Abstract
Epidermal growth factor receptor (EGFR), an aberrantly overexpressed or activated receptor-tyrosine kinase in many cancers, plays a pivotal role in cancer progression and has been an attractive target for cancer therapy. Gefitinib and erlotinib, two EGFR-tyrosine kinase inhibitors, have been approved for non-small cell lung cancer. However, durable clinical efficacy of these EGFR inhibitors is severely limited by the emergence of acquired resistance. For example, the expression of breast cancer-resistant protein (BCRP/ABCG2) has been shown to confer acquired resistance of wild-type EGFR (wtEGFR)-expressing cancer cells to gefitinib. However, the underlying molecular mechanisms still remain unclear. Here, we show that wtEGFR expression is elevated in the nucleus of acquired gefitinib-resistant cancer cells. Moreover, nuclear translocation of EGFR requires phosphorylation at Ser-229 by Akt. In the nucleus, EGFR then targets the proximal promoter of BCRP/ABCG2 and thereby enhances its gene transcription. The nuclear EGFR-mediated BCRP/ABCG2 expression may contribute at least in part to the acquired resistance of wtEGFR-expressing cancer cells to gefitinib. Our findings shed light on the role of nuclear EGFR in the sensitivity of wtEGFR-expressing cancer cells to EGFR tyrosine kinase inhibitors and also deciphered a putative molecular mechanism contributing to gefitinib resistance through BCRP/ABCG2 expression.
Keywords: Drug Resistance, Gene Regulation, Nuclear Translocation, Protein Phosphorylation, Receptor-tyrosine Kinase, Akt, BCRP/ABCG2, Gefitinib, Nuclear EGFR
Introduction
The receptor tyrosine kinase EGFR4 (also known as ErbB1 or HER1) of the ErbB (HER) family, plays pivotal roles in the etiology of cancer and is frequently overexpressed or aberrantly activated in many cancers and has been as an attractive target for cancer therapy (1). Two small molecule tyrosine kinase inhibitors, gefitinib (ZD1839, Iressa) and erlotinib (OSI-774, Tarceva), specifically and reversibly bind to the ATP binding pocket of EGFR and thereby inhibit tyrosine kinase activity and downstream survival signals of EGFR. Although EGFR is overexpressed in many cancer types, these two agents showed more dramatic efficacy and clinical benefits for non-small cell lung cancer (NSCLC) patients, particularly those characterized as East Asian, non-smoker, adenocarcinoma histological type, and female gender. The encouraging responses in these selected NSCLC patients to EGFR inhibitors show strong association with specific activating mutations within the EGFR-tyrosine kinase domain (2–4). However, these patients would ultimately become resistant to gefitinib or erlotinib through development of a secondary mutation in EGFR that reduces its binding affinity for gefitinib (5, 6) or amplification of the MET gene to raise the compensatory survival signals (7, 8).
Although the response rates are not as high compared with patients with EGFR mutations, about 20–30% of NSCLC patients with amplified wild-type EGFR (wtEGFR) treated with gefitinib and erlotinib still demonstrate a significant survival benefit (9–11). No identifiable EGFR mutations were found in ∼10–20% of gefitinib responders (4, 10–15). These observations indicate that EGFR mutations may not be the only determinant for the sensitivity to EGFR tyrosine kinase inhibitors and that using these mutations as single criteria for receiving EGFR tyrosine kinase inhibitor therapy may exclude a significant population of patients who may otherwise receive clinical benefit. Unlike the well characterized studies between EGFR mutation and gefitinib sensitivity (5–8), a few studies have addressed the molecular determinants accounting for the cellular sensitivity to gefitinib in wtEGFR-expressing cancer cells. In a cell culture system with acquired resistance to gefitinib, an increased activity of insulin-like growth factor receptor by down-regulating insulin-like growth factor-binding proteins has been found to maintain the PI3K/Akt-mediated survival signaling in response to acquired gefitinib resistance in gefitinib-sensitive and wtEGFR-expressing cancer cells (16, 17). In addition, it has also been reported that a non-smoking female NSCLC patient with wtEGFR expression developed acquired gefitinib resistance without any identifiable EGFR mutations (18). Further examination showed that breast cancer-resistant protein (BCRP)/ATP binding cassette subfamily G member 2 (ABCG2) was detected in this patient's recurrent tumor (18). Aside for these studies, the underlying mechanisms of the sensitivity to gefitinib in wtEGFR-expressing cancer cells are still largely unknown.
In addition to its downstream signaling, EGFR has been identified in the nucleus and associates with specific functions, including gene transcription (19–22), DNA repair (23), radioresistance (24–26), and chemoresistance (26). A study recently showed that increased nuclear expression of EGFR conferred acquired resistance to EGFR antibody cetuximab in NSCLC cancer cells (27), bolstering the nuclear functions of EGFR in drug resistance. Importantly, EGFR was reported to be internalized and located in the perinuclear region of gefitinib-resistant cancer cells (13, 28). However, it still remains unclear whether nuclear localization of EGFR plays a role in the development of acquired gefitinib resistance.
In this study, using wtEGFR-expressing and gefitinib-sensitive A431 and its derived gefitinib-resistant (A431/GR) cell lines as the assay model (16), we observed an increased accumulation of EGFR in the nucleus of A431/GR and other gefitinib-treated cell lines, and this required Akt-mediated EGFR phosphorylation at Ser-229. Moreover, nuclear EGFR (nEGFR) in A431/GR cells targeted the BCRP/ABCG2 promoter and enhanced its transcriptional expression. As expression of BCRP/ABCG2 has been implicated in gefitinib resistance in breast cancer cells harboring wtEGFR, our findings here suggest that nEGFR-mediated activation of BCRP/ABCG2 gene expression is one of the mechanisms through which cells acquire gefitinib resistance.
EXPERIMENTAL PROCEDURES
Materials
Commercially available gefitinib was used for in vitro and in vivo studies. Cells were transfected with siRNA oligo (5′-AAAUCCAGACUCUUUCGAU-3′) targeting EGFR 3′-UTR or non-targeting control siRNA (5′-UGGUUUACAUGUCGACUAA-3′) with DharmaFECT 1 (Dharmacon) and used for experiments 72 h after transfection. siRNAs against Akt1 (M-003000-03-0005), Akt2 (M-003001-02-0005), and Akt3 (M-003002-02-0005) were purchased from Dharmacon. EGFR cDNA was constructed into a pCDNA3.1 vector, and the S229A and S229D mutations were generated by using the QuikChange site-directed mutagenesis kit according to the manufacturer's protocol (Stratagene). Anti-EGFR (Ab-13) antibody purchased from Thermo Scientific and anti-EGFR (SC-03) antibody from Santa Cruz were used for EGFR immunoprecipitation and EGFR immunoblotting, respectively. For detection of Akt-dependent EGFR phosphorylation, antibody against phosphorylated Akt substrate (PAS) (#9611) from Cell Signaling was used. Anti-Akt and anti-phospho-Akt antibody were purchased from Cell Signaling. For detection of BCRP/ABCG2 protein levels by immunoblotting, anti-BCRP/ABCG2 antibody from Santa Cruz (SC58222) was used. Epidermal growth factor (EGF) was purchased from Sigma. The following peptides were chemically synthesized from LTK Biolaboratories (Taiwan) for anti-phospho-EGFR Ser-229 antibody production in mice and the peptide competition assay: unmodified peptide, RGKSPSDC; keyhole limpet hemocyanin-conjugated phosphorylated peptide, RGKSPpSDC.
