Abstract
Signal transducer and activator of transcription 2 (STAT2), the critical component of type I interferons signaling, is a prototype latent cytoplasmic signal-dependent transcription factor. Activated tyrosine-phosphorylated STAT2 associates with STAT1 and IRF9 to bind the ISRE elements in the promoters of a subset of IFN-inducible genes (ISGs). In addition to activate hundreds of ISGs, IFNα also represses numerous target genes but the mechanistic basis for this dual effect and transcriptional repression is largely unknown. We investigated by ChIP-chip the binding dynamics of STAT2 and “active” phospho(P)-STAT2 on 113 putative IFNα direct target promoters before and after IFNα induction in Huh7 cells and primary human hepatocytes (PHH). STAT2 is already bound to 62% of our target promoters, including most “classical” ISGs, before IFNα treatment. 31% of STAT2 basally bound promoters also show P-STAT2 positivity. By correlating in vivo promoter occupancy with gene expression and changes in histone methylation marks we found that: 1) STAT2 plays a role in regulating ISGs expression, independently from its phosphorylation; 2) P-STAT2 is involved in ISGs repression; 3) “activated” ISGs are marked by H3K4me1 and H3K4me3 before IFNα; 4) “repressed” genes are marked by H3K27me3 and histone methylation plays a dominant role in driving IFNα-mediated ISGs repression.
Keywords: Chromatin Immunoprecipitation (ChiP), Chromatin Histone Modification, Interferon, Liver, STAT Transcription Factor, Transcription Regulation, Interferon α, Phospho-STAT2, STAT2
Introduction
Interferons are pleiotropic cytokines induced upon virus infection and other stimuli to modulate host immune response and are classified as type I interferon α and β (IFNα and IFNβ),3 type II (IFNγ), and the recently discovered type III (IFNλ) (1–3)). Interferons exert their function by phosphorylating latent transcription factors belonging to the signal transducer and activator of transcription (STAT) family after a signaling cascade, which begins with the binding of interferon to their membrane receptors and involves Janus kinases (JAKs). Receptor dimerization or oligomerization leads to Jak apposition and the trans-phosphorylation on tyrosine residues that releases their intrinsic catalytic activity. Tyrosine-phosphorylated cytokine-receptor cytoplasmic domains then provides binding sites for the Src homology-2 (SH2) domain of the STAT proteins, which are recruited to the JAKs and phosphorylated on a single tyrosine residue (Tyr-689 in the case of STAT2). The interaction between phosphorylated-SH2 domains on STAT proteins leads to homo- or hetero-dimerization and nuclear translocation (4–6). STAT2 Tyr-689 and STAT1 Tyr-701 can also be phosphorylated by non-receptor TKs, including SRC and ABL in the absence of ligand-induced receptor signaling (7). STAT dimers directly activate genes containing the IFNγ activation site (GAS) element, while the association of STATs with the DNA-binding protein interferon regulatory factor (IRF) 9 expands the range of DNA response elements that can be targeted by the JAK-STAT pathway to interferon-stimulate response element (ISRE) and IRF response element (IRE) (8). Un-phosphorylated STAT2 binds IRF9 and constitutively shuttles in and out of the nucleus (9). STAT2 tyrosine phosphorylation promotes the association of STAT2/IRF9 with STAT1 to form the IFN-stimulate gene factor 3 (ISGF3). ISGF3 enters the nucleus, binds to the ISRE elements present in the promoter regions of a subset of IFN-inducible genes and triggers transcription (10). In the ISGF3 complex, STAT1 and IRF9 are essential to mediate the DNA binding, whereas STAT2 provides a potent TAD (11). Although all STATs have been found to be activated by different type I IFNs in specific cell types and to participate in the formation of complexes with different intrinsic DNA binding specificities to GAS, ISRE, or IRE sites (11–13), STAT2 is absolutely required for a fully effective response to IFNα. Indeed, STAT2 may form stable homodimers in response to IFNα that interact with IRF9 and activate transcription of ISRE-containing genes (14) and cells lacking STAT2 do not respond at all to IFNα activation of ISGs whose transcription is ISRE-dependent (15). In addition, a hybrid IRF9-STAT2 protein is able to recapitulate type I IFN-stimulated gene expression and antiviral response in the absence of STAT1 (16).
