Abstract
A diverse array of external stimuli, including most hormones and neurotransmitters, bind to cell surface receptors that activate G proteins. Mating pheromones in yeast Saccharomyces cerevisiae activate G protein-coupled receptors and initiate events leading to cell cycle arrest in G1 phase. Here, we show that the Gα subunit (Gpa1) is phosphorylated and ubiquitinated in response to changes in the cell cycle. We systematically screened 109 gene deletion strains representing the non-essential yeast kinome and identified a single kinase gene, ELM1, as necessary and sufficient for Gpa1 phosphorylation. Elm1 is expressed in a cell cycle-dependent manner, primarily at S and G2/M. Accordingly, phosphorylation of Gpa1 in G2/M phase leads to polyubiquitination in G1 phase. These findings demonstrate that Gpa1 is dynamically regulated. More broadly, they reveal how G proteins can simultaneously regulate, and become regulated by, progression through the cell cycle.
Keywords: Cell Cycle, G Proteins, Protein Phosphorylation, Serine Threonine Protein Kinase, Ubiquitination
Introduction
G protein coupled receptors and heterotrimeric G proteins are the predominant components through which cells receive and transduce extracellular signals. G protein signal transduction is highly conserved throughout eukaryotes, including the yeast Saccharomyces cerevisiae. In yeast, haploid a-type cells secrete an a-factor pheromone that binds to receptors on the surface of α-type cells, whereas α-type cells secrete an α-factor that acts exclusively on a-type cells (1). Consequently, the haploid cells fuse to form an a/α diploid cell.
As with other G protein systems, activation of the yeast pheromone receptor stimulates exchange of GDP for GTP on the G protein α subunit (Gpa1),2 which promotes its dissociation from the Gβγ (Ste4/18) heterodimer (1, 2). In yeast, Gβγ is primarily responsible for transmission and amplification of the signal to effector proteins, whereas the Gα subunit serves primarily to regulate the levels of free Gβγ. As a consequence, cells are highly sensitive to small changes in the stoichiometry of Gα and Gβγ (3–5). The signal is terminated by hydrolysis of GTP to GDP on the Gα subunit, which promotes reassociation of the heterotrimeric G protein complex. Further regulation is imposed by accelerating the GTPase activity of Gα via regulators of G protein signaling proteins (6).
Propagation of the G protein signal requires components of a MAPK cascade (Ste20, Ste11, Ste7, Fus3, or Kss1), a MAPK scaffold (Ste5), as well as a transcription factor (Ste12) (1). Consequently, the pheromone initiates changes in gene expression and cell morphology that prepare the cell to undergo cell-cell and nuclear fusion. Critical to this process is the initiation of cell cycle arrest in G1 phase, which ensures that each haploid cell contains exactly one copy of every chromosome before fusion (7). Pheromone-induced G1 arrest necessarily prevents fusion during DNA replication (S phase) or before mitosis (M phase), during which nuclear fusion could lead to missegregation of genetic material, aneuploidy, and other proliferative disadvantages (8, 9). Consequently, yeast mating and cell cycle progression must be highly coordinated processes.
The mating pathway is well known to regulate the cell cycle by stimulating expression of Far1, a cyclin-dependent kinase inhibitor that arrests cells at START in late G1 phase (10–14). Reciprocally, Far1 is itself regulated in a cell cycle-dependent manner and is degraded after cells pass through START and exit G1 phase (10, 11). Degradation of Far1 is initiated in G1 by cyclin-dependent kinase-mediated phosphorylation, which promotes its ubiquitination by the SCF (Skp1/Cullin/F-box) ubiquitin ligase (13, 15, 16).
Emerging evidence suggests that multiple mating pathway components are regulated by ubiquitination. Targets of ubiquitin-mediated degradation include the regulator of G protein signaling protein Sst2 (17), the MAPK kinase Ste7 (18), the MAPK scaffold protein Ste5 (19), and the G protein α subunit Gpa1 (20–23). Of these, the SCF has been shown to be necessary and sufficient for ubiquitination of Gpa1 (4). While SCF substrates are generally phosphorylated prior to ubiquitination (24, 25), as yet the kinase that phosphorylates Gpa1 has not been identified.
Here we identify a novel G protein kinase, Elm1. We show that the G protein is phosphorylated directly by Elm1 and that phosphorylation occurs in coordination with Elm1 expression during the cell cycle. G protein phosphorylation in G2/M phase leads to ubiquitination during the following G1 phase. In the absence of Elm1, the G protein is neither phosphorylated nor ubiquitinated. Taken together, these findings show how G proteins can simultaneously regulate, and become regulated by, progression through the cell cycle.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
Standard methods for growth, maintenance, and transformation of yeast and bacteria were used throughout. To ensure accurate cell density comparisons, a diluted fraction of each cell culture was lightly sonicated before measurement at an A600 nm to break apart cell clusters that commonly form between elm1Δ cells. Strains used in this study were BY4741 (MATa leu2Δ met15Δ his3Δ ura3Δ), elm1Δ (BY4741 elm1Δ::KanMX4), ELM1-TAP (Yeast TAP-fusion library, Open Biosystems), cdc6-1 (BY4741 cdc6-1 hph) (provided by Jean Cook, University of North Carolina), 15Dau (MATa ade1 his2 leu2–3, trp1–112, ura3Δ), cdc28-1 (15Dau, cdc28-1) (provided by Beverly Errede, University of North Carolina), LHY488 (MATa his3-Δ200 leu2Δ1 ura3–52 lys2–801 trp1Δ63 ade2–101), LHY489 (LHY488 cim3-1) (provided by Linda Hicke, Northwestern University) (26), MTY235 (MATa ade2-1 his3–11,15 leu2–3,112 trp1-1 ura3-1 can1–100), MTY670 (MTY235 cdc34-2), and MTY668 (MTY235 cdc4-1) (provided by Mike Tyers, Samuel Lunenfield Research Institute) (27). Integrated yeast strains were constructed by plasmid integration of pRS406-GPA1-(81-1539 bp) or pRS406-GPA1-(81-1539 bp)S200A at the naturally occurring HindIII site within GPA1, resulting in a single full-length copy of GPA1 at its endogenous locus. Integration was validated by immunoblotting with wild-type cells to confirm loss of the phosphorylation dependent mobility shift (in the case of S200A). All experiments were done in BY4741 unless otherwise specified. Table of yeast strains used in this study are shown in supplemental Table 1.