Cell Lines and Cell Culture
A431 and A431/GR cell lines were gifts from Dr. Carlos L. Arteaga (Vanderbilt-Ingram Cancer Center, Nashville, TN). Other acquired gefitinib-resistant cancer cells were established by selecting with gradually elevated concentrations of gefitinib for two months as described previously (16). Insensitivity to gefitinib treatment was tested in these established resistant cancer cell lines, which were cultured in the presence of 1 μm gefitinib.
Cellular Fractionation
Cells were washed twice with 1× PBS and then lysed in Nori buffer (20 mm HEPES, pH 7.0, 10 mm KCl, 2 mm MgCl2, 0.5% Nonidet P-40, 1 mm Na3VO4, 10 mm NaF, 1 mm phenylmethanesulfonyl fluoride (PMSF), 2 μg/ml aprotinin) followed by incubation on ice for 10 min. Then cells were homogenized by 40–70 strokes in a tightly fitting Dounce homogenizer. Homogenates were centrifuged at 1500 × g for 5 min. The supernatant was further centrifuged at 16,100 × g for 20 min, which formed as nonnuclear fraction. The former pellet was then washed in Nori buffer without protease inhibitors and centrifuged at 1500 × g for 5 min. The washing step was repeated three times. Then the pellet was resuspended and sonicated in NETN buffer (150 mm NaCl, 1 mm EDTA, 20 mm Tris-Cl, pH 8.0, 0.5% Nonidet P-40, 1 mm Na3VO4, 10 mm NaF, 1 mm PMSF, and 2 μg/ml aprotinin) and then centrifuged at 16,100 × g for 20 min, which formed as the nuclear fraction.
Immunofluorescence Staining
Cells were grown in Lab-Tek chamber slides followed by transfection of the indicated amounts of plasmids described in each experiment. Cells were then washed twice with 1× PBS, fixed with 4% paraformaldehyde for 15 min, and permeabilized with 0.5% Triton X-100 for 15 min followed by blocking with 10% bovine serum albumin for at least 1 h. Dilution of the primary antibodies were used as follows: rabbit polyclonal EGFR antibodies (1:100, Santa Cruz Biotechnology) and mouse monoclonal HA antibody (1:300, Roche Applied Science). Respective secondary antibodies tagged with Texas Red and Cy5 were then used (1:500). The fluorescence of Texas Red and Cy5 were visualized and captured by using Leica TCS SP5 confocal microscopy.
In Vitro Kinase Assay
Briefly, the GST-fused EGFR extracellular domain was incubated with PreScission protease to remove GST and then used as a substrate for the purified Akt protein in the presence of ATP. Phosphorylation was detected by using anti-PAS antibody.
Cell Proliferation Assay
In vitro cell viability was characterized by (3-(4,5-dimethylthiazol-2-yl)-2,5 -diphenyltetrazolium bromide (MTT) colorimetric assay. Briefly, cells (8 × 103 cells per well) with transfection of specific EGFR mutants were seeded in 96-well plates for 24 h and subsequently subjected to pretreatments as indicated. After treatment of gefitinib for 72 h, relative cell viability was determined by measuring absorbance at 570 nm after a 3-h incubation of 1 mg/ml MTT in each well and solubilized in 100 μl of dimethyl sulfoxide (DMSO).
Reporter Gene Assay
A431 and A431/GR cells that reached 60–70% confluence were transfected with the indicated plasmids as described in each experiment as well as BCRP/ABCG2 promoter-luciferase plasmid containing EGFR binding regions. Forty-eight hours later, the luciferase activities in cell lysates were measured by the luciferase assay system. Luciferase activity was normalized per μg of protein extract.
DNA Affinity Precipitation Assay
An in vitro DNA binding assay was performed by mixing whole cell lysates with 10 μg of biotinylated BCRP/ABCG2 promoter-containing DNA probes for 1 h followed by adding 30 μl of streptavidin-agarose beads (4%) with a 50% slurry. The mixture was incubated at room temperature for another hour with rotation. Beads were pelleted and washed three times with 1× PBS plus 0.1% Tween 20. The binding proteins were eluted by 2× SDS loading buffer and separated by SDS-PAGE followed by Western blot analysis. The probes were synthesized by PCR amplification of genomic DNA with the following primers: 5′-CCCGTTTCCTGAACATGCGC-3′ (forward) and Biotin-5′-TTTTTTTTTTTTGCCCAGTCACAAGCGCTG-3′ (reverse) for AT1, 5′-GCTGTCACTCCTTGCCCAGC-3′ (forward) and Biotin-5′-TTTTTTTTTTTTGCCCAGTCACAAGCGCTG-3′ (reverse) for AT2, and 5′-CGTGTCACGGCAGGGTGACC-3′ (forward) and Biotin-5′-TTTTTTTTTTGCGGCTGGAGGTCACGATGG-3′ (reverse) for non-AT.
Chromatin Immunoprecipitation (ChIP)
An in vivo DNA binding assay was performed by using EZ-ChIPTM kit (Millipore) according to the manufacturer's instructions. Briefly, cells were cross-linked, lysed, and sonicated to shear the size of DNA to 500–1000 bp followed by immunoprecipitation with EGFR antibodies (Ab-13). DNA was then purified, and specific sequences in the immunoprecipitates were detected by PCR amplification. The PCR product was separated by 1.5% agarose gel electrophoresis and visualized by ethidium bromide staining. Primers for PCR amplification were 5′-CCCGTTTCCTGAACATGCGC-3′ (forward) and 5′-GCCCAGTCACAAGCGCTG-3′ (reverse) for AT1 and 5′-CGTGTCACGGCAGGGTGACC-3′ (forward) and 5′-GCGGCTGGAGGTCACGATGG-3′ (reverse) for non-AT.
RESULTS
Nuclear Localization of EGFR Is Increased in Gefitinib-treated Cells
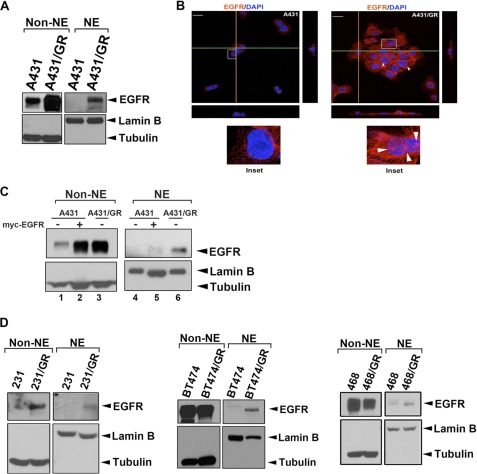
To address whether nEGFR plays a role in acquired gefitinib resistance in wtEGFR-expressing cancer cells, we first isolated the cytoplasmic (non-NE) and nuclear (NE) fractions in both gefitinib-sensitive A431 and its derived gefitinib-resistant (A431/GR) cell lines (16) and then examined the level of EGFR in these fractions (Fig. 1A). We found that both cytoplasmic and nuclear levels of EGFR increased in A431/GR cells compared with the parental A431 cells (Fig. 1A). The increase in nuclear localization of EGFR in A431/GR cells (right panel, Fig. 1B) was further demonstrated by confocal microscope analysis. To ensure that the enhanced level of EGFR in the nucleus in A431/GR cells is not a result of an increase in the overall level of EGFR, we enforced the expression of EGFR in the parental cell line by transient transfection of myc-tagged EGFR to a level comparable with that we observed in the A431/GR cell line. As shown in Fig. 1C, although the level of EGFR increased in the cytoplasm in A431 cells, the EGFR level in the nucleus remained unchanged, indicating that the EGFR nuclear import is regulated by other unexplored mechanisms and not as a result of enhanced expression level of EGFR in response to acquired gefitinib resistance.