Significantly less progress has been made toward identifying the specific STAT-regulated genes responsible for the different biological effects of IFNs and determining how these target genes are selected and regulated at the chromatin level. The repertoire of ISGs whose expression is modulated by the different IFNs has been greatly expanded by DNA microarrays studies (17–19). However, very few of these genes were originally validated as ISGF3 direct target genes (20, 21). Hartman et al. (20) have detected by ChIP-chip a large number of genomic sites in chromosome 22 to which STAT1 and/or STAT2 bind after IFNγ and/or IFNα stimulation and identified a number of new candidate direct target genes for STAT1 homodimers and STAT1:STAT2 heterodimers. Similarly, STAT1 binding sites have been located on the whole genome using several approaches, including ChIPSeq (22–24). Expression profiling studies have also shown that, in addition to activate hundreds of genes, IFNα is also able to repress numerous target genes (21, 25–27). The basis for this dual effect, and in particular for transcriptional repression, is still unclear as the composition of STAT containing complexes in genes activated or repressed in response to IFNs is largely unknown. To generate further mechanistic knowledge on ISREs-bound protein complexes, we investigated the binding dynamics of STAT2 and its “active” phospho-STAT2 form to a large panel of putative IFNα direct target promoters by ChIP-chip on a custom oligonucleotide promoter array. We found that Stat2 is already bound to 62% of investigated target promoters, including most “classical” ISGs, before IFNα treatment. Only a proportion of STAT2 basally bound promoters also show P-STAT2 positivity before and after IFNα. Combined ChIP and expression analysis indicates that STAT2 plays a role in regulating ISGs expression, independently from its phosphorylation. By correlating in vivo promoter occupancy and gene expression with RNA Pol II recruitment and changes in histone methylation, we could show that “activated” ISGs are marked by H3K4me and H3K4me3 before IFNα whereas IFNα “repressed” genes either have STAT2 bound before IFNα or recruit both STAT2 and P-STAT2, thus linking STAT2 with ISGs repression. Finally, “repressed” genes are consistently marked by H3K27me3, suggesting a dominant role of histone methylation in driving IFNα-mediated ISGs repression.
EXPERIMENTAL PROCEDURES
IFNα/STAT2 Array Design, Probe Generation, and Microarray Hybridization
We aimed at designing an oligonucleotide-based array containing a large repertoire of genes targeted by IFN-α/β, but not IFN-γ (21). Selection criteria and detail about the oligos can be found in supplemental materials. Probe generation was done essentially as previously described in the Whole Genome Amplification Method (28) using 10 ng of the input and the whole ChIP samples. For a detailed description of the amplification and the array hybridization see supplemental materials.
Chromatin Immunoprecipitation (ChIP)
ChIP analysis was performed as described in (29). Briefly, cells (0.5/1 × 108) were washed in phosphate-buffered saline (PBS) and incubated for 10 min with 1% formaldehyde; after quenching the reaction with glycine 0.125 m, cells were sonicated, and chromatin fragments of an average length of 1 kb recovered by centrifugation. Immunoprecipitations were performed with ProtG-Sepharose (KPL, 2235101) and 3–5 μg of the indicated antibodies. After immunoprecipitation, washes, and reverse cross-linking, the samples were extracted twice with phenol/chloroform, once with chloroform and ethanol precipitated in the presence of 30 μg of glycogen. In the Re-ChIp protocol, after the first immunoprecipitation, the samples were eluted in 10 mm DTT at 37 °C for 30′, the supernatant was collected, diluted 1:20 in Re-ChIP buffer (1% Triton, 2 mm EDTA, 150 mm NaCl, 20 mm Tris-HCl, pH 8) and incubated with the second antibody. Washes, reverse crosslinking, and extraction were the same as previously described. Quantitative PCR was performed using the SYBR-Green ROX mix, and an ABI Prism 7900 Sequence Detection System according to manufacturer's instructions (Applied Biosystems). Primers used are listed in supplemental Table S1. Dissociation curves after amplification showed that all primer pairs generated single products. The amount of PCR product amplified was calculated as % of a standard curve of the input. A genomic region between the GAPDH gene and the chromosome condensation-related SMC-associated protein (CNAP1) gene was used as negative internal control (CTL) (ChIP-IT kit -Active Motif) and the % of input values on target sites were divided for % of input values on the CTL region to obtain the specific enrichment.
Antibodies Used in ChIP
Phospho-STAT2 (Tyr-689) (07-224, Upstate); STAT2 (sc-476, Santa Cruz Biotechnology), H3K4me1, H3K4me3, and H3K27me3 (ab8895, ab1012, and ab6002, Abcam).
