Abstract
Familial encephalopathy with neuroserpin inclusion bodies is a neurodegenerative disorder characterized by the accumulation of neuroserpin polymers in the endoplasmic reticulum (ER) of cortical and subcortical neurons in the CNS because of neuroserpin point mutations. ER-associated degradation (ERAD) is involved in mutant neuroserpin degradation. In this study, we demonstrate that two ER-associated E3 ligases, Hrd1 and gp78, are involved in the ubiquitination and degradation of mutant neuroserpin. Overexpression of Hrd1 and gp78 decreases the mutant neuroserpin protein level, whereas Hrd1 and gp78 knockdown increases mutant neuroserpin stability. Moreover, ERAD impairment by mutant valosin-containing protein increases the mutant neuroserpin protein level and aggregate formation. Thus, these findings identify mutant neuroserpin as an ERAD target and show that Hrd1 and gp78 mediate mutant neuroserpin turnover through the ERAD pathway.
Keywords: Neurochemistry, Neurodegeneration, Neurological Diseases, Proteasome, Protein Degradation, Ubiquitin Ligase
Introduction
Misfolding of specific proteins can lead to the formation of aggregates, which is associated with most neurodegenerative disorders. Protein aggregates are extracellular in Alzheimer's disease, cytosolic in Parkinson's disease and amyotrophic lateral sclerosis, both cytosolic and intranuclear in polyglutamine (polyQ)3 diseases, and localize to the endoplasmic reticulum (ER) in familial encephalopathy with neuroserpin inclusion bodies (FENIB), which is caused by neuroserpin mutations (1). Although the role of the protein aggregates in the pathogenesis of these diseases is still poorly understood, a common feature of these aggregates is that they are all marked by ubiquitin. Neurodegenerative protein aggregation is closely related to the ubiquitin-proteasome system, a major cellular turnover system (2). Mutant neuroserpin aggregates have been observed in brain and cultured cells (3, 4). Neuroserpin is a secretory glycoprotein (5) that is a member of the serine protease inhibitor serpin family, and it is mainly expressed in the CNS (6). Mutations in neuroserpin result in its misfolding and accumulation in the ER (3, 4, 7). To date, four mutants of neuroserpin, S49P, S52R, H338R, and G392E, have been identified in association with FENIB (8). In FENIB-diseased brains, G392E, the most disruptive mutant, forms more inclusion bodies and at an earlier age of onset (13 years) than the other neuroserpin mutants (9). Because the earlier onset of FENIB is strongly correlated with the level of intracellular neuroserpin accumulation, neuroserpin accumulation appears to play a central role in FENIB pathology.
Many accumulated proteins that are caused by mutations or ER stress are targeted by a protein quality control system, termed ER-associated degradation (ERAD). ERAD is a degradation system in which proteins that are misfolded in the ER are exported to the cytosol for proteasomal degradation (10, 11). ER-associated E3 ubiquitin ligases, together with the VCP-Ufd1-Npl4 complex, are key components of the ERAD machinery. During the ERAD process, selected proteins are ubiquitinated by E3 ubiquitin ligases and then retrotranslocated from the ER to the cytosol by the VCP-Ufd1-Npl4 complex for degradation (12). Hrd1 and gp78 are the two best-characterized mammalian ERAD E3 ubiquitin ligases and are responsible for ubiquitinating a subset of misfolded substrates (13–16). The interactions of VCP with Hrd1 and gp78 play a central role in ERAD (12, 17, 18). The proteasome inhibitors lactacystin and MG132 block neuroserpin degradation in the ER, suggesting that ERAD may be involved in neuroserpin degradation (19). However, the mechanism by which neuroserpin is regulated by ERAD and the E3(s) that is/are involved in its degradation are still largely unknown.
In this study, we show that the cellular turnover and aggregation of mutant neuroserpin are regulated by ERAD. Our results suggest that central factors in the ERAD machinery, such as AAA (ATPases associated with diverse cellular activities) ATPase, VCP/p97, and the ubiquitin ligases Hrd1 and gp78, are involved in the regulation of mutant neuroserpin aggregation and degradation.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
Full-length human neuroserpin cDNA was first amplified using PCR from a human fetal brain cDNA library (Clontech) using the primers 5′-GAAGATCTATATGGCTTTCCTTGGAC-3′ and 5′-CGGAATTCTTAAAGTTCTTCGAAATC-3′. The PCR product was subsequently inserted into the pEGFP-C2 (Clontech) vector at the BglII and EcoRI sites. FLAG-neuroserpin was constructed by subcloning the amplified PCR product using the primers 5′-CCCAAGCTTATGGCTTTCCTTGGAC-3′ and 5′-GCTCTAGAAAGTTCTTCGAAATC-3′ from the pEGFP-C2-neuroserpin construct and inserting it into the p3xFLAG-Myc-CMV-24 (Sigma) vector at the HindIII and XbaI sites. To create the HA-, EGFP-, and Myc-tagged neuroserpin constructs, we subcloned neuroserpin from p3xFLAG-Myc-CMV-24 into the pKH3-HA vector at the HindIII and XbaI sites or into the pEGFP-N3 or pCS2-MT-Myc vector at the HindIII and BamHI sites. Pathogenic neuroserpin mutants were generated by site-directed mutagenesis using the MutanBEST kit (Takara) with the following primers: S49P neuroserpin, 5′-CCTCCATTGAGTATTGCTC-3′ and 5′-GAAGAGAATATTTTCATC-3′; S52R neuroserpin, 5′-TTGCGTATTGCTCTTGCA-3′ and 5′-TGGAGAGAAGAGAATATT-3′; and G392E neuroserpin, 5′-GAACGAGTCATGCATCCTGAA-3′ and 5′-CATGAATAGAATTGTACCAGT-3′. FLAG-tagged parkin was constructed by subcloning full-length parkin cDNA from pGEX-5x-1-parkin and inserting it into p3xFLAG-Myc-CMV-24 at the BamHI and SalI sites.