FIGURE 1.
Nuclear EGFR is involved in drug resistance to EGFR-tyrosine kinase inhibitor gefitinib. A, A431 and A431/GR cells were subjected to cellular fractionation followed by WB analysis of cellular localization of EGFR. Levels of tubulin and lamin B were used as markers for cytosolic and nuclear fractions, respectively. B, immunofluorescence staining of EGFR (red) and DAPI (blue) was analyzed by confocal microscopy with z-stacks. Yellow and green lines represented corresponding points in the orthogonal planes, which confirmed distribution of the labels within the pictured cells after the summation of serial optical sections. The scale bar represents 10 μm. C, shown is EGFR overexpression in A431 cells followed by cellular fractionation. The cellular localization of EGFR was analyzed by WB. NE, nuclear extract. D, nuclear localization of EGFR in several gefitinib-resistant cell line pairs was analyzed as described in A.
To determine whether the increase in nEGFR also occurs in other wtEGFR-expressing cancer cell lines, we first established gefitinib-resistant clones of breast cancer cells, MDA-MB-231, BT474, and MDA-MB-468, by culturing and selecting them with increasing concentrations of gefitinib as previously described (16). In MDA-MB-231/GR cells (left panel, Fig. 1D), we observed a similar pattern of increased EGFR expression level in both cytoplasmic and nuclear fractions compared with A431/GR cells. In contrast, even though the level of cytoplasmic EGFR in BT474 and MDA-MB-468 gefitinib-resistant cells did not change significantly (center and right panels, Fig. 1D, respectively), we continued to see increased EGFR in the nuclear fraction. These observations suggest that the nuclear translocation of EGFR may be a general phenomenon in response to long term treatment of gefitinib.
Akt Phosphorylates EGFR at Ser-229 in Response to Both Gefitinib Resistance and EGF Treatment
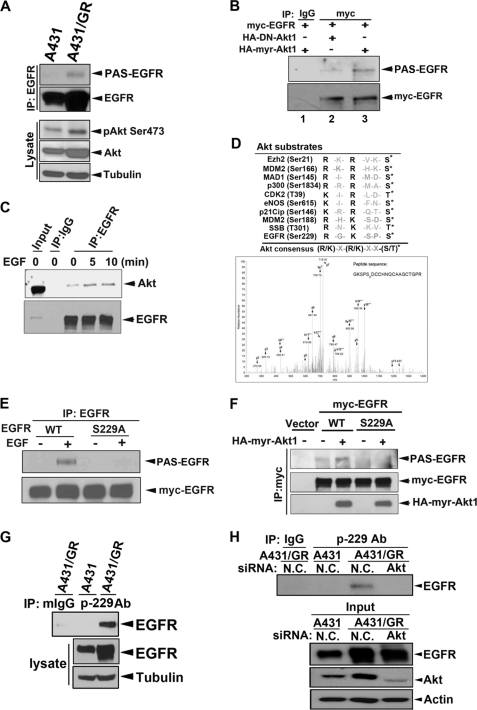
Next, we investigated the underlying mechanisms of EGFR nuclear translocation in the gefitinib-resistant cancer cells. The status of Akt activity is a critical feature in determining the responsiveness of cancer cells to gefitinib and erlotinib (29, 30). Tumor cells resistant to these drugs are characterized by the failure of Akt suppression (31, 32). ErbB2, ErbB3, IGF-1 receptor, and MET have all been proposed to maintain the continuous activation of Akt in the gefitinib-resistant cells (33). Consistent with the previous study (16), Akt activity was significantly increased in A431/GR compared with parental A431 cells (Fig. 2A). We also confirmed the down-regulation of insulin-like growth factor-binding protein-3 and -4 in A431/GR cells (supplemental Fig. S1A), which was reported to maintain this continued Akt activity in A431/GR cells (16). Therefore, we first asked whether the activated Akt phosphorylates EGFR. Using an anti-phospho-Akt substrate (anti-PAS) antibody, which recognizes the phosphorylated consensus motif ((R/K)X(R/K)XX(pS/T)) of Akt substrates, we found that the Akt-dependent phosphorylation of EGFR (PAS-EGFR) was detected in A431/GR but not in A431 cells. Moreover, this phosphorylation can be inhibited by specific Akt inhibitor API-2 (supplemental Fig. S1B). Similarly, PAS-EGFR was also induced by EGF and can be inhibited by both specific Akt inhibitor API-2 (supplemental Fig. S1C) and PI3K inhibitor LY294002 but not by another inhibitor that is not involved in the PI3K signaling pathway such as the p38 inhibitor, SB202190 (supplemental Fig. S1D), which indicates that this anti-PAS antibody specifically recognized Akt-dependent phosphorylated motifs. To further validate that Akt phosphorylates EGFR, we overexpressed either a constitutively active (HA-myr-Akt) or a kinase-dead mutant of Akt (HA-DN-Akt) and showed that PAS-EGFR was enhanced only under Akt activation (lane 2 versus lane 3, Fig. 2B). The increased interaction between EGFR and Akt in response to EGF stimulation as indicated by IP/Western blot (WB) analysis (Fig. 2C) further supports our hypothesis that Akt binds to and phosphorylates EGFR.
FIGURE 2.
Akt phosphorylates EGFR at Ser-229. A, whole cell lysates prepared from cells were subjected to IP/WB analysis by using indicated antibodies. PAS, anti-phospho-Akt-substrate antibody. B, HEK293 cells were transfected with the indicated constructs and then subjected to IP/WB analysis. C, MDA-MB-468 cells were treated with EGF for indicated times, and IP/WB analysis was performed to assess the physical interaction between Akt and EGFR. D, in vivo EGFR Ser-229 phosphorylation was detected in anti-EGFR immunoprecipitates from EGF-treated MDA-MB-468 cells by mass spectrometry. E and F, substitution of Ser-229 to Ala abolished EGF- or Akt-induced EGFR phosphorylation detected by anti-PAS antibody in anti-EGFR (E) or anti-myc (F) immunoprecipitates from transfected HEK293 cells. G, endogenous EGFR Ser-229 phosphorylation was detected in A431/GR cells by using anti-phospho-EGFR Ser-229 for IP and anti-EGFR antibody for subsequent WB. H, Akt expression in A431/GR cells was deprived by Akt siRNA. Then endogenous EGFR Ser-229 phosphorylation was detected by using anti-phospho-EGFR Ser-229 for IP and anti-EGFR antibody for subsequent WB. N.C., non-specific control.
