Abstract
Pyridomycin is a structurally unique antimycobacterial cyclodepsipeptide containing rare 3-(3-pyridyl)-l-alanine and 2-hydroxy-3-methylpent-2-enoic acid moieties. The biosynthetic gene cluster for pyridomycin has been cloned and identified from Streptomyces pyridomyceticus NRRL B-2517. Sequence analysis of a 42.5-kb DNA region revealed 26 putative open reading frames, including two nonribosomal peptide synthetase (NRPS) genes and a polyketide synthase gene. A special feature is the presence of a polyketide synthase-type ketoreductase domain embedded in an NRPS. Furthermore, we showed that PyrA functioned as an NRPS adenylation domain that activates 3-hydroxypicolinic acid and transfers it to a discrete peptidyl carrier protein, PyrU, which functions as a loading module that initiates pyridomycin biosynthesis in vivo and in vitro. PyrA could also activate other aromatic acids, generating three pyridomycin analogues in vivo.
Keywords: Antibiotics, Bacterial Genetics, Bacterial Metabolism, Coenzyme A, Enzyme Mechanisms, Mass Spectrometry (MS), Peptide Biosynthesis, Non-ribosomal Peptide Synthetase, Polyketide Synthase, Biosynthesis
Introduction
For more than half of the twentieth century, natural products and derivatives thereof have been essential products of the pharmaceutical industry because of the diversity of their structures and biological activities (1). Many of these natural products, such as cyclosporins, vancomycin, erythromycin, and FK506, are synthesized by multifunctional megasynthase molecular “assembly lines” (2–5). There are two types of megasynthases: nonribosomal peptide synthetases (NRPSs)2 and polyketide synthases (PKSs). They are composed of multiple large peptides, each of which contains several catalytic modules. They assemble in a stepwise fashion two distinct classes of secondary metabolites. The precursors, amino acids and short carboxylic acids, are selected and attached as thioesters to the long phosphopantetheinyl arms of carrier proteins. They are then linked by the condensation (C) domains in NRPSs via peptide bonds or by ketosynthase (KS) domains of PKSs that form C–C bonds by Claisen-like condensation. The mature chain is usually released from the synthase by cyclization or hydrolysis catalyzed by a thioesterase (TE) domain in the terminal NRPS or PKS modules of the assembly line.
NRPSs and PKSs are remarkably similar in their architectures, which made it possible for nature to evolve PKS/NRPS hybrid systems that combine NRPSs and PKSs in the same assembly line and are capable of incorporating both carboxylic acids and nonproteinogenic amino acids into the final products. Thus, the NRPS/PKS hybrid biosynthetic paradigm generates enormous structural diversity (6, 7).
Pyridomycin is an antimycobacterial antibiotic produced by Streptomyces pyridomyceticus NRRL B-2517 (Fig. 1) (8). Pyridomycin is an unusual 12-membered ring depsipeptide composed of four moieties in the following order: N-3-hydroxypicolinyl-l-threonine, 3-(3-pyridyl)-l-alanine, propionic acid, and 2-hydroxy-3-methylpent-2-enoic acid, which is probably epimerized from α-keto-β-methylvaleric acid (9, 10). To the best of our knowledge, it is the only known depsipeptide that contains an enolic acid. Isotope labeling studies (11) indicated that the biosynthesis of pyridomycin might involve the assembly of the backbone by a hybrid NRPS/PKS system using 3-hydroxypicolinic acid (3-HPA) as the starter unit. Feeding experiments confirmed that l-Asp is a precursor of the 3-hydroxypicolinyl moiety, whereas lysine was not incorporated into the compound, indicating that the formation of the 3-hydroxypicolinyl moiety follows a different pathway from that involved in the biosynthesis of streptogramin B antibiotics, such as pristinamycin I, virginiamycin S (Fig. 1), and etamycin, which also use 3-HPA as the starter unit (12–14). Chemical synthesis schemes have been devised for pyridomycin (15), but little was known about the pathway of pyridomycin biosynthesis.
FIGURE 1.
Structures of natural products containing 3-hydroxypicolinic acid (boldface type). A, pyridomycin; B, pristinamycins IA (where R represents N(CH3)2) and IB (where R represents NHCH3) and virginiamycin S1 (where R represents H).
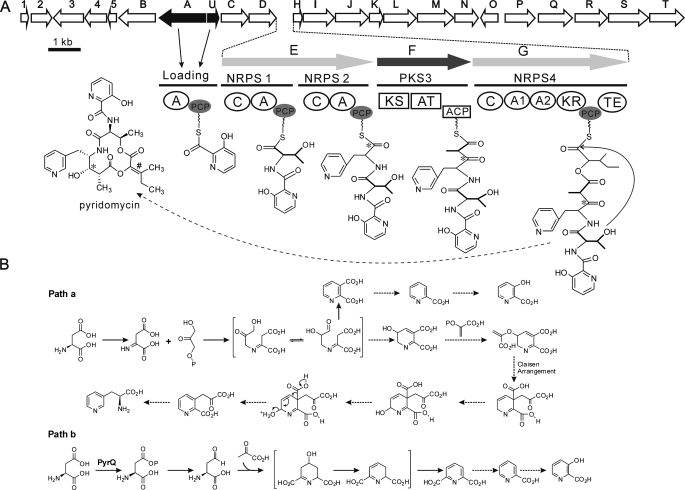
Here we report the cloning and sequencing of the pyridomycin biosynthetic gene cluster within a 42.5-kb DNA region containing 26 open reading frames (ORFs) (Fig. 2A). At the center of this region is a hybrid NRPS/PKS system (NRPS/PKS/NRPS). The last NRPS contains an essential KR domain in an unusual position. Based on targeted mutagenesis, in vitro enzyme studies, and sequence analysis, we propose a putative pathway for pyridomycin biosynthesis. In vivo and in vitro characterization of PyrA and PyrU demonstrated that PyrA and PyrU constituted the loading module that initiated the assembly of the pyridomycin backbone. Together with the observed broad substrate specificity of PyrA, these findings provide opportunities to generate pyridomycin derivatives with novel or enhanced bioactivities by rational engineering of the biosynthetic pathway or combinatorial biosynthesis. Given its special molecular architecture (9, 16), pyridomycin also offers an opportunity to discover new chemistry for natural product biosynthesis.
FIGURE 2.
