Abstract
The inhibitor of DNA binding 2, dominant negative helix-loop-helix protein, ID2, acts as an oncogene and elevated levels of ID2 have been reported in several malignancies. Whereas some inducers of the ID2 gene have been characterized, little is known regarding the proteins capable to repress its expression. We developed siRNA microarrays to perform a large scale loss-of-function screen in human adult keratinocytes engineered to express GFP under the control of the upstream region of ID2 gene. We screened the effect of siRNA-dependent inhibition of 220 cancer-associated genes on the expression of the ID2::GFP reporter construct. Three genes NBN, RAD21, and p63 lead to a repression of ID2 promoter activity. Strikingly NBN and RAD21 are playing on major role in cell cycle progression and mitosis arrest. These results underline the pregnant need to silence ID2 expression at transcript level to promote cell cycle exit. Central to this inhibitory mechanism we find p63, a key transcription factor in epithelial development and differentiation, which binds specific cis-acting sequence within the ID2 gene promoter both in vitro and in vivo. P63 would not suppress ID2 expression, but would rather prevent excessive expression of that protein to enable the onset of keratinocyte differentiation.
Keywords: Epithelial Cell, Gene Silencing, Microarray, RNA Interference (RNAi), Transcription Regulation, Functional Screen
Introduction
ID2 is a member of the helix-loop-helix (HLH)3 family of transcription factors (1–3). Proteins of the HLH family positively regulate transcription by binding to DNA via a basic domain, as either homodimers or heterodimers, and often drive cell lineage commitment and differentiation in various cell types. In contrast, the ID proteins (ID1, ID2, ID3, and ID4) lack the basic domain and associate instead with other members of the HLH family, preventing them from binding DNA. Thus, the ID proteins act as dominant negative regulators of bHLH. For instance, ID proteins interact with the E transcription factors, including E12, E47, and E2-2 (4), as well as members of the Ets family (5). Generally, ID proteins function as positive regulators of cell growth and as negative regulators of cell differentiation (6).
ID2 is the only member of the ID family which physically interacts with the retinoblastoma protein (RB). RB is actually an upstream regulator of the ID2 gene, which in turn restrains the activity of ID2 (7). Although ID2 is not a bona fide oncogene triggering transformation of normal cells after a genetic alteration, its overexpression seems to contribute to tumorigenesis by inhibiting cell differentiation and stimulating proliferation. Elevated levels of ID2 have been reported in several malignancies, such as pancreas carcinomas (8), breast cancer (9), neuroblastomas (10), prostate cancer (11), and lung cancer (12).
ID genes play an important role in controlling epidermal homeostasis and cell fate in human keratinocytes. We demonstrated that overexpression of ID2 in HaCaT cells induced their proliferation, while the siRNA-mediated depletion of ID2 resulted in cell cycle arrest (13). The anti-proliferative effect of retinoids on human keratinocytes seems to result from the down-regulation of ID2 gene expression through a transcriptional convergence between Wnt and retinoid signaling (14). Transforming growth factor β (TGFβ) also inhibits growth of epithelial cells, including keratinocytes, through long term repression of ID2 and ID3 (15, 16). Similarly ID2 promotes tumor cell proliferation via control of cyclin D1 protein level (17). Finally, small enhancement of ID1 expression, but likely of other ID proteins as well, affects proliferation, differentiation, and apoptosis of keratinocytes grown in organotypic cultures (16).
Several genes, including some transcription factors, have been involved in the regulation of ID2 expression. For example, FSH (18), IGF1R (19), β-catenin (20), and MYC (7) are inducers of ID2. TGFβ seems to exert more subtle regulation, as it leads to repression of ID2 in epithelial cells (15), while it acts as an inducer of ID2 in immune cells (21). As ID2 overexpression seems to affect key oncogenic pathways in cells, we sought to characterize more repressors of ID2 expression.
RNA interference (RNAi) is a powerful approach to perform systematic loss-of-function screens. Several high-throughput phenotypic screens have been done in human cells using small interfering RNA (siRNA) libraries in a 96-well plate format. For instance, genes involved in TRAIL-induced apoptosis (22), cancer cell chemoresponsiveness (23), cell division (24), and haploinsufficiency diseases (25) have been recently characterized by large scale RNAi screens. Other groups have rather used large pools of small hairpin RNA (shRNA) using barcoded microarrays to analyze systematic loss-of-function in human cells (26–28). Here, we used siRNA microarrays (29) to perform a large-scale RNAi screen and characterize genes involved in the regulation of ID2 expression. We monitored the effect of siRNA-dependent specific inhibition of 220 genes involved in cancer, in human keratinocytes stably expressing an ID2 promoter::GFP reporter construct. We identified three new repressors of ID2. Specifically, we report that NBN and RAD21, two key actors of cell cycle checkpoint control (30–33) indirectly lead to repression of ID2, and that p63, a major regulator of skin development (34, 35), is a direct ID2 repressor.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
The non-tumorigenic, spontaneously transformed human keratinocyte cell line HaCaT (36) was obtained from CLS (Cell Line Service, Eppelheim, Germany). HaCaT cells were maintained at 37 °C in a 5% CO2 humidified atmosphere in Dulbecco's modified Eagle's medium (DMEM) without calcium chloride containing GLUTAMAX, 4.5 g/liter glucose (Invitrogen), 10% fetal calf serum (Hyclone, Perbio Sciences, Erembodegem-Aalst, Belgium), 100,000 units/liter penicillin, and 50 mg/liter streptomycin (Invitrogen).
For the forward transfection experiments, keratinocytes were plated in 6-well plates and transfected the following day in complete medium containing 10 nm siRNA complexed to interferin (Polyplus Transfection, Illkirch, France). Cells were assayed 72 h after transfection. siRNA duplexes (control siRNA, AllStars Negative Control siRNA; p63 siRNA, SI00055118) were obtained from Qiagen (Hilden, Germany). TAp63γ (Addgene plasmid 14575), ΔNp63α (Addgene plasmid 14574) and the control plasmid pBABE-puro (Addgene plasmid 1764) were obtained from Addgene. The pRc/RSV plasmid was obtained from Invitrogen, and the plasmid pRc-Id2 was constructed as described in Ref. 13. Stable cell HaCaT cell lines expressing pRc-Id2 were generated through antibiotic selection.
Human primary keratinocytes were obtained from human mammary skin biopsy. Briefly, keratinocytes were isolated by overnight trypsinization in 0.5% trypsin (Invitrogen)/5% penicillin/streptomycin (Eurobio) in phosphate-buffered saline at 4 °C, followed by scraping with a scalpel; then cells were cultured in semi-defined KGM-2 medium (Clonetics) on flasks coated with collagen type I (Falcon Biocoat) at 37 °C and 5% CO2. The differentiation program of secondary cultures was induced at confluency by a switch to the following differentiation medium: KBM-2 (Clonetics) supplemented with 0.5 mg/ml hydrocortisone, 5 mg/ml insulin, 5 ng/ml EGF, and 1.85 mm calcium.
ID2 Promoter and Plasmid Cloning
A DNA fragment of the ID2 promoter region, spanning from −2046 bp upstream to +13 bp downstream of the TSS, was amplified by PCR on genomic DNA from HaCaT cells using flanking BamHI and KpnI restriction sequences for the forward and reverse primers, respectively (see supplemental Table S3 for primer sequences). The PCR was performed using high-fidelity DNA polymerase (Platinum HiFi Taq Polymerase, Invitrogen) according to the manufacturer's recommendations.