Next, we wanted to determine the specific residue(s) of EGFR that is phosphorylated by Akt. Mass spectrometry analysis of the anti-EGFR immunoprecipitates showed several Ser/Thr sites of EGFR that were phosphorylated in the EGF-treated MDA-MB-468 breast cancer cells (data not shown). Among these phosphorylation sites, only Ser-229 phosphorylation fits the consensus site for Akt substrate when aligned with other well established Akt substrates (Fig. 2D) (34–42). Substitution of Ser-229 to Ala also blocked EGF-induced or active Akt-mediated PAS-EGFR (Figs. 2, E and F, respectively). Direct phosphorylation of EGFR at Ser-229 by Akt was further demonstrated by in vitro kinase assay (supplemental Fig. S1E). To specifically detect phosphorylation status of EGFR at Ser-229 in A431/GR cells in vivo, we generated and characterized a p-229 antibody that recognizes phosphorylated EGFR at Ser-229. As shown in supplemental Fig. S1F, the p-229 antibody selectively immunoprecipitated wtEGFR (lanes 5 and 6) in the presence of active Akt but was unable to pull down the S229A mutant (lane 7). The immunoprecipitates can be reduced by phospho-peptide but not by non-phospho-peptide (lanes 1 and 2), suggesting that this antibody more preferentially recognizes the natural form of Ser-229-phosphorylated EGFR. The antibody does not recognize the denatured form of the phosphorylated EGFR by WB analysis. As expected, phosphorylation of EGFR at Ser-229 was detected primarily in A431/GR but not in A431 cells by IP/WB analysis (Fig. 2G). Importantly, EGFR Ser-229 phosphorylation was blocked when Akt expression was knocked down by siRNA in A431/GR cells (Fig. 2H), demonstrating that EGFR is a substrate of Akt phosphorylation at Ser-229.
Phosphorylation of EGFR at Ser-229 by Akt Is Critical for EGFR Nuclear Translocation and Gefitinib Resistance
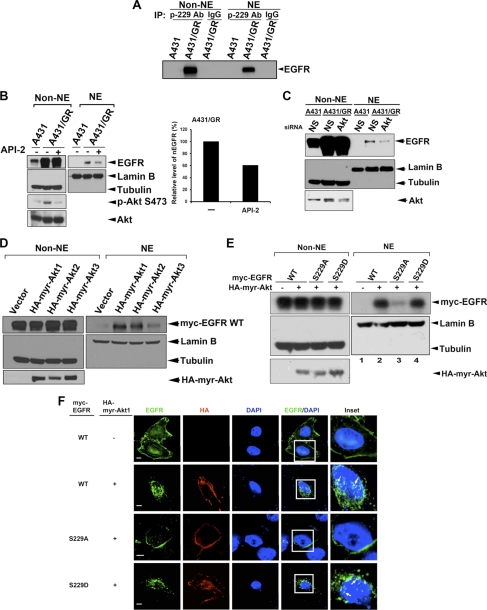
From the above results, we found that Akt could phosphorylate EGFR at Ser-229. We then determined if EGFR phosphorylation by Akt regulates EGFR nuclear translocation. We looked at the phosphorylation status in nuclear and cytoplasmic fractions of A431/GR cells and detected Ser-229 phosphorylation of EGFR in both nuclear and cytoplasmic fractions (Fig. 3A), suggesting that phosphorylation at this site of EGFR might be one possible mechanism for nuclear accumulation of EGFR. Indeed, targeting Akt by pharmacological inhibitor API-2 (Fig. 3B) or by Akt1/2/3 siRNA (Fig. 3C) attenuated the nuclear transport of EGFR in A431/GR cells. Similarly, EGF-induced nuclear import of EGFR was reduced by PI3K inhibitor LY294002 (supplemental Fig. S2A). Ectopic expression of PTEN, a negative regulator of PI3K/Akt activation, also attenuated EGFR nuclear transport (supplemental Fig. S2B). In contrast, when we overexpressed constitutively active Akt1 (HA-myr-Akt1), the nuclear level of EGFR was increased in a dose-dependent manner. This effect was not seen in cells that expressed HA-DN-Akt1 (supplemental Fig. S2C).
FIGURE 3.
Phosphorylation by Akt increases EGFR nuclear translocation. A, endogenous EGFR Ser-229 phosphorylation in both cytoplasm (Non-NE) and nucleus (NE) of A431/GR cells was detected by using anti-phospho-EGFR Ser-229 for IP and subsequently anti-EGFR antibody for WB. B, Western blot analysis and quantification of nuclear EGFR expression in A431 and A431/GR cells treated with or without the Akt inhibitor, API-2, is shown. C, shown is a Western blot analysis of nuclear EGFR expression in A431 and A431/GR cells transfected with control (N.S., non-specific) or Akt siRNA. D, shown is WB analysis of nuclear EGFR expression in HEK293 cells transfected with three Akt isoforms individually. E, shown is Western blot analysis of nuclear expressions of EGFR-WT and EGFR-Ser-229 mutants in HEK293 cells co-transfected with or without HA-myr-Akt1. F, nuclear localization of EGFR-WT and EGFR-Ser-229 mutants in HeLa cells co-transfected with or without HA-myr-Akt1 was examined by confocal microscopy. Bar, 5 μm.
Because the Akt family is composed of three members, we also examined the effect of other two isoforms of Akt on the nuclear transport of EGFR. As shown in Fig. 3D, all three isoforms of Akt were able to increase the nuclear accumulation of EGFR, although the effect of Akt3 was less compared with Akt1 or Akt2, suggesting that the nuclear translocation of EGFR is concordantly regulated by all three Akt isoforms and that Akt1 and Akt2 do so more dominantly. Consistently, when the individual Akt isoforms were silenced by siRNA, only Akt1 and Akt2 siRNA significantly attenuated the EGF-induced nuclear translocation of EGFR (supplemental Fig. S2D). Furthermore, substitution of Ser-229 to Ala significantly attenuated Akt-mediated EGFR nuclear accumulation (lane 2 versus lane 3, Fig. 3E). In contrast, when Ser-229 was substituted by glutamic acid to mimic the phosphorylated status, the Akt-mediated nuclear import of EGFR S229D mutant was comparable with that of wtEGFR (lane 2 versus lane 4, Fig. 3E). These effects were further confirmed by confocal microscope analysis (Fig. 3F). Our findings indicate that phosphorylation of Ser-229 by Akt enhanced the nuclear import of EGFR.
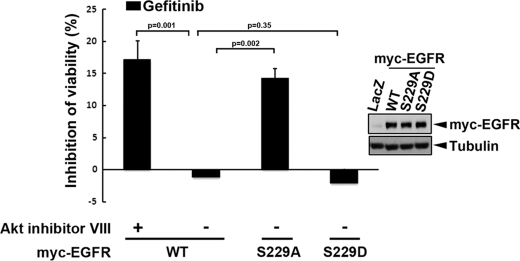
After we showed that Ser-229 phosphorylation by Akt is essential for regulating nuclear translocation of EGFR, we next examined whether this phosphorylation is critical for the development of gefitinib resistance. We expressed EGFR S229A and S229D mutants, mimicking the unphosphorylated and phosphorylated forms of EGFR, respectively, in A431/GR cells to determine the role of this phosphorylation in gefitinib resistance. As shown in Fig. 4, EGFR S229A is more sensitive than EGFR S229D mutant to gefitinib-mediated growth suppression. Likewise, Akt inhibitor also rendered wtEGFR more sensitive to gefitinib. Together, these findings indicate that Akt mediates EGFR trafficking to the nucleus by phosphorylating EGFR at Ser-229 in response to EGF stimulation and likely plays an important role in gefitinib resistance.
FIGURE 4.