Organization of the pyridomycin biosynthetic gene cluster and model for the biosynthesis of pyridomycin. A, at the top are ORFs 1–5 and PyrA to -U predicted from the 42.5-kb DNA sequence. Horizontal lines below the ORFs indicate the multifunctional NRPS and PKS modules. Below the module designations are the functional domains: NRPS adenylation domains (A, A1, and A2), peptidyl carrier protein (PCP), condensation domain (C), thioesterase (TE), PKS ketosynthase (KS), acyltransferase (AT), acyl carrier protein (ACP), and ketoreductase (KR). The broken arrow to pyridomycin indicates that two additional enzymatic steps are required to form the hydroxyl at the position indicated by an asterisk and the double bond at the position indicated by the number sign. B, alternative putative biosynthetic pathways for pyridyl moieties. Solid arrows, confirmed reactions (39, 58); dashed arrows, predicted reactions.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Culture Conditions
Strains and plasmids used in the study are summarized in supplemental Tables S1 and S2. S. pyridomyceticus NRRL B-2517 and its derivatives were cultivated at 30 °C in YEME liquid medium for growth of mycelia, on COM medium (1% corn starch, 1% oat flour, 0.1% malt extract, 0.1% yeast extract, 0.1% tryptone, 1.2% agar, pH 7.2) for sporulation, and on 2CM medium (17) (1% soluble starch, 0.2% tryptone, 0.1% NaCl, 0.2% (NH4)2SO4, 0.1% K2HPO4, 0.1% MgSO4, 0.2% CaCO3, 1.2% agar with 1 ml of inorganic salt solution/liter and adjusted to pH 7.2) for conjugation. Luria-Bertani (LB) broth and agar were used for culturing Escherichia coli strains. All plasmid subcloning experiments were performed in E. coli DH10B using standard protocols (18). The final antibiotic concentrations used for selection of E. coli and Streptomyces were as follows: 5 μg/ml thiostrepton and 15 μg/ml apramycin for S. pyridomyceticus; 30 μg/ml apramycin, 50 μg/ml kanamycin, 100 μg/ml ampicillin, and 12.5 μg/ml chloramphenicol for E. coli.
DNA Isolation and General Manipulations
DNA isolation and manipulation in E. coli and Streptomyces were carried out following the standard procedures (18, 19). PCR amplifications were performed on a Veriti thermal cycler (Applied Biosystems, Carlsbad, CA) using TaqDNA polymerase or KOD-plus high fidelity PCR polymerase. Synthetic PCR primers were obtained from Invitrogen, and DNA sequencing was accomplished at Shanghai Majorbio Biotech Co. Ltd. (Shanghai, China).
Genomic Library Construction and Screening
A pOJ446-derived genomic cosmid library of S. pyridomyceticus NRRL B-2517 was constructed according to a standard protocol (18, 19). Recombinant clones were selected on LB agar plates supplemented with apramycin and screened by PCR using degenerate primers for NRPS and KS that were designed according to the CODENHOP algorithm (20).
DNA Sequence Analysis
ORFs were analyzed and identified using the Frame Plot 3.0 beta online program (21), and the deduced proteins were compared with other known proteins in the databases using the NCBI BLAST server (22). Multiple nucleotide sequence alignments and analysis were performed using the BioEdit Sequence Alignment Editor (available on the World Wide Web) or Vector NTI Advance 11.0 (Invitrogen). The NRPS-PKS architecture was predicted by NRPS-PKS (available on the World Wide Web) (23).
Inactivating PyrG-KR in S. pyridomyceticus NRRL B-2517 by Point Mutation
The mutated sequence on pJTU4676PSY (ThioR; supplemental Fig. S1) was introduced into S. pyridomyceticus by interspecific conjugation. Thiostrepton-sensitive (ThioS) clones were selected and confirmed to contain the desired chromosomal point mutation (HTT19PSY) using PCR and sequencing.
Genetic Manipulation of pyrA and pyrU in S. pyridomyceticus
See the supplemental material.
Pyridomycin Fermentation, Isolation, Feeding Experiments, and HPLC-MS Analysis
See the supplemental material.
Cloning of pyrU and pyrA
pyrU and pyrA were amplified using KOD-plus high fidelity PCR polymerase (Toyobo) and cosmid 9A3 as template (primers listed in supplemental Table S3). The purified PCR products pyrU (255-bp NdeI/BamI fragment) and pyrA (1632-bp NdeI/EcoRI fragment) were ligated into pET28a digested with the same restriction enzymes to generate plasmids pJTU4655 and pJTU4637. The correct sequences of pyrU in pJTU4655 and pyrA in pJTU4637 were confirmed by DNA sequencing.
Construction of pJTU4655(S47A) and pJTU4652(S47A)
To achieve the mutation of serine to alanine at position 47 in PyrU, pJTU4655(S47A) and pJTU4652(S47A) were constructed by site-directed mutagenesis using the primers PyrUS47AF and PyrUS47AR (supplemental Table S3). pJTU4655 and pJTU4652 served as templates. The mutations were verified by DNA sequencing. pJTU4655(S47A) was used for PyrU(S47A) overproduction, whereas pJTU4652(S47A) was used to complement the ΔpyrU mutant.
Overproduction and Purification of Recombinant Proteins
See the supplemental material.
Phosphopantetheinylation of PyrU and PyrU(S47A)
The in vitro assays for phosphopantetheinylation of PyrU or PyrU(S47A) were carried out in a 100-μl reaction containing 30 μm apo-PyrU, 0.5 mm CoA, 3 μm Sfp, 10 mm MgCl2, 2 mm DTT, and 20 mm Tris·HCl, pH 8.0, at 37 °C for 45 min. Reactions were started by adding Sfp (24) and quenched by flash freezing at −80 °C. The in vivo assays were performed by coexpression of PyrU and PyrU(S47A) with Sfp in E. coli BL21 (DE3), followed by purification of the recombinant proteins.
The assays were analyzed by HPLC on an Agilent HPLC series 1100 with a ZORBAX 300SB-C18 column (4.6 × 250 mm, 5 μm, 300 Å, Agilent), using a 10–90% (v/v) gradient of acetonitrile/water containing 0.1% (v/v) trifluoroacetic acid (TFA) for 30 min at a flow rate of 0.5 ml/min and UV detection at 280 nm.
The LC-MS analyses were performed on a 6530 Accurate-Mass QTOF spectrometer coupled to an Agilent HPLC 1200 series (Agilent Technologies) that was developed for 30 min using a 5–95% linear gradient of acetonitrile/water containing 0.1% TFA at a flow rate of 0.2 ml/min. Data were obtained in the positive mode, and the mass scan range was set between 600 and 2500 m/z. The resulting spectra were analyzed using the software Mass Hunter, which calculated the masses of the intact proteins.
Determination of Substrate Specificity of PyrA
PyrA activity was measured by monitoring PPi release at 360 nm continuously for 30 min, using the EnzChek pyrophosphate assay kit according to the manufacturer's instructions (MicroProbes) on a Synergy 2 multimode microplate reader (BioTek). PyrA (1.9 μg and 0.3 μm) was used to determine the initial velocity with 3-HPA or its analogs (4 mm) as the substrates. The standard curve was created using the pyrophosphate standard from the kit. Reactions were carried out in triplicate with boiled PyrA as control. Kinetic analysis of PyrA for 3-HPA, 2,3-dihydroxybenzoic acid (2,3-DHBA), and 4-amino-2-hydroxybenzoic acid (4A2HBA) were performed by varying each substrate concentration (0.01–0.32 mm 3-HPA, 0.4–4.8 mm 2,3-DHBA, and 0.4–4.0 mm 4A2HBA) in the presence of 2 mm ATP. The reactions were initiated, adding 1.2, 0.3, and 0.6 μm PyrA, respectively. The velocity was calculated based on the increase in absorbance at 360 nm. The Michaelis-Menten equation was fitted to plots of velocity of PPi release versus substrate concentration to extract values for Km and kcat using the program GraphPad Prism 5.