The PCR products were inserted into a modified pENTRTM5′ (Gateway plasmid, Invitrogen) via TOPO-TA cloning technology (Invitrogen). Gateway recombination using pENTR5′-[ID2 promoter], pENTR-EGFP and the lentiviral backbone plasmid pLenti6/R4R2/V5-DEST was used to insert the ID2 promoter region upstream of EGFP coding sequence following the manufacturer instructions (Invitrogen). The resulting lentiviral construct was verified by sequencing.
The promoter region (−2574 to ∼+100 bp) was cloned into pENTRTM5′ vector. Truncated ID2 promoters were PCR amplified and cloned into pENTR5′-TOPO vectors (PCR primers listed in supplemental Table S3). The pID2-Luc2P (pID2–2.7k, pID2–980, and pID2–329) vector was obtained using the MultiSite Gateway technology. Truncated ID2 promoters were PCR amplified and cloned into pENTR5′-TOPO vectors (PCR primers listed in supplemental Table S3). The luciferase coding sequence (Luc2P ORF) was PCR-amplified from the pGL4.24 vector (Promega Madison, WI) using primer with attB flanking cassette, and inserted into the pDONR221 vector through a BP recombination. The reporter vector pID2-Luc2P was generated through LR recombination with pENTR5′-promID2, pENTR-Luc2P, and plenti6-R4R2-V5-DEST (Invitrogen).
Viral Production in HEK293-FT
Recombinant lentiviruses were produced by transient transfection of HEK 293FT cells according to the manufacturer's protocol (Invitrogen) with minor modifications. Lentivirus-containing supernatant was collected 48 h after transfection, 0.22 μm-filtered, and ultracentrifuged at 22,000 rpm for 90 min. The pellet was resuspended in 200 μl of phosphate-buffered saline and stored at −80 °C until the day of use.
Transduction and Cell Sorting
HaCaT cells were plated in 24-well plates and infected 12 h later in complete medium supplemented with 8 μg/ml protamine sulfate (Sigma-Aldrich) and diluted viral preparation to achieve the estimated range of multiplicity of infection (MOI) required. After 24 h, fresh medium was added and cells were cultured and passaged for several days. A subpopulation of pID2-HaCaT cells exhibiting homogeneous GFP expression was selected by two consecutive runs of FACS (MoFlo, Dako, Denmark) to increase sensitivity and allow measurement of subtle changes in GFP expression, in both direction.
siRNA Microarrays
siRNA were transfected into HaCaT cells using a reverse-transfection format as previously described (29) with slight modifications. Briefly, 0.5 μl of a 20 μm siRNA solution in 100 mm KoAc, 30 mm Hepes-KOH, 2 mm MgOAC pH 7.4 (Qiagen, Hilden, Germany) was mixed with 10 μl of PBS 1× (Invitrogen), 2 μl of 1.5 m sucrose solution (Sigma-Aldrich), and then 2 μl of Interferin transfection reagent (PolyPlus transfection, Strasbourg, France) and incubated 10 min at room temperature to allow for the formation of siRNA-Interferin complexes. Three microliters of a 1% (w/v) type B-gelatin solution (Sigma #G-1393) diluted in RNase-free water and 3 μl of Matrigel (BD Biosciences, Erembodegem, Belgium) were added. After lipid/DNA complex formation with the transfection reagent, the mixture was arrayed in quadruplicate on Superfrost glass slides (Menzel-Gläser, Braunschweigm Germany) using a MicroGrid Compact microarrayer (Genomic Solutions, Cambridgeshire, UK) with 400 μm solid pins at 19 °C and 60% humidity. Each slide comprised 640 features, mainly corresponding to 150 distinct siRNA printed in quadruplicate, 16 negative controls (scrambled siRNA) and 16 positive controls (siGFP), ordered in twelve 8 × 8 blocks. Positive and negative control spots were scattered all over the microarray to take into account cell-seeding variability or local artifacts, to quality control the transfection efficacy and to establish a reference for GFP expression in transfected cells. After printing, the slides were stored at 19 °C in a desiccator until the day of use. Slides were transferred to fresh sterile 10-cm dishes, and cells were plated onto the slides in 10 ml of DMEM at ∼3 × 105 cells/cm2. Cell microarrays were assayed 72 h after cell seeding. A minimum of three independent biological replicate experiments were carried out for each of the three batches of printed slides.
Immunofluorescence
Cells were fixed for 15 min in 4% (w/v) paraformaldehyde solution, washed 3 times in PBS and permeabilized for 5 min in a 0.1% (v/v) Triton X-100 solution (Sigma-Aldrich). Slides were then blocked in PBS-BSA 1% (w/v) for 30 min. EGFP expression was probed by immunostaining using Alexa647-coupled anti-GFP antibodies (1:250 dilution, Invitrogen) for 1 h at room temperature to increase the GFP signal. After several washes in PBS, nuclear DNA was then stained in a 0.25 μm Sytox Orange solution (Invitrogen) for 10 min. Slides were washed and dried, and independent fluorescence images of arrays were immediately captured using a Genepix 4000D microarray scanner (Molecular Devices) at 532 nm (PMT 200V) and 635 nm (PMT 550V) for DNA content and GFP expression staining, respectively.
siRNA Microarray Analysis
The fluorescence intensity values for each feature on the array were extracted using Genepix 4.0 (Molecular Devices). A .gal file (Genepix Array list) containing the coordinates of the different siRNA was generated before transfection and aligned on a pre-scan of the arrays to determine the precise localization of spots. After reverse-transfection, this grid was overlaid on the fluorescence images and used to extract the total intensity values corresponding to GFP (Ftotal 635) and DNA staining (Ftotal 532) of all cells in each feature (supplemental Fig. S1). A manual flagging of marginal spots (i.e. features comprising too few cells or dust) and regions of the slides with staining artifacts was performed and greatly improved the quality of the data (data not shown). For each spot, the ratio (Ftotal 635/Ftotal 532) or rGFP was computed. This ratio corresponds to the relative GFP expression (F635, represented in green in Fig. 1) related to the DNA content (F532, represented in red in Fig. 1). Thus, the ratio yields a GFP-fluorescence value normalized by the number of cells present in the spot. Data were then summarized to take into account intra-array replicates, and also replicate slides (inter-array replicates) (supplemental Fig. S1). Indeed, each siRNA was arrayed in quadruplicate on each slide, and three independent experiments were carried out. A total of 12 independent measures of the effect of a particular siRNA on ID2 expression were obtained, revealing the reliability of the screening process. The median of rGFP was calculated and ranked for each array. siRNA having at least 3 unflagged replicates out of 4 were further considered in the analysis.