Phosphorylation of EGFR at Ser-229 by Akt plays a role in the development of gefitinib resistance. The cytostatic effect of gefitinib on the A431/GR cells expressing adenoviral-derived EGFR was measured by MTT assay. Error bars denote S.E. (n = 3).
Nuclear EGFR Regulates BCRP/ABCG2 Expression in A431/GR Cells
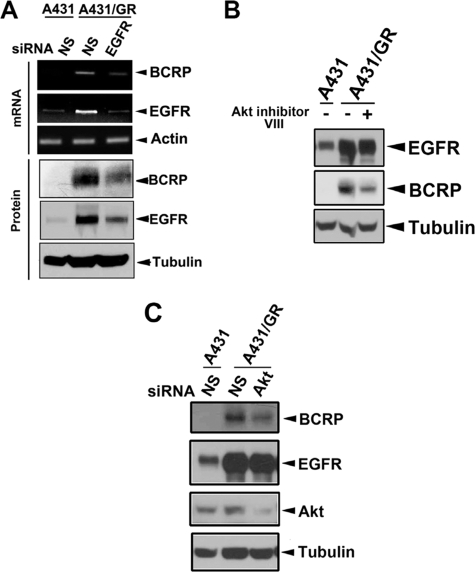
BCRP/ABCG2, a well known ATP binding cassette (ABC) transporter, has been shown to mediate chemoresistance by pumping out anti-cancer drugs, such as doxorubicin (22, 43). In fact, several studies have shown that gefitinib is also a BCRP/ABCG2 substrate (44–46), and stably enforced BCRP/ABCG2 expression in A431 cells conferred gefitinib resistance (47). Furthermore, as mentioned above, a case report showed that a wtEGFR-expressing NSCLC patient developed acquired gefitinib resistance without any identifiable EGFR mutations. Rather, BCRP/ABCG2 expression was detected in the recurrent tumor of this patient (18). Therefore, this raises a possibility that Akt-dependent nuclear translocation of EGFR might contribute to acquired gefitinib resistance through regulation of BCRP/ABCG2 expression in wtEGFR-expressing cancer cells. As shown in Fig. 5A, both mRNA and protein levels of BCRP/ABCG2 were increased in A431/GR cells compared with those in parental A431 cells. However, the increase in mRNA and protein level can be attenuated when EGFR expression was down-regulated by siRNA. Moreover, reduction of nEGFR levels by silencing importin β1, an essential regulator for its nuclear trafficking, or by substituting the EGFR nuclear localization signal (48) also decreased the protein level of BCRP/ABCG2 (supplemental Fig. S3, A and B, respectively), suggesting that nEGFR is important in regulating BCRP/ABCG2 expression. Because we showed earlier that Akt activates EGFR nuclear translocation, we hypothesized that inhibition of Akt in A431/GR cells would reduce nuclear localization of EGFR, which would in turn block EGFR-mediated BCRP/ABCG2 expression. Indeed, when we added a pharmacological Akt inhibitor VIII (Fig. 5B) or silenced Akt expression by siRNA (Fig. 5C), we found that BCRP/ABCG2 expression was also decreased, supporting that EGFR-mediated BCRP/ABCG2 expression requires activation of EGFR phosphorylation at Ser-229 by Akt.
FIGURE 5.
Nuclear EGFR regulates BCRP/ABCG2 expression in A431/GR cells. A, mRNA and protein expressions of BCRP/ABCG2 in A431 and A431/GR cells transfected with control or EGFR siRNA were analyzed by RT-PCR and WB, respectively. B and C, effects of Akt inhibitor VIII (B) and Akt siRNA (C) on the BCRP/ABCG2 protein expression in A431/GR cells were examined by WB. N.S., non-specific siRNA.
Nuclear EGFR Regulates BCRP/ABCG2 Expression Transcriptionally in A431/GR Cells
Next, we examined the regulatory mechanisms of BCRP/ABCG2 gene expression by nEGFR in A431/GR cells. As a transcription factor, nEGFR complex is known to target the AT-rich minimal consensus sequences (ATRSs) (49). BCRP/ABCG2 promoter contains multiple ATRSs as putative EGFR-targeting sequences (Fig. 6A). We asked if nEGFR might also target these sites to mediate BCRP/ABCG2 expression in A431/GR cells. To further address this issue, we performed DNA affinity precipitation assay to examine the association of nEGFR with the BCRP/ABCG2 promoter. Three different biotinylated probes containing three various regions in BCRP/ABCG2 promoter as illustrated in Fig. 6B were used to pull down the promoter-associated EGFR. As predicted, we observed positive binding activity of nEGFR to BCRP/ABCG2 promoter in A431/GR with AT1 but not with non-AT probe (Fig. 6C). Furthermore, the binding activity of nEGFR to the AT2 probe, which contains several more ATRSs within the BCRP/ABCG2 promoter, was similar to the AT1 probe (supplemental Fig. S3C), suggesting that the AT1 region (from −637 to −365 bp) may be sufficient for the association of nEGFR with the BCRP/ABCG2 promoter. This nEGFR binding activity to BCRP/ABCG2 promoter in vivo was also observed by ChIP analysis (Fig. 6D). To further support the transcriptional regulation of BCRP/ABCG2 by nEGFR, we performed BCRP/ABCG2 promoter luciferase reporter assays and found that BCRP/ABCG2 promoter activity was higher in A431/GR cells compared with A431 parental cells (Fig. 6E). Moreover, transfection of wtEGFR and its S229D mutant enhanced BCRP/ABCG2 promoter activity in A431/GR cells, whereas transfection of EGFR S229A mutant did not significantly affect the promoter activity (Fig. 6F). Similarly, BCRP/ABCG2 protein expression was also increased by EGFR S229D but not by S229A mutant in HEK293 cells (Fig. 6G). These results provide strong evidence linking BCRP/ABCG2 up-regulation to Akt-mediated Ser-229 phosphorylation of nEGFR in gefitinib-resistant cells.
FIGURE 6.
Nuclear EGFR enhances transcriptional activation of BCRP/ABCG2 in A431/GR cells via recruitment to the BCRP/ABCG2 promoter. A, the DNA binding consensus sites of EGFR in BCRP/ABCG2 promoter are shown. B, three probes for DNA affinity precipitation assay were designed according to the sequence of human BCRP/ABCG2 promoter. C, the binding of EGFR to BCRP/ABCG2 promoter was analyzed by DNA affinity precipitation assay (left). The inputs of probes were shown in at the right. D, the binding of EGFR to BCRP/ABCG2 promoter was analyzed by chromatin immunoprecipitation (ChIP). E, transcriptional activities of BCRP/ABCG2 in A431 and A431/GR cells were analyzed by luciferase reporter assay. F, effects of EGFR Ser-229 mutations on EGFR-induced BCRP/ABCG2 promoter activity were analyzed in A431/GR cell by luciferase reporter assay. Error bars in E and F denote S.E. (n = 3). G, protein expression of BCRP/ABCG2 in HEK293 cells co-transfected with EGFR Ser-229 mutants. H, the proposed model for the mechanism underlying nuclear EGFR-mediated BCRP/ABCG2 expression conferring gefitinib resistance.