Acylation of Holo-PyrU
Loading of 3-HPA and its analogs onto holo-PyrU catalyzed by PyrA was performed in standard reactions containing 50 mm Tris·HCl (pH 8.0), 10 mm MgCl2, 2 mm DTT, 5 mm ATP, 2 mm substrate, 5 μm PyrA, and 30 μm holo-PyrU. Reactions were initiated by the addition of PyrA, incubated at 37 °C for 30 min, and quenched by flash freezing at −80 °C. After centrifugation, the clarified supernatant was separated by HPLC, and the new peak for candidate enzymatic products was collected and concentrated using a Savant SPD111V SpeedVac concentrator (Thermo Scientific). The identities of products were determined by HPLC and QTOF MS analysis as for the analysis of PyrU.
RESULTS
Cloning, Sequencing, and Analysis of the pyr Gene Cluster
To identify the pyridomycin biosynthetic gene cluster, a genomic library of S. pyridomyceticus NRRL B-2517 was constructed in pOJ446 (about 2,000 cosmid clones). Degenerate PCR primers (supplemental Table S3) designed according to the conserved core regions A3 and A7 of NRPS adenylation (A) domains (23, 25) were used for the initial screening of the cosmid library, and more than 100 A domain positive cosmids were isolated. These were then tested using degenerate primers designed according to the conserved regions that are unique to KS domains of hybrid NRPS/PKS (7). More than 20 cosmids hybridizing to both the A domain and the KS domain probes were isolated. Restriction mapping produced two separate contigs. In order to identify the contig containing the pyridomycin biosynthesis gene cluster, additional degenerate primers specific for conserved motifs in 3-HPA:AMP ligases (responsible for activation of 3-HPA in the biosynthesis of pristinamycin, virginiamycin, and etamycin) (13, 14, 26) were used. The deduced protein sequence of the amplified fragment from contig 2 resembled the 3-HPA:AMP ligase from Streptomyces pristinaespiralis (supplemental Fig. S2). From this, we concluded that contig 2 may be involved in pyridomycin biosynthesis. To confirm this hypothesis, a 20-kb sequence in cosmid 9A3 of contig 2 was replaced by a kan cassette. This mutation was then introduced into the S. pyridomyceticus chromosome. The resulting strain, HTT7, no longer produced pyridomycin (supplemental Fig. S3). Thus, cosmid 9A3 was sequenced to yield a continuous 42.5-kb DNA region, the GC content of which is 73.56%, typical for Streptomyces (19), and 26 ORFs were predicted (Fig. 2 and Table 1).
TABLE 1.
ORFs of the pyr gene cluster, closest homologues, and proposed functions
| ORF | Sizea | Homologous protein, species | Identity/Similarity | Accession number | Proposed functionb |
|---|---|---|---|---|---|
| aa | % | ||||
| Pyr1 | 49 | Ccel_0989, Clostridium cellulolyticm H10 | 44/68 | YP_002505331 | FkbH-like protein |
| Pyr2 | 246 | SCO7266, Streptomyces coelicolor A3(2) | 55/68 | NP_631322 | Ketoreductase |
| Pyr3 | 334 | Bthur0003_63860, Bacillus thuringiensis serovar thuringiensis str. T01001 | 29/53 | ZP_04137149 | Transport/self-resistance |
| Pyr4 | 210 | Shewmr7_2680, Shewanella sp. MR-7 | 33/47 | YP_738722 | Unknown |
| Pyr5 | 72 | SSDG_07478, S. pristinaespiralis ATCC 25486 | 71/87 | EFH32214 | Unknown |
| PyrB | 418 | SSDG_07479, S. pristinaespiralis ATCC 25486 | 66/75 | EFH32215 | Aminotransferase |
| PyrA | 543 | SSDG_07480, S. pristinaespiralis ATCC 25486 | 68/76 | EFH32216 | 3-Hydroxypicolinic acid:AMP ligase |
| PyrU | 84 | SSDG_05103, S. pristinaespiralis ATCC 25486 | 36/59 | ZP_05014213 | Phosphopantetheine binding |
| PyrC | 384 | SSDG_05104, S. pristinaespiralis ATCC 25486 | 67/76 | ZP_05014214 | Oxidoreductase |
| PyrD | 381 | SSDG_05106, S. pristinaespiralis ATCC 25486 | 60/71 | ZP_05014216 | Oxidase |
| PyrE | 2147 | SgriT_010100025868, Streptomyces griseoflavus Tu4000 | 49/61 | ZP_05541578 | Dimodular NRPS (C-A-T-C-A-T) |
| PyrF | 1411 | Ava_1612, Anabaena variabilis ATCC 29413 | 38/55 | YP_322130 | PKS (KS-AT-ACP) |
| PyrG | 2451 | SAML0376, S. ambofaciens ATCC 23877 | 45/55 | CAJ89363 | NRPS (C-A1-A2-KR-T-TE) |
| PyrH | 71 | Sros_3469, S. fungicidicus | 63/74 | YP_003339148 | MbtH-like protein |
| PyrI | 402 | SBI_09258, Streptomyces bingchenggensis BCW-1 | 52/66 | ADI12376 | Cytochrome P450 |
| PyrJ | 372 | SBI_05317, S. bingchenggensis BCW-1 | 65/76 | ADI08437 | Aminotransferase |
| PyrK | 173 | ROP_53460, Rhodococcus opacus B4 | 53/65 | YP_002782538 | Oxidoreductase |
| PyrL | 402 | SSAG_05882, Streptomyces sp. Mg1 | 74/83 | ZP_05001560 | Dehydrogenase |
| PyrM | 488 | nfa46290, Nocardia farcinica IFM 10152 | 53/64 | YP_120844 | Dehydrogenase |
| PyrN | 239 | SBI_07222, S. bingchenggensis BCW-1 | 49/60 | ADI10342 | Esterase |
| PyrO | 212 | SAV_2270, Streptomyces avermitilis MA-4680 | 45/64 | NP_823446 | TetR family regulator |
| PyrP | 370 | Francci3_4206, Frankia sp. CcI3 | 47/66 | YP_483283 | 3-Dehydroquinate synthase |
| PyrQ | 454 | SCAB_41651, Streptomyces scabiei 87.22 | 39/56 | YP_003489785 | Aspartate kinase |
| PyrR | 408 | MXAN_4919, Myxococcus xanthus DK 1622 | 37/53 | YP_633075 | Cytochrome P450 |
| PyrS | 513 | BTH_II2161, Burkholderia thailandensis TXDOH | 43/59 | YP_440349 | Dehydrogenase |
| PyrT | 497 | nfa32650, Nocardia farcinica IFM 10152 | 68/82 | YP_119476 | Aldehyde dehydrogenase |
a Number of amino acids in the ORF predicted by Frame Plot 3.0.
b Functions of the most similar proteins from the NCBI database and from the predicted conserved motifs. NRPS and PKS domains are abbreviated as follows: A, adenylation; C, condensation; T, thiolation; KS, ketoacyl synthase; AT, acyltransferase; ACP, acyl carrier protein; KR, ketoreductase; TE, thioesterase.