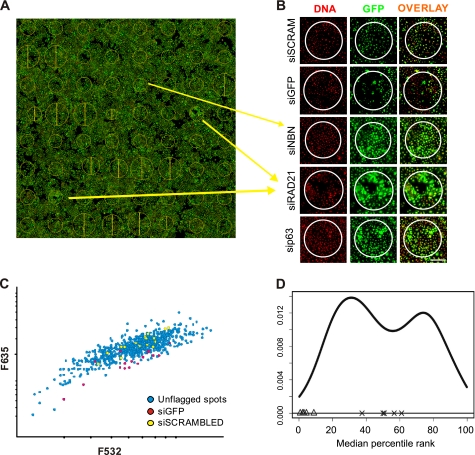
FIGURE 1.
siRNA microarray screen. A, scan of a transfected arrays is presented, with cellular GFP appearing in green, and the DNA content stained in red. The yellow circles represent areas where specific siRNA was printed and where reverse transfection of cells occurred. Position and size of the circles were automatically calculated by the Genepix software that was used to analyze the data B, close-up view of the image presented in A. The green/red channels as well as the overlay are presented for the two controls and new putative ID2 regulators. A decrease in GFP can be observed with the GFP-targeted siRNA compared with cells transfected with the scrambled siRNA. By contrast, NBN- (NBS1), p63-, and RAD21-targeting siRNA induced up-regulation of ID2-dependent GFP expression. Scale bar indicates 200 μm. C, scatter plot displaying distribution of F635 and F532 signal intensities for each spot in a given siRNA microarrays showed a linear increase of the GFP signal proportional to the number of cells estimated from DNA content. siGFP-targeted siRNA (red) and scrambled siRNA (yellow) are also indicated on the plot. D, distribution of the median of rankings among all replicated siRNA microarrays. The graph depicts the ranking-based non-parametric approach output for a batch of 4 replicate siRNA microarrays. The GFP-targeted siRNA spots (Δ) are consistently classified at the lower end of the distribution while scrambled siRNA (×) transfected cells are consistently ranked in the middle of the distribution, validating the strategy used for the analysis.
Semi-quantitative PCR (qRT-PCR)
One microgram of total RNA isolated with the RNeasy isolation kit (Qiagen) was reverse-transcribed using random primers and a SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Quantitative PCR was carried out with the Platinum® qPCR SuperMix (Invitrogen) in an ABI 7500 Real-Time PCR System (Applied Biosystems) with 1/20th of the cDNA for each reaction (supplemental Table S3 for primer sequences). Assays were performed in triplicate. Data were transformed using the 2−ΔΔCt formula (37). Changes in abundance of the tested (target) gene were normalized to the 18 S ribosomal RNA level (reference gene) and compared with the relative expression of a calibrator sample (control, siRNA scrambled transfection).
Western Blotting
Cells were lysed in RIPA buffer (Pierce) (25 mm Tris-HCl, pH 7.6, 150 mm NaCl, 1% sodium pyrophosphate, 1% sodium deoxycholate, 1% SDS) containing protease and phosphatase inhibitors (1 mm PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 1 mm Na3VO4, 50 mm NaF). One hundred micrograms of each protein sample were separated by SDS-PAGE (12% (w/v) acrylamide) and then blotted onto Hybond-P PVDF membranes (Amersham Biosciences, Freiburg, Germany) using a semi-dry transfer system. Membranes were blocked for 1 h at room temperature in 5% nonfat dry milk TBST (140 mm NaCl, 10 mm Tris, pH 7.6, 0.05% Tween-20) and probed overnight at 4 °C with primary antibodies against ID2 (C-20, sc-489, Santa Cruz Biotechnology) or p63 (4A4, sc-8431, Santa Cruz Biotechnology), which were diluted 200 times in 1% nonfat dry milk TBST solution. An HRP-conjugated antibody was used to probe β-actin (1/20000 dilution, A3854, Sigma-Aldrich) as a loading control on the same membranes. The blots were then washed and incubated with the respective HRP-conjugated secondary antibody (1/2000 dilution, anti-rabbit, 12–348; 1/2000 dilution, anti-mouse, 12–349, Millipore, CA) at room temperature for 1 h. Detection was carried out with the SuperSignal chemiluminescent system (Pierce).
Expression Profiles
The GEO database to analyze ID2 expression in p63 knockdown expression profiles were obtained by colleagues in the world (38).
Luciferase Reporter Assay
HaCaT cells were seeded in 96-well plates the day before transfection at a density of 5000 cells/cm2. Using jetSI-ENDO (PolyPlus Transfection), 120 ng of pID2-Luc2P plasmid was transfected in each well along with 10 nm siRNA and 5 ng of phRL-TK plasmid (Promega), which was used as an internal control of transfection. Luciferase activity was measured 48 h post-transfection using a dual-luciferase reporter assay system (Promega).
Chromatin Immunoprecipitation Experiment
ChIP experiments were carried out as described previously (39, 40). Briefly, growing HaCaT cells were fixed with formaldehyde and cross-linked chromatin was extracted, and subsequently fragmented by sonication into 1–2-kb fragments. The chromatin was then immunoprecipitated with a polyclonal antibody against p63α, or a control anti-FLAG antibody. After washing and reverse crosslinking, enrichment of ID2 promoter loci compared with a control loci were assessed on agarose gel after PCR amplification (see supplemental Table S3 for primer sequences). A Bio-Rad MyIQ single color thermal cycler and a SYBR Green PCR Master mix was used in all Q-PCR experiments. Specificity of products was monitored with a heat dissociation curve. Fold enrichment was calculated with the formula 2−ΔΔCt where the Ct represented the threshold cycles of the input, the specific antibody and the negative antibody; a further normalization with the enrichment obtained on a negative genomic region (centromeric satellite 11) was applied. This double normalization reduced extent of enrichment but increased reproducibility.
Skin Sections Immunofluorescence
Skin sections were derived from biopsies of healthy donors. They were either fixed in formalin and embedded in paraffin wax or frozen prior to sectioning. Paraffin sections were dewaxed with two xylene washes, two 100% ethanol washes, two 70% ethanol washes and two distilled water washes (5 min at room temperature for each step). Samples were then unmasked in a 10 mm Tris, 1 mm EDTA, and 0.05% Tween 20 solution for 5 min at 85 °C. After blocking in 1× PBS and 10% goat serum, skin sections were incubated overnight with the primary antibodies: mouse anti-p63 (4A4, Dako) and rabbit anti-ID2 (Zymed Laboratories Inc.). After a 1-h incubation with Alexa-conjugated secondary antibodies (Invitrogen) and Hoechst dye, slides were mounted with 100 mm Tris and 70% glycerol.
Frozen skin sections were stored in optimal cutting temperature compound. Five microns skin sections were generated with a cryostat and frozen on polylysine glass slides (Menzel-Gläser) until used. After blocking in 1× PBS and 1% bovine serum albumin, skin sections were incubated overnight with the primary antibodies as for paraffin sections. IF with ID2 alone were performed with a Cy3-conjugated secondary antibodies (Jackson Immunoresearch) and mounted with Vectashield containing 1.5 μg/ml DAPI. Fluorescence was analyzed with a Leica confocal microscope.
RESULTS
siRNA Microarray Screen
To characterize repressors of ID2 gene expression, we performed a large-scale RNAi-based loss-of-function screen using reverse transfection on siRNA microarrays coupled to a GFP reporter assay. A 2-kb fragment from the ID2 promoter region was cloned upstream of the GFP coding sequence, and the resulting construct was transduced into the HaCaT human keratinocyte cell line. HaCaT cells stably expressing the ID2-promoter::GFP reporter construct (pID2-HaCaT cells) were reverse transfected on siRNA microarrays. The expression of GFP under control of the ID2 promoter was monitored 72h after transfection by analysis of rGFP (the ratio of GFP-specific/DNA-specific fluorescence) (supplemental Fig. S1). We used a subset of 440 siRNA targeting 220 human genes; oncogenes, tumor suppressors and cell cycle regulators, from the Human Cancer siRNA Set v2 (Qiagen) targeting (supplemental Table S1).
