mutant protein into the thin filaments is sufficient to recapitulate the functional phenotype seen in the presence of 100% of the mutant protein. These data support the findings that RCM is an autosomal dominant disease. Additionally, the cTnT-RCM double deletion ΔN100/ΔE101 shows changes in the cooperativity of thin filament activation in skinned fibers (see the Hill coefficient data in Table 3) similar to what was previously seen with the other cTnT-RCM mutation (17). These changes could help to explain the timing of onset and severity of the disease once they were identified in a pediatric patient; in fact, fetal TnI isoform expression may have afforded protection against the deleterious effects of the cTnT-RCM double deletion ΔN100/ΔE101 during early childhood (22). However, this concept will be further refined by additional studies that explore the contribution of mutations toward the development of RCM.
Since the first publication of a cardiomyopathy-associated mutation in TNNT2, more than 30 distinct mutations in TNNT2 have been identified in patients with HCM, and several TNNT2 mutations have also been described in DCM patients (8). Recently, a mutation in TNNT2 (p.96delE) was reported in an infant with RCM (39). Functional analysis of this mutation revealed a large increase in Ca2+ sensitivity and an impaired ability to inhibit ATPase activity and to undergo relaxation in skinned fibers. This is consistent with the functional anomalies in muscle relaxation associated with RCM (17, 49–53). Patients with RCM present severe diastolic dysfunction, and mutations associated with RCM generally cause drastically increased Ca2+ sensitivity of force development in cTnT-substituted skinned fibers (54). Contraction of cardiac muscles is regulated by Ca2+ through its binding to the ternary complex of cardiac TnT, TnI, and TnC. Cardiac TnT links cTnI to the thin filament due to simultaneous binding to both cTnI and Tm. This allows the inhibition of cardiomyocyte contraction by cTnI only at low [Ca2+]i and enables the ternary troponin complex to exert its Ca2+-dependent regulation on the thin filament (55, 56). Studies of cTnT fragments have identified a critical tropomyosin binding region corresponding to residues 70–170 in the N-terminal domain. Mutations within this region may impair tropomyosin-dependent functions of cTnT and most likely affect the cooperativity of the thin filament (57). Subsequently, three reported TNNT2 mutations associated with RCM (I79N, ΔE96, and E136K) occur within this region. These findings may indicate the presence of a mutational hotspot region as three previous mutations associated with RCM occur in this portion of TNNT2. Although the two residues appear conserved across species (Fig. 6B), the results suggest a predominant functional effect of Asn-100 over Glu-101. Additionally, the Asn-100 deletion may impact the overall cTnT structure or interactions between cTnT and Tm more drastically than that of the Glu-101 deletion.
Co-sedimentation analysis using a 7 actin:1 Tm:1 Tn ratio was performed to determine whether the RCM cTnT mutant had altered the affinity of cTn for the components of the thin filament. The results suggest that the RCM cTnT mutation either does not interfere with the association between cTn and the thin filament or that this method may not be sensitive enough to detect small affinity changes. Further biochemical studies may help to elucidate whether any modifications of the thin filament occur in the presence of RCM cTnT mutation.
A functional phenotype for cTn mutations linked to RCM is emerging, and it is possible that the molecular mechanism that governs the manifestation of this disease arises from increased myofilament Ca2+ sensitivity along with an impaired ability to relax (52, 53, 58–60). In a more physiological scenario we speculate that increased affinity of the myofilament for Ca2+ would slow the rate of Ca2+ dissociation from TnC and consequently delay the Ca2+ and force transients that consequently could result in diastolic dysfunction (49, 51). In addition, clinical, genetic, and functional data placed together provide a valuable tool to strengthen our knowledge of the causes of cardiomyopathic disease (27). Additionally, the data suggest that this RCM mutation did not cause lethality during gestation as no aberrant changes in the Ca2+ sensitivity of force development and cooperativity of thin filament activation were observed in the fetal environment where fetal troponin isoforms are present.
Supplementary Material
Acknowledgment
We gratefully acknowledge the help of Dr. C. Boutin in the preparation of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant HL-042325 (to J. D. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- DCM
- dilated cardiomyopathy
- HCM
- hypertrophic cardiomyopathy
- RCM
- restrictive cardiomyopathy
- TnT
- troponin
- cTnT
- cardiac TnT
- cTnI
- cardiac TnI
- cTnC
- cardiac TnC
- Tm
- tropomyosin
- SSTnI
- slow skeletal troponin I.