Plasmid Construction
Yeast shuttle plasmids pRS315-ELM1 and pRS316-ELM1 were constructed by PCR amplification of ELM1 ± 500 bp flanking the open reading frame (primers SalI-ELM1-F and SacI-ELM1-R) and directional cloning into the SalI and SacI sites of pRS315 or pRS316. Single point mutations (Gpa1S200A, Gpa1S200E, and Elm1K117R) were constructed by QuikChange (Stratagene) mutagenesis using the indicated primers (supplemental Table 2). Plasmids containing gpa115S/T-A were constructed by chemical synthesis of a 413-bp fragment of GPA1 (base pairs 392–786 of the open reading frame starting from the naturally occurring HindIII restriction site of GPA1) within which all serine and threonine codons were mutated to alanine, and a silent BglII site was introduced by mutation of base pairs 781 and 783 (GenScript). The synthesized fragment was cloned into GPA1 plasmids in which the same silent BglII mutation was introduced by QuikChange (using primer GPA1-BglII-F/R), followed by restriction digestion with HindIII and BglII. pYEX 4T-1-GST-ELM1 was purified from the yeast GST-fusion library host strain EJ 758 (28) using a yeast plasmid miniprep kit (Zymo Research) and then retransformed into BY4741 elm1Δ cells for expression of GST-Elm1. All plasmids were verified by DNA sequencing.
Growth Arrest and Release
Log-phase cell cultures were arrested in G1 phase with a synthetic α-factor peptide (30 μm final concentration, CHI Scientific 53424), in S phase by addition of hydroxyurea (added in powder form to 10 mg/ml final concentration, Sigma Aldrich H8627), and in G2/M phase by addition of nocodazole (15 μg/ml final concentration, Sigma Aldrich M1404). Note that for G2/M phase arrest, cells were first treated with 1% dimethyl sulfoxide for 30 min at 30 °C, followed by addition of nocodazole (100× stock at 1.5 mg/ml in dimethyl sulfoxide), resulting in a 2% final concentration of dimethyl sulfoxide in the cell culture. Each arrest was allowed to proceed for 2.5 h at 30 °C unless otherwise noted. Cells were released from arrest by centrifugation and washing with 3 × 100 ml of sterile water followed by resuspension in fresh medium to an A600 nm of 0.7 and growth at 30 °C.
Growth of Temperature-sensitive Mutants
Temperature-sensitive mutants (cdc6-1, cdc28-1, cdc4-1, or cdc34-2) and the isogenic wild type counterstrains were grown at a permissive temperature (25 °C) to early log-phase and then shifted to the restrictive temperature (37 °C) for 2.5 h to induce cell cycle arrest in G1 phase (cdc28-1 cells), or early S phase (cdc6-1 cells) and for 1 h to inactivate the SCF ubiquitin ligase (cdc4-1 or cdc34-2 cells).
In Vivo Ubiquitination Assays
Gpa1 polyubiquitination was detected by constitutive (ADH1 promoter) expression of Gpa1 in yeast harboring a temperature-sensitive proteasome mutation (cim3-1) or by coexpression of Gpa1 and Myc-ubiquitin as described previously (22). For cim3-1 and isogenic wild type cells, log-phase cultures were grown at the permissive temperature (25 °C) to an A600 nm of 0.5–0.6, followed by transition to the restrictive temperature (37 °C) for 3 h. Inducible Myc-ubiquitin strains were grown at 30 °C to an A600 nm of 0.5–0.6, followed by addition of CuSO4 to 100 μm for 3 h at 30 °C as described previously (29). Detection of Gpa1 ubiquitination at different cell cycle stages was accomplished by arresting log-phase cells followed by induction of Myc-ubiquitin expression for 3 h.
Protein Detection
Unless otherwise noted, cell pellets were harvested by addition of 100% trichloroacetic acid (5% final concentration), centrifugation at 4000 × g for 1 min, washing with 1 ml of 10 mm NaN3, and then stored as a frozen cell pellet at −80 °C. Protein extracts were generated by glass bead lysis in trichloroacetic acid as described previously (30) and resolved by 10% SDS-PAGE and immunoblotting. Membranes were probed with anti-Gpa1 at 1:1000 (31), anti-FLAG at 1:1000 (F1804, Sigma Aldrich), anti-Clb2 at 1:350 (sc-9071, Santa Cruz Biotechnology), anti-p44/42 at 1:500 (9101L, Cell Signaling Technology), anti-Kss1 at 1:350 (sc-6775, Santa Cruz Biotechnology), anti-Fus3 at 1:350 (sc-6773, Santa Cruz Biotechnology), anti-G6PDH at 1:50,000 (A9521, Sigma Aldrich), anti-Myc at 1:100 (9E10 mouse monoclonal antibody), and anti-protein A at 1:50,000 (P3775, Sigma Aldrich). Immunoreactive species were visualized by chemiluminescent detection (PerkinElmer Life Sciences) of horseradish peroxidase-conjugated anti-rabbit (170-5046) or anti-mouse IgG (170-5047) (Bio-Rad). Where indicated, image densitometry was conducted using National Institutes of Health ImageJ software (32). Statistical analysis was conducted using GraphPad Prism 4 for a minimum of three independent experiments unless noted otherwise.
Phosphatase Assays
Cells grown to an A600 nm of 1.0 were harvested by centrifugation at 2000 × g and stored at −80 °C. Cell pellets were resuspended in 1× λ phosphatase buffer (New England Biolabs) containing 50 mm HEPES (pH 7.5), 100 mm NaCl, 2 mm DTT, 0.01% Brij 35, 1 mm MnCl2, and 1× EDTA-free protease inhibitors (Roche). Each resuspended pellet was split in half and subjected to glass bead lysis in the presence or absence of phosphatase inhibitors (50 mm NaF and 1.3 mm sodium orthovanadate) (S6508, Sigma Aldrich). Lysates were centrifuged at 21,000 × g for 15 min, and the supernatant fraction was then collected into a fresh tube with or without 60 units (2.25 units/μl final concentration) of λ protein phosphatase (New England Biolabs) for 30 min at 30 °C. The reaction was stopped by addition of 6× SDS-PAGE loading buffer, and the samples were immediately subjected to SDS-PAGE and immunoblot analysis. Alternatively, phosphatase assays were conducted on purified protein. Briefly, 1 μl of yeast-purified Gpa1-FLAG was diluted in 1× λ-phosphatase buffer with or without 20 units (2 units/μl final concentration) λ protein phosphatase.
GST-Elm1 Expression, Purification, and in Vitro Kinase Assay
1 liter cultures of elm1Δ cells harboring pYEX-4T1-GST-ELM1 or elm1K117R were grown to an A600 nm of 0.7 and treated with 500 μm CuSO4 for 2 h to induce expression of GST-Elm1. Cells were harvested, washed with water, and stored at −80 °C. The cell pellets were subjected to glass bead lysis in buffer containing 20 mm Tris-HCl (pH 8.0), 400 mm NaCl, 2 mm MgCl2, 5% glycerol, 0.1% Triton X-100 (Sigma Aldrich), 1 mm DTT, 1× protease inhibitor mixture tablets, and 500 μm PMSF. The lysate mixture was subjected to microcentrifugation at 21,000 × g for 10 min, and GST-Elm1 was purified from the soluble extract using glutathione-SepharoseTM 4 Fast-flow (GE Healthcare) followed by washing with buffer containing 20 mm Tris-HCl (pH 8.0), 400 mm NaCl, 2 mm MgCl2, 5% glycerol, and 1 mm DTT and elution with dialysis buffer containing 20 mm glutathione (pH 7.5). Eluted protein was dialyzed with a slide-a-lyzer mini cartridge (Pierce) into kinase storage buffer containing 20 mm Tris-HCl (pH 8.0), 100 mm NaCl, 2 mm MgCl2, and 5% glycerol. In vitro kinase assays were conducted by incubating 0.08–0.16 pmol of purified GST-Elm1 (3.2–6.4 nm final concentration) and 12.5–25 pmol of recombinant purified Gpa1 or indicated mutants (0.5–1 μm final concentration) in 1× kinase reaction buffer previously described for in vitro Elm1 kinase assays and containing 50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 5 mm DTT, 2 mm EGTA, 200 mm sodium orthovanadate, and 10 μCi of [γ-32P]ATP (PerkinElmer Life Sciences) for 1 h at 30 °C (33). Reactions were stopped by addition of 6× SDS-PAGE loading buffer and immediately subjected to SDS-PAGE. The resulting gel was dehydrated and exposed to autoradiography film (HyBlot CL, Denville Scientific).