The screening procedure is illustrated in supplemental Fig. S1. The images obtained contained both positive (GFP-targeted siRNA) and negative (scrambled siRNA) control spots, and as an example show three siRNA targeting new negative regulators of ID2, NBN, p63, and RAD21 (Fig. 1A). A closer view of the spots clearly showed changes in GFP expression (Fig. 1B). Despite slight variations in cell number between spots, a significant decrease in the GFP-specific signal was observed within spots containing a GFP-targeted siRNA compared with spots containing scrambled siRNA. In contrast, a significant increase in GFP expression was observed in spots containing siRNA targeting either NBN, p63 or RAD21 (Fig. 1B). Moreover, the use of scrambled siRNA as a negative control showed that cell viability and GFP expression were not affected by reverse transfection of nonspecific siRNA, as indicated by the equal number of cells growing within spots and in the “transfection-free” area.
The distribution of fluorescence intensities over the siRNA microarrays typically showed proportionality between 532 nm and 635 nm, indicating that the GFP signal increased with cell number (Fig. 1C). The distribution of the negative control (scrambled siRNA) spots followed the global trend of all spots, dispersed along the x axis as a function of cell number. However, the 635 nm fluorescence signal in these spots was well-centered in the distribution, suggesting that GFP expression was not modulated in these cells. In contrast, the distribution of all but two positive controls (GFP-targeted siRNA) was shifted toward low fluorescence intensities (635 nm values), reflecting an efficient knockdown of GFP expression. The remainders of the spots were dispersed along the x axis of fluorescence intensity values, illustrating the necessity of normalizing GFP signal to cell number.
Each siRNA was processed independently, even when they targeted the same gene. siRNA having at least 3 unflagged features out of the quadruplicate on a given array were considered in the analysis. For each array, siRNA were ranked according to the median of the rGFP measurements. The reproducibility of this ranking was assessed across 3 independent microarrays for each siRNA, by calculating the median of percentile ranks per array (supplemental Fig. S1). The distribution of the median of percentile ranks for each siRNA exhibited a bi-modal shape (Fig. 1D). Effective siRNA were expected to rank at both tails of this distribution. The analysis procedure was validated with both negative and positive controls. Cells transfected with the GFP-targeted siRNA were systematically classified at the left side of the distribution, while negative control siRNA were scattered in the middle of this bi-modal distribution, as expected (Fig. 1D). A list of the top 6 ranking siRNA which consistently led to the induction of GFP after transfection, were considered to target potential repressors of ID2 promoter activity (supplemental Table S2). Among them we identified 4 genes, AURKB, NBN (NBS1), p63, and RAD21 that we choose to further validate.
Validation of the Putative Repressors of ID2 Promoter Activity
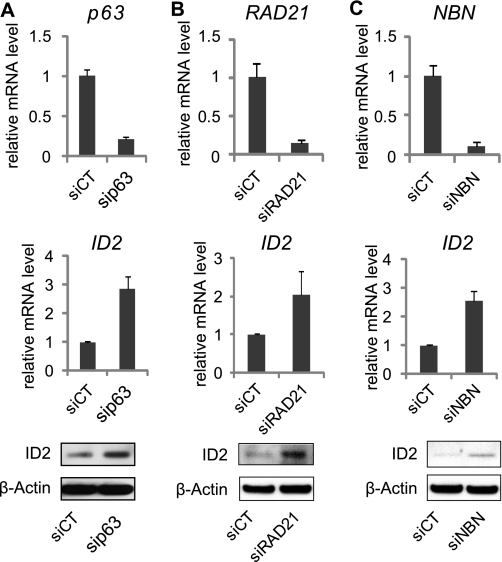
Candidate ID2 repressors were further validated by “forward” transfection in regular cell cultures. Semi-quantitative real-time PCR (qRT-PCR) analysis was first used to determine the extent of gene knock-down upon siRNA transfection. P63-, RAD21-, and NBN-targeted siRNA efficiently reduced the expression of corresponding gene (Fig. 2, A–C). Concomitantly, we validated effects of the siRNA on endogenous ID2 gene expression in HaCaT cells. The knockdown of p63, NBN, and RAD21 increased the expression of endogenous ID2 both at mRNA and protein levels (Fig. 2, A–C), consistent with the ID2 promoter-dependent GFP modulations observed on siRNA microarrays. On the contrary we were not able to validate the effect of AURKB depletion on ID2 expression (data not shown). Concordant results between the siRNA microarrays, qRT-PCR and Western blot analyses were obtained for the selected siRNA. This siRNA microarrays-mediated loss-of-function screen enabled the characterization of three new putative repressors of ID2. Interestingly two of them are involved in cell cycle checkpoint control. NBN participates in S-phase checkpoint pathway, while RAD21 acts in establishing normal spindle-kinetochore interaction and is part of spindle checkpoint activation. Both genes are important to promote cell cycle progression and mitosis arrest. This result suggests that silencing ID2 at transcript level would be necessary to promote cell cycle exit. However none of these proteins are transcription factor and probably act indirectly on ID2 expression. Only p63, a member of the p53 gene family, is a well-known transcription factor that could act as a direct repressor of ID2.
FIGURE 2.
Validation of the putative repressors of ID2. Inhibition of p63 (A), RAD21 (B), and NBN (C) by siRNA, was quantified by RT-qPCR. Endogenous expression of ID2 was measured by RT-qPCR and immunoblot. Samples were extracted 48 h post-transfection from HaCaT cells, and error bars represent ± S.D.