Hrd1, gp78, the FLAG-ubiquitin constructs, pEGFP-Mito, and ER markers were described previously (20). pCMV-VCP-Myc (Myc-tagged VCP/p97) was obtained from Dr. Baoliang Song (Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, China), and pcDNA3.1-His-p97 WT and pcDNA3.1-His-p97 QQ (His-tagged VCP/p97) was obtained from Dr. Yihong Ye (National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health). Myc-His-CHIP (Myc-tagged CHIP), was obtained from Dr. Nihar Ranjan Jana (National Brain Research Centre, India).
Cell Culture, Transfection and Chemicals
Human embryonic kidney 293 (293) cells or mouse neuroblastoma (N2a) cells were cultured in DMEM (Invitrogen) containing 10% newborn calf serum (Invitrogen). Cells were transfected with either expression vectors or siRNAs using Lipofectamine 2000 reagent (Invitrogen) at ∼50% confluence in DMEM without serum. Double-stranded oligonucleotides against gp78 named si-gp78–1 and si-gp78–2 were described previously (20). Double-stranded oligonucleotides designed against nucleotides 175–195 of human Hrd1 cDNA or 883–903 of human VCP cDNA were synthesized by Shanghai GenePharma (Shanghai, China). A non-targeting oligonucleotide was used as a negative control. To establish each stable cell line expressing EGFP-tagged WT, S49P, S52R, or G392E neuroserpin, 293 cells were transfected with the EGFP vector containing the indicated neuroserpin variant, and individual clones stably expressing each of the neuroserpin variants were selected using 200 μg/ml G418 (Invitrogen). Cycloheximide (CHX), lactacystin, and pepstatin A were purchased from Sigma, and MG132 was purchased from Calbiochem.
Immunoblot Analysis and Antibodies
Cells were first lysed in a buffer containing 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.5% sodium deoxycholate, 1% Nonidet P-40, and protease inhibitors (Roche), and the proteins were then separated by 10 or 12% SDS-PAGE and transferred onto PVDF membranes (Millipore).
The following primary antibodies were used: mouse monoclonal antibodies against GFP, HA, Tom20, His, Myc, and ubiquitin, and rabbit polyclonal antibodies against GFP and HA (Santa Cruz Biotechnology). Mouse monoclonal GAPDH antibody was from Chemicon. Mouse monoclonal anti-FLAG, anti-FLAG conjugated with HRP, and anti-tubulin and rabbit polyclonal anti-calnexin antibodies were all from Sigma. Rabbit polyclonal anti-gp78 antibodies were described previously (20). The secondary antibodies sheep anti-mouse IgG-HRP and anti-rabbit IgG-HRP (Amersham Biosciences) were used. The proteins were visualized using an ECL detection kit (Amersham Biosciences).
Immunofluorescence
Transfected 293 cells grown on coverslips were washed with PBS and fixed in 4% paraformaldehyde for 5 min at room temperature. The cells were treated with 0.25% Triton X-100 for 10 min, blocked with 0.5% FBS in PBS, and then incubated with the primary antibody followed by Rho (red)- or FITC (green)-conjugated donkey anti-mouse or anti-rabbit secondary antibodies (Santa Cruz Biotechnology) or Alexa Fluor 350 (blue)-labeled goat anti-mouse IgG (Invitrogen). Nuclei were stained with DAPI (Sigma).
Immunoprecipitation
Crude cell lysates were sonicated in lysis buffer. Cellular debris was removed by centrifugation at 15,000 × g for 30 min at 4 °C. The supernatants were incubated with rabbit polyclonal anti-GFP antibodies in 0.1% BSA for 4 h at 4 °C. After incubation, protein G-Sepharose (Roche) was used for precipitation. The beads were washed with lysis buffer five times, and proteins were eluted with SDS sample buffer for immunoblot analysis.
Cycloheximide Chase Analysis
Twenty-four hours after transfection, the 293 cells expressing neuroserpin were treated with 150 μg/ml CHX to inhibit protein synthesis. The cells were harvested at 0, 3, and 6 h after CHX treatment. The same lysate volumes were analyzed by immunoblot analysis.
Pulse-chase Analysis
293 cells stably expressing EGFP-tagged G392E neuroserpin were starved in methionine-free medium and incubated for 30 min. The cells were then labeled with 200 μCi/ml [35S]methionine (PerkinElmer Life Sciences) for 1 h. Pulse labeling was terminated by incubating cells in fresh medium containing an excess of unlabeled methionine (300 mg/liter), and the cells were harvested at 0, 3, and 6 h. Cells lysates were immunoprecipitated as described above and analyzed by SDS-PAGE and autoradiography.
Cell Fractionation
293 cells were transfected with the indicated plasmids and homogenized in buffer containing 50 mm Tris-HCl (pH 8.0), 1 mm 2-mercaptoethanol, 1 mm EDTA, 0.32 mm sucrose, and 0.1 mm PMSF. Lysates were initially centrifuged at 9,000 × g for 10 min. Microsomal and cytosolic fractions were further separated from the supernatant by centrifugation at 105,000 × g for 60 min, and the fractions were further processed for immunoblot analysis.