Affinity Purification of Gpa1-FLAG
Yeast harboring pRS316-ADH-GPA1-FLAG was grown to early log-phase and then harvested by centrifugation. The resulting cell pellet was lysed, and Gpa1-FLAG was purified as described previously (4).
Gene Transcription Assay
Pheromone-dependent transcription reporter assays were conducted as described previously (34). Briefly, cell cultures at an A600 nm of 0.8 were dispensed (90 μl into each of 48 wells of a 96-well plate) and mixed with 10 μl of α-factor peptide at the indicated concentration for 90 min at 30 °C. Next, each well was mixed with 20 μl of FDG solution (130 mm PIPES (pH 7.2), 0.25% Triton-X100, 0.5 mm fluorescein di-β-galactopyranoside (Marker Gene Technologies, M0250)) for 1 h at 37 °C. The reaction was stopped by the addition of 20 μl of 1 m sodium bicarbonate followed by fluorescence quantification using a fluorescence plate reader (SpectraMax M5, Molecular Devices).
Escherichia coli Expression of His6 Fusion Proteins
Recombinant His6-Gpa1 was expressed by autoinduction as described previously (35). All procedures were conducted at 4 °C unless noted otherwise. Briefly, competent BL21-RIPL cells (Stratagene) were transformed with the indicated expression vector. 5 ml of saturated starter cultures (containing ZY medium (10 g/l tryptone, 5 g/l yeast extract) with 15 mg of glucose, 50 μg/ml carbenicillin, 25 μg/ml chloramphenicol) were used to inoculate 800 ml of ZY medium containing 1× M solution (25 mm Na2HPO4, 25 mm KH2PO4, 50 mm NH4Cl, 5 mm Na2SO4), 1× 5052 solution (0.5% (w/v) glycerol, 0.5% (w/v) glucose, 10% (w/v) α-lactose), 50 mm MgSO4, 50 μg/ml carbenicillin, and 25 μg/ml chloramphenicol. Cultures were grown for 8 h at 37 °C and then shifted to 18 °C overnight. The G protein was purified by nickel-affinity chromatography as described previously (23) but without cleavage of the N-terminal His6 tag. The resultant protein was dialyzed by slide-a-lyzer (Pierce) in 20 mm Tris (pH 8.0), 100 mm NaCl, 2 mm MgCl2, 20 μm GDP, and 0.5 mm tris(2-carboxyethyl)phosphine.
RESULTS
Gpa1 Is a Phosphoprotein
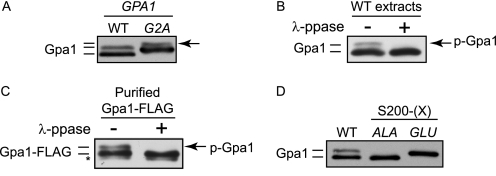
Gpa1 undergoes a variety of posttranslational modifications, including myristoylation, palmitoylation, and ubiquitination (21, 36). The myristoylation state of Gpa1 can be distinguished by an electrophoretic mobility shift following SDS-PAGE and immunoblotting. Because only a fraction of Gpa1 is mobility-shifted under these conditions, the prevailing view has been that Gpa1 exists in both myristoylated and non-myristoylated states (37). However, upon close inspection of overexpressed protein, we found that even non-myristoylated Gpa1G2A exhibits differential mobility by immunoblotting, indicating the presence of another modification (Fig. 1A). Given that phosphorylation can likewise alter the electrophoretic mobility of proteins, we asked whether phosphorylation rather than myristoylation might account for the second form of Gpa1. Consistent with this hypothesis, phosphatase treatment of whole cell extracts (Fig. 1B) or of purified Gpa1 (C) resulted in complete loss of the slower migrating form of Gpa1. We also found that mutation of serine 200, which was identified as a phosphorylation site by mass spectrometry (38), altered the mobility of Gpa1. Substitution of Ser-200 with alanine (S200A) or glutamate (S200E) replicated the mobility of dephosphorylated and phosphorylated Gpa1, respectively (Fig. 1D). We conclude that Gpa1 is a phosphorylated protein.
FIGURE 1.
Phosphorylation induces an electrophoretic mobility shift in Gpa1. Immunoblot analysis of Gpa1 using Gpa1-specific antibodies. A, cells over-expressing wild-type GPA1 (pAD4M-GPA1) (WT) or the myristoylation-site mutant GPA1G2A (G2A) analyzed by immunoblotting. The arrow marks the position of the presumptive phosphorylated form of Gpa1. B, soluble protein extracts from cells overexpressing wild-type Gpa1 were split into two separate aliquots, and one half was treated (+) with λ-protein phosphatase. p-Gpa1, phosphorylated Gpa1. C, Gpa1-FLAG purified from nocodazole-arrested cells and treated (+) with λ-protein phosphatase (ppase). D, wild type cells overexpressing wild-type Gpa1 (WT) or phosphorylation site mutants (S200A, ALA or S200E, GLU). *, nonspecific immunoblot band.
A Yeast Kinome Screen Reveals Elm1 as a G Protein Kinase
To identify the Gpa1 kinase, we monitored the phosphorylation-dependent mobility shift of endogenously expressed Gpa1 in gene deletion strains representing the majority of the yeast kinome. Of the 109 strains tested, deletion of ELM1 alone resulted in a significant observable loss of phosphorylated Gpa1. Phosphorylation of Gpa1 was restored by plasmid-borne expression of ELM1 (Fig. 2A). We observed no such differences in the absence of kinases that act downstream of Elm1 or that are functionally similar to Elm1 (see discussion, supplemental Fig. 1A) (33, 39). In addition to testing kinase deletions, we also monitored Gpa1 phosphorylation in each of 31 phosphatase-deletion strains, including all those involved in the pheromone response, but did not observe any difference in the abundance of phosphorylated Gpa1 (supplemental Fig. 1C). We conclude that Elm1 phosphorylates Gpa1 in vivo.
FIGURE 2.