DISCUSSION
EGFR nuclear localization is induced rapidly and transiently within 2 h of EGF stimulation (48). The coat protein complex I (COPI)-mediated retrograde trafficking from the Golgi to the ER has been shown to regulate EGF-induced EGFR nuclear transport (50). Unlike the transient nuclear localization by EGF stimulation, EGFR is steadily present in the nucleus under conditions such as chemoresistance (51), radioresistance (26), or cetuximab insensitivity (27, 52), which all share a common mechanism of resistance that is mediated by elevated or continuous activation of Akt survival signaling (53). Although EGFR is suppressed by gefitinib, the compensatory and continuous activation of PI3K/Akt by enhancing insulin-like growth factor receptor signaling has also been shown to contribute to the acquired gefitinib resistance in wtEGFR-expressing cancer cells (16). Specifically, increased activity of insulin-like growth factor receptor through down-regulation of insulin-like growth factor-binding proteins maintains PI3K/Akt-mediated survival signaling in response to acquired gefitinib resistance in gefitinib-sensitive and wtEGFR-expressing cancer cells (16, 17).
In the current study, we identified EGFR Ser-229 as a novel Akt substrate and demonstrated that this phosphorylation is required for EGFR nuclear translocation, which plays a role in the development of acquired gefitinib resistance. These findings suggest that Ser-229 phosphorylation by continuously activated Akt may function as a common mechanism to regulate EGFR nuclear transport and likely contribute to resistance to chemotherapy, radiotherapy, cetuximab, and gefitinib. Interestingly, whereas nuclear accumulation of EGFR in response to both cetuximab (27) and gefitinib resistance is observed in the wtEGFR-expressing cell lines, changes in the level of nEGFR was not observed in EGFR mutant-expressing cell lines in response to acquired gefitinib resistance and irradiation (supplemental Fig. S4) (26, 54).
Nuclear EGFR has been implicated in DNA repair through its interaction with DNA-proliferating cell nuclear antigen (23, 55) and DNA-dependent protein kinase (26) in the resistance to cisplatin treatment (51) and ionizing radiation (26). An increased level of nEGFR has been proposed to provide survival signals through induction of cyclin D1, proliferating cell nuclear antigen, and B-myb expressions in cetuximab-resistant cells (27). Here, we demonstrate that nEGFR targets BCRP/ABCG2 promoter, enhances its expression transcriptionally, and contributes to gefitinib resistance (a proposed model shown in Fig. 6H). Although nEGFR can function as a transcription regulator by targeting ATRSs (49), it does not contain a DNA binding domain, and thus, it likely targets BCRP/ABCG2 promoter indirectly by interacting with transcription factors. Nuclear EGFR has been demonstrated to interact with STAT3, STAT5, E2F1, and RHA to regulate gene expressions (21, 56–58, 60). The human BCRP/ABCG2 promoter contains several potential binding sites for STAT5 and E2F1 that overlap with the ATRSs within the AT1 regions, suggesting that nEGFR might be recruited to BCRP/ABCG2 promoter through interaction with STAT5 or E2F1.
In response to EGF stimulation, we also found that activated Akt is able to induce the EGFR phosphorylation at Ser-229 and subsequently promotes EGFR nuclear translocation to activate BCRP/ABCG2 expression in A431/GR but not in A431 cells. In addition, transient transfection of A431 cells with wtEGFR or its phosphorylation-mimicking S229D mutant did not induce BCRP/ABCG2 promoter activity (data not shown). Overexpression of EGFR S229D mutant only slightly increased the BCRP/ABCG2 level in HEK293 cells (Fig. 6G). These data suggest that transient Akt and nuclear EGFR activities may not be sufficient to induce BCRP/ABCG2 gene expression and that other mechanisms elicited by chronic gefitinib treatment remain to be explored. It is worthwhile to note that Akt has been reported to regulate BCRP/ABCG2 activity via enhancing its cell-surface display (61, 62), which could indicate that continuously activated Akt in A431/GR cells likely increases BCRP/ABCG2 activity through up-regulation of its transport to the plasma membrane in addition to the nEGFR-mediated gene expression.
BCRP/ABCG2 is a well recognized determinant for various types of chemoresistance (22, 43). Several studies have demonstrated that gefitinib is also a substrate of BCRP/ABCG2 at low concentrations (44–46). Stable transfection of A431 with BCRP/ABCG2 cDNA resulted in insensitivity of cells to gefitinib (47). BCRP/ABCG2 expression by immunohistochemical staining was detected in 46% of treatment-naive NSCLC patients (59). Our current study further indicated that chronic treatment with gefitinib induced BCRP/ABCG2 expression through the Akt/nEGFR pathway, leading to the acquired gefitinib resistance. Consistent with our findings, Usuda et al. (18) observed an elevated BCRP/ABCG2 expression level in a NSCLC patient with wtEGFR expression who received gefitinib therapy. Moreover, our unpublished results also indicated that blockage of BCRP/ABCG2 activity can re-sensitize A431/GR cells to gefitinib in vitro and potentiate the therapeutic effects of gefitinib in the A431/GR xenograft mice. It would be of interest to determine the clinical implication of BCRP/ABCG2 expression in tumor tissue samples from patients with wtEGFR expressing NSCLC.
In summary, our findings demonstrate that continuously activated Akt, in addition to delivering the survival signals in gefitinib-resistant cancer cells with wtEGFR expression, also phosphorylates EGFR and facilitates its nuclear transport to mediate BCRP/ABCG2 expression. Although further investigations would be required to demonstrate the clinical relevance, the link between BCRP/ABCG2 expression and nEGFR could serve as a predictor for gefitinib sensitivity, and targeting BCRP/ABCG2 may have important implications for the treatment of wtEGFR-expressing cancer types with gefitinib.
Supplementary Material
Acknowledgments
We are grateful to Dr. Carlos L. Arteaga for providing the A431 and A431/GR cell lines and Dr. Jennifer L. Hsu for reviewing and editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants PO1 CA099031 and RO1 CA109311. This work was supported by the following grants: National Institutes of Health (NIH PO1 CA099031 and NIH RO1 CA109311), National Breast Cancer Foundation, Inc., Cancer Center Research of Excellence DOH-99-TDC-111-05 (to M.-C. H.), NSC-2632-B-039-001-MY3 (to M.-C. H.), NSC-3111-B-039-002 (to M.-C. H. and L.-Y. L.), NSC-97-2320-B-039-033-MY3 (to W.-C. H.), NSC-98-3112-B-039-002 (to W.-C. H.), NSC-99-3112-B-039-002 (to W.-C. H.), NSC-96-2917-I-006-004 (to Y.-J. C.), NHRI-EX-98-9812BC (to W.-C. H.), NHRI-EX98-9603BC (to L.-Y. L.), DOH97-TD-G-111-027 (to M.-C. H.), and The University of Texas M.D. Anderson-China Medical University and Hospital Sister Institution Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- EGFR
- EGF receptor
- nEGFR
- nuclear EGFR
- NSCLC
- non-small cell lung cancer
- BCRP
- breast cancer-resistant protein
- ABCG2
- ATP binding cassette subfamily G member 2
- PAS
- phosphorylated Akt substrate
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5 -diphenyltetrazolium bromide
- ABC
- ATP binding cassette
- ATRSs
- AT-rich minimal consensus sequences
- WB
- Western blot
- IP
- immunoprecipitate.