RESULTS
Regulation of Aggregate Formation and Cellular Turnover of Mutant Neuroserpin by the Proteasome
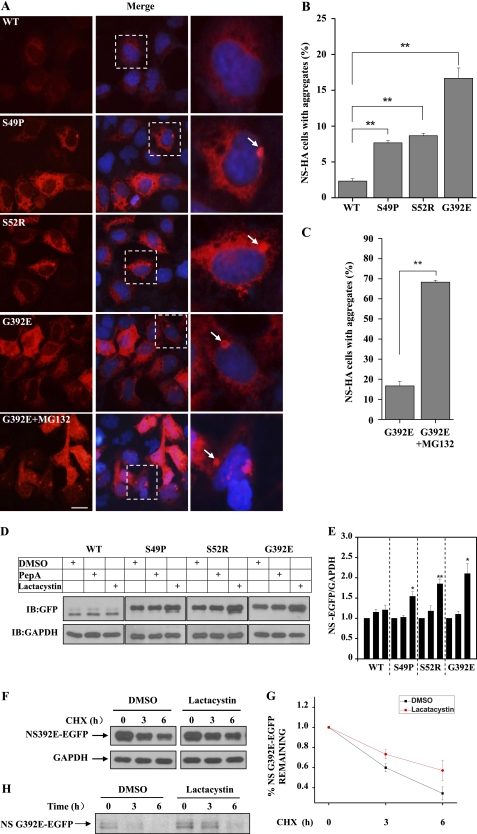
To investigate whether mutant neuroserpin is degraded by the proteasome, we constructed HA-tagged WT, S49P, S52R, and G392E mutant neuroserpins and transfected each into 293 cells. When neuroserpin was expressed in 293 cells, it was reticularly distributed throughout the cytoplasm (Fig. 1A). Although all forms of neuroserpin were mainly diffusively distributed, aggregates were observed in cells transfected with mutant neuroserpins (Fig. 1A). Notably, G392E neuroserpin, which is associated with the most severe form of FENIB (7), formed the most aggregates, consistent with previous findings that this mutant easily forms accumulated “polymers” (21). The quantitative data are shown in Fig. 1B. We next examined the effects of the proteasome on mutant neuroserpin aggregate formation using the proteasome inhibitor MG132. The number of G392E neuroserpin aggregates in transfected cells was significantly increased after MG132 treatment (Fig. 1, A and C). To further investigate the cellular turnover of neuroserpin, we established 293 cell lines that stably expressed each of the neuroserpin variants. These cells were treated with either proteasome or lysosome inhibitor, and the neuroserpin variant protein levels were examined by immunoblot analysis. As shown in Fig. 1, D and E, the levels of S49P, S52R, and G392E neuroserpin increased in the presence of the proteasome inhibitor lactacystin but not the lysosome inhibitor pepstatin A. However, the WT neuroserpin levels were not affected by either of the inhibitors. CHX chase analysis also showed that lactacystin treatment increased the stability of G392E neuroserpin (Fig. 1, F and G). To further confirm the involvement of the proteasome in neuroserpin degradation, we performed a pulse-chase analysis using the G392E neuroserpin stable cell line. Consistent with the observations in the CHX chase analysis, lactacystin treatment stabilized G392E neuroserpin (Fig. 1H). These data suggest that inhibition of the proteasome results in an abnormal accumulation of mutant neuroserpin.
FIGURE 1.
The proteasome regulates aggregation and turnover of neuroserpin. A, 293 cells were transfected with HA-tagged WT, S49P, S52R, or G392E neuroserpin. Twelve hours later, the cells were subjected to immunofluorescence analysis using anti-HA antibody (red), and they were visualized using an inverted system microscope IX71 (Olympus). Blue fluorescence shows DAPI nuclear staining. Scale bar = 10 μm. B, experiments were performed as in A to quantify neuroserpin aggregates. Quantification of the mean percentage of positive-transfected cells with neuroserpin aggregates from three independent transfections is plotted as a bar graph. The results are indicated as the mean ± S.E. **, p < 0.01, one-way ANOVA. C, G392E neuroserpin-transfected cells were treated with DMSO or MG132 (10 μm) 12 h after transfection and the mean percentage of cells with aggregates are plotted as a bar graph. **, p < 0.01, one-way ANOVA. D, 293 cells stably expressing EGFP-tagged WT, S49P, S52R, or G392E neuroserpin were incubated with DMSO, pepstatin A (10 μg/ml), or lactacystin (10 μm) for 12 h. Cells were harvested, lysed, and immunoblotted (IB) for GFP or GAPDH. E, quantification of the data presented in D, representing three independent experiments. The neuroserpin band density relative to that of GAPDH is plotted as a bar graph. The results are indicated as mean ± S.E. *, p < 0.05; **, p < 0.01; one-way ANOVA. F, 293 cells stably expressing G392E neuroserpin were treated with DMSO or lactacystin (10 μm) in the presence of CHX (150 μg/ml), collected at the indicated times after the incubation, and immunoblotted with GFP or GAPDH antibody. G, quantification of the data in F is shown. The results are indicated as the mean ± S.E. H, 293 cells stably expressing G392E neuroserpin were labeled with 200 μCi/ml [35S]methionine for 1 h and then chased in the presence of DMSO or 10 μm lactacystin at the indicated times. Cells lysates were subjected to immunoprecipitation using anti-GFP antibodies, and the immunoprecipitates were separated by SDS-PAGE and analyzed using autoradiography.
Involvement of ERAD in Mutant Neuroserpin Degradation
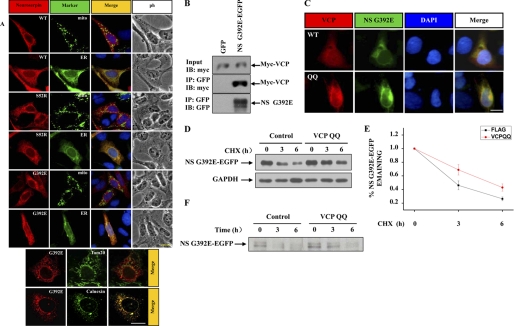
To examine the subcellular localization of neuroserpin, we cotransfected 293 cells with HA-tagged wild-type or mutant neuroserpin and EGFP-ER or EGFP-Mito, the specific organelle markers that labeled ER or mitochondria. WT, S52R, and G392E neuroserpin all colocalized with the ER but not mitochondria (Fig. 2A). ERAD is a well known degradation system for ER-retained proteins (10, 11). We therefore examined whether VCP/p97, a crucial component of ER retrotranslocation for proteasomal degradation, is involved in mutant neuroserpin degradation. We cotransfected 293 cells with EGFP-tagged G392E neuroserpin and Myc- or His-tagged VCP/p97. Using immunoprecipitation with anti-GFP antibodies, we found that VCP/p97 was coimmunoprecipitated with G392E neuroserpin (Fig. 2B), suggesting that ERAD may be involved in mutant neuroserpin degradation. Moreover, VCP/p97 QQ, a VCP/p97 mutant form that has no function in ERAD because of its deficiency in ATP hydrolysis (22), was recruited to mutant neuroserpin aggregates (C). CHX chase and pulse-chase analysis showed that in the G392E neuroserpin stable cell line transfected with VCP QQ, G392E neuroserpin was stabilized (Fig. 2, D–F), suggesting that the loss of VCP function significantly stabilizes mutant neuroserpin. These data further suggest the involvement of ERAD in mutant neuroserpin degradation.