Yeast kinome screen reveals Elm1 as a Gpa1 kinase. The abundance of phosphorylated Gpa1 was determined by immunoblotting 109 different kinase gene deletion strains. A single kinase, Elm1, is required for detection of phosphorylated Gpa1. A, validation of kinome screen results. Wild-type cells (WT) or cells expressing GPA1S200A (by gene replacement) or elm1Δ cells harboring empty vector or plasmid-borne ELM1 (ELM1). B, analysis of pheromone-dependent (+) MAP kinase activation (top panels, different exposures) and total expression (bottom panels) in WT, elm1Δ, and elm1Δ kss1Δ cells. IB, immunoblot. C, differential interference contrast (DIC) images of yeast strains used in the pheromone signaling assays shown in B and D. D, analysis of pheromone-dependent gene transcription in elm1Δ cells with the indicated plasmid containing ELM1, elm1K117R, or an empty vector (EV). Indicated are yeast strains expressing a FUS1-lacZ reporter treated with the indicated concentrations of mating pheromone. Results show the mean ± S.E. for four individual experiments, each performed in quadruplicate. RFU, relative fluorescence units.
Elm1 Is Required for Maximal Pheromone-induced Gene Transcription
Elm1 is best known as a regulator of cell morphology during G2/M phase of the cell cycle. During G2/M, Elm1 phosphorylates proteins required for the morphogenesis checkpoint that coordinates bud emergence and mitosis (40, 41) as well as for organization of septins during cytokinesis (33, 39, 42). Yeast harboring elm1 mutations exhibit a morphologically distinct growth pattern in which cells delay cytokinesis and undergo elongated bud growth, a process that also occurs under conditions of nitrogen starvation and filamentous growth (43). Elm1 has been proposed to inhibit the filamentous growth response that includes multiple signaling pathway branches, including (but not limited to) the MAP kinases Ste20, Ste11, Ste7, and Kss1 (43, 44), all of which participate as well in the pheromone response pathway. Accordingly, we found that Kss1 was more highly expressed and activated in elm1Δ compared with wild-type cells (Fig. 2B). In addition, deletion of KSS1 in elm1Δ cells reduced the filamentous-like phenotype (elongated buds and flocculation) typical of elm1Δ cells (Fig. 2C). In contrast, the other pheromone-responsive MAP kinase, Fus3, was largely unaffected by the absence of Elm1 (Fig. 2B).
We next determined whether Elm1 regulates pheromone-dependent gene transcription using a β-galactosidase reporter fused with the promoter of the mating-specific gene FUS1. We found that elm1Δ cells exhibit a significantly reduced maximum level of pheromone-induced gene transcription. Signaling was restored upon plasmid-borne expression of wild-type Elm1 but not kinase-inactive Elm1K117R (Fig. 2D). Thus, Elm1 represses activation of the MAP kinase branch of the filamentous growth response, including Kss1, and is required for maximum response to the pheromone.
Gpa1 Is Phosphorylated Directly by Elm1
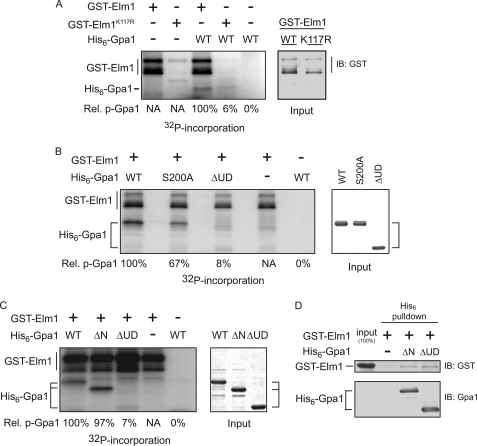
Elm1 clearly plays a role in multiple signaling pathways, including the filamentous growth and pheromone response pathways, any of which could be indirectly responsible for the diminished Gpa1 phosphorylation in elm1Δ cells. Therefore, we asked whether Elm1 acts directly on Gpa1. We established that purified Elm1 can bind to and phosphorylate recombinant Gpa1 in vitro (Fig. 3). Elm1 is capable of autophosphorylation (43), and we show that this activity is exhibited as well by GST-Elm1 (Fig. 3, A–C). Both Elm1 autophosphorylation and Gpa1 transphosphorylation were blocked when catalytically inactive GST-Elm1K117R was substituted for the wild-type kinase (Fig. 3A). Consistent with our findings in vivo, Ser-200 is required for maximum phosphorylation of Gpa1 (Fig. 3B). Similar results were observed upon mutation of all 15 Ser and Thr residues in the ubiquitination domain (data not shown). Finally, Elm1 was able to bind to, but not effectively phosphorylate, Gpa1ΔUD, a mutant that lacks the ubiquitination domain of the protein (residues 129–236) (Fig. 3, C and D). We conclude that Elm1 phosphorylates Gpa1 directly at Ser-200 and multiple other sites throughout the protein.
FIGURE 3.
Elm1 is necessary and sufficient for Gpa1 phosphorylation. The association between Elm1 and Gpa1 as determined by copurification and by in vitro kinase assay. A, in vitro kinase assay with purified GST-Elm1 or catalytically inactive GST-Elm1K117R and purified recombinant Gpa1 in the presence of [γ32P]ATP (left panel) and GST-Elm1 input detected by immunoblot with anti-GST (right panel). B, in vitro kinase assay with GST-Elm1 and purified recombinant Gpa1 (WT), Gpa1S200A (S200A), or Gpa1Δ1-36,Δ129-236 (ΔUD) in the presence of [γ32P]ATP. Shown is the incorporation of radioactive phosphate (left panel) and Gpa1 input detected by Coomassie staining (right panel). C, in vitro kinase assay with GST-Elm1 and with purified recombinant His6-tagged Gpa1 (WT), Gpa1Δ1-36 (ΔN), or Gpa1Δ1-36, Δ129-236 (ΔUD) lacking the ubiquitinated subdomain. Shown is the incorporation of radioactive phosphate (left panel) and Gpa1 input detected by Coomassie gel staining (right panel). D, in vitro affinity purification using nickel affinity matrix and immunodetection of bound GST-Elm1 described in C. Relative 32P incorporation (Rel. p-Gpa1) is shown for Gpa1 and was calculated as follows: [32P signal/Coomassie signal]mutant/[32P signal/Coomassie signal]WT.