REFERENCES
- 1. Ammash N. M., Seward J. B., Bailey K. R., Edwards W. D., Tajik A. J. (2000) Circulation 101, 2490–2496 [DOI] [PubMed] [Google Scholar]
- 2. Zangwill S. D., Naftel D., L'Ecuyer T., Rosenthal D., Robinson B., Kirklin J. K., Stendahl G., Dipchand A. I. (2009) J. Heart Lung Transplant. 28, 1335–1340 [DOI] [PubMed] [Google Scholar]
- 3. Dipchand A. I., Naftel D. C., Feingold B., Spicer R., Yung D., Kaufman B., Kirklin J. K., Allain-Rooney T., Hsu D. (2009) J. Heart Lung Transplant. 28, 1312–1321 [DOI] [PubMed] [Google Scholar]
- 4. Weller R. J., Weintraub R., Addonizio L. J., Chrisant M. R., Gersony W. M., Hsu D. T. (2002) Am. J. Cardiol. 90, 501–506 [DOI] [PubMed] [Google Scholar]
- 5. Chen S. C., Balfour I. C., Jureidini S. (2001) J. Heart Lung Transplant. 20, 90–92 [DOI] [PubMed] [Google Scholar]
- 6. Morimoto S. (2008) Cardiovasc. Res. 77, 659–666 [DOI] [PubMed] [Google Scholar]
- 7. Kaski J. P., Syrris P., Burch M., Tomé-Esteban M. T., Fenton M., Christiansen M., Andersen P. S., Sebire N., Ashworth M., Deanfield J. E., McKenna W. J., Elliott P. M. (2008) Heart 94, 1478–1484 [DOI] [PubMed] [Google Scholar]
- 8. Willott R. H., Gomes A. V., Chang A. N., Parvatiyar M. S., Pinto J. R., Potter J. D. (2010) J. Mol. Cell. Cardiol. 48, 882–892 [DOI] [PubMed] [Google Scholar]
- 9. Parvatiyar M. S., Pinto J. R., Dweck D., Potter J. D. (2010) J. Biomed. Biotechnol. 2010, 350706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu Q., Dewey S., Nguyen S., Gomes A. V. (2010) J. Mol. Cell. Cardiol. 48, 899–909 [DOI] [PubMed] [Google Scholar]
- 11. Alcalai R., Seidman J. G., Seidman C. E. (2008) J. Cardiovasc. Electrophysiol. 19, 104–110 [DOI] [PubMed] [Google Scholar]
- 12. Gordon A. M., Homsher E., Regnier M. (2000) Physiol. Rev. 80, 853–924 [DOI] [PubMed] [Google Scholar]
- 13. Potter J. D., Sheng Z., Pan B. S., Zhao J. (1995) J. Biol. Chem. 270, 2557–2562 [DOI] [PubMed] [Google Scholar]
- 14. Ahmad F., Banerjee S. K., Lage M. L., Huang X. N., Smith S. H., Saba S., Rager J., Conner D. A., Janczewski A. M., Tobita K., Tinney J. P., Moskowitz I. P., Perez-Atayde A. R., Keller B. B., Mathier M. A., Shroff S. G., Seidman C. E., Seidman J. G. (2008) PLoS ONE 3, e2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nishii K., Morimoto S., Minakami R., Miyano Y., Hashizume K., Ohta M., Zhan D. Y., Lu Q. W., Shibata Y. (2008) Dev. Biol. 322, 65–73 [DOI] [PubMed] [Google Scholar]
- 16. Sehnert A. J., Huq A., Weinstein B. M., Walker C., Fishman M., Stainier D. Y. (2002) Nat. Genet. 31, 106–110 [DOI] [PubMed] [Google Scholar]
- 17. Pinto J. R., Parvatiyar M. S., Jones M. A., Liang J., Potter J. D. (2008) J. Biol. Chem. 283, 2156–2166 [DOI] [PubMed] [Google Scholar]
- 18. Anderson P. A., Malouf N. N., Oakeley A. E., Pagani E. D., Allen P. D. (1991) Circ. Res. 69, 1226–1233 [DOI] [PubMed] [Google Scholar]
- 19. Townsend P. J., Barton P. J., Yacoub M. H., Farza H. (1995) J. Mol. Cell. Cardiol. 27, 2223–2236 [DOI] [PubMed] [Google Scholar]
- 20. Anderson P. A., Greig A., Mark T. M., Malouf N. N., Oakeley A. E., Ungerleider R. M., Allen P. D., Kay B. K. (1995) Circ. Res. 76, 681–686 [DOI] [PubMed] [Google Scholar]
- 21. Gomes A. V., Guzman G., Zhao J., Potter J. D. (2002) J. Biol. Chem. 277, 35341–35349 [DOI] [PubMed] [Google Scholar]