REFERENCES
- 1. Hynes N. E., Lane H. A. (2005) Nat. Rev. Cancer 5, 341–354 [DOI] [PubMed] [Google Scholar]
- 2. Lynch T. J., Bell D. W., Sordella R., Gurubhagavatula S., Okimoto R. A., Brannigan B. W., Harris P. L., Haserlat S. M., Supko J. G., Haluska F. G., Louis D. N., Christiani D. C., Settleman J., Haber D. A. (2004) N. Engl. J. Med. 350, 2129–2139 [DOI] [PubMed] [Google Scholar]
- 3. Paez J. G., Jänne P. A., Lee J. C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F. J., Lindeman N., Boggon T. J., Naoki K., Sasaki H., Fujii Y., Eck M. J., Sellers W. R., Johnson B. E., Meyerson M. (2004) Science 304, 1497–1500 [DOI] [PubMed] [Google Scholar]
- 4. Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., Singh B., Heelan R., Rusch V., Fulton L., Mardis E., Kupfer D., Wilson R., Kris M., Varmus H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13306–13311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balak M. N., Gong Y., Riely G. J., Somwar R., Li A. R., Zakowski M. F., Chiang A., Yang G., Ouerfelli O., Kris M. G., Ladanyi M., Miller V. A., Pao W. (2006) Clin. Cancer Res. 12, 6494–6501 [DOI] [PubMed] [Google Scholar]
- 6. Pao W., Miller V. A., Politi K. A., Riely G. J., Somwar R., Zakowski M. F., Kris M. G., Varmus H. (2005) PLoS. Med. 2, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Engelman J. A., Zejnullahu K., Mitsudomi T., Song Y., Hyland C., Park J. O., Lindeman N., Gale C. M., Zhao X., Christensen J., Kosaka T., Holmes A. J., Rogers A. M., Cappuzzo F., Mok T., Lee C., Johnson B. E., Cantley L. C., Jänne P. A. (2007) Science 316, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 8. Engelman J. A., Jänne P. A. (2008) Clin. Cancer Res. 14, 2895–2899 [DOI] [PubMed] [Google Scholar]
- 9. Tsao M. S., Sakurada A., Cutz J. C., Zhu C. Q., Kamel-Reid S., Squire J., Lorimer I., Zhang T., Liu N., Daneshmand M., Marrano P., da Cunha, Santos G., Lagarde A., Richardson F., Seymour L., Whitehead M., Ding K., Pater J., Shepherd F. A. (2005) N. Engl. J. Med. 353, 133–144 [DOI] [PubMed] [Google Scholar]
- 10. Cappuzzo F., Hirsch F. R., Rossi E., Bartolini S., Ceresoli G. L., Bemis L., Haney J., Witta S., Danenberg K., Domenichini I., Ludovini V., Magrini E., Gregorc V., Doglioni C., Sidoni A., Tonato M., Franklin W. A., Crino L., Bunn P. A., Jr., Varella-Garcia M. (2005) J. Natl. Cancer. Inst. 97, 643–655 [DOI] [PubMed] [Google Scholar]
- 11. Bell D. W., Lynch T. J., Haserlat S. M., Harris P. L., Okimoto R. A., Brannigan B. W., Sgroi D. C., Muir B., Riemenschneider M. J., Iacona R. B., Krebs A. D., Johnson D. H., Giaccone G., Herbst R. S., Manegold C., Fukuoka M., Kris M. G., Baselga J., Ochs J. S., Haber D. A. (2005) J. Clin. Oncol. 23, 8081–8092 [DOI] [PubMed] [Google Scholar]
- 12. Takano T., Ohe Y., Sakamoto H., Tsuta K., Matsuno Y., Tateishi U., Yamamoto S., Nokihara H., Yamamoto N., Sekine I., Kunitoh H., Shibata T., Sakiyama T., Yoshida T., Tamura T. (2005) J. Clin. Oncol. 23, 6829–6837 [DOI] [PubMed] [Google Scholar]
- 13. Nishimura Y., Yoshioka K., Bereczky B., Itoh K. (2008) Mol. Cancer 7, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim E. S., Hirsh V., Mok T., Socinski M. A., Gervais R., Wu Y. L., Li L. Y., Watkins C. L., Sellers M. V., Lowe E. S., Sun Y., Liao M. L., Osterlind K., Reck M., Armour A. A., Shepherd F. A., Lippman S. M., Douillard J. Y. (2008) Lancet 372, 1809–1818 [DOI] [PubMed] [Google Scholar]
- 15. Huang S. F., Liu H. P., Li L. H., Ku Y. C., Fu Y. N., Tsai H. Y., Chen Y. T., Lin Y. F., Chang W. C., Kuo H. P., Wu Y. C., Chen Y. R., Tsai S. F. (2004) Clin. Cancer Res. 10, 8195–8203 [DOI] [PubMed] [Google Scholar]
- 16. Guix M., Faber A. C., Wang S. E., Olivares M. G., Song Y., Qu S., Rinehart C., Seidel B., Yee D., Arteaga C. L., Engelman J. A. (2008) J. Clin. Invest. 118, 2609–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cappuzzo F., Toschi L., Tallini G., Ceresoli G. L., Domenichini I., Bartolini S., Finocchiaro G., Magrini E., Metro G., Cancellieri A., Trisolini R., Crino L., Bunn P. A., Jr., Santoro A., Franklin W. A., Varella-Garcia M., Hirsch F. R. (2006) Ann. Oncol. 17, 1120–1127 [DOI] [PubMed] [Google Scholar]
- 18. Usuda J., Ohira T., Suga Y., Oikawa T., Ichinose S., Inoue T., Ohtani K., Maehara S., Imai K., Kubota M., Tsunoda Y., Tsutsui H., Furukawa K., Okunaka T., Sugimoto Y., Kato H. (2007) Lung Cancer 58, 296–299 [DOI] [PubMed] [Google Scholar]
- 19. Xie Y., Hung M. C. (1994) Biochem. Biophys. Res. Commun. 203, 1589–1598 [DOI] [PubMed] [Google Scholar]
- 20. Wang S. C., Lien H. C., Xia W., Chen I. F., Lo H. W., Wang Z., Ali-Seyed M., Lee D. F., Bartholomeusz G., Ou-Yang F., Giri D. K., Hung M. C. (2004) Cancer Cell 6, 251–261 [DOI] [PubMed] [Google Scholar]
- 21. Lo H. W., Hsu S. C., Ali-Seyed M., Gunduz M., Xia W., Wei Y., Bartholomeusz G., Shih J. Y., Hung M. C. (2005) Cancer Cell 7, 575–589 [DOI] [PubMed] [Google Scholar]
- 22. Kuo M. T. (2007) Adv. Exp. Med. Biol. 608, 23–30 [DOI] [PubMed] [Google Scholar]
- 23. Wang S. C., Nakajima Y., Yu Y. L., Xia W., Chen C. T., Yang C. C., McIntush E. W., Li L. Y., Hawke D. H., Kobayashi R., Hung M. C. (2006) Nat. Cell Biol. 8, 1359–1368 [DOI] [PubMed] [Google Scholar]
- 24. Dittmann K., Mayer C., Kehlbach R., Rodemann H. P. (2008) Radiother. Oncol. 86, 375–382 [DOI] [PubMed] [Google Scholar]
- 25. Dittmann K., Mayer C., Kehlbach R., Rodemann H. P. (2008) Mol. Cancer 7, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dittmann K., Mayer C., Fehrenbacher B., Schaller M., Raju U., Milas L., Chen D. J., Kehlbach R., Rodemann H. P. (2005) J. Biol. Chem. 280, 31182–31189 [DOI] [PubMed] [Google Scholar]