Gpa1 Phosphorylation Is Cell Cycle-dependent
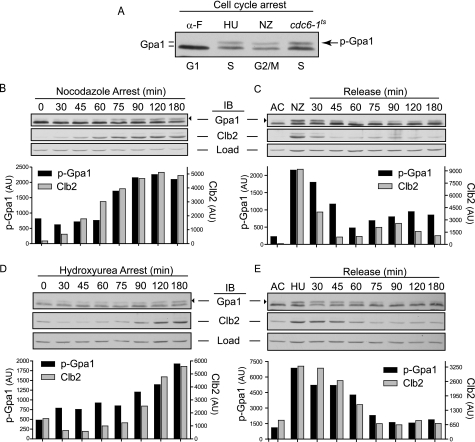
We next determined how phosphorylation of Gpa1 is regulated. Elm1 is expressed primarily during S and G2/M phases of the cell cycle (42, 43, 45), a phenotype that we corroborated (supplemental Fig. 2). Further, longstanding evidence indicates that the mobility-shifted (phosphorylated) form of Gpa1 is significantly reduced upon pheromone stimulation (31), which induces cell cycle arrest in G1 phase. Thus, we considered whether Gpa1 phosphorylation is regulated during the cell cycle. We directly compared Gpa1 from cells arrested in G1 phase with α-factor (α-F), in S phase with hydroxyurea, or in G2/M phase with nocodazole. Hydroxyurea inhibits deoxyribonucleotide synthesis, inducing a DNA replication checkpoint arrest in S phase (46). Nocodazole inhibits microtubule polymerization and induces a checkpoint arrest at the metaphase/anaphase transition of mitosis (47). We found that phosphorylated Gpa1 is most abundant in cells arrested in S and G2/M phase but lowest in cells arrested in G1 phase (Fig. 4A). To determine the dynamics of phosphorylation during the cell cycle, we monitored Gpa1 during arrest and release in cells treated with nocodazole or hydroxyurea. To validate the arrest and release protocol, we monitored changes in the mitotic cyclin Clb2 (48). We found that phosphorylated Gpa1 accumulates in a coordinated fashion with Clb2 as cells arrest in either G2/M or S phase (Fig. 4, B and D). Upon release from nocodazole, phosphorylated Gpa1 and Clb2 rapidly decrease in abundance and then increase concomitantly as cells reenter the cell cycle (Fig. 4C and supplemental Fig. 3). Similar coordination between phosphorylated Gpa1 and Clb2 was evident in cells released from hydroxyurea-induced cell cycle arrest (Fig. 4E). Finally, we observed temperature-dependent accumulation of phosphorylated Gpa1 in cdc6–1ts cells, which are incapable of DNA replication licensing, resulting in a post-START arrest in late G1/early S phase (Fig. 4A) (49, 50). We conclude that Gpa1 phosphorylation is Elm1-dependent and cell cycle-regulated. Although phosphorylated Gpa1 accumulates throughout the S and G2/M phases, it is rapidly eliminated from cells during G1 phase.
FIGURE 4.
Gpa1 phosphorylation is cell cycle-dependent. Cells treated for 2.5 h with α-factor pheromone (α-F, arrests cells in G1 phase), hydroxyurea (HU, arrests cells in S phase) or nocodazole (NZ, arrests cells in G2/M phase) followed by release into fresh medium where indicated. Shown are aliquots taken at the indicated time points followed by immunoblotting. The abundance of phosphorylated Gpa1 (black arrowhead) and Clb2 were quantified by densitometry from the immunoblot shown above. AU, arbitrary units; AC, asynchronous cells; Load, G6PDH loading control. A, direct comparison of Gpa1 from arrested cells. Cells were also arrested in late G1/early S phase by temperature shift of cdc6-1 cells. B, nocodazole arrest. C, nocodazole release. D, hydroxyurea arrest. E, hydroxyurea release.
Elm1 Is Required for Gpa1 Polyubiquitination
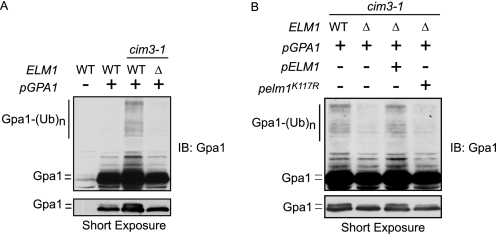
We have shown previously that Gpa1 is polyubiquitinated by the SCFCdc4 ubiquitin ligase (4) and that ubiquitination occurs primarily at lysine 165 within the ubiquitinated subdomain of Gpa1 (21). Typically, SCF recruits phosphorylated proteins as substrates for ubiquitination (24, 25). Therefore, we asked whether Elm1 phosphorylation promotes Gpa1 polyubiquitination. We compared Gpa1 polyubiquitination in the presence and absence of ELM1 using a proteasome-deficient yeast strain, cim3–1 (22, 23). For these experiments, Gpa1 was overexpressed to allow detection of the minor ubiquitinated species. Growth of cim3–1 cells at the restrictive temperature inactivates Cim3, an essential protein component of the 26 S proteasome (51), thereby stabilizing polyubiquitinated proteins and further enabling their detection by immunoblotting. Consistent with the hypothesis, Gpa1 polyubiquitination is considerably diminished in the absence of Elm1 or in the presence of plasmid-borne Elm1K117R (Fig. 5). To validate the observations made in cim3–1 cells, we also monitored Gpa1 polyubiquitination in wild-type or elm1Δ cells expressing myc-ubiquitin (proteins conjugated to myc-ubiquitin are degraded slowly) (supplemental Fig. 4) (52, 53). Once again Gpa1 polyubiquitination was diminished in the absence of Elm1. We conclude that Elm1 is required for ubiquitination, as well as for phosphorylation, of Gpa1.
FIGURE 5.
Elm1 is required for Gpa1 polyubiquitination. Temperature-sensitive proteasome-deficient (cim3-1 or cim3-1/elm1Δ) or isogenic wild-type cells overexpressing the indicated form of plasmid-borne GPA1 (pAD4M-GPA1, GPA1) grown at the restrictive temperature. A, accumulation of Gpa1 polyubiquitination in cim3-1 versus cim3-1/elm1Δ. B, accumulation of Gpa1 polyubiquitination in cim3-1 or cim3-1/elm1Δ cells with or without plasmid expression of ELM1 or elm1K117R.
Gpa1 Ubiquitination Is Regulated during the Cell Cycle
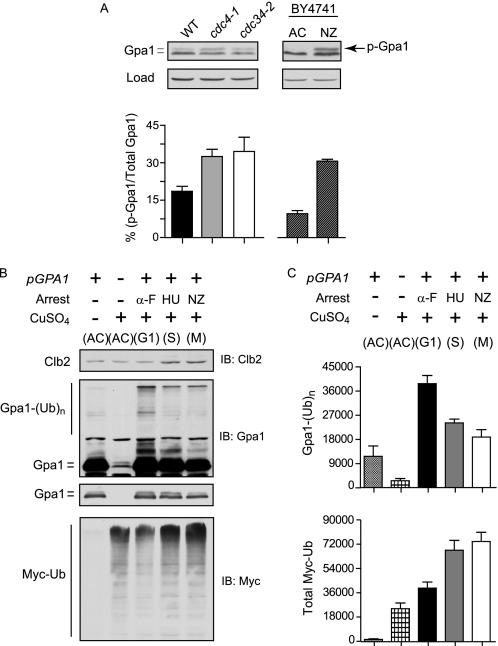
Phosphorylation is a well established precursor to ubiquitination by the SCF (25) and serves as a signal for recruitment of target substrates by F-box proteins (54). Our previous findings indicate that Gpa1 is ubiquitinated by SCFCdc4. The data presented above indicate that Gpa1 is phosphorylated by Elm1 and that Elm1 is required for ubiquitination. Therefore, we postulated that phosphorylation by Elm1 precedes ubiquitination by SCF. To test this hypothesis, we compared the relative proportion of phosphorylated Gpa1 in cells lacking functional Cdc4 (F-box protein) or Cdc34 (ubiquitin-conjugating enzyme), which we previously identified as responsible for polyubiquitination of Gpa1 (4). Accordingly, we monitored Gpa1 in temperature-sensitive cdc4-1 and cdc34-2 mutants (27). Consistent with the hypothesis, we found the proportion of phosphorylated Gpa1 to be higher in asynchronous cells lacking active forms of either Cdc4 or Cdc34 but comparatively lower in identically treated wild-type cells (Fig. 6A, left panel). Similar results were obtained using Tet-repressible versions of CDC4 and CDC34 (data not shown). Thus, phosphorylated Gpa1 accumulates in the absence of functional SCFCdc4 ubiquitin ligase.