Data Analysis
Data analysis was conducted essentially as described in Ref. 30. For each array, background-subtracted signal intensities for ChIP Cy5 and the Input Cy3 channels were obtained. The STAT2 (or the STAT2+P-STAT2 in the re-ChIP experiments) ChIP-derived Cy5 values were analyzed both by considering the enrichment versus the Input Cy3 and the NoAb (or the Stat2+NoAb in the re-ChIP experiments) ChIP Cy5 channels. In the first analysis, as Cy5-ChIPed chromatin is hybridized together with Cy3-Input chromatin on the same array, values corrected for the corresponding background signals of the NoAb controls were used to calculate the ratio between Cy5 and Cy3 channels. A value >2 was considered significant. Moreover, to compare the Cy5 intensities of the same spot on different arrays (STAT2 versus NoAb, STAT2+P-STAT2 versus STAT2+NoAb), raw background-subtracted data were normalized to mean value of elements present in the second quartile of lowest intensities in the Cy5 channel. As previously reported, this normalization approach equalizes negative array elements without dampening the enrichment of signals in positive array elements (31). After this normalization, the Cy5/Cy3 intensity ratio of each spot in the STAT2 or STAT2+P-STAT2 slides were divided by the Cy5/Cy3 intensity ratio of the corresponding spot of the NoAb or STAT2+NoAb slide. An enrichment ratio of 1.4 was scored positive. The three replicate experiments for both STAT2 and STAT2+P-STAT2 were scaled to one another using the ExpressYourself data processing platform. More than 90% of oligonucleotides with a Cy5/Cy3 ratio considered “positive” (>2) were confirmed by the enrichment analysis over the corresponding NoAb. Entire list of values for the 339 oligos is available upon request.
Cell Cultures and IFNα Treatment
Huh7 hepatoma cells were maintained in DMEM supplemented with 10% fetal bovine serum (Invitrogen), 1% penicillin/streptomycin and 1% glutamine (Sigma). IFNα (Roferon-A, Roche, Basel) was used at a final concentration of 1000 IU/ml and added directly to the culture medium.
Liver Tissue Samples and Primary Culture of Human Hepatocytes
Primary human hepatocytes (PHH) were prepared from adult patients undergoing lobectomy or segmental liver resection for medically required purposes unrelated to this research program. The use of these human hepatic specimens for scientific purposes has been approved by the French National Ethic Committee. The PHH used in this study were obtained from two patients: FT304, 52-year-old female transplant donor and FT310, 60-year-old female, undergoing liver resection for a cystadenoma. Both patients were negative for HBV, HCV, and HIV serologic markers. PHH were prepared and cultured as described elsewhere (32). The cells were plated into collagen-coated dishes (BD Biosciences) at 1.7 × 105 cells/cm2 in a hormonally and chemically defined medium (32). Forty-eight hours after plating, PHH were exposed to IFNα at 1000 units/ml for the indicated times.
RNA Extraction and Taqman® Low Density Arrays
Subconfluent Huh7 cells were treated with 1000 IU/ml of IFNα and/or 300 nm TSA and total mRNAs were extracted at the indicated time points using the RNAeasy mini kit (Qiagen). Reverse transcription was performed using the random examers method (Superscript II kit, Invitrogen). RNA quality and quantity were monitored by ethidium bromide staining and by UV absorbance. Custom real-time PCR liquid arrays (Taqman® Low Density Arrays or TLDAs - Applied Biosystems) were loaded with 200 ng of cDNA per lane and run in a 7900HT Fast Real-Time apparatus (Applied Biosystems), according to manufacturer's instructions. Data were analyzed using the SDS 2.2.2 program. 18 S RNA was used as internal control for normalizing equal loading of samples.
RESULTS
In Vivo Binding of STAT2 to Target Promoters in Huh7 Cells
Aiming to get insights on IFNα/STAT2 trascriptome regulation at the chromatin level, we designed a custom oligonucleotide array to probe by ChIP-chip 113 gene promoters known to be modulated by type I IFNs (and not IFNγ) in expression profiling experiments and/or to be specifically bound by STAT2 in vivo in ChIP assays (see supplemental Table S2 and “Experimental Procedures” for selection criteria). For each promoter, three 50-mers were designed, the first one located in the upstream regulatory region (Oligo 1; between −1500 and −500 from tss), the second in the proximal promoter (Oligo 2; between −500 and +0) and the third downstream of the tss (oligo 3, between +0 and +1000). A complete description of the ISREs and 50-mers probes positions is given in supplemental Table S3. Cross-linked chromatin was prepared from untreated and IFNα treated Huh7 cells and immunoprecipitated with an α-STAT2-specific antibody. Amplified and Cy5-labeled anti-STAT2 and control NoAb ChIPed-chromatins were hybridized on the array together with the Cy3-labeled Input chromatin. Both the enrichment of STAT2 Cy5 channel over the NoAb Cy5 channel hybridized in parallel and of Cy5-ChIPed chromatin over the corresponding Cy3-Input chromatin were considered for data analysis (see “Experimental Procedures”). As expected, positive spots corresponded to the ISRE-positive oligonucleotides for each promoter set, the flanking negative oligonucleotides serving as internal controls for hybridization specificity (see supplemental Table S4). As shown in supplemental Fig. S1a (S1A) when oligonucleotides are divided into classes according to their enrichment over the Input before (left panel) and after (right panel) IFNα treatment, the negative internal control oligos fall into the group with Cy5/Cy3 ratios between 1 and 2 (therefore scored “negative”) and the frequency of negative internal control oligos scored “negative” is not significantly affected by IFN treatment. Notably, the vast majority of oligonucleotides scored as “positive” [i.e. with a Cy5/Cy3 ratio >2.0 (blue colums)] and display enrichment ratios >2,5 and only a marginal number of them falls near the cut-off values (2.0 to 2,5).