FIGURE 2.
Impairment of ERAD promotes neuroserpin aggregation and blocks neuroserpin degradation. A, 293 cells were cotransfected with HA-tagged wild-type or mutant neuroserpin and EGFP-tagged mitochondrial or ER marker. Cells were subjected to immunofluorescence analysis using anti-HA antibody (red), anti-Tom20 antibody (green) representing mitochondria, or anti-calnexin antibody (green) representing the ER. Scale bars = 10 μm. B, 293 cells were cotransfected with EGFP or EGFP-tagged G392E neuroserpin, along with Myc-tagged VCP/p97. After 24 h, the cells were immunoprecipitated (IP) with rabbit polyclonal GFP antibodies. The immunoprecipitates were immunoblotted (IB) with the indicated antibodies. C, HA-tagged G392E neuroserpin was cotransfected with His-tagged VCP/p97 or VCP/p97 QQ for 12 h and subjected to immunofluorescence analysis using polyclonal HA antibodies (red) and monoclonal His antibody (green). Blue fluorescence shows nuclear DAPI. Scale bar = 10 μm. D and F, CHX chase and pulse-chase analyses were performed. His-tagged VCP/p97 QQ or control empty vector were transfected into the G392E neuroserpin stable cell line. After 24 h, the cells were subjected to CHX chase and pulse-chase analysis. E, quantification of the data in D. The results are indicated as mean ± S.E.
Increased Ubiquitination and Degradation of Mutant Neuroserpin by ERAD-associated E3s Hrd1 and gp78
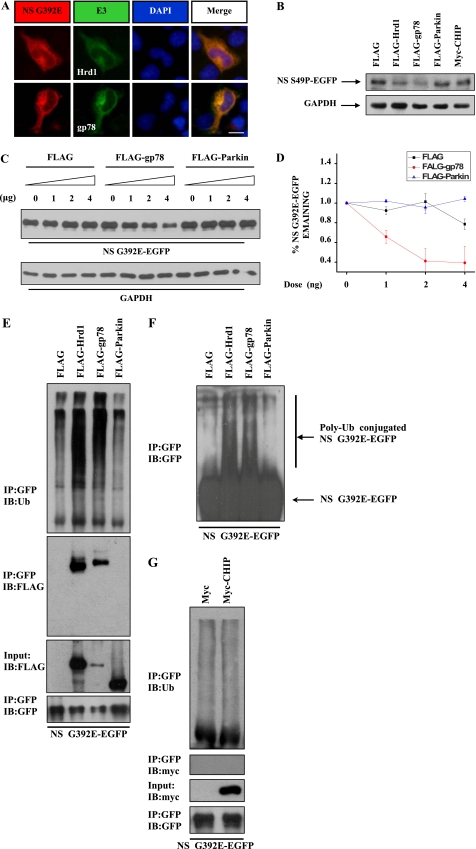
To further confirm whether ERAD is involved in the degradation of neuroserpin, we examined whether ERAD-associated E3s could promote mutant neuroserpin degradation. First, we examined G392E neuroserpin localization using 293 cells that were cotransfected with mutant neuroserpin and the ER-resident E3s Hrd1 and gp78. G392E neuroserpin colocalized with Hrd1 and gp78 (Fig. 3A). Cell fractionation assays also demonstrated that Hrd1, gp78, and G392E neuroserpin were isolated in the ER-containing microsomal membrane fraction (m) (supplemental Fig. S1). Next, we examined whether ER-associated E3s promote mutant neuroserpin degradation. We cotransfected 293 cells with S49P neuroserpin and several mammalian E3s, including parkin and CHIP, two cytoplasmic E3s reported to be associated with ERAD (11), and Hrd1 and gp78. The level of S49P neuroserpin was markedly decreased in the presence of Hrd1 or gp78, but it was not changed in the presence of parkin or CHIP (Fig. 3B). These data suggest that Hrd1 and gp78, but not CHIP or parkin, are involved in mutant neuroserpin degradation.
FIGURE 3.
Hrd1 and gp78 are involved in ERAD of neuroserpin. A, HA-tagged G392E neuroserpin was cotransfected with FLAG-tagged Hrd1 or gp78 into 293 cells. Cells were subjected to immunofluorescent staining with polyclonal HA antibodies (red) and monoclonal FLAG antibody (green). Blue fluorescence shows nuclear DAPI staining. Scale bar = 10 μm. B, 293 cells were cotransfected with EGFP-tagged S49P neuroserpin and FLAG-tagged Hrd1, gp78, gp78 Rm, parkin, or Myc-tagged CHIP. After 12 h, the cell lysates were subjected to immunoblot analysis. GAPDH served as a loading control. C, 293 cells stably expressing G392E neuroserpin were transfected with increasing amounts of FLAG tag, FLAG-gp78, or FLAG-parkin. Cell lysates were immunoblotted with the indicated antibodies. D, quantification of the data in C. The results are indicated as mean ± S.E. E, N2a cells were cotransfected with EGFP-tagged G392E neuroserpin and FLAG-tagged Hrd1, gp78, or parkin. After 24 h, the cells were treated with 5 μm MG132 for 6 h and immunoprecipitated (IP) with rabbit polyclonal GFP antibodies. The immunoprecipitates were immunoblotted (IB) for Ubiquitin (Ub), FLAG, or GFP. F, polyubiquitinated EGFP-tagged G392E neuroserpin from the experiment in E was detected by GFP antibody. G, EGFP-tagged G392E neuroserpin was cotransfected with Myc or Myc-CHIP in 293 cells for 24 h. After incubating with 5 μm MG132 for 6 h, the cell lysates were immunoprecipitated and immunoblotted with the indicated antibodies.