FIGURE 6.
Gpa1 polyubiquitination by SCFCdc4 is cell cycle-regulated. Gpa1 phosphorylation in SCF mutant strains or Gpa1 polyubiquitination under different cell cycle arrested conditions. A, accumulation of phosphorylated Gpa1 (p-Gpa1) in cells lacking functional SCFCdc4. Gpa1 immunoblot analysis of whole cell extracts from temperature-sensitive cdc4-1 or cdc34-2 mutant strains, or the isogenic wild-type strain. The bar graph represents the ratio of phosphorylated Gpa1 (top band) to the sum total of Gpa1 (top and bottom bands) expressed as a percentage (left panel). Shown is the same analysis conducted for Gpa1 from asynchronous (AC) or nocodazole-arrested (NZ) wild-type cells (right panel). Results are the mean ± S.E. for three independent experiments analyzed in triplicate. B, accumulation of Gpa1 polyubiquitination (from plasmid pAD4M, top panel) or total Myc-ubiquitin (bottom panel) in wild type BY4741 cells harboring copper-inducible myc-ubiquitin and arrested in G1 phase with α-factor (α-F), in S phase with hydroxyurea (HU), or in G2/M phase with nocodazole (NZ), followed by CuSO4 induction of Myc-ubiquitin. C, quantitation of absolute immunoblot intensity for ubiquitinated Gpa1 and Myc-ubiquitin (Myc-Ub) from B. Results are the mean ± S.E. for three independent experiments analyzed in triplicate.
Finally, we considered whether ubiquitination, in addition to phosphorylation, might be regulated by the cell cycle. In support of this model, accumulation of phosphorylated Gpa1 in SCF-deficient cells is comparable with that observed in G2/M-arrested cells (Fig. 6A, right panel). Moreover, we found that Gpa1 polyubiquitination is significantly higher in cells arrested in G1 phase with α-factor mating pheromone or by temperature inactivation of cdc28-1 when compared with cells arrested in S phase or in G2/M phase (Fig. 6, B and C, and supplemental Fig. 5). Although treatment with α-factor results in Far1-mediated G1 arrest, cells expressing cdc28-1 undergo G1 arrest independent of pheromone pathway activation at the restrictive temperature (55). In either case, Gpa1 polyubiquitination is highest in cells arrested in G1 phase. Taken together, the data presented here reveal that Gpa1 is dynamically regulated. Phosphorylation and ubiquitination are independent of the pheromone stimulus and therefore not the result of feedback regulation. Rather, these modifications occur in conjunction with the cell cycle. Thus, the G protein is simultaneously a regulator of, and regulated by, cell cycle progression.
DISCUSSION
It is now well established that the G protein α subunit Gpa1 is ubiquitinated. Here we have begun to discern how this ubiquitination event is regulated. Our investigation began by showing that Gpa1 is phosphorylated and that phosphorylation results in an electrophoretic mobility shift of the protein. This property allowed us to determine that a single kinase (Elm1) is necessary for proper phosphorylation in vivo. Using purified components, we showed that Elm1 is also sufficient for Gpa1 phosphorylation in vitro. Elm1 is expressed primarily in S and G2/M phases of the cell cycle. Correspondingly, we found that the G protein is phosphorylated in a cell cycle-dependent manner. Gpa1 is polyubiquitinated by SCF, and phosphorylation is typically required for SCF-mediated ubiquitination. Thus, we investigated whether phosphorylation leads to ubiquitination of Gpa1 and whether ubiquitination also occurs in a cell cycle-dependent manner. Indeed, although Gpa1 phosphorylation peaks in G2/M phase, ubiquitination occurs in the subsequent G1 phase. These findings establish that G proteins can be regulated by progression through the cell cycle. More broadly, they raise the possibility that other pathway components may also be subject to cell cycle regulation.
The participation of Elm1 in G protein signaling was unexpected. Elm1 is best known as a protein kinase that coordinates events leading to cell division, including bud emergence, mitosis, and cytokinesis (33, 39–43). In this capacity, Elm1 phosphorylates protein kinases involved in septin organization and cytokinesis (Gin4 and Cla4) (33, 39, 42), another kinase that phosphorylates and deactivates the morphogenesis checkpoint protein Swe1 (Hsl1) (40, 41), and a fourth kinase that inhibits the mitotic exit network when the spindle position checkpoint is activated (Kin4) (56). It is unlikely that Gpa1 is regulated by any of these Elm1 substrates, given that deletion of those genes has no effect on Gpa1 phosphorylation or pheromone responsiveness. Thus, Gpa1 represents a new target of Elm1.
Whereas elm1Δ was the only mutant that exhibited a loss of Gpa1 phosphorylation, some residual phosphorylation could still be detected in vivo (Fig. 2A). Therefore, we speculate that Gpa1 is phosphorylated by another kinase. Possibilities include kinases that are absent from the gene deletion array, kinases essential for cell viability, and kinases that have functions related to Elm1. Elm1 is one of three closely related kinases, including Tos3 and Sak1. All three proteins are known to phosphorylate and activate Snf1 (the yeast homolog of human AMPK (5′ AMP-activated protein kinase), primarily under conditions of glucose starvation (57). Under the normal growth conditions used to assess mating pheromone responses however, deletion of TOS3, SAK1, or SNF1 does not appear to affect Gpa1 phosphorylation or pheromone signaling (supplemental Fig. 1A). Taken together, our findings indicate that Elm1 is largely responsible for Gpa1 phosphorylation, is uniquely able to regulate the pheromone response pathway, and does so in a cell cycle-dependent manner.
Other lines of evidence support the model that Elm1 phosphorylates Gpa1. First, Gpa1 is neither phosphorylated nor ubiquitinated when Elm1 is absent. In contrast, we did not observe any such differences in the absence of 108 other kinases. Second, we did not observe any differences in the absence of 31 different protein phosphatases. Thus it is unlikely that Elm1 acts indirectly by inhibiting the function of a Gpa1-phosphatase. Third, we detected a substantial accumulation of phosphorylated Gpa1 in the absence of the SCF function. Finally, we detected a substantial enrichment of Gpa1 polyubiquitination in G1-arrested cells as compared with S or G2/M-arrested cell cultures. These data confirm that phosphorylation and ubiquitination occur in a cell cycle-dependent manner and that phosphorylation precedes ubiquitination.