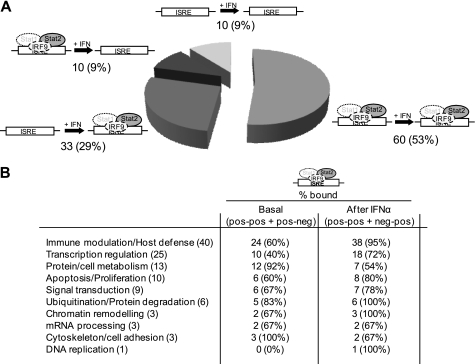
We found STAT2 already bound to 70 promoters, corresponding to 62% of total, before IFNα stimulation, and to 90 promoters, corresponding to 80% of total, after treatment (Fig. 1A). In particular, 60 (53%) promoters display STAT2 binding before and after IFNα stimulation, 10 (9%) lose a pre-existing binding after IFNα stimulation, while 33 (29%) acquire STAT2 binding only after IFNα treatment. 10 promoters (9%) show no STAT2 binding in both conditions (Fig. 1A). This latter observation may be related to the specific cell line used in the study or might reflect a binding kinetic different from the time points under investigation. The Gene Ontology categories most enriched in basal STAT2 binding are Immune modulation/Host defense (24 out of 40 genes), Transcription Regulation (10/25), Protein/Cell metabolism (12/13), Ubiquitination/Protein degradation (5/6) cytoskeleton/cell adhesion (3/3). After IFNα treatment, while cytoskeleton/cell adhesion and protein/cell metabolism genes tend to lose STAT2 binding, the immune modulation/host defense and transcription regulation categories dramatically increase the association with STAT2 (Fig. 1B). These data indicate that STAT2 binds in vivo to a number of target promoters, among which the large majority of “classical” ISGs before IFNα treatment and that the cytokine stimulation triggers a STAT2 re-distribution in addition to its well characterized recruitment on responsive genes. The notion of STAT2 occupancy on target promoters in unstimulated cells is indirectly supported by the observation that STAT2 shuttles in and out the nucleus without being phosphorylated (9), although the biological significance and the consequences on gene transcription were not established.
FIGURE 1.
ISRE occupancy by STAT2 before and after IFNα treatment. A, STAT2-ChIPped DNAs from untreated and IFNα-treated Huh7 cells were labeled and hybridized to the STAT2/IFNα array as described under “Experimental Procedures.” Genes were divided into four categories depending on whether they: 1) already display STAT2 binding to their ISRE site before IFNα treatment and maintain it after the treatment (pos-pos; 53%) 2) acquire STAT2 binding after treatment (neg-pos; 29%) 3) STAT2 is bound prior to IFNα treatment and detaches after treatment (pos-neg; 9%) 4) display no STAT2 binding before and after IFNα treatment (neg-neg; 9%). B, dynamics of ISRE/STAT2 occupancy according to Gene Ontology categories (within brackets the number of genes belonging to each category). The figures represent the percentage of genes for each GO category displaying ISRE STAT2 binding before (pos-pos and pos-neg groups, as defined in A) and after IFNα stimulation (pos-pos or neg-pos groups), respectively. The schemes representing the ISGF3 include all the known components of the complex (STAT1, STAT2, and IRF9). However, because our ChIP-chip experiments investigated only STAT2 direct binding to the ISREs, STAT1, and IRF9 are depicted as shaded with dotted line (STAT1) and as only dotted line (IRF9) (see the Introduction section for a detailed discussion of IRF9 requirement for STAT2 activity).
Phosphorylated STAT2 Binding to Target Promoters
Next, we investigated the binding of the putative “active” phosphorylated STAT2 (P-STAT2) to the target promoters before and after IFNα treatment. To this aim, we performed ChIP-chip experiments using chromatin sequentially chipped with α-STAT2 and then with α-P-STAT2 antibodies (see “Experimental Procedures”). Preliminary experiments showed that this approach permits to minimize false positive signals. supplemental Fig. S1B shows that, likewise for STAT2, also for P-STAT2 most oligonucleotides with a Cy5/Cy3 ratio evaluated as “positive” in our analysis (blue colums) display higher enrichment ratios (>2.5). The frequency of the negative internal control oligonucleotides is comparable to that found after STAT2 hybridization and is, again, not affected by IFNα stimulation.