To further characterize the effects of Hrd1 and gp78 on mutant neuroserpin degradation, we transfected G392E neuroserpin stable cell lines with different amounts of FLAG, FLAG-tagged gp78, or FLAG-tagged parkin. Increased amounts of FLAG-gp78, but not FLAG or FLAG-parkin, resulted in a greater decrease in S49P neuroserpin (Fig. 3, C and D), suggesting that gp78 promotes neuroserpin degradation in a dose-dependent manner. As Hrd1 and gp78 are well known E3 ubiquitin ligases, we wondered if they affect neuroserpin ubiquitination. We cotransfected N2a and 293 cells with EGFP-tagged G392E neuroserpin and FLAG, FLAG-tagged Hrd1, gp78, or parkin and performed immunoprecipitation assays using polyclonal GFP antibodies. G392E neuroserpin ubiquitination was strikingly increased in the presence of Hrd1 and gp78 but was not changed in the presence of parkin (Fig. 3, E and F) or CHIP (G). Moreover, Hrd1 and gp78, but not parkin or CHIP, were coimmunoprecipitated with G392E neuroserpin (Fig. 3, E and G). Taken together, these data suggest that the degradation of mutant neuroserpin is specifically driven by the ER-resident E3s Hrd1 and gp78 but not by the non-ER-resident E3s parkin and CHIP.
E3 Activity and Proteasome-dependent Degradation of Mutant Neuroserpin
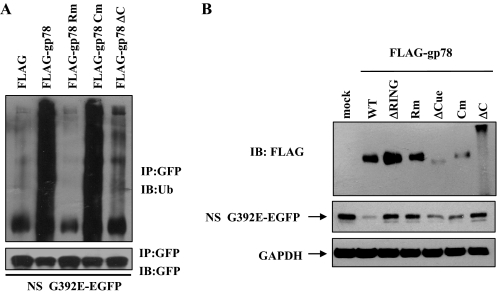
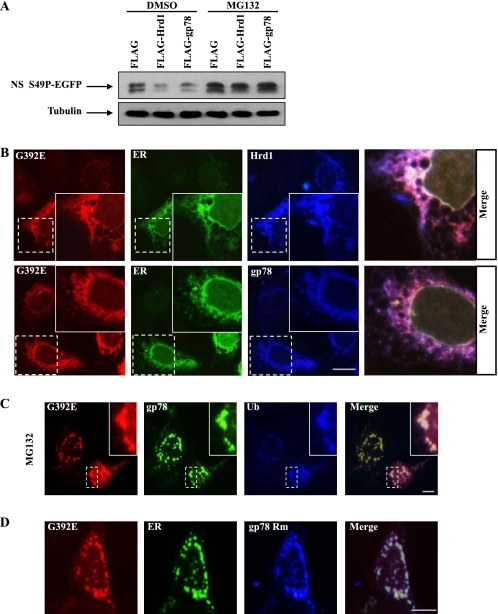
Hrd1 and gp78 both contain a RING finger E3 activity domain, and there is a ubiquitin-binding Cue domain in the C terminus of gp78, but not in Hrd1 (23, 24). To explore whether E3 activity is responsible for neuroserpin degradation, we generated the following FLAG-tagged gp78 mutants: a deletion mutant (ΔC) that lacked the C terminus, which includes the RING finger and Cue domains; a RING finger deletion mutant (ΔRING); a Cue domain deletion mutant (ΔCue); a RING domain point mutant (Rm); and a Cue domain point mutant (Cm). We cotransfected 293 cells with EGFP-tagged G392E neuroserpin along with each of the above mutants. Gp78 Rm and gp78 ΔC failed to promote the ubiquitination of G392E neuroserpin, whereas wild-type gp78 and gp78 Cm strikingly increased the ubiquitination of G392E neuroserpin (Fig. 4A). Consistent with these observations, gp78 ΔC, ΔRING, and Rm, but not wild-type gp78, ΔCue, or Cm, failed to decrease the levels of G392E neuroserpin (Fig. 4B), further suggesting that the RING finger, which has E3 ligase activity but not the Cue domain of gp78 is necessary for neuroserpin ubiquitination and degradation. We next examined whether inhibition of the proteasome affects neuroserpin degradation and aggregation. In MG132-treated cells, Hrd1- or gp78-driven mutant neuroserpin degradation was completely blocked (Fig. 5A). Using immunocytochemical staining, we found that G392E neuroserpin colocalized with Hrd1 and gp78 in the ER (Fig. 5B) and that MG132 treatment (C) or overexpression of gp78 Rm (D) significantly increased G392E neuroserpin aggregate formation in the ER, suggesting that impairments of proteasome function and ERAD increase mutant neuroserpin aggregation.
FIGURE 4.
E3 ligase activity is necessary for ERAD of neuroserpin. A, EGFP-tagged G392E neuroserpin was cotransfected with FLAG-tagged gp78, gp78 Rm, gp78 Cm, or gp78 ΔC. After 24 h, the cells were treated with 5 μm MG132 for 6 h, subjected to immunoprecipitation (IP), and immunoblotted (IB) with the indicated antibodies. B, 293 cells were cotransfected with EGFP-tagged G392E neuroserpin and different FLAG-gp78 constructs, as indicated. The cell lysates were immunoblotted for EGFP, FLAG, or GAPDH.
FIGURE 5.