p63 Represses ID2 Gene Expression in Adult Human Keratinocytes
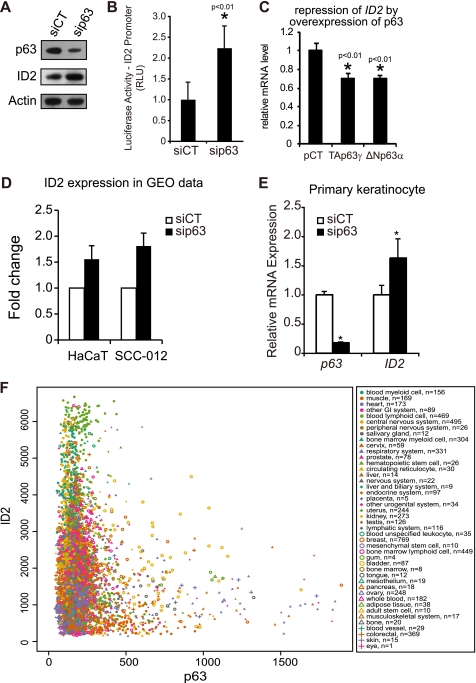
Transfection of p63-targeted siRNA in HaCaT cells led to an effective knockdown of p63 both at the transcript (Fig. 2A) and protein levels (Fig. 3A), and with an induction of endogenous ID2 (Fig. 3A). Using an ID2 promoter assay based on luciferase activity instead of GFP, we confirmed that p63 could regulate the ID2 gene promoter activity in HaCaT cells (Fig. 3B). Two isoforms, TAp63 and ΔNp63, are functionally active in human keratinocytes. Although the specific role of these isoforms in epidermis development remains mostly unclear, ΔNp63 seems to be the main mediator of keratinocyte proliferation and differentiation, as this isoform is largely predominant in human keratinocytes (41). Interestingly, overexpression of either TAp63γ or ΔNp63α only slightly down-regulated ID2 transcript level (Fig. 3C). This result suggests that, in physiological conditions, p63 would not suppress ID2 expression, but would rather prevent excessive transcription of it. We also referenced the GEO database to analyze ID2 expression in p63 knockdown expression profiles obtained by colleagues in the world. p63 knockdown-dependent regulation of ID2 was confirmed in HaCaT and other epithelial cells (Fig. 3D). The p63 knockdown-dependent up-regulation of ID2 was also observed in primary cultures of human keratinocytes (Fig. 3E). Finally, we studied the co-expression of ID2 and p63 mRNAs using a database containing normalized gene expression data on most of the human genes across 9783 tissue samples (42). If p63 were an inducer of ID2, one would observe a positive linear regression between the two mRNA expression levels. On the contrary, if p63 were a repressor of ID2, one would observe a negative linear regression. Actually, as demonstrated in Fig. 3F, when the relative expression level of p63 is above 600, that of ID2 rarely exceeds 2000; i.e. the lower third. These expression data exquisitely confirm our molecular data and suggest that p63 prevent excessive expression of ID2. Furthermore these data suggest that the regulation of ID2 by p63 occurs in many tissues. The maximum amplitude of variation observed for ID2 expression in the skin expression profiles in the database was from 550 to 2,300 relative units of expression (supplemental Fig. S2), i.e. up to 4.2-fold, at most. The quartile distribution (25–75%) range of expression was from 900 to 1600, i.e. up to 1.8-fold, at most. Strikingly, our ex vivo molecular data fit well within these ranges of expression. Taken together, these results demonstrate that p63 acts as a repressor of ID2 not completely suppressing ID2 expression, but rather controlling it to prevent excessive expression of that gene in adult human keratinocytes.
FIGURE 3.
p63 is a repressor of ID2 in adult human keratinocytes. A, an efficient knockdown of the p63 at protein level was observed by immunoblot and a concomitant up-regulation of ID2 when compared with control siRNA-transfected cells (siCT, control siRNA; siP63, p63-targeted siRNA). Actin was used as loading control. B, quantification of ID2 promoter-luciferase reporter activity in p63-deficient HaCaT cells. Renilla luciferase was used as internal control. Error bars indicate the ± S.D. (*, t test, p < 0.01, n = 6). C, overexpression of TAp63γ or ΔNp63α represses endogenous ID2 expression. Error bars indicate the ± S.D. (*, t test, p < 0.01, n = 3). pCT was the control plasmid. D, data were analyzed from a comprehensive study including expression profiling in HaCaT and SCC-012 cells transfected with p63 siRNA using the algorithm of the expression profiling database GEO (GSE4975) (38). The p63 depletion-dependent up-regulation of ID2 was reported with three different probes in two cell lines, HaCaT and SCC-012. E, endogenous ID2 is up-regulated consecutive to p63 silencing in primary human keratinocytes. Error bars represent ± S.D. (*, t test, p < 0.01, n = 3). F, in silico co-expression analysis of ID2 and p63 mRNAs in normal and cancer human tissues present in the in silico transcriptomics database (42). The normalized expression levels of ID2 are presented on y axis as a function of p63 expression levels on x axis. The tissues and number of samples (n) are indicated in the box.
ID2 Gene Repression by p63 Is Mediated by Direct Binding of p63 on the ID2 Promoter
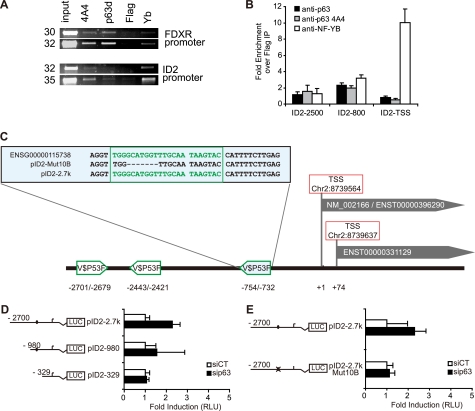
To determine if p63 binds onto the ID2 promoter, chromatin immunoprecipitation (ChIP) experiments were performed in growing HaCaT cells using two different α-p63-specific antibodies (43). P63 binding onto the ID2 promoter was detected by PCR (Fig. 4A). Two distinct regions of the ID2 promoter, located 800 and 2500 bp upstream from the TSS, were enriched after p63 immunoprecipitation (IP) compared with the control (Fig. 4B). This suggests a direct binding of p63 onto ID2 promoter in these two regions. Potential p63 binding sites within the promoter region of ID2 were assessed using Genomatix software, which indicated three putative p63 binding sites at −754 bp, −2443 bp, and −2701 bp upstream of the first TSS (Fig. 4C). To study the function of each of these putative binding sites, increasing lengths of the promoter region that controls luciferase activity were deleted, and this deletion analysis showed that the −754 bp site was the most efficient in down-regulating ID2 gene promoter activity (Fig. 4D). Finally, we engineered a short deletion of 7 nucleotides within the −754 p63 binding site and showed that this site mediated the p63-dependent repression of ID2 gene promoter activity (Fig. 4E).
FIGURE 4.
p63 directly binds onto the promoter region of ID2. A, ChIP was performed in growing HaCaT cells with polyclonal (p63D) or monoclonal (4A4) antibodies raised against p63, a flag (Flag) antibody as a negative control and NF-YB (NF-YB) directed antibody as a positive control. FDXR promoter was used as a positive control of p63 binding. Immunoprecipitated DNA was then further analyzed by quantitative-PCR. The set of primers used for ID2 the promoter amplification were the ID2–2500 (supplemental Table S3) B, a major enrichment was observed in a region around 800-bp upstream of the TSS with both anti-p63 antibodies, whereas another weaker binding occurred at around 2500 bp upstream of TSS. As expected, NF-YB binding was observed near the TSS. C, map of the ID2 locus is displayed with two alternate TSS. Potential p63 binding sites (V$P53F) were identified within the ID2 promoter sequence using Genomatix MatInspector tool at −2701, −2443, and −754 bp from the first TSS, respectively. The −754 binding site, present in both the wild-type gene (ENSG00000115738) and the reporter construct pID2–2.7k is detailed in the upper box. The deletion of this site obtained by directed mutagenesis of the reporter construct is also indicated (pID2Mut10B). D and E, HaCaT cells were cotransfected with ID2 promoter-luciferase reporter of various lengths (pId2–2.7k, pId2–980, pId2–329) or mutant ID2 promoter-luciferase reporter (pId2–2.7k Mut10B) together with a control reporter pRL-TK and either scrambled or p63-targeted siRNA. On schematic representations of the ID2 promoter-reporter constructs, a black circle represents the putative −754 nt p63 binding sites, and a crossed circle is the mutated sequence. Luciferase activity (RLU) was defined as the ratio of firefly luciferase activity from ID2 promoter-reporters relative to Renilla luciferase activity as the transfection control. Results are displayed as changes to induction of the luciferase activity after p63-targeted siRNA transfection compared with control siRNA. Bars and brackets indicate mean and standard deviation, respectively.