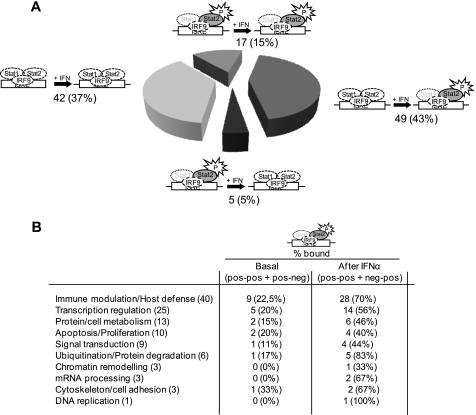
The overall view of the results shows that the majority of the P-STAT2 positive promoters (49; 43%) acquire P-STAT2 binding after IFNα treatment, while only a small subset of targets loses (5; 5%) or displays P-STAT2 occupancy before IFNα stimulation (17; 15%), respectively (Fig. 2A). 42 out of 113 promoters (37%) are not occupied by P-STAT2 at the investigated time points. All the promoters that are P-STAT2 positive also show STAT2 positivity. Conversely, all the promoters that are STAT2 negative do not show any P-STAT2 recruitment, thus confirming the reliability of the sequential ChIP protocol. It is worth to note that most well-known target genes belonging to the Immune modulation/Host defense Gene Ontology category acquire, as expected, P-STAT2 binding after IFNα stimulation. A similar behavior is shared by many target genes belonging to ubiquitination/protein degradation and transcription regulation GO categories (Fig. 2B).
FIGURE 2.
ISRE occupancy by P-STAT2 before and after IFNα treatment. A, cross-linked chromatin from untreated and IFNα-treated Huh7 cells was sequentially ChIPed first with α-STAT2 and then with α-P-STAT2, labeled and hybridized to the STAT2/IFNα array as described under “Experimental Procedures.” Genes were divided into 4 categories as in Fig. 1A: 1) [pos-pos]: 15%; 2) [neg-pos]: 43%; 3) [pos-neg]: 5%; 4) [neg-neg]: 37%. B, dynamics of ISRE/P-STAT2 occupancy according to Gene Ontology categories (within brackets the number of genes belonging to each category). The figures represent the percentage of genes for each GO category displaying ISRE P-Stat2 binding before (pos-pos or pos-neg groups, see Fig. 1A) and after IFNα stimulation (pos-pos or neg-pos groups), respectively. The ISGF3 cartoons are represented as in Fig. 1. Moreover, as a fraction of the promoters that are negative for P-STAT2 binding could be occupied by un-phosphorylated STAT2 in this categories the whole ISGF3 complex is drawn with dotted lines.
Next, we crossed the STAT2 and P-STAT2 occupancy data sets obtained in all the six ChIP-chip experiments. The analysis of STAT2 and P-STAT2 dynamic recruitment on chromatin before and after IFNα revealed a rather complex scenario. On the whole, 22 of the 72 (30%) target promoters that are STAT2 positive before IFNα treatment are also P-STAT2 positive (i.e. STAT2 and P-STAT2 pos-pos + pos-neg). This percentage raises to 74% after IFN treatment (i.e. STAT2 and P-STAT2 pos-pos + neg-pos) (Table 1). Among the genes that recruit STAT2 after IFN treatment (i.e. STAT2 neg-pos), about two-thirds of them (23/33) display a concomitant recruitment of P-STAT2 (i.e. P-STAT2 neg-pos). Similarly, between the gene targets that are STAT2 positive before and after IFN stimulation (i.e. STAT2 pos-pos), STAT2 occupation is accompanied by a P-STAT2 binding in 19 out of 60 (i.e. P-STAT2 pos-pos + pos-neg) before treatment, and in 46 out of 60 (i.e. P-STAT2 pos-pos + neg-pos) after treatment. Finally, the 12 promoters that possess STAT2 binding before IFN stimulation and lose it after treatment show a concomitant P-STAT2 occupancy only in three cases. From this data, it is evident that STAT2 and P-STAT2 binding is not always concomitant. In particular, P-STAT2 can be recruited to promoters that already show a pre-existing STAT2 binding (i.e. STAT2 pos-pos and P-STAT2 neg-pos); P-STAT2 could not be found on a number of STAT2 positive targets (i.e. P-STAT2 neg-neg) either before or after IFN, and, finally, it may be lost from regions that continue to display a STAT2 binding (i.e. STAT2 pos-pos and P-STAT2 pos- neg). Although some of these observations might be due to kinetic changes of P-STAT2 recruitment that we unable to reveal at the investigated time points, with mobile complexes that continuously bind and detach from DNA, they could also reflect a role of un-phosphorylated STAT2 in transcription regulation.