Hrd1 and gp78 regulate the degradation and aggregation of mutant neuroserpin in a proteasome-dependent manner. A, EGFP-tagged S49P neuroserpin was cotransfected with FLAG or FLAG-tagged Hrd1 or gp78 into 293 cells. Cells were cultured in medium containing DMSO or MG132 (10 μm) for 16 h, and the cell lysates were immunoblotted for GFP or tubulin. B, 293 cells were cotransfected with HA-tagged G392E neuroserpin and FLAG-tagged Hrd1 or gp78 as well as EGFP-tagged ER marker. Cells were subjected to immunofluorescence analysis with polyclonal HA antibodies (red) and monoclonal FLAG antibody (blue). Scale bar = 10 μm. C, 293 cells were cotransfected with HA-tagged G392E neuroserpin and FLAG-tagged ubiquitin (Ub) along with EGFP-tagged gp78. After 24 h, the cells were treated with 10 μm MG132 for 6 h. Cells were subjected to immunofluorescent staining with polyclonal HA antibody (red) and monoclonal FLAG antibody (blue). Scale bar = 10 μm. D, 293 cells were cotransfected with HA-tagged G392E neuroserpin and FLAG-tagged gp78 Rm as well as EGFP-tagged ER marker. After 24 h, the cells were subjected to immunofluorescent staining with polyclonal HA antibodies (red) and monoclonal FLAG antibody (blue). Scale bar = 10 μm.
Hrd1 or gp78 Knockdown Results in Increased Mutant Neuroserpin Stability
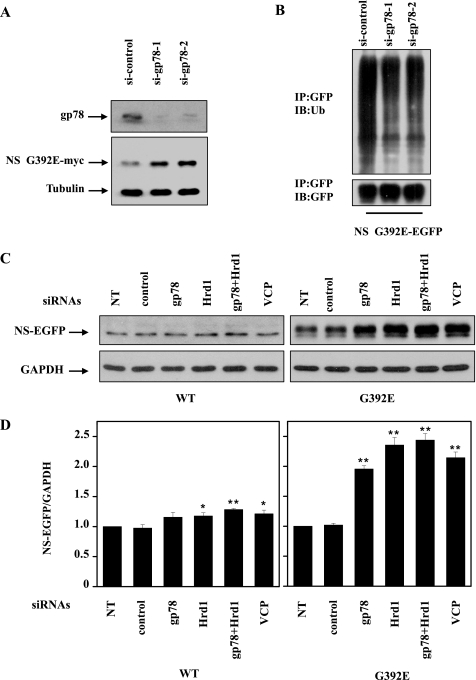
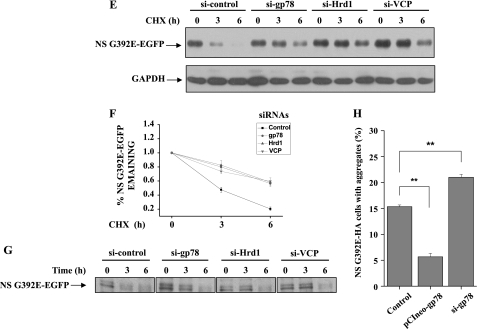
To further explore the role of ERAD E3s in neuroserpin degradation, we cotransfected siRNA oligonucleotides against gp78 (si-gp78–1 or si-gp78–2) or a control non-targeting oligonucleotide (si-control) with Myc-tagged G392E neuroserpin into 293 cells. These siRNAs were also transfected into the G392E neuroserpin stable cell line. In gp78-knockdown cells, the G392E neuroserpin level was increased (Fig. 6A). Moreover, G392E neuroserpin ubiquitination was decreased in gp78-knockdown cells (Fig. 6B). In the G392E neuroserpin stable cell line, knockdown of gp78, Hrd1, gp78 plus Hrd1, or VCP increased the mutant neuroserpin level. As a comparison, the level of WT neuroserpin was not increased to the same extent (Fig. 6, C and D). The knockdown effects of Hrd1 and VCP are shown in supplemental Fig. S2. CHX chase as well as pulse-chase experiments demonstrated that knockdown of the above genes stabilized G392E neuroserpin (Fig. 6, E, F, and G). Furthermore, gp78 overexpression decreased the number of G392E neuroserpin aggregates, whereas gp78 knockdown increased its aggregates (Fig. 6H).
FIGURE 6.
Effects of endogenous Hrd1 and gp78 on mutant neuroserpin degradation. A, 293 cells were transfected with Myc-tagged G392E neuroserpin and siRNAs targeting two regions of gp78 or a negative control siRNA. The indicated proteins were detected by immunoblot analysis 72 h after transfection. Endogenous gp78 was detected with anti-gp78 antibodies. B, 293 cells stably expressing G392E neuroserpin were transfected with the indicated siRNAs. After 48 h, the cells were treated with MG132 (10 μm) and incubated for 16 h. The cells were immunoprecipitated (IP) and immunoblotted (IB) with the indicated antibodies. C, 293 cells stably expressing G392E neuroserpin were transfected with the indicated siRNAs for 72 h, and the cell lysates were immunoblotted with the indicated antibodies. NT, untransfected. D, quantification of the data in C. The results are indicated as mean ± S.E. *, p < 0.05; **, p < 0.01; one-way ANOVA. E and G, CHX chase and pulse-chase analyses of EGFP-tagged G392E neuroserpin were performed. 293 cells stably expressing G392E neuroserpin were transfected with control siRNA or siRNA targeting gp78, Hrd1, or VCP. After 72 h, the cells were subjected to CHX chase and pulse-chase analyses. F, quantification the data in E. The results are indicated as mean ± S.E. H, HA-tagged neuroserpin was cotransfected with control siRNA, pCIneo-gp78, or siRNA targeting gp78 in 293 cells. The quantitative data represent the mean percentage of positive-transfected cells with neuroserpin aggregates from three independent transfections and are plotted as a bar graph. The results are indicated as mean ± S.E. **, p < 0.01, one-way ANOVA.