p63 and ID2 Proteins Expression in Human Epidermis and in Differentiating Primary Keratinocytes
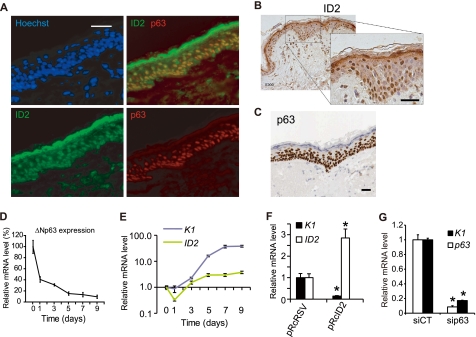
The expression of p63 and ID2 proteins was monitored in situ in human skin sections. ID2 protein was detected in the epidermal basal layer, with very little presence in the spinous layer corresponding to the first stage of keratinocyte differentiation and high quantities in the granular layer, the terminal differentiation layer (Fig. 5, A and B and supplemental Fig. S3). Interestingly, ID2 was mainly nuclear in the basal layer and cytoplasmic in the granular layer (Fig. 5B). This is consistent with the subcellular location of ID2 in human skin cells both in the nucleus but not nucleoli and the cytoplasm (supplemental Fig. S3). p63 was expressed in the nuclei, very strongly in the basal layer, to a smaller extent in the spinous layer and could not be detected in the granular layer (Fig. 5, A and C). In the basal layer both proteins were expressed, while in the granular layer, the absence of p63 resulted in very high expression of ID2, thus confirming our molecular data (Fig. 5A). These in vivo results are again consistent with the role of p63 as a repressor of ID2, not abolishing its expression, but rather regulating it to prevent excessive expression of ID2.
FIGURE 5.
Expression of ID2 and p63 in differentiating primary keratinocytes and in human skin. A, expression of p63 and ID2 proteins in human skin. Sections of paraffin-embedded breast skin biopsies were stained with fluorescent p63-specific mAb (red), ID2-specific pAb (green), and Hoechst (blue). Scale bar is 100 μm. B, expression of ID2 in paraffin-embedded skin section was monitored by immunohistochemistry using DAB-stained ID2-specific pAb. Nuclei are stained by Hemalun and appeared in blue. Scale bar is 100 μm. C, p63-specific immunohistochemistry in paraffin-embedded skin sections extracted from the Human Protein Atlas database (57). Scale bar is 100 μm. D, expression of ΔNp63 in differentiating primary keratinocytes. qRT-PCR analyses were perfomed with isoform-specific primers for ΔNp63 (supplemental Table S3). GAPDH was used as an endogenous standard. E, ID2 and keratin 1 (K1) expression were monitored by qRT-PCR in differentiating human primary keratinocytes. F, expression of K1 and ID2 was monitored after 2 days in differentiating HaCaT cell line with an overexpression of ID2. HaCaT Were also transformed with pRcRSV the control empty plasmid. (*, t test, p < 0.01, n = 3). G, Expression of K1 and p63 at 6 days post-transfection in differentiating HaCaT (*, t test, p < 0.01, n = 3). Error bars in all qRT-PCR graphs represent the standard deviation (S.D.) of triplicates.
We next monitored over 9 days in vitro, the p63 and ID2 transcript levels in differentiating primary keratinocytes. While the expression of ΔNp63 constantly decreased over that period (Fig. 5D), ID2 expression was more complex. It decreased at day 1, then slowly increased and finally remained constant after 5 days, when ΔNp63 was almost no longer expressed (Fig. 5E). We also monitored the expression of keratin 1 (K1), a well characterized marker of early differentiation and of the basal-spinous switch in epidermis. K1 expression level started to increase at day 2 and then continuously increased (Fig. 5E). Interestingly, p63 and ID2 expression patterns in differentiating keratinocytes modeled in a way the in vivo protein expression patterns in skin sections. Moreover, it confirmed once again that high ΔNp63 expression level contains excessive ID2 expression. Finally, we investigated the role of p63 and ID2 in the keratinocyte commitment to differentiation. Functional studies were performed at day 2 in in vitro differentiation experiments as it is a key time in differentiation commitment, functionally equivalent to the transition between basal and spinous layers in vivo. While a 2.8-fold overexpression of ID2 resulted in the inhibition of differentiation (Fig. 5F), knockdown of p63 in HaCaT cells also inhibited differentiation as demonstrated by the reduced expression of K1 (Fig. 5G). After 7 days the over-expression of ID2 no longer inhibited K1 expression (data not shown). Taken together our results suggest that the p63-dependent repression of ID2 would be necessary to favor the keratinocyte cell cycle withdrawal and the onset of differentiation that occurs in vivo during the transition from basal to spinous layers. However, later in the differentiation process, in the granular layer, the expression of p63 seems too low to contain ID2 expression. The role of this high cytosolic expression of ID2 in the granular layer remains to be discovered.
DISCUSSION
To date, reports of ID2 repressors are very scarce; only TGFβ had been identified as a partial and indirect repressor of ID2 in epithelial cells (15). The RNAi/reporter screen in the present study enabled the characterization of 3 new direct and indirect repressors of ID2. Strikingly, we demonstrated that two genes playing a major role in cell cycle checkpoints, NBN in S-phase checkpoint and RAD21 in spindle checkpoint, are leading to repression of ID2, which further highlighted the role for ID2 in cell cycle exit. Indeed, ID2 is a very unstable protein that is eliminated as cells withdraw from cycle (44). The anaphase promoting complex/cyclosome and its activator CDH1 (APC/CCdh1) targets ID2 for degradation through a destruction box motif (D box) that is conserved in ID1 and ID4 to couple cell cycle exit and axonal growth (44). We found that in addition to this ID2-targeted protein degradation mechanism, two genes, involved in the regulation of cell cycle withdrawal, NBN and RAD21, lead to a repression of ID2 at the transcript level. These results emphasize the need to silence expression, at both the transcript and protein levels to prevent keratinocytes from re-entering the cell cycle. Indeed, due to the short half-life of ID2 protein (17), this dual level of repression might be particularly necessary. This finding is in agreement with our previous results, which show that ID2 was necessary to revert double strand break-dependent cell-cycle arrest in human keratinocytes (13), but also with a recent report about the central role of ID2 within a generic transcription network regulating cell cycle in mouse and man (45). Together these results suggest a new potential role for ID2 in response to DNA damage and checkpoints exit.
We also show for the first time a p63-dependent direct repression of ID2. Based on p63 knock-out mouse models, p63 was found to play an essential role in epidermal-mesenchymal interactions during embryonic development (34, 35). It is required for the stratification of the apical ectodermal ridge, for limb development and craniofacial development, and hence for the establishment and maintenance of stratified epithelia (46). p63-depleted keratinocytes exhibited impaired stratification and differentiation (41). Furthermore, p63 is crucial for the activation of the epithelial cell adhesion program (47) or to maintain the proliferative potential of stem cells (48). Taking into account the well-established roles of ID2 in controlling epidermal homeostasis and cell fate in human keratinocytes (13–16), including its capacity to inhibit differentiation (6, 32, 49) the regulatory link that we observe between p63 and ID2 seems particularly interesting. Indeed, as it was recently suggested in an excellent review on epidermal homeostasis (50) that the identification of key genes downstream of p63 would provide important new insights into its roles in dynamic equilibrium of differentiation and proliferation. Our results show that ID2 is one of these potential key genes acting downstream of p63 to control keratinocytes cell fate.