TABLE 1.
Combined analysis of STAT2 and P-STAT2 occupation ChIP-chip data
STAT2 and P-STAT2 ChIP-chip data sets are from six (3 + 3) experiments.
| STAT2 occupancya | n° genes | P-STAT2 occupancyb | n° genes |
|---|---|---|---|
| Neg-pos | 33 | Neg-pos | 23 |
| Neg-neg | 10 | ||
| Pos-pos | 60 | Neg-pos | 29 |
| Pos-pos | 17 | ||
| Pos-neg | 2 | ||
| Neg-neg | 12 | ||
| Pos-neg | 12 | Pos-neg | 3 |
| Neg-neg | 9 |
a STAT2 occupancy: genes are grouped according to STAT2 recruitment on their promoters before and after IFNα treatment.
b P-STAT2 occupancy: genes in each STAT2 category are subclassified according to P-STAT2 binding before and after IFNα.
ChIP-chip Results Validation
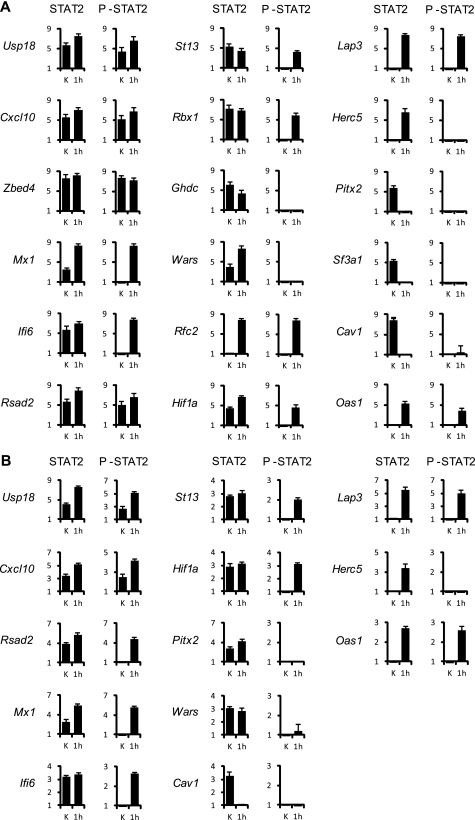
We selected 18 gene promoters representative of the different combinations of STAT2/P-STAT2 occupation status before and after IFNα to independently test the array data generated in Huh7 cells by performing quantitative PCR reactions (Fig. 3A). USP18, CXCL10 and the zinc finger transcription factor ZBED4 are positive for both STAT2 and P-STAT2 before and after IFNα treatment. USP18 was originally identified as a protease cleaving ISG15 from its conjugated proteins, but recent works attribute to USP18 also a role in the long-term desensitization of IFN signal transduction pathway in the mouse liver (33, 34). CXCL10 is a chemokine induced by both IFNα and IFNγ responses. Among the gene promoters that are STAT2 positive before and after IFN treatment, but recruit P-STAT2 only after the stimulus, we chose the typical ISGs MX1 (35), IFI6 (36), and RSAD2 (VIPERIN; (37)), the ubiquitin ligase RBX1 (38) and “Suppressor of tumorigenity 13” (ST13; (39)), encoding for the HIP1α protein that interacts with and potentiates the chaperone functions of HSP70 in protein folding and repair. Whereas MX1, IFI6, and RSAD2 increase STAT2 enrichment at their promoters after IFN treatment, ST13 and RBX1 display a global reduction of STAT2 binding after treatment that is accompanied by the recruitment of P-STAT2. The tryptophanyl-tRNA synthetase WARS (40) and the histone deacetylase inhibitor GHDC (LGP1; (41)) promoters are bound by STAT2 but do not display any P-STAT2 recruitment before or after IFN treatment. RFC2, HIF1A, and the interleukin-1β-converting enzyme (ICE) family member LAP3 show STAT2 and P-STAT2 recruitment only after IFN stimulation. RFC2 encodes the third-largest subunit of the replication factor C complex, involved in clamp loading and DNA replication (42). HIF1a, in addition to its well known role in the cell response to hypoxia, has been recently involved in innate immune responses: IFNα induces HIF1A in endothelial cells under normoxic conditions (43) and the administration of double-stranded nucleic acids results in an increase of HIF1α protein levels and HIF1α target genes expression (44). HERC5 (also referred to as CEB1), a E3 ligase that mediates type I IFN induced ISGylation of protein targets (45), shows STAT2 recruitment after IFN treatment and no binding of P-STAT2 at any investigated time points. The splicing factor subunit SF3A1, the caveolae structural protein CAV1 (46) and the transcription factor involved in β-catenin/Wnt signaling PITX2 (47) display STAT2 binding before IFN stimulation and no STAT2 and P-STAT2 binding after the treatment. Finally, we performed additional ChIP experiments to confirm STAT2 and P-STAT2 occupancy on the same set of promoters in primary human hepatocytes (PHH) (Fig. 3B). Altogether the validation step fully confirmed the ChIP-chip results, not only in the hepatocellular carcinoma cell line Huh7, but also in the context of non transformed primary cells.