DISCUSSION
Misfolded protein aggregation or inclusion body formation is a key feature of neurodegenerative disorders. Many pathogenic proteins involved in neurodegenerative diseases aggregate in brain of affected humans and in cultured mammalian cells (25, 26). Mutant neuroserpin was first identified as the major component of inclusion bodies found in the brains of FENIB patients (4). Subsequent studies showed that mutant neuroserpin is retained in the ER as accumulated polymers in a cellular model (3). Interestingly, ERAD E3s, such as Hrd1 and gp78, suppress the neurodegeneration mediated by aggregate-prone proteins, including Alzheimer's disease-linked amyloid precursor proteins (27), Parkinson's disease-linked α-synuclein (28), amyotrophic lateral sclerosis-linked SOD1 (20), polyglutamine disease-linked Nhtt and ataxin-3 (20, 29, 30), and prion disease-linked prion protein (31, 32). In this study, we found that Hrd1 and gp78 are involved in mutant neuroserpin ubiquitination and degradation and that the ERAD pathway contributes to the neuroserpin degradation and aggregation machinery, as indicated by the observations that impairment of ERAD by defective VCP and gp78 significantly increases mutant neuroserpin stability and aggregation.
Our findings demonstrate that mutant neuroserpin is targeted by ERAD with the aid of VCP/p97 is polyubiquitinated by the two ER-resident E3s Hrd1 and gp78 and is subsequently degraded by the proteasome. This mechanism is essentially the same as that involved in the quality control of other misfolded and aberrantly synthesized proteins in the ER (10, 11). Because VCP/p97 is an essential regulator in ERAD that facilitates the retrotranslocation of ubiquitinated ER proteins to the cytosol for degradation (12), forms a complex with the relevant E3s and functions in classical ERAD (17, 18), the activity of VCP/p97 involved in the degradation of mutant neuroserpin strongly suggests that VCP/p97 cooperates with Hrd1 and gp78 for neuroserpin degradation. The N-terminal fragments of polyQ-expanded huntingtin (Nhtt) and ataxin-3 inhibit the function of ERAD and retrotranslocation (33, 34). In our observations, we also found that the polyQ-expanded Nhtt or ataxin-3 significantly stimulates mutant neuroserpin aggregation and perturbs its proteasomal degradation (data not shown). As the polyQ-expanded Nhtt and ataxin-3 affect ERAD by influencing the VCP complex (33, 34), and VCP deficiency significantly decreases mutant neuroserpin degradation rates (Fig. 2D), these data further suggest that the ERAD machinery is involved in mutant neuroserpin degradation. Interestingly, consistent with previous studies (3), we observed that MG132 treatment does not significantly up-regulate the WT neuroserpin protein level as much as it does for mutant neuroserpins, suggesting a higher proteasomal turnover for the mutant. These data provide further evidence that mutant neuroserpin is misfolded and targeted by the proteasome.
Hrd1 and gp78 are localized in the ER membrane, whereas parkin and CHIP are localized in the cytoplasm (10, 11, 35). Therefore, ER-resident Hrd1 and gp78, but not non-ER-resident parkin and CHIP, may directly interact with ER-resident neuroserpin to promote neuroserpin ubiquitination, as indicated by strong Hrd1/gp78 binding to neuroserpin but no interaction between parkin/CHIP and neuroserpin (Figs. 3E and G). In our observations, parkin and CHIP have no effect on neuroserpin degradation, further suggesting that gp78 and Hrd1 are neuroserpin-specific E3s.
Hrd1 and gp78 serve divergent ERAD substrate populations (20, 24, 36, 37). gp78 differs from Hrd1 in its complex domain structure, which, in addition to its RING finger, includes the ubiquitin-binding Cue domain (23, 24, 36). However, studies have also shown that Hrd1 and gp78 share many common substrates, such as the “classic” ERAD substrate T cell antigen receptor CD3δ subunit, 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMG-CoA reductase), polyQ-expanded huntingtin and α-1-antitrypsin (AAT)-deficiency disease protein Z variant of α-1-antitrypsin (14–16, 29, 30, 38). Overlapping gp78 and Hrd1 substrates in ERAD could be explained by the high identity of the N-terminal and RING finger E3 activity domains of Hrd1 and gp78, which are the most closely related ERAD E3s (14). In addition, both proteins use UBC7 as the E2 ubiquitin-conjugating enzyme (14, 15). In our study, we show that the RING finger, but not the Cue domain is necessary for mutant neuroserpin ubiquitination (Fig. 4A) and degradation (B), suggesting that the E3 RING finger activity in Hrd1 and gp78 is specifically involved in regulating mutant neuroserpin degradation.
In summary, the two ER-associated E3s Hrd1 and gp78 regulate mutant neuroserpin ubiquitination and degradation. The degradation of mutant neuroserpin is controlled by the ERAD machinery in association with VCP. Thus, ERAD may play a role in the regulation of mutant neuroserpin.