Six p63 proteins have been described, resulting from two promoters and alternative splicing (α, β, γ) at the 3′ end of the gene, with or without a sterile alpha motif (SAM) domain. The TA isoforms are structurally more like p53 and contain a transactivating domain while the ΔN isoforms lack this domain (51). The major isoform present in keratinocytes is ΔNp63α, and the genomic targets of this isoform have been partially described (52). Here we have shown that the ΔNp63α isoform was the most highly expressed in human keratinocytes and both isoform were able to slightly repress ID2 gene promoter activity. Interestingly several studies recently demonstrated that ΔN isoforms are capable of transactivating genes through a proline-rich transactivation domain (53, 54).
Here we show that, siRNA-mediated knockdown of endogenous p63 represses ID2-inducible reporter gene activity in human cells. However, in apparent contradiction to these observations, upon transient overexpression of ΔNp63α ex vivo in human keratinocyte, we only observed a slight repression of the ID2-reporter construct. To explain these results we propose two hypotheses.
First, ΔNp63α could have a function in recruiting transcriptional repressors to ID2 promoter region. Overexpression of p63 may lead to sequestration of such repressors, preventing excessive repression, as recently proposed on the role of p63 as a negative regulator of Wnt-induced transcription (55).
Second, it was recently shown that in cycling cells, p73 and p63 are bound to the p53-responsive elements (RE) present in the regulatory region of cell cycle regulating genes and compete with p53 to attach to these sites during the different phases of the cycle (56). Similarly, the binding of p63 to p53 RE upstream of ID2 would not directly represses ID2 but rather prevents action of positive regulators of ID2.
Nevertheless, the role of p63 as a negative ID2-regulator that we report here matches with the frequently observed down-regulation of p63 during tumor progression, when cancer cells adopt a more mesenchymal, invasive phenotype.
Supplementary Material
This work was funded by CEA (to N. W. and D. C.), a fellowship from the “Fondation pour la Recherche Médicale” (to N. W.), and an Association pour la Recherche sur le Cancer (ARC) Fellowship (to D. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1–S3.
- HLH
- helix-loop-helix
- RB
- retinoblastoma protein
- TSS
- transcriptional start site.
REFERENCES
- 1. Yokota Y., Mori S. (2002) J. Cell Physiol. 190, 21–28 [DOI] [PubMed] [Google Scholar]
- 2. Ruzinova M. B., Benezra R. (2003) Trends Cell Biol. 13, 410–418 [DOI] [PubMed] [Google Scholar]
- 3. Perk J., Iavarone A., Benezra R. (2005) Nat. Rev Cancer 5, 603–614 [DOI] [PubMed] [Google Scholar]
- 4. Langlands K., Yin X., Anand G., Prochownik E. V. (1997) J. Biol. Chem. 272, 19785–19793 [DOI] [PubMed] [Google Scholar]
- 5. Ohtani N., Zebedee Z., Huot T. J., Stinson J. A., Sugimoto M., Ohashi Y., Sharrocks A. D., Peters G., Hara E. (2001) Nature 409, 1067–1070 [DOI] [PubMed] [Google Scholar]
- 6. Norton J. D., Deed R. W., Craggs G., Sablitzky F. (1998) Trends Cell Biol. 8, 58–65 [PubMed] [Google Scholar]
- 7. Lasorella A., Noseda M., Beyna M., Yokota Y., Iavarone A. (2000) Nature 407, 592–598 [DOI] [PubMed] [Google Scholar]
- 8. Kleeff J., Ishiwata T., Friess H., Büchler M. W., Israel M. A., Korc M. (1998) Cancer Res. 58, 3769–3772 [PubMed] [Google Scholar]
- 9. Stighall M., Manetopoulos C., Axelson H., Landberg G. (2005) Int. J. Cancer 115, 403–411 [DOI] [PubMed] [Google Scholar]
- 10. Alaminos M., Gerald W. L., Cheung N. K. (2005) Pediatr. Blood Cancer 45, 909–915 [DOI] [PubMed] [Google Scholar]
- 11. Asirvatham A. J., Carey J. P., Chaudhary J. (2007) Prostate 67, 1411–1420 [DOI] [PubMed] [Google Scholar]
- 12. Rollin J., Bléchet C., Régina S., Tenenhaus A., Guyétant S., Gidrol X. (2009) PLoS ONE 4, e4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baghdoyan S., Lamartine J., Castel D., Pitaval A., Roupioz Y., Franco N., Duarte M., Martin M. T., Gidrol X. (2005) J. Biol. Chem. 280, 15836–15841 [DOI] [PubMed] [Google Scholar]
- 14. Memezawa A., Takada I., Takeyama K., Igarashi M., Ito S., Aiba S., Kato S., Kouzmenko A. P. (2007) Oncogene 26, 5038–5045 [DOI] [PubMed] [Google Scholar]
- 15. Kowanetz M., Valcourt U., Bergström R., Heldin C. H., Moustakas A. (2004) Mol. Cell Biol. 24, 4241–4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rotzer D., Krampert M., Sulyok S., Braun S., Stark H. J., Boukamp P., Werner S. (2006) Oncogene 25, 2070–2081 [DOI] [PubMed] [Google Scholar]
- 17. Tokuriki A., Iyoda T., Inaba K., Ikuta K., Fujimoto S., Kumakiri M., Yokota Y. (2009) Carcinogenesis 30, 1645–1650 [DOI] [PubMed] [Google Scholar]
- 18. Scobey M. J., Fix C. A., Walker W. H. (2004) J. Biol. Chem. 279, 16064–16070 [DOI] [PubMed] [Google Scholar]
- 19. Belletti B., Prisco M., Morrione A., Valentinis B., Navarro M., Baserga R. (2001) J. Biol. Chem. 276, 13867–13874 [DOI] [PubMed] [Google Scholar]
- 20. Rockman S. P., Currie S. A., Ciavarella M., Vincan E., Dow C., Thomas R. J., Phillips W. A. (2001) J. Biol. Chem. 276, 45113–45119 [DOI] [PubMed] [Google Scholar]
- 21. Hacker C., Kirsch R. D., Ju X. S., Hieronymus T., Gust T. C., Kuhl C., Jorgas T., Kurz S. M., Rose-John S., Yokota Y., Zenke M. (2003) Nat. Immunol. 4, 380–386 [DOI] [PubMed] [Google Scholar]
- 22. Aza-Blanc P., Cooper C. L., Wagner K., Batalov S., Deveraux Q. L., Cooke M. P. (2003) Mol. Cell 12, 627–637 [DOI] [PubMed] [Google Scholar]
- 23. Whitehurst A. W., Bodemann B. O., Cardenas J., Ferguson D., Girard L., Peyton M., Minna J. D., Michnoff C., Hao W., Roth M. G., Xie X. J., White M. A. (2007) Nature 446, 815–819 [DOI] [PubMed] [Google Scholar]
- 24. Kittler R., Pelletier L., Heninger A. K., Slabicki M., Theis M., Miroslaw L., Poser I., Lawo S., Grabner H., Kozak K., Wagner J., Surendranath V., Richter C., Bowen W., Jackson A. L., Habermann B., Hyman A. A., Buchholz F. (2007) Nat. Cell Biol. 9, 1401–1412 [DOI] [PubMed] [Google Scholar]