As the primary negative regulator of the mating pathway, Gpa1 is a logical target for regulation by posttranslational modifications. We have shown that pheromone-dependent gene transcription is diminished in cells that lack Elm1 function. Similarly, pheromone-dependent gene transcription is diminished in cells expressing Gpa1ΔUD. Gpa1ΔUD lacks a domain (UD, residues 129–236) that is required for ubiquitination by SCF as well as phosphorylation by Elm1 (4, 21, 22). The Gpa1 ubiquitination domain cannot be the only target of phosphorylation, however, because mutation of all 15 serine and threonine residues within this region failed to diminish phosphorylation in vitro (data not shown) or ubiquitination in vivo (supplemental Fig. 4). Thus, alternate phosphorylation sites are likely to exist, and these may likewise target the protein for polyubiquitination, at least under some circumstances. Taken together, these data indicate that phosphorylation can occur at multiple sites throughout the protein, whereas ubiquitination is restricted to a specific subdomain of the protein.
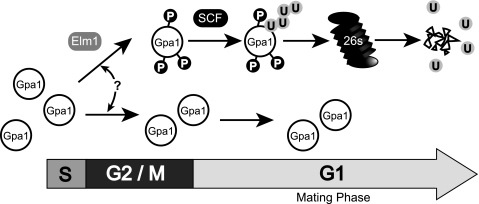
We have now identified the primary components necessary for Gpa1 phosphorylation (Elm1) and polyubiquitination (SCF). While much has been learned, substantive questions remain. For instance, we have yet to establish how Elm1 and SCF work together to modulate G protein function. All existing data indicate redundancy within this process because perturbations to phosphorylation or ubiquitination have modest effects on G protein stability. Selective pressure in yeast may have instilled this property because cells lacking Gpa1 cannot grow as a result of G1 cell cycle arrest. Alternatively phosphorylation and ubiquitination may affect G protein signaling in other ways. For example, ubiquitination could affect G protein catalytic activity. Ubiquitination could also serve to restrict Gpa1 localization to specific signal transduction complexes. Considering that Elm1 is localized predominately to the bud neck between dividing cells, Gpa1 phosphorylation and subsequent polyubiquitination may occur only during cell division or within a specialized subdomain of the plasma membrane (Fig. 7). Indeed, Gpa1 was recently shown to concentrate to the bud neck during G2/M phase, where Elm1 is located (58).
FIGURE 7.
Model of cell cycle G protein regulation. Elm1 phosphorylates a subpopulation of Gpa1 during S and G2/M phase. Phosphorylated Gpa1 is stable until entrance into the following G1 phase, when it is targeted for polyubiquitination by the SCFCdc4 ubiquitin ligase. The selective mechanism responsible for initiating phosphorylation of a fraction of the total G protein population is unknown (?) but may be due to cellular localization of signal transduction complexes near the bud neck of a dividing cell where Elm1 is localized during the S and G2/M phases. Localization-specific cell cycle-dependent ubiquitination of Gpa1 may provide a mechanism to optimize the local stoichiometry of Gα relative to Gβγ subunits and restrict signaling competency to G1 phase of the cell cycle.
The yeast mating response is perhaps the best-characterized of any signal transduction system, yet it continues to reveal new mechanisms of signal regulation. It has long been known that pheromone stimulation activates the G protein and promotes cell cycle arrest in G1. We now find that the G protein α subunit is phosphorylated and ubiquitinated in a manner that is contingent on cell cycle progression. The abundance of phosphorylated Gpa1 increases as cells progress through the S and G2/M phases and decreases rapidly after cells divide and enter G1 phase. Taken together, these data show that the G protein can be dynamically regulated. More broadly, these findings reveal a previously unsuspected degree of coordination between G protein signaling and cell division.
Supplementary Material
Acknowledgments
We thank Daniel Isom for constructive discussions and technical assistance with analysis of G protein point mutants, Candice Carlile and Jean Cook for the cdc6-1 yeast strain, and Rachael Baker and Jeffrey Duffy for plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM059167 and R01 GM080739 (to H. G. D.) and R01 GM080739-S1 and K99 GM094533-01 (to M. P. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2, Figs. 1–5, and additional references.
- Gpa1
- G protein α subunit
- SCF
- Skp1/Cullin/F-box
- PIPES
- 1,4-piperazinediethanesulfonic acid
- UD
- ubiquitination domain.
REFERENCES
- 1. Dohlman H. G., Thorner J. W. (2001) Annu. Rev. Biochem. 70, 703–754 [DOI] [PubMed] [Google Scholar]
- 2. Sprang S. R. (1997) Annu. Rev. Biochem. 66, 639–678 [DOI] [PubMed] [Google Scholar]
- 3. Cole G. M., Stone D. E., Reed S. I. (1990) Mol. Cell. Biol. 10, 510–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cappell S. D., Baker R., Skowyra D., Dohlman H. G. (2010) Mol. Cell 38, 746–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spain B. H., Koo D., Ramakrishnan M., Dzudzor B., Colicelli J. (1995) J. Biol. Chem. 270, 25435–25444 [DOI] [PubMed] [Google Scholar]
- 6. Hepler J. R. (2003) Mol. Pharmacol. 64, 547–549 [DOI] [PubMed] [Google Scholar]
- 7. Wittenberg C., La Valle R. (2003) BioEssays 25, 856–867 [DOI] [PubMed] [Google Scholar]
- 8. Torres E. M., Sokolsky T., Tucker C. M., Chan L. Y., Boselli M., Dunham M. J., Amon A. (2007) Science 317, 916–924 [DOI] [PubMed] [Google Scholar]
- 9. Torres E. M., Williams B. R., Amon A. (2008) Genetics 179, 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McKinney J. D., Chang F., Heintz N., Cross F. R. (1993) Genes Dev. 7, 833–843 [DOI] [PubMed] [Google Scholar]
- 11. Peter M., Gartner A., Horecka J., Ammerer G., Herskowitz I. (1993) Cell 73, 747–760 [DOI] [PubMed] [Google Scholar]
- 12. Tyers M., Futcher B. (1993) Mol. Cell. Biol. 13, 5659–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oehlen L. J., Cross F. R. (1994) Genes Dev. 8, 1058–1070 [DOI] [PubMed] [Google Scholar]
- 14. Peter M., Herskowitz I. (1994) Science 265, 1228–1231 [DOI] [PubMed] [Google Scholar]