FIGURE 3.
Validation of STAT2/P-STAT2 occupancy of ISRE sites in selected array genes. Chromatin from untreated (K) and IFNα-treated (1000 UI/ml for 1 h) Huh7 cells was immunoprecipitated with either α-STAT2 or α-P-STAT2 (right column) antibodies and analyzed by real-time PCR with primers amplifying the regions spanning the oligonucleotides specifically enriched according to the ChIP-chip arrays data analysis (see supplemental Tables S1 and S4 for the complete list of primers). Results are expressed as specific enrichment (% of input values on selected regions divided for % of input values on the control (CTL) region), (as detailed in the “Experimental Procedures”). The figure shows mean values derived from three independent ChIPs experiments. Bars indicate standard deviation.
Correlation between STAT2/P-STAT2 Promoter Occupation and IFNα-dependent Gene Expression Regulation
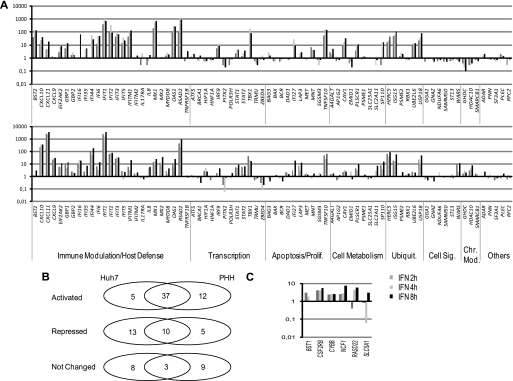
Next we investigated how STAT2/P-STAT2 promoter occupation and changes in ISRE-bound STAT2 complexes translated into gene expression in untreated and IFNα-treated Huh7 cells and PHH. More in detail we wanted to assess: 1) the function of STAT2 and P-STAT2 bound to promoters under basal conditions 2) whether un-phosphorylated STAT2 could play a role in directing ISGs transcription independent from P-STAT2, and 3) whether transcriptionally activated and repressed genes cluster in different STAT2/P-STAT2 binding categories. To this aim, we correlated the expression profile of 76 genes belonging to the ChIP-chip array with the STAT2/P-STAT2 occupancy at their promoters. Total RNAs were prepared from Huh7 cells and PHH left untreated or treated with 1000 IU/ml of IFNα for 2, 4, and 8 h and the cDNA loaded onto custom TLDAs (see “Experimental Procedures” for details). The results of the real time transcripts quantification in both cell lines are shown in Fig. 4 and in supplemental Fig. S6. Six genes that are not expressed in Huh7 cells, but are modulated in PHH are shown separately in panel C. Four of them are genes involved in metabolism of cells belonging to the immunomodulatory system (BST1, CSF2RB, CYBB, and NCF1), while RASD2 is a Ras protein homolog probably involved in schizophrenia and SLC5A1 is a sodium/glucose co-transporter (see supplemental Table S2 for gene annotations). Genes displaying a fold enrichment over control (untreated cells) >2 were considered as “activated” and those which had more than 40% reduction respect to the control were labeled as “repressed.”
FIGURE 4.
ISGs expression profiles in Huh7 cells and PHH after IFNα treatment. A, custom real-time PCR liquid arrays (TLDAs, Applied Biosystems) designed to include 76 genes from the STAT2/IFNα ChIP-chip array (see “Experimental Procedures” for more details) were loaded with 200 ng of cDNA obtained from total RNAs extracted from Huh7 cells (upper panel) or PHH (lower panel) treated with 1000 IU/ml for the indicated time points. Results are plotted as log10 enrichment over the basal level of expression in untreated cells. Genes that are not expressed in Huh7 cells, but are expressed and regulated in PHH by IFNα are shown in C. The complete list of expression values obtained from the average of three independent experiments is shown in supplemental Table S5. Standard deviation is always lower than 5% B, Venn representation of gene expression overlaps between Huh7 cells and PHH.