Assembly of the Pyridomycin Core
Among the 26 ORFs, two typical NRPS genes, pyrE and pyrG, and a PKS gene, pyrF, were identified, and their functional domains matched the chemical structure of the pyridomycin core (Fig. 2A).
PyrE consists of two minimal NRPS modules and is most similar (45% identity) to DhbF, involved in the biosynthesis of the catecholic siderophore bacillibactin from Bacillus subtilis (27). The two A domains of PyrE are very similar to known A domains, and they feature all 10 highly conserved motifs. Module 1 of PyrE is similar to the pristinamycin I synthetase 2, which forms a 3-hydroxypicolinic acid-threonine moiety as was predicted for pyridomycin biosynthesis. Also, the 10 residues in the aa binding pocket predict incorporation of threonine by module 1 (28), consistent with the chemical structure and the feeding experiments (11). PyrE module 2 probably incorporates 3-(3-pyridyl)-l-alanine, which is similar to the weakly predicted phenylalanine.
PyrF is a minimal PKS module. The KS domain features the highly conserved catalytic Cys-His-His triad (29). The KS domain is most similar to typical NRPS/PKS hybrid KSs from blmIII (36% identity) or epoD (34% identity) (30, 31). The acyltransferase domain of PyrF contains the highly conserved active site GHSXG and is similar to methylmalonyl-CoA acyltransferases.
Special features of the PyrG (2451 aa) architecture are the location of a PKS KR domain and two A domains. PyrG is therefore an NRPS/PKS hybrid protein. The C domain in PyrG probably forms a C–O (ester) link as has been found in fumonisin (32) and antibiotic C-1027 (33).
The KR domain in PyrG was predicted to be functional because it contains a Rossmann fold for NAD(P)H binding and conserved Lys, Ser, and Tyr residues (34) (supplemental Fig. S4). Mutation of conserved active sites of the KR domain S163A/Y176F resulted in complete loss of pyridomycin production, and no intermediate product was detected in the supernatant or in the mycelium of three independent mutant clones. Analysis of the aa binding pockets of the two NRPS A domains gave no firm prediction for the type of extender unit. The PyrG-A1 domain lost the conserved A3 motif, which is critical for adenylate formation, whereas the A6 and A8 motifs in PyrG-A2 are not conserved (35) (supplemental Fig. S5). A TE domain at the C-terminal end of PyrG is probably responsible for lactone formation, as shown in Fig. 2A.
Biosynthesis of the Pyridyl Moieties
PyrB is an l-lysine 2-aminotransferase similar to VisA (61% identity and 70% similarity) involved in 3-HPA formation for virginiamycin biosynthesis in Streptomyces virginiae (14, 36) and to NikC (56% identity and 69% similarity) that catalyzes the initial reaction for converting lysine to the pyridyl residue in nikkomycin D in Streptomyces tendae (37). However, l-aspartic acid instead of l-lysine (along with glycerol or pyruvic acid) was incorporated into the two pyridyl residues in pyridomycin. This suggested that pyrB should not be involved in the pyridomycin biosynthesis and that the formation of the pyridyl residues may follow either the NAD biosynthetic pathway (path a in Fig. 2B) (38, 39) or the aspartate family of amino acids biosynthetic pathway (path b in Fig. 2B) (40).
Five genes (pyrP to -T) are transcribed in the same orientation and may constitute an operon (Table 1 and Fig. 2A). They initially seemed to be involved in the biosynthesis of pyridyl moieties. PyrQ is a putative aspartate kinase (40). PyrP resembles 3-dehydroquinate synthase (41). The other genes encode oxidoreductases. However, the inactivation of all of these genes did not affect pyridomycin production (supplemental Tables S2 and S3). These findings indicated that the biosyntheses of the pyridyl moieties would follow path a in Fig. 2B, using some genes from the primary metabolism.
Unknown or Tentative Role in Pyridomycin Biosynthesis
Immediately downstream of the NRPS/PKS genes, pryH to -N may form an operon (Fig. 2A and Table 1). PyrH (71 aa) is 74% identical to the MbtH-like protein from Streptomyces fungicidicus, and it contains the three conserved Trp residues that may be important for moderating protein-protein interactions (42, 43). Similar proteins are integral parts of several NRPSs that stimulate specific A domains (44). PyrJ is probably a histidinol-phosphate aminotransferase catalyzing the formation of the enol carboxylic acid moiety α-keto-β-methylvaleric acid (module 4, Fig. 2). PyrI, -K, -L, and -M are putative oxidoreductases (cytochrome P450 or dehydrogenases), and PyrN is similar to an esterase. The roles of these enzymes in pyridomycin biosynthesis remain unclear.
pyrC and pyrD, located upstream of NRPS/PKS genes, encode a flavin-dependent oxidoreductase and a sarcosine oxidase, respectively. Similar gene pairs of unknown functions (snaO (67% aa identity) and snaN (58% aa identity), and virN (65% aa identity) and virM (58% aa identity)) occur in the pristinamycin II (45) and in the virginamycin M biosynthesis gene clusters (14).
Genes Involved in Regulation and Self-resistance
Pyr3, a putative membrane protein, could be involved in a transport system for pyridomycin export and self-resistance. PyrO resembles TetR regulators containing a conserved helix-turn-helix DNA-binding domain (Pfam00440) (46). The inactivation of pyr2 (HTT12; supplemental Table S1) had no effect on pyridomycin production (confirmed by LC-MS analysis; data not shown), and it may be outside the biosynthetic gene cluster.
3-HPA Activation by PyrA and PyrU
The above analysis of PyrE to -G found no loading module. PyrA (543 aa), upstream of the NRPS/PKS/NRPS locus, is a probable 3-HPA:AMP ligase because it is similar to SnbA involved in pristinamycin I biosynthesis by S. pristinaespiralis (68% identity) and VisB involved in virginamycin S biosynthesis by S. virginiae (65% identity) (14, 26). Presumably, 3-HPA is activated by PyrA and incorporated into the pyridomycin assembly line (Fig. 2A). To confirm the role of pyrA in pyridomycin biosynthesis, an internal 1101-bp DNA fragment (encoding the entire catalytic triad) was replaced in the S. pyridomyceticus genome using the PCR targeting method (Fig. 3, A and B). HPLC and bioassay showed that pyridomycin production was completely abolished in the pyrA deletion mutant HTT6, but it was restored to near wild-type level by in trans complementation by a full-length pyrA gene expressed from PermE* (promoter of the erythromycin resistance gene) in the integrating plasmid pJTU4637b (Fig. 3, C and D).
FIGURE 3.