- 25. Ebert B. L., Pretz J., Bosco J., Chang C. Y., Tamayo P., Galili N., Raza A., Root D. E., Attar E., Ellis S. R., Golub T. R. (2008) Nature 451, 335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berns K., Hijmans E. M., Mullenders J., Brummelkamp T. R., Velds A., Heimerikx M., Kerkhoven R. M., Madiredjo M., Nijkamp W., Weigelt B., Agami R., Ge W., Cavet G., Linsley P. S., Beijersbergen R. L., Bernards R. (2004) Nature 428, 431–437 [DOI] [PubMed] [Google Scholar]
- 27. Paddison P. J., Silva J. M., Conklin D. S., Schlabach M., Li M., Aruleba S., Balija V., O'Shaughnessy A., Gnoj L., Scobie K., Chang K., Westbrook T., Cleary M., Sachidanandam R., McCombie W. R., Elledge S. J., Hannon G. J. (2004) Nature 428, 427–431 [DOI] [PubMed] [Google Scholar]
- 28. Schlabach M. R., Luo J., Solimini N. L., Hu G., Xu Q., Li M. Z., Zhao Z., Smogorzewska A., Sowa M. E., Ang X. L., Westbrook T. F., Liang A. C., Chang K., Hackett J. A., Harper J. W., Hannon G. J., Elledge S. J. (2008) Science 319, 620–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baghdoyan S., Roupioz Y., Pitaval A., Castel D., Khomyakova E., Papine A., Soussaline F., Gidrol X. (2004) Nucleic Acids Res. 32, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lim D. S., Kim S. T., Xu B., Maser R. S., Lin J., Petrini J. H., Kastan M. B. (2000) Nature 404, 613–617 [DOI] [PubMed] [Google Scholar]
- 31. Pati D., Zhang N., Plon S. E. (2002) Mol. Cell Biol. 22, 8267–8277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Toyoda Y., Furuya K., Goshima G., Nagao K., Takahashi K., Yanagida M. (2002) Curr. Biol. 12, 347–358 [DOI] [PubMed] [Google Scholar]
- 33. Zhang Y., Zhou J., Lim C. U. (2006) Cell Res. 16, 45–54 [DOI] [PubMed] [Google Scholar]
- 34. Mills A. A., Zheng B., Wang X. J., Vogel H., Roop D. R., Bradley A. (1999) Nature 398, 708–713 [DOI] [PubMed] [Google Scholar]
- 35. Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R. T., Tabin C., Sharpe A., Caput D., Crum C., McKeon F. (1999) Nature 398, 714–718 [DOI] [PubMed] [Google Scholar]
- 36. Boukamp P., Petrussevska R. T., Breitkreutz D., Hornung J., Markham A., Fusenig N. E. (1988) J. Cell Biol. 106, 761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 38. Barbieri C. E., Tang L. J., Brown K. A., Pietenpol J. A. (2006) Cancer Res. 66, 7589–7597 [DOI] [PubMed] [Google Scholar]
- 39. Viganò M. A., Lamartine J., Testoni B., Merico D., Alotto D., Castagnoli C., Robert A., Candi E., Melino G., Gidrol X., Mantovani R. (2006) EMBO J. 25, 5105–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Viganò M. A., Mantovani R. (2007) Cell Cycle 6, 233–239 [DOI] [PubMed] [Google Scholar]
- 41. Truong A. B., Kretz M., Ridky T. W., Kimmel R., Khavari P. A. (2006) Genes Dev. 20, 3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kilpinen S., Autio R., Ojala K., Iljin K., Bucher E., Sara H., Pisto T., Saarela M., Skotheim R. I., Björkman M., Mpindi J. P., Haapa-Paananen S., Vainio P., Edgren H., Wolf M., Astola J., Nees M., Hautaniemi S., Kallioniemi O. (2008) Genome Biol. 9, R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Beretta C., Chiarelli A., Testoni B., Mantovani R., Guerrini L. (2005) Cell Cycle 4, 1625–1631 [DOI] [PubMed] [Google Scholar]
- 44. Lasorella A., Stegmüller J., Guardavaccaro D., Liu G., Carro M. S., Rothschild G., de la Torre-Ubieta L., Pagano M., Bonni A., Iavarone A. (2006) Nature 442, 471–474 [DOI] [PubMed] [Google Scholar]
- 45. Ravasi T., Suzuki H., Cannistraci C. V., Katayama S., Bajic V. B., Tan K., Akalin A., Schmeier S., Kanamori-Katayama M., Bertin N., Carninci P., Daub C. O., Forrest A. R., Gough J., Grimmond S., Han J. H., Hashimoto T., Hide W., Hofmann O., Kawaji H., Kubosaki A., Lassmann T., van Nimwegen E., Ogawa C., Teasdale R. D., Tegnér J., Lenhard B., Teichmann S. A., Arakawa T., Ninomiya N., Murakami K., Tagami M., Fukuda S., Imamura K., Kai C., Ishihara R., Kitazume Y., Kawai J., Hume D. A., Ideker T., Hayashizaki Y. (2010) Cell 140, 744–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McKeon F. (2004) Genes Dev. 18, 465–469 [DOI] [PubMed] [Google Scholar]
- 47. Carroll D. K., Carroll J. S., Leong C. O., Cheng F., Brown M., Mills A. A., Brugge J. S., Ellisen L. W. (2006) Nat. Cell Biol. 8, 551–561 [DOI] [PubMed] [Google Scholar]
- 48. Senoo M., Pinto F., Crum C. P., McKeon F. (2007) Cell 129, 523–536 [DOI] [PubMed] [Google Scholar]
- 49. Simbulan-Rosenthal C. M., Trabosh V., Velarde A., Chou F. P., Daher A., Tenzin F., Tokino T., Rosenthal D. S. (2005) Oncogene 24, 5443–5458 [DOI] [PubMed] [Google Scholar]
- 50. Fuchs E. (2009) Cell Stem Cell 4, 499–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Irwin M. S., Kaelin W. G. (2001) Cell Growth Differ. 12, 337–349 [PubMed] [Google Scholar]
- 52. Pozzi S., Zambelli F., Merico D., Pavesi G., Robert A., Maltère P., Gidrol X., Mantovani R., Vigano M. A. (2009) PLoS ONE 4, e5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Helton E. S., Zhu J., Chen X. (2006) J. Biol. Chem. 281, 2533–2542 [DOI] [PubMed] [Google Scholar]
- 54. Liu G., Nozell S., Xiao H., Chen X. (2004) Mol. Cell Biol. 24, 487–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Drewelus I., Göpfert C., Hippel C., Dickmanns A., Damianitsch K., Pieler T., Dobbelstein M. (2010) Cell Cycle 9, 580–587 [DOI] [PubMed] [Google Scholar]
- 56. Lefkimmiatis K., Caratozzolo M. F., Merlo P., D'Erchia A. M., Navarro B., Levrero M., Sbisa E., Tullo A. (2009) Cancer Res. 69, 8563–8571 [DOI] [PubMed] [Google Scholar]
- 57. Berglund L., Björling E., Oksvold P., Fagerberg L., Asplund A., Al-khalili Szigyarto C., Persson A., Ottosson J., Wernérus H., Nilsson P., Lundberg E., Sivertsson A., Navani S., Wester K., Kampf C., Hober S., Pontén F., Uhlén M. (2008) Mol. Cell. Proteomics 10, 2019–2027 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.