- 27. Li C., Iida M., Dunn E. F., Ghia A. J., Wheeler D. L. (2009) Oncogene 28, 3801–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kwak E. L., Sordella R., Bell D. W., Godin-Heymann N., Okimoto R. A., Brannigan B. W., Harris P. L., Driscoll D. R., Fidias P., Lynch T. J., Rabindran S. K., McGinnis J. P., Wissner A., Sharma S. V., Isselbacher K. J., Settleman J., Haber D. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 7665–7670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moasser M. M., Basso A., Averbuch S. D., Rosen N. (2001) Cancer Res. 61, 7184–7188 [PubMed] [Google Scholar]
- 30. Anderson N. G., Ahmad T., Chan K., Dobson R., Bundred N. J. (2001) Int. J. Cancer 94, 774–782 [DOI] [PubMed] [Google Scholar]
- 31. Sordella R., Bell D. W., Haber D. A., Settleman J. (2004) Science 305, 1163–1167 [DOI] [PubMed] [Google Scholar]
- 32. Engelman J. A., Jänne P. A., Mermel C., Pearlberg J., Mukohara T., Fleet C., Cichowski K., Johnson B. E., Cantley L. C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3788–3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arteaga C. L. (2007) Nat. Med. 13, 675–677 [DOI] [PubMed] [Google Scholar]
- 34. Zhou B. P., Liao Y., Xia W., Zou Y., Spohn B., Hung M. C. (2001) Nat. Cell Biol. 3, 973–982 [DOI] [PubMed] [Google Scholar]
- 35. Michell B. J., Harris M. B., Chen Z. P., Ju H., Venema V. J., Blackstone M. A., Huang W., Venema R. C., Kemp B. E. (2002) J. Biol. Chem. 277, 42344–42351 [DOI] [PubMed] [Google Scholar]
- 36. Maddika S., Ande S. R., Wiechec E., Hansen L. L., Wesselborg S., Los M. (2008) J. Cell Sci. 121, 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Y., Dowbenko D., Lasky L. A. (2002) J. Biol. Chem. 277, 11352–11361 [DOI] [PubMed] [Google Scholar]
- 38. Huang W. C., Chen C. C. (2005) Mol. Cell. Biol. 25, 6592–6602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Feng J., Tamaskovic R., Yang Z., Brazil D. P., Merlo A., Hess D., Hemmings B. A. (2004) J. Biol. Chem. 279, 35510–35517 [DOI] [PubMed] [Google Scholar]
- 40. Chou C. K., Lee D. F., Sun H. L., Li L. Y., Lin C. Y., Huang W. C., Hsu J. M., Kuo H. P., Yamaguchi H., Wang Y. N., Liu M., Wu H. Y., Liao P. C., Yen C. J., Hung M. C. (2009) Mol. Carcinog. 48, 1048–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cha T. L., Zhou B. P., Xia W., Wu Y., Yang C. C., Chen C. T., Ping B., Otte A. P., Hung M. C. (2005) Science 310, 306–310 [DOI] [PubMed] [Google Scholar]
- 42. Brenet F., Socci N. D., Sonenberg N., Holland E. C. (2009) Oncogene 28, 128–139 [DOI] [PubMed] [Google Scholar]
- 43. Takara K., Sakaeda T., Okumura K. (2006) Curr. Pharm. Des. 12, 273–286 [DOI] [PubMed] [Google Scholar]
- 44. Shi Z., Peng X. X., Kim I. W., Shukla S., Si Q. S., Robey R. W., Bates S. E., Shen T., Ashby C. R., Jr., Fu L. W., Ambudkar S. V., Chen Z. S. (2007) Cancer Res. 67, 11012–11020 [DOI] [PubMed] [Google Scholar]
- 45. Shi Z., Parmar S., Peng X. X., Shen T., Robey R. W., Bates S. E., Fu L. W., Shao Y., Chen Y. M., Zang F., Chen Z. S. (2009) Oncol. Rep. 21, 483–489 [PMC free article] [PubMed] [Google Scholar]
- 46. Nakamura Y., Oka M., Soda H., Shiozawa K., Yoshikawa M., Itoh A., Ikegami Y., Tsurutani J., Nakatomi K., Kitazaki T., Doi S., Yoshida H., Kohno S. (2005) Cancer Res. 65, 1541–1546 [DOI] [PubMed] [Google Scholar]
- 47. Sugimoto Y., Tsukahara S., Ishikawa E., Mitsuhashi J. (2005) Cancer Sci. 96, 457–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hsu S. C., Hung M. C. (2007) J. Biol. Chem. 282, 10432–10440 [DOI] [PubMed] [Google Scholar]
- 49. Lin S. Y., Makino K., Xia W., Matin A., Wen Y., Kwong K. Y., Bourguignon L., Hung M. C. (2001) Nat. Cell Biol. 3, 802–808 [DOI] [PubMed] [Google Scholar]
- 50. Wang Y. N., Wang H., Yamaguchi H., Lee H. J., Lee H. H., Hung M. C. (2010) Biochem. Biophys. Res. Commun. 399, 498–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hsu S. C., Miller S. A., Wang Y., Hung M. C. (2009) Am. J. Transl. Res. 1, 249–258 [PMC free article] [PubMed] [Google Scholar]
- 52. Wheeler D. L., Huang S., Kruser T. J., Nechrebecki M. M., Armstrong E. A., Benavente S., Gondi V., Hsu K. T., Harari P. M. (2008) Oncogene 27, 3944–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang W. C., Hung M. C. (2009) J. Formos. Med. Assoc. 108, 180–194 [DOI] [PubMed] [Google Scholar]
- 54. Das A. K., Chen B. P., Story M. D., Sato M., Minna J. D., Chen D. J., Nirodi C. S. (2007) Cancer Res. 67, 5267–5274 [DOI] [PubMed] [Google Scholar]
- 55. Wang S. C., Hung M. C. (2009) Clin. Cancer Res. 15, 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hanada N., Lo H. W., Day C. P., Pan Y., Nakajima Y., Hung M. C. (2006) Mol. Carcinog. 45, 10–17 [DOI] [PubMed] [Google Scholar]
- 57. Hung L. Y., Tseng J. T., Lee Y. C., Xia W., Wang Y. N., Wu M. L., Chuang Y. H., Lai C. H., Chang W. C. (2008) Nucleic Acids. Res. 36, 4337–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lo H. W., Cao X., Zhu H., Ali-Osman F. (2010) Mol. Cancer Res. 8, 232–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yoh K., Ishii G., Yokose T., Minegishi Y., Tsuta K., Goto K., Nishiwaki Y., Kodama T., Suga M., Ochiai A. (2004) Clin. Cancer Res. 10, 1691–1697 [DOI] [PubMed] [Google Scholar]
- 60. Huo L., Wang Y. N., Xia W., Hsu S. C., Lai C. C., Li L. Y., Chang W. C., Wang Y., Hsu M. C., Yu Y. L., Huang T. H., Ding Q., Chen C. H., Tsai C. H., Hung M. C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 16125–16130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Takada T., Suzuki H., Gotoh Y., Sugiyama Y. (2005) Drug Metab. Dispos. 33, 905–909 [DOI] [PubMed] [Google Scholar]
- 62. Bleau A. M., Hambardzumyan D., Ozawa T., Fomchenko E. I., Huse J. T., Brennan C. W., Holland E. C. (2009) Cell Stem Cell 4, 226–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.