Construction and fermentation of the ΔpyrA mutant HTT6. A, schematic representation of the pyrA gene replacement in HTT6. B, confirmation of the inactivation of pyrA by insertion of an apramycin resistance-oriT cassette (aac(3)IV + oriT) by PCR using the primers pyrAidx1-f and pyrAidx1-r; the wild type gave a 2710 bp band, whereas all mutants yielded the expected 2994 bp band. M, 1 Kb plus DNA ladder (Invitrogen). C, HPLC profiles. a, pyridomycin standard; b–d, extracts from the wild-type strain (WT), non-producing mutant HTT6, and HTT6 complemented in trans by a functional pyrA gene. The slanted arrows point to the pyridomycin peak. D, bioassay with M. smegmatis. a–d, equal amounts of the samples labeled as for the HPLC profiles.
Next to pyrA but reading in the opposite direction is pyrU, which encodes a putative carrier protein. PSI-BLAST of its deduced amino acid sequence showed 42% identity to isochorismatase from Vibrio harveyi HY01, 37% to enterobactin synthetase component B in Azotobacter vinelandii DJ, and 36% to a conserved hypothetical protein from S. pristinaespiralis ATCC 25486. Sequence alignment with homologous proteins revealed an LGXXS motif in PyrU, which is a conserved motif in carrier proteins (47) (supplemental Fig. S6). Thus, PyrU is proposed to function as a PCP for tethering 3-HPA. No similar protein has been found among the pristinamycin I and virginamycin S biosynthetic genes.
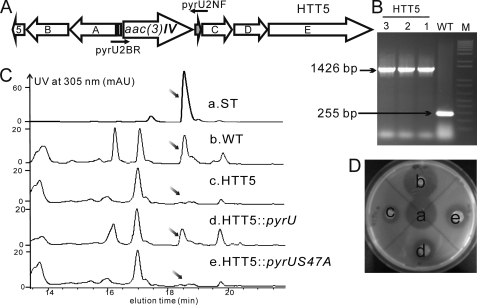
To confirm that pyrU is critical to the biosynthesis of pyridomycin, the ΔpyrU mutant HTT5 was constructed (Fig. 4, A and B) and lost the production of pyridomycin, as analyzed by HPLC. Complementation by introducing pJTU4655 containing pyrU constitutively expressed from the ermE* promoter partially restored pyridomycin production of HTT5 (Fig. 4C). As expected, the mutant gene pyrUS47A failed to restore normal pyridomycin production. The bioassay using Mycobacterium smegmatis mc2155 indicated that a trace amount of pyridomycin was produced by HTT5, and this was confirmed by LC-MS analysis ([M + H]+541.3 m/z, identical to the pyridomycin standard; Fig. 4, C and D). All of these findings clearly demonstrated that the putative PCP PyrU was essential for pyridomycin biosynthesis and that the serine 47 residue is the active site for the proposed phosphopantetheinylation.
FIGURE 4.
Construction and fermentation of ΔpyrU mutant HTT5. A, schematic representation of the pyrU mutant HTT5 by double crossover gene replacement. B, conformation of the replacement of pyrU by PCR using the primers pyrU2NF and pyrU2BR; the wild-type gave a 255 bp band, whereas all mutants gave the expected 1426 bp band. M, 1 Kb plus DNA ladder (Invitrogen). C, HPLC profiles of fermentation. a, pyridomycin standard; b–e, extracts from the WT strain, mutant HTT5, and HTT5 complemented by pyrU or pyrUS47A. The slanted arrows point to the pyridomycin peak. D, bioassay with M. smegmatis. a–d, equal amounts of the samples labeled as for the HPLC profiles. AU, absorbance units.
PyrU Is a Peptidyl Carrier Protein for Loading 3-HPA in Vivo and in Vitro
To test whether PyrU functions as a PCP for loading 3-HPA, His6-apo-PyrU, His6-holo-PyrU, and, as a control, the presumably inactive mutant His6-PyrUS47A were overproduced separately in E. coli (supplemental Fig. S7).
Phosphopantetheinylation in vivo was tested by coexpression of PyrU and Sfp (B. subtilis phosphopantetheinyl transferase expressed from pSV20) (48). Expression of PyrU without Sfp was used as a control. Protein purification and HPLC analysis showed that coexpression produced a new peak at 27.8 min compared with the peak eluting at 28.3 min from PyrU alone (Fig. 5A, a and b). The identity of the two peaks was confirmed by ESI-QTOF-MS analysis, giving 11,294.90 Da (calcd: 11,294.77 Da for apo-PyrU) and 11,635.21 Da (calcd: 11,635.86 Da for holo-PyrU), which is a +340 mass shift, consistent with a 4′-phosphopantetheine cofactor covalently attached to the Ser residue of apo-PyrU (Fig. 5B) (49). However, PyrU with the S47A mutation coexpressed with pSV20 only gave one peak at 28.8 min on HPLC analysis, showing that it was inactive (Fig. 5A, d and e).
FIGURE 5.
Phosphopantetheinylation of PyrU. A, HPLC profiles for phosphopantetheinylation. a, apo-PyrU (♦) from E. coli BL21(DE3)/pJTU4655; b, holo-PyrU (▾) biosynthesized by coexpression of PyrU and Sfp; c, in vitro conversion of apo-PyrU to holo-PyrU catalyzed by Sfp (●); d, apo-PyrUS47A (◊) from E. coli BL21(DE3)/pJTU4655(S47A); e, coexpression of PyrUS47A and Sfp. B, QTOF-MS analysis of apo-PyrU and holo-PyrU. The mass shift of 340 Da is consistent with the phosphopantetheinyl moiety transferred from CoA to apo-PyrU. AU, absorbance units; amu, atomic mass units.
Subsequently, we monitored the incorporation of the 4′-phosphopantetheine cofactor in vitro. When incubated with CoA and Sfp, apo-PyrU was quantitatively converted to holo-PyrU (Fig. 5A, c), as demonstrated by HPLC analysis, whereas no change was observed for PyrUS47A. Together, the data provide direct evidence that PyrU is a PCP with an active site Ser47.
Substrate Specificity of PyrA
To determine the substrate specificity in vitro, the 58.9-kDa N-terminally His6-tagged PyrA was produced in E. coli (supplemental Fig. S8). The reaction velocity of PPi release from ATP, catalyzed by PyrA with different aromatic acids and amino acids at 4 mm concentrations, was determined using a continuous spectrophotometric assay (see “Experimental Procedures”). The results depicted in Fig. 6A showed that 2,3-DHBA and 4A2HBA were activated with even higher reaction velocity than 3-HPA. The Km and kcat values for these three substrates were determined and are shown in Fig. 6C. Although higher turnover numbers (kcat) were indeed found for 2,3-DHBA and 4A2HBA, the catalytic efficiency (kcat/Km) of PyrA for 3-HPA was 22- and 40-fold higher than that for 2,3-DHBA and 4A2HBA, respectively. Taken together, the results are consistent with the assignment of 3-HPA as the native substrate of PyrA. However, the capability of PyrA to accept a range of different substrates if supplied at sufficient concentration offers the prospect to generate pyridomycin derivatives by feeding alternative starter units to a strain that has lost by mutation the ability to synthesize 3-HPA.