- 22. Sasse S., Brand N. J., Kyprianou P., Dhoot G. K., Wade R., Arai M., Periasamy M., Yacoub M. H., Barton P. J. (1993) Circ. Res. 72, 932–938 [DOI] [PubMed] [Google Scholar]
- 23. Huang X., Pi Y., Lee K. J., Henkel A. S., Gregg R. G., Powers P. A., Walker J. W. (1999) Circ. Res. 84, 1–8 [DOI] [PubMed] [Google Scholar]
- 24. Perry S. V. (1999) Mol. Cell. Biochem. 190, 9–32 [PubMed] [Google Scholar]
- 25. Metzger J. M., Michele D. E., Rust E. M., Borton A. R., Westfall M. V. (2003) J. Biol. Chem. 278, 13118–13123 [DOI] [PubMed] [Google Scholar]
- 26. Eidem B. W., McMahon C. J., Cohen R. R., Wu J., Finkelshteyn I., Kovalchin J. P., Ayres N. A., Bezold L. I., O'Brian Smith E., Pignatelli R. H. (2004) J. Am. Soc. Echocardiogr. 17, 212–221 [DOI] [PubMed] [Google Scholar]
- 27. Hershberger R. E., Pinto J. R., Parks S. B., Kushner J. D., Li D., Ludwigsen S., Cowan J., Morales A., Parvatiyar M. S., Potter J. D. (2009) Circ. Cardiovasc. Genet. 2, 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hernandez O. M., Szczesna-Cordary D., Knollmann B. C., Miller T., Bell M., Zhao J., Sirenko S. G., Diaz Z., Guzman G., Xu Y., Wang Y., Kerrick W. G., Potter J. D. (2005) J. Biol. Chem. 280, 37183–37194 [DOI] [PubMed] [Google Scholar]
- 29. Szczesna D., Zhang R., Zhao J., Jones M., Guzman G., Potter J. D. (2000) J. Biol. Chem. 275, 624–630 [DOI] [PubMed] [Google Scholar]
- 30. Dweck D., Reyes-Alfonso A., Jr., Potter J. D. (2005) Anal. Biochem. 347, 303–315 [DOI] [PubMed] [Google Scholar]
- 31. Pinto J. R., Parvatiyar M. S., Jones M. A., Liang J., Ackerman M. J., Potter J. D. (2009) J. Biol. Chem. 284, 19090–19100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ommen S. R., Nishimura R. A., Appleton C. P., Miller F. A., Oh J. K., Redfield M. M., Tajik A. J. (2000) Circulation 102, 1788–1794 [DOI] [PubMed] [Google Scholar]
- 33. Soldin S. J., Soldin O. P., Boyajian A. J., Taskier M. S. (2006) Clin. Chim. Acta 366, 304–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chimenti C., Hamdani N., Boontje N. M., DeCobelli F., Esposito A., Bronzwaer J. G., Stienen G. J., Russo M. A., Paulus W. J., Frustaci A., van der Velden J. (2008) Am. J. Pathol. 172, 1482–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karam S., Raboisson M. J., Ducreux C., Chalabreysse L., Millat G., Bozio A., Bouvagnet P. (2008) Congenit. Heart Dis. 3, 138–143 [DOI] [PubMed] [Google Scholar]
- 36. Kostareva A., Gudkova A., Sjöberg G., Mörner S., Semernin E., Krutikov A., Shlyakhto E., Sejersen T. (2009) Int. J. Cardiol. 131, 410–412 [DOI] [PubMed] [Google Scholar]
- 37. Menon S. C., Michels V. V., Pellikka P. A., Ballew J. D., Karst M. L., Herron K. J., Nelson S. M., Rodeheffer R. J., Olson T. M. (2008) Clin. Genet. 74, 445–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mogensen J., Kubo T., Duque M., Uribe W., Shaw A., Murphy R., Gimeno J. R., Elliott P., McKenna W. J. (2003) J. Clin. Invest. 111, 209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peddy S. B., Vricella L. A., Crosson J. E., Oswald G. L., Cohn R. D., Cameron D. E., Valle D., Loeys B. L. (2006) Pediatrics 117, 1830–1833 [DOI] [PubMed] [Google Scholar]