Supplementary Material
This work was supported in part by National Natural Sciences Foundation of China Grants 30970921 and 31000473 and by National High-Tech Research and Development Program of China 973 Projects Grant 2011CB504102.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- polyQ
- polyglutamine
- ER
- endoplasmic reticulum
- FENIB
- familial encephalopathy with neuroserpin inclusion bodies
- ERAD
- endoplasmic reticulum-associated degradation
- VCP
- valosin-containing protein
- CHIP
- C terminus of Hsc70-interacting protein
- CHX
- cycloheximide
- ANOVA
- analysis of variance
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1. Rubinsztein D. C. (2006) Nature 443, 780–786 [DOI] [PubMed] [Google Scholar]
- 2. Ciechanover A., Brundin P. (2003) Neuron 40, 427–446 [DOI] [PubMed] [Google Scholar]
- 3. Miranda E., Römisch K., Lomas D. A. (2004) J. Biol. Chem. 279, 28283–28291 [DOI] [PubMed] [Google Scholar]
- 4. Davis R. L., Shrimpton A. E., Holohan P. D., Bradshaw C., Feiglin D., Collins G. H., Sonderegger P., Kinter J., Becker L. M., Lacbawan F., Krasnewich D., Muenke M., Lawrence D. A., Yerby M. S., Shaw C. M., Gooptu B., Elliott P. R., Finch J. T., Carrell R. W., Lomas D. A. (1999) Nature 401, 376–379 [DOI] [PubMed] [Google Scholar]
- 5. Osterwalder T., Contartese J., Stoeckli E. T., Kuhn T. B., Sonderegger P. (1996) EMBO J. 15, 2944–2953 [PMC free article] [PubMed] [Google Scholar]
- 6. Hastings G. A., Coleman T. A., Haudenschild C. C., Stefansson S., Smith E. P., Barthlow R., Cherry S., Sandkvist M., Lawrence D. A. (1997) J. Biol. Chem. 272, 33062–33067 [DOI] [PubMed] [Google Scholar]
- 7. Lomas D. A., Carrell R. W. (2002) Nat. Rev. Genet. 3, 759–768 [DOI] [PubMed] [Google Scholar]
- 8. Carrell R. W. (2005) Trends Cell Biol. 15, 574–580 [DOI] [PubMed] [Google Scholar]
- 9. Davis R. L., Shrimpton A. E., Carrell R. W., Lomas D. A., Gerhard L., Baumann B., Lawrence D. A., Yepes M., Kim T. S., Ghetti B., Piccardo P., Takao M., Lacbawan F., Muenke M., Sifers R. N., Bradshaw C. B., Kent P. F., Collins G. H., Larocca D., Holohan P. D. (2002) Lancet 359, 2242–2247 [DOI] [PubMed] [Google Scholar]
- 10. Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 11. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ye Y., Shibata Y., Kikkert M., van Voorden S., Wiertz E., Rapoport T. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14132–14138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cao J., Wang J., Qi W., Miao H. H., Wang J., Ge L., DeBose-Boyd R. A., Tang J. J., Li B. L., Song B. L. (2007) Cell Metab. 6, 115–128 [DOI] [PubMed] [Google Scholar]
- 14. Kikkert M., Doolman R., Dai M., Avner R., Hassink G., van Voorden S., Thanedar S., Roitelman J., Chau V., Wiertz E. (2004) J. Biol. Chem. 279, 3525–3534 [DOI] [PubMed] [Google Scholar]
- 15. Fang S., Ferrone M., Yang C., Jensen J. P., Tiwari S., Weissman A. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shen Y., Ballar P., Fang S. (2006) Biochem. Biophys. Res. Commun. 349, 1285–1293 [DOI] [PubMed] [Google Scholar]
- 17. Zhong X., Shen Y., Ballar P., Apostolou A., Agami R., Fang S. (2004) J. Biol. Chem. 279, 45676–45684 [DOI] [PubMed] [Google Scholar]
- 18. Lilley B. N., Ploegh H. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14296–14301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kroeger H., Miranda E., MacLeod I., Pérez J., Crowther D. C., Marciniak S. J., Lomas D. A. (2009) J. Biol. Chem. 284, 22793–22802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ying Z., Wang H., Fan H., Zhu X., Zhou J., Fei E., Wang G. (2009) Hum. Mol. Genet. 18, 4268–4281 [DOI] [PubMed] [Google Scholar]
- 21. Miranda E., MacLeod I., Davies M. J., Pérez J., Römisch K., Crowther D. C., Lomas D. A. (2008) Hum. Mol. Genet. 17, 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Q., Li L., Ye Y. (2006) J. Cell Biol. 174, 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen B., Mariano J., Tsai Y. C., Chan A. H., Cohen M., Weissman A. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morito D., Hirao K., Oda Y., Hosokawa N., Tokunaga F., Cyr D. M., Tanaka K., Iwai K., Nagata K. (2008) Mol. Biol. Cell 19, 1328–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bossy-Wetzel E., Schwarzenbacher R., Lipton S. A. (2004) Nat. Med. 10, S2–9 [DOI] [PubMed] [Google Scholar]
- 26. Ross C. A., Poirier M. A. (2004) Nat. Med. 10, S10–17 [DOI] [PubMed] [Google Scholar]
- 27. Kaneko M., Koike H., Saito R., Kitamura Y., Okuma Y., Nomura Y. (2010) J. Neurosci. 30, 3924–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gitler A. D., Chesi A., Geddie M. L., Strathearn K. E., Hamamichi S., Hill K. J., Caldwell K. A., Caldwell G. A., Cooper A. A., Rochet J. C., Lindquist S. (2009) Nat. Genet. 41, 308–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang H., Liu C., Zhong Y., Luo S., Monteiro M. J., Fang S. (2010) PLoS ONE 5, e8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang H., Zhong X., Ballar P., Luo S., Shen Y., Rubinsztein D. C., Monteiro M. J., Fang S. (2007) Exp. Cell Res. 313, 538–550 [DOI] [PubMed] [Google Scholar]
- 31. Yedidia Y., Horonchik L., Tzaban S., Yanai A., Taraboulos A. (2001) EMBO J. 20, 5383–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Apodaca J., Kim I., Rao H. (2006) Biochem. Biophys. Res. Commun. 347, 319–326 [DOI] [PubMed] [Google Scholar]
- 33. Duennwald M. L., Lindquist S. (2008) Genes Dev. 22, 3308–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhong X., Pittman R. N. (2006) Hum. Mol. Genet. 15, 2409–2420 [DOI] [PubMed] [Google Scholar]
- 35. Ron I., Rapaport D., Horowitz M. Hum. Mol. Genet. 19, 3771–3781 [DOI] [PubMed] [Google Scholar]
- 36. Tsai Y. C., Mendoza A., Mariano J. M., Zhou M., Kostova Z., Chen B., Veenstra T., Hewitt S. M., Helman L. J., Khanna C., Weissman A. M. (2007) Nat. Med. 13, 1504–1509 [DOI] [PubMed] [Google Scholar]
- 37. Pabarcus M. K., Hoe N., Sadeghi S., Patterson C., Wiertz E., Correia M. A. (2009) Arch. Biochem. Biophys. 483, 66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Christianson J. C., Shaler T. A., Tyler R. E., Kopito R. R. (2008) Nat. Cell Biol. 10, 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.