FIGURE 6.
Substrate specificity of PyrA in vitro. A, activation of different acyl substrates (4 mm) by PyrA. PPi release was determined by a continuous spectrophotometric assay with 30-min reaction. Error bars, S.D. values from three independently performed experiments. B, structures of the tested substrates. C, determination of Km and kcat values for 3-HPA (left), 2,3-DHBA (middle), and 4A2HBA (right).
In Vitro Reconstitution of the Loading Module for the Pyridomycin Biosynthesis
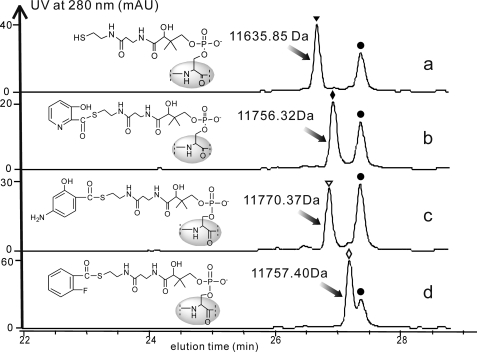
The functions and the location of PyrA and PyrU led to the expectation that PyrA may activate 3-HPA and tether it to PyrU, followed by transfer to the NRPS/PKS assembly line. To confirm this hypothesis, His6-holo-PyrU, His6-PyrA, and the substrate 3-HPA were incubated in a buffer containing Mg2+, DTT, and ATP. HPLC analysis and high resolution ESI-QTOF-MS showed a new peak at m/z 11,756.32 Da, consistent with the expected 3-HPA-holo-PyrU (calcd: 11,756.87 Da) (Fig. 7, a and b).
FIGURE 7.
HPLC profiles for acyl-PyrU formation catalyzed by PyrA. a, control performed with PyrA (●) and holo-PyrU (▾) without the acyl substrate; b, 3-HPA-PyrU (♦) formation with 3-HPA as a substrate; c, 4A2HBA-PyrU (▿) formation with 4A2HBA as a substrate; d, 2-FBA-PyrU (◊) formation with 2-FBA as a substrate. AU, absorbance units.
The 3-HPA analogs 4A2HBA and 2-fluorobenzoic acid (2-FBA) also yielded the expected products at m/z 11,770.37 and 11,757.40 Da (calcd: 11770.89 and 11,757.87 Da, respectively) (Fig. 7, c and d). Therefore, PyrA tethers 3-HPA and alternative aromatic molecules to PyrU, which acts as the loading module for pyridomycin assembly.
Production of New Pyridomycin Analogues
Inspired by the broad substrate specificity of PyrA and PyrU, we created new pyridomycin analogs by feeding picolinic acid, 2,3-DHBA, 4A2HBA, 2-FBA, and 2-chlorobenzoic acid into the wild-type strain. LC-QTOF-MS revealed three analogs derived from picolinic acid, 2,3-DHBA, and 4A2HBA (Table 2 and supplemental Fig. S9). No new product was generated from 2-FBA or 2-chlorobenzoic acid.
TABLE 2.
Aromatic compounds tested for precursor-directed feeding
| Feeding compounds | Expected m/z [M + H]+ for pyridomycin analogs | Found m/z [M + H]+ for pyridomycin analogs |
|---|---|---|
| 2,3-DHBA | 556.2290 | 556.2314 |
| 4A2HBA | 555.2449 | 555.2461 |
| Picolinic acid | 525.2344 | 525.2377 |
| 2-Chlorobenzoic acid | 558.2002 | NDa |
| 2-FBA | 542.2297 | ND |
a ND, not detected.
DISCUSSION
In this work, we report the cloning and characterization of a 42.5-kb DNA fragment of S. pyridomyceticus NRRL B-2517, which contains a gene cluster that encodes the enzymes for the assembly of the core structure of the antimycobacterial antibiotic pyridomycin.
The gene cluster represents an unusual NRPS/PKS hybrid system. PyrE, an NRPS elongation module containing two minimal modules (C-A-PCP), activates and tethers threonine and 3-(3-pyridyl)-l-alanine to the PCPs of NRPS1 and NRPS2, respectively, and forms an amide bond. Next, PyrF, a typical PKS elongation module, probably activates and transfers a methylmalonyl CoA to the PKS3 acyl carrier protein and elongates the chain.
Surprisingly, the PKS3, PyrF, lacks a KR domain that was thought to be responsible for the reduction of the β-keto group (*) to the hydroxyl group (*) of pyridomycin (Fig. 2A).
A probably functional KR domain is embedded in the subsequent NRPS4 (PyrG). Based on the structural analysis of pyridomycin, the KR in PyrG was initially predicted to catalyze the reduction of the ω-keto group (*) of the biosynthetic intermediate transferred from PKS3. Disruption by nonpolar point mutations should thus have produced a keto group at the position in pyridomycin (indicated by asterisk in Fig. 2A). However, no product was detected, indicating that chain assembly was prematurely aborted.
Similar NRPS modules have been described for cereulide, valinomycin, cryptophycin, and hectochlorin biosynthesis (50–53). They all catalyze the reduction of the α-keto carboxyl acid that is tethered directly to the PCP by a cognate A domain. As predicted from the in vitro biochemical investigation (54), the KR point mutant HTT19PSY (S163A/Y176F) did not produce any pyridomycin intermediate. The conservative point mutations were unlikely to change the overall structure of the NRPS4. It therefore seemed likely that the KR domain needs to be functional and is essential for pyridomycin production.
Probably, α-keto-β-methylvaleric acid (derived from isoleucine) is activated by the tandem A domains of NRPS4 and tethered to PCP. Then the KR domain may reduce the α-keto group of α-keto-β-methylvaleric acid to hydroxy, ready for ester bond formation catalyzed by the C domain (Fig. 2A). The TE domain then detaches and circularizes the chain. This would generate a hypothetical precursor that needs to be reduced at the position indicated by the asterisk in Fig. 2A and oxidized to form a double bond (#) in the structure of pyridomycin. The enzymes catalyzing these steps remain to be identified.
The tandem A domains of PyrG may act together to activate the substrate and tether it to PCP because PyrG-A1 lacks the conserved A3 motif for adenylate formation, and the A6 and A8 motifs in PyrG-A2 are not conserved. The TE domain at the C terminus of PyrG was proposed to catalyze the cyclization of the mature pyridomycin linear chain to form the lactone.
PyrE, PyrF, and PyrG constitute the hybrid NRPS/PKS that synthesizes the pyridomycin ring structure. However, an enterobactin EntB-like loading module containing an ArL-ArCP didomain was still missing for the pyridomycin assembly line (55).
PyrA, a predicted 3-HPA:AMP ligase, was shown to link ATP and 3-HPA, releasing PPi. 3-HPA:AMP ligases were proved to be involved in the biosynthesis of streptogramin B antibiotics, such as pristinamycin I, etamycin, and virginiamycin S (12–14). Therefore, PyrA was envisioned to activate 3-HPA using ATP. Indeed, the in vivo and in vitro experiments proved that PyrA selected and activated 3-HPA in the presence of ATP, a function that is normally performed by NRPS A domains.