- 15. Blondel M., Galan J. M., Chi Y., Lafourcade C., Longaretti C., Deshaies R. J., Peter M. (2000) EMBO J. 19, 6085–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Henchoz S., Chi Y., Catarin B., Herskowitz I., Deshaies R. J., Peter M. (1997) Genes Dev. 11, 3046–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hao N., Yildirim N., Wang Y., Elston T. C., Dohlman H. G. (2003) J. Biol. Chem. 278, 46506–46515 [DOI] [PubMed] [Google Scholar]
- 18. Wang Y., Ge Q., Houston D., Thorner J., Errede B., Dohlman H. G. (2003) J. Biol. Chem. 278, 22284–22289 [DOI] [PubMed] [Google Scholar]
- 19. Garrenton L. S., Braunwarth A., Irniger S., Hurt E., Künzler M., Thorner J. (2009) Mol. Cell. Biol. 29, 582–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Madura K., Varshavsky A. (1994) Science 265, 1454–1458 [DOI] [PubMed] [Google Scholar]
- 21. Marotti L. A., Jr., Newitt R., Wang Y., Aebersold R., Dohlman H. G. (2002) Biochemistry 41, 5067–5074 [DOI] [PubMed] [Google Scholar]
- 22. Wang Y., Marotti L. A., Jr., Lee M. J., Dohlman H. G. (2005) J. Biol. Chem. 280, 284–291 [DOI] [PubMed] [Google Scholar]
- 23. Torres M. P., Lee M. J., Ding F., Purbeck C., Kuhlman B., Dokholyan N. V., Dohlman H. G. (2009) J. Biol. Chem. 284, 8940–8950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deshaies R. J. (1999) Annu. Rev. Cell Dev. Biol. 15, 435–467 [DOI] [PubMed] [Google Scholar]
- 25. Willems A. R., Goh T., Taylor L., Chernushevich I., Shevchenko A., Tyers M. (1999) Philos Trans. R. Soc. Lond. B Biol. Sci. 354, 1533–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ghislain M., Udvardy A., Mann C. (1993) Nature 366, 358–362 [DOI] [PubMed] [Google Scholar]
- 27. Tang X., Orlicky S., Liu Q., Willems A., Sicheri F., Tyers M. (2005) Methods Enzymol. 399, 433–458 [DOI] [PubMed] [Google Scholar]
- 28. Martzen M. R., McCraith S. M., Spinelli S. L., Torres F. M., Fields S., Grayhack E. J., Phizicky E. M. (1999) Science 286, 1153–1155 [DOI] [PubMed] [Google Scholar]
- 29. Roth A. F., Sullivan D. M., Davis N. G. (1998) J. Cell Biol. 142, 949–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee M. J., Dohlman H. G. (2008) Curr. Biol. 18, 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dohlman H. G., Goldsmith P., Spiegel A. M., Thorner J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9688–9692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rasband W. S. (1997–2009) U.S. National Institutes of Health, Bethesda, Maryland [Google Scholar]
- 33. Asano S., Park J. E., Yu L. R., Zhou M., Sakchaisri K., Park C. J., Kang Y. H., Thorner J., Veenstra T. D., Lee K. S. (2006) J. Biol. Chem. 281, 27090–27098 [DOI] [PubMed] [Google Scholar]
- 34. Hoffman G. A., Garrison T. R., Dohlman H. G. (2002) Methods Enzymol. 344, 617–631 [DOI] [PubMed] [Google Scholar]
- 35. Studier F. W. (2005) Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 36. Wedegaertner P. B., Wilson P. T., Bourne H. R. (1995) J. Biol. Chem. 270, 503–506 [DOI] [PubMed] [Google Scholar]
- 37. Stone D. E., Cole G. M., de Barros Lopes M., Goebl M., Reed S. I. (1991) Genes Dev. 5, 1969–1981 [DOI] [PubMed] [Google Scholar]
- 38. Li X., Gerber S. A., Rudner A. D., Beausoleil S. A., Haas W., Villén J., Elias J. E., Gygi S. P. (2007) J. Proteome Res. 6, 1190–1197 [DOI] [PubMed] [Google Scholar]
- 39. Sreenivasan A., Kellogg D. (1999) Mol. Cell. Biol. 19, 7983–7994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Szkotnicki L., Crutchley J. M., Zyla T. R., Bardes E. S., Lew D. J. (2008) Mol. Biol. Cell 19, 4675–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Crutchley J., King K. M., Keaton M. A., Szkotnicki L., Orlando D. A., Zyla T. R., Bardes E. S., Lew D. J. (2009) Mol. Biol. Cell 20, 1926–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bouquin N., Barral Y., Courbeyrette R., Blondel M., Snyder M., Mann C. (2000) J. Cell Sci. 113, 1435–1445 [DOI] [PubMed] [Google Scholar]
- 43. Koehler C. M., Myers A. M. (1997) FEBS Lett. 408, 109–114 [DOI] [PubMed] [Google Scholar]
- 44. Palecek S. P., Parikh A. S., Kron S. J. (2000) Genetics 156, 1005–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Manderson E. N., Malleshaiah M., Michnick S. W. (2008) PLoS ONE 3, e1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weinert T. A., Kiser G. L., Hartwell L. H. (1994) Genes Dev. 8, 652–665 [DOI] [PubMed] [Google Scholar]
- 47. Jacobs C. W., Adams A. E., Szaniszlo P. J., Pringle J. R. (1988) J. Cell Biol. 107, 1409–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Deshaies R. J. (1997) Curr. Opin. Genet. Dev. 7, 7–16 [DOI] [PubMed] [Google Scholar]
- 49. Liang C., Weinreich M., Stillman B. (1995) Cell 81, 667–676 [DOI] [PubMed] [Google Scholar]
- 50. Liang C., Stillman B. (1997) Genes Dev. 11, 3375–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Finley D., Tanaka K., Mann C., Feldmann H., Hochstrasser M., Vierstra R., Johnston S., Hampton R., Haber J., McCusker J., Silver P., Frontali L., Thorsness P., Varshavsky A., Byers B., Madura K., Reed S. I., Wolf D., Jentsch S., Sommer T., Baumeister W., Goldberg A., Fried V., Rubin D. M., Toh-e A. (1998) Trends Biochem. Sci 23, 244–245 [DOI] [PubMed] [Google Scholar]
- 52. Ellison M. J., Hochstrasser M. (1991) J. Biol. Chem. 266, 21150–21157 [PubMed] [Google Scholar]
- 53. Hochstrasser M., Ellison M. J., Chau V., Varshavsky A. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 4606–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Skowyra D., Craig K. L., Tyers M., Elledge S. J., Harper J. W. (1997) Cell 91, 209–219 [DOI] [PubMed] [Google Scholar]
- 55. Neiman A. M., Chang F., Komachi K., Herskowitz I. (1990) Cell Regul. 1, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Caydasi A. K., Kurtulmus B., Orrico M. I., Hofmann A., Ibrahim B., Pereira G. (2010) J. Cell Biol. 190, 975–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hedbacker K., Carlson M. (2008) Front. Biosci. 13, 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Suchkov D. V., DeFlorio R., Draper E., Ismael A., Sukumar M., Arkowitz R., Stone D. E. (2010) Mol. Biol. Cell 21, 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.