- 40. Ware S. M., Quinn M. E., Ballard E. T., Miller E., Uzark K., Spicer R. L. (2008) Clin. Genet. 73, 165–170 [DOI] [PubMed] [Google Scholar]
- 41. Solaro R. J., Lee J. A., Kentish J. C., Allen D. G. (1988) Circ. Res. 63, 779–787 [DOI] [PubMed] [Google Scholar]
- 42. Varnava A. M., Elliott P. M., Baboonian C., Davison F., Davies M. J., McKenna W. J. (2001) Circulation 104, 1380–1384 [DOI] [PubMed] [Google Scholar]
- 43. Kranias E. G., Bers D. M. (2007) Subcell. Biochem. 45, 523–537 [DOI] [PubMed] [Google Scholar]
- 44. Baudenbacher F., Schober T., Pinto J. R., Sidorov V. Y., Hilliard F., Solaro R. J., Potter J. D., Knollmann B. C. (2008) J. Clin. Invest. 118, 3893–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lompré A. M., Hajjar R. J., Harding S. E., Kranias E. G., Lohse M. J., Marks A. R. (2010) Circulation 121, 822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peña J. R., Szkudlarek A. C., Warren C. M., Heinrich L. S., Gaffin R. D., Jagatheesan G., del Monte F., Hajjar R. J., Goldspink P. H., Solaro R. J., Wieczorek D. F., Wolska B. M. (2010) J. Mol. Cell. Cardiol. 49, 993–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Layland J., Cave A. C., Warren C., Grieve D. J., Sparks E., Kentish J. C., Solaro R. J., Shah A. M. (2005) FASEB J. 19, 1137–1139 [DOI] [PubMed] [Google Scholar]
- 48. Urboniene D., Dias F. A., Peña J. R., Walker L. A., Solaro R. J., Wolska B. M. (2005) Circ. Res. 97, 70–77 [DOI] [PubMed] [Google Scholar]
- 49. Wen Y., Xu Y., Wang Y., Pinto J. R., Potter J. D., Kerrick W. G. (2009) J. Mol. Biol. 392, 1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Davis J., Wen H., Edwards T., Metzger J. M. (2007) Circ. Res. 100, 1494–1502 [DOI] [PubMed] [Google Scholar]
- 51. Du J., Zhang C., Liu J., Sidky C., Huang X. P. (2006) Arch. Biochem. Biophys. 456, 143–150 [DOI] [PubMed] [Google Scholar]
- 52. Gomes A. V., Liang J., Potter J. D. (2005) J. Biol. Chem. 280, 30909–30915 [DOI] [PubMed] [Google Scholar]
- 53. Li Y., Charles P. Y., Nan C., Pinto J. R., Wang Y., Liang J., Wu G., Tian J., Feng H. Z., Potter J. D., Jin J. P., Huang X. (2010) J. Mol. Cell. Cardiol. 49, 402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chang A. N., Parvatiyar M. S., Potter J. D. (2008) Biochem. Biophys. Res. Commun. 369, 74–81 [DOI] [PubMed] [Google Scholar]
- 55. Murakami K., Stewart M., Nozawa K., Tomii K., Kudou N., Igarashi N., Shirakihara Y., Wakatsuki S., Yasunaga T., Wakabayashi T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7200–7205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Takeda S., Yamashita A., Maeda K., Maéda Y. (2003) Nature 424, 35–41 [DOI] [PubMed] [Google Scholar]
- 57. Palm T., Graboski S., Hitchcock-DeGregori S. E., Greenfield N. J. (2001) Biophys. J. 81, 2827–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Davis J., Wen H., Edwards T., Metzger J. M. (2008) J. Mol. Cell. Cardiol. 44, 891–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kobayashi T., Solaro R. J. (2006) J. Biol. Chem. 281, 13471–13477 [DOI] [PubMed] [Google Scholar]
- 60. Yumoto F., Lu Q. W., Morimoto S., Tanaka H., Kono N., Nagata K., Ojima T., Takahashi-Yanaga F., Miwa Y., Sasaguri T., Nishita K., Tanokura M., Ohtsuki I. (2005) Biochem. Biophys. Res. Commun. 338, 1519–1526 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.