The search for a carrier protein identified PyrU, a small 84-aa protein that has not been observed in the streptogramin B antibiotics biosynthesis (14, 45). Although it shows low homology with known PCPs or aryl carrier proteins (ArCPs), it features the conserved LGXXS motif for phosphopantetheinylation (supplemental Fig. S6). Therefore, PyrU was proposed to function as a PCP or ArCP receiving the activated 3-HPA from PyrA (55–57). To confirm the function of PyrU, the ΔpyrU mutant HTT5 was constructed. It almost completely lost pyridomycin production, which was restored by trans complementation, demonstrating the involvement of PyrU in pyridomycin biosynthesis. To obtain clear evidence that PyrU functions as a PCP/ArCP for loading 3-HPA, PyrU and its mutant PyrUS47A were overproduced and purified for biochemical characterization. Phosphopantetheinylation of PyrU but not of PyrUS47A was observed in vivo and in vitro, clearly identifying PyrU as a PCP/ArCP. With the characterized PyrA and PyrU in hand, we successfully reconstituted the loading module for pyridomycin assembly in vitro.
Pyridomycin and pristinamycin I use the same two initial building blocks. A predicted PCP gene (pyrU orthologue; SSDG_07480; Table 1) was found in the S. pristinaespiralis ATCC 25486 genome sequence, outside the pristinamycin biosynthetic gene cluster (45). We thus predict that the separate PCP may participate in the biosynthesis of pristinamycin I and other streptogramin B compounds.
Streptogramin B and pyridomycin contain 3-HPA starter units. The streptogramin 3-HPA is derived from lysine (14, 36, 37), but the labeling experiment indicated that both pyridyl moieties of pyridomycin originate from l-aspartic acid, glycerol, and/or pyruvate, but lysine is not incorporated (11). Pyridyl ring formation from aspartate is known from the primary metabolism NAD biosynthetic pathway (39) (path a in Fig. 2B). The three initial steps of path b in Fig. 2B (aspartate kinase, aspartate semialdehyde dehydrogenase, and dihydropicolinate synthase) are also used in the biosynthesis of lysine (40). For pyridyl ring formation, dehydrogenation produces pyridine 2,6-dicarboxylic acid (58), which can be envisioned to be converted to picolinic acid by decarboxylation. However, inactivation of the predicted aspartate kinase pyrQ did not reduce pyridomycin production. We therefore propose that both pyridyl moieties are synthesized by the reactions shown in Fig. 2, path a, similar to NAD biosynthesis in the primary metabolism and the shikimate pathway (38).
PyrA activated a series of aromatic acids, including two aromatic amino acids, and transferred them to PyrU. Precursor feeding of S. pyridomycetus yielded three pyridomycin analogs (Figs. 6 and 7 and supplemental Fig. S9), but no product was generated from 2-chlorobenzoic acid or 2-fluorobenzoic acid. A combination of rational engineering of the biosynthetic pathway and precursor feeding will provide opportunities to produce novel pyridomycin derivatives. This work sets the stage for ongoing in depth investigations of pyridomycin biosynthesis.
Supplementary Material
Acknowledgments
We are grateful to Dr. Tobias Kieser for critical reading of the manuscript and valuable comments and Prof. Lutz Heide for helpful discussions. We thank Dr. Wen Liu from the Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, for kindly providing plasmid pSV20.
This work was supported by the Ministry of Science and Technology of China 973 and 863 programs and the National Science Foundation of China.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Methods,” Tables S1–S3, and Figs. S1–S9.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) HM436809.
- NRPS
- nonribosomal peptide synthetase
- 3-HPA
- 3-hydroxypicolinic acid
- 4A2HBA
- 4-amino-2-hydroxybenzoic acid
- 2,3-DHBA
- 2,3-dihydroxybenzoic acid
- 2-FBA
- 2-fluorobenzoic acid
- QTOF
- quadrupole-time of flight
- A domain
- adenylation domain
- PCP
- peptidyl carrier protein
- C domain
- condensation domain
- TE
- thioesterase
- PKS
- polyketide synthase
- KS
- ketosynthase
- KR
- ketoreductase
- ArCP
- aryl carrier protein
- aa
- amino acid(s)
- contig
- group of overlapping clones.
REFERENCES
- 1. Koehn F. E., Carter G. T. (2005) Nat. Rev. Drug Discov. 4, 206–220 [DOI] [PubMed] [Google Scholar]
- 2. Cane D. E. (1997) Chem. Rev. 97, 2463–2464 [DOI] [PubMed] [Google Scholar]
- 3. Staunton J., Weissman K. J. (2001) Nat. Prod. Rep. 18, 380–416 [DOI] [PubMed] [Google Scholar]
- 4. Grünewald J., Marahiel M. A. (2006) Microbiol. Mol. Biol. Rev. 70, 121–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fischbach M. A., Walsh C. T. (2006) Chem. Rev. 106, 3468–3496 [DOI] [PubMed] [Google Scholar]
- 6. Du L., Shen B. (2001) Curr. Opin. Drug Discov. Dev. 4, 215–228 [PubMed] [Google Scholar]
- 7. Du L., Sánchez C., Shen B. (2001) Metab. Eng. 3, 78–95 [DOI] [PubMed] [Google Scholar]
- 8. Maeda K., Kosaka H., Okami Y., Umezawa H. (1953) J. Antibiot. 6, 140. [PubMed] [Google Scholar]
- 9. Maeda K. (1957) J. Antibiot. 10, 94–106 [PubMed] [Google Scholar]
- 10. Ogawara H., Maeda K., Koyama G., Naganawa H., Umezawa H. (1968) Chem. Pharm. Bull. 16, 679–687 [DOI] [PubMed] [Google Scholar]
- 11. Ogawara H., Maeda K., Umezawa H. (1968) Biochemistry 7, 3296–3302 [DOI] [PubMed] [Google Scholar]
- 12. de Crécy-Lagard V., Blanc V., Gil P., Naudin L., Lorenzon S., Famechon A., Bamas-Jacques N., Crouzet J., Thibaut D. (1997) J. Bacteriol. 179, 705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schlumbohm W., Keller U. (1990) J. Biol. Chem. 265, 2156–2161 [PubMed] [Google Scholar]
- 14. Namwat W., Kamioka Y., Kinoshita H., Yamada Y., Nihira T. (2002) Gene 286, 283–290 [DOI] [PubMed] [Google Scholar]
- 15. Kinoshita M., Nakata M., Takarada K., Tatsuta K. (1989) Tetrahedron Lett. 30, 7419–7422 [Google Scholar]
- 16. Waters B., Saxena G., Wanggui Y., Kau D., Wrigley S., Stokes R., Davies J. (2002) J. Antibiot. 55, 407–416 [DOI] [PubMed] [Google Scholar]
- 17. Yuan L. (1983) Antibiotics 8, 380–387 [Google Scholar]
- 18. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 19. Kieser T., Bibb M. J., Chater K. F., Butter M. J., Hopwood D. (2000) Practical Streptomyces Genetics: A Laboratory Manual, John Innes Foundation, Norwich, UK [Google Scholar]
- 20. Rose T. M., Henikoff J. G., Henikoff S. (2003) Nucleic Acids Res. 31, 3763–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ishikawa J., Hotta K. (1999) FEMS Microbiol. Lett. 174, 251–253 [DOI] [PubMed] [Google Scholar]
- 22. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ansari M. Z., Yadav G., Gokhale R. S., Mohanty D. (2004) Nucleic Acids Res. 32, W405–W413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lambalot R. H., Gehring A. M., Flugel R. S., Zuber P., LaCelle M., Marahiel M. A., Reid R., Khosla C., Walsh C. T. (1996) Chem. Biol. 3, 923–936 [DOI] [PubMed] [Google Scholar]
- 25. Marahiel M. A., Stachelhaus T., Mootz H. D. (1997) Chem. Rev. 97, 2651–2674 [DOI] [PubMed] [Google Scholar]
- 26. Thibaut D., Bisch D., Ratet N., Maton L., Couder M., Debussche L., Blanche F. (1997) J. Bacteriol. 179, 697–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. May J. J., Wendrich T. M., Marahiel M. A. (2001) J. Biol. Chem. 276, 7209–7217 [DOI] [PubMed] [Google Scholar]
- 28. Rausch C., Weber T., Kohlbacher O., Wohlleben W., Huson D. H. (2005) Nucleic Acids Res. 33, 5799–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Olsen J. G., Kadziola A., von Wettstein-Knowles P., Siggaard-Andersen M., Lindquist Y., Larsen S. (1999) FEBS Lett. 460, 46–52 [DOI] [PubMed] [Google Scholar]
- 30. Molnár I., Schupp T., Ono M., Zirkle R., Milnamow M., Nowak-Thompson B., Engel N., Toupet C., Stratmann A., Cyr D. D., Gorlach J., Mayo J. M., Hu A., Goff S., Schmid J., Ligon J. M. (2000) Chem. Biol. 7, 97–109 [DOI] [PubMed] [Google Scholar]
- 31. Shen B., Du L., Sanchez C., Edwards D. J., Chen M., Murrell J. M. (2001) J. Ind. Microbiol. Biotechnol. 27, 378–385 [DOI] [PubMed] [Google Scholar]
- 32. Zaleta-Rivera K., Xu C., Yu F., Butchko R. A., Proctor R. H., Hidalgo-Lara M. E., Raza A., Dussault P. H., Du L. (2006) Biochemistry 45, 2561–2569 [DOI] [PubMed] [Google Scholar]
- 33. Lin S., Van Lanen S. G., Shen B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4183–4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reid R., Piagentini M., Rodriguez E., Ashley G., Viswanathan N., Carney J., Santi D. V., Hutchinson C. R., McDaniel R. (2003) Biochemistry 42, 72–79 [DOI] [PubMed] [Google Scholar]
- 35. Stachelhaus T., Mootz H. D., Marahiel M. A. (1999) Chem. Biol. 6, 493–505 [DOI] [PubMed] [Google Scholar]
- 36. Namwat W., Kinoshita H., Nihira T. (2002) J. Bacteriol. 184, 4811–4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bruntner C., Bormann C. (1998) Eur. J. Biochem. 254, 347–355 [DOI] [PubMed] [Google Scholar]
- 38. Dewick P. M. (2002) Medicinal Natural Products: A Biosynthetic Approach, pp. 307–314, John Wiley & Sons, Ltd., West Sussex, UK [Google Scholar]
- 39. Foster J. W., Moat A. G. (1980) Microbiol. Rev. 44, 83–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Umbarger H. E. (1978) Annu. Rev. Biochem. 47, 532–606 [DOI] [PubMed] [Google Scholar]
- 41. Dewick P. M. (1995) Nat. Prod. Rep. 12, 579–607 [DOI] [PubMed] [Google Scholar]
- 42. Zhang W., Heemstra J. R., Jr., Walsh C. T., Imker H. J. (2010) Biochemistry 49, 9946–9947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Felnagle E. A., Barkei J. J., Park H., Podevels A. M., McMahon M. D., Drott D. W., Thomas M. G. (2010) Biochemistry 49, 8815–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wolpert M., Gust B., Kammerer B., Heide L. (2007) Microbiology 153, 1413–1423 [DOI] [PubMed] [Google Scholar]
- 45. Mast Y., Weber T., Gölz M., Ort-Winklbauer R., Gondran A., Wohlleben W., Schinko E. (2011) Microb. Biotechnol. 4, 192–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ramos J. L., Martínez-Bueno M., Molina-Henares A. J., Terán W., Watanabe K., Zhang X., Gallegos M. T., Brennan R., Tobes R. (2005) Microbiol. Mol. Biol. Rev. 69, 326–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weber T., Baumgartner R., Renner C., Marahiel M. A., Holak T. A. (2000) Structure 8, 407–418 [DOI] [PubMed] [Google Scholar]
- 48. Nakano M. M., Corbell N., Besson J., Zuber P. (1992) Mol. Gen. Genet. 232, 313–321 [DOI] [PubMed] [Google Scholar]
- 49. Quadri L. E., Weinreb P. H., Lei M., Nakano M. M., Zuber P., Walsh C. T. (1998) Biochemistry 37, 1585–1595 [DOI] [PubMed] [Google Scholar]
- 50. Magarvey N. A., Beck Z. Q., Golakoti T., Ding Y., Huber U., Hemscheidt T. K., Abelson D., Moore R. E., Sherman D. H. (2006) ACS Chem. Biol. 1, 766–779 [DOI] [PubMed] [Google Scholar]
- 51. Ehling-Schulz M., Vukov N., Schulz A., Shaheen R., Andersson M., Märtlbauer E., Scherer S. (2005) Appl. Environ. Microbiol. 71, 105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cheng Y. Q. (2006) Chembiochem 7, 471–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ramaswamy A. V., Sorrels C. M., Gerwick W. H. (2007) J. Nat. Prod. 70, 1977–1986 [DOI] [PubMed] [Google Scholar]
- 54. Magarvey N. A., Ehling-Schulz M., Walsh C. T. (2006) J. Am. Chem. Soc. 128, 10698–10699 [DOI] [PubMed] [Google Scholar]
- 55. Gehring A. M., Bradley K. A., Walsh C. T. (1997) Biochemistry 36, 8495–8503 [DOI] [PubMed] [Google Scholar]
- 56. Schmoock G., Pfennig F., Jewiarz J., Schlumbohm W., Laubinger W., Schauwecker F., Keller U. (2005) J. Biol. Chem. 280, 4339–4349 [DOI] [PubMed] [Google Scholar]
- 57. Keller U., Schlumbohm W. (1992) J. Biol. Chem. 267, 11745–11752 [PubMed] [Google Scholar]
- 58. Orsburn B. C., Melville S. B., Popham D. L. (2010) Mol. Microbiol. 75, 178–186 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.