Supplemental Fig. S1C (S1C) shows the kinetics of IFNα effect on gene expression in Huh7 cells. Interestingly, activated genes (red columns) show a fold induction several folds higher than cut-off (>2), and their number reaches the maximum after 4 h of stimulation and remains the same also after 8 h post-IFNα. Repressed genes show a different kinetics, in that their highest frequency is found 2 h after treatment and then diminishes progressively.
42 genes (55,3% of total) in Huh7 and 49 genes (64% of total) in PHH are activated in response to IFNα, whereas repressed genes represent 30% of the total in Huh7 cells (23 genes) and 20% in PHH (15 genes) (Fig. 4A). The overlap between the two cell lines is about 80% for the activated genes and 70% for the repressed ones (Fig. 4B). In Fig. 4A, to allow a better visual comparison of the results in Huh7 cells (upper panel) and PHH (lower panel) genes were arranged according to their Gene Ontology category. Notably, immune modulation genes show little differences among the two cell lines, the more relevant being represented by IL8, which is, as expected, activated by IFNα in PHH but has an opposite behavior in Huh7 cells and MX2, a “typical” ISG that is unexpectedly only marginally activated by IFNα in Huh7. Transcription, cell metabolism, ubiquitination, and cell signaling categories show a very conserved expression profile in Huh7 and PHH. The few exceptions are the tryptophanyl tRNA synthetase WARS, the transporters SLC25A1 and SLC2A11, the E3 ligase RBX1 and the transcription factor HIF1a. Interestingly, the categories that display more differences in gene expression between Huh7 and PHH are apoptosis/cell proliferation and chromatin remodeling. The 3 chromatin modifiers HDAC10, GHDC, and SMARCB1 are all activated in PHH and repressed in Huh7 cells. Interestingly, HDAC10 C>T polymorphism, associated to an augmented HDAC10 expression, has been linked to an accelerated HCC development (48). GHDC functions as an HDAC inhibitor and it has been shown to sensitize U937 cells to TRAIL-induced apoptosis (41). SMARCB1 encodes for the key subunit of the SWI/SNF-like complex BAF, which has been involved in the rapid induction of ISGs by IFNα (49). The apoptosis/growth suppression promoting genes BAX, DAD1, and SGSM3 are all repressed in Huh7 cells and activated in PHH. Conversely, BAG3, a member of the BAG co-chaperone protein family which sustains cell survival (50), shows an opposite behavior. These differences also account for most of the differences existing between repressed genes in Huh7 with respect to PHH. Because most of the genes displaying a differential behavior are involved in cell proliferation and survival, it is likely that these differences might be related to the transformed nature of Huh7 cells.
Next, we crossed the expression profile data with the ChIP-chip results. As shown in Table 2, among the 42 activated genes, 25 are STAT2 positive before and after IFNα treatment and almost all of them display also a P-STAT2 binding, either before and after IFNα (P-STAT2 pos-pos, 10 genes) or only after IFNα (P-STAT2 neg-pos, 12 genes). Except a few exceptions (i.e. IFIT1, IFIT3, IFITM1), all the “typical” immunomodulated ISGs present in our custom TLDA enter this category. A smaller number of activated genes show the recruitment of STAT2 alone (STAT2 neg-pos and P-STAT2 neg-neg, 4 genes) or of both STAT2 and P-STAT2 only after IFNα stimulation (STAT2 and P-STAT2 neg-pos, 8 genes). On the other hand, half of the repressed genes display a STAT2 binding before IFN treatment (STAT2 pos-pos, 10 genes, and STAT2 pos-neg, 2 genes. Finally, (6) of the 23 repressed genes show the recruitment of both STAT2 and P-STAT2 after IFN stimulation.
TABLE 2.
Correlation between gene expression and STAT2/P-STAT2 occupation of ISRE sites
| STAT2 occupancyb | P-STAT2 occupancyb | n°genes | % | |||
|---|---|---|---|---|---|---|
| Activateda | n° genes | % total | ||||
| 42 | 55.3% | Pos-pos | Pos-pos | 10 | 23.8% | |
| Pos-pos | Neg-pos | 12 | 28.6% | |||
| Pos-pos | Pos-neg | 1 | 2.4% | |||
| Pos-pos | Neg-neg | 2 | 4.8% | |||
| Neg-pos | Neg-pos | 8 | 19% | |||
| Neg-pos | Neg-neg | 4 | 9.5% | |||
| Pos-neg | Neg-neg | 2 | 4,8% | |||
| Neg-neg | Neg-neg | 3 | 7.1% | |||
| Represseda | n° genes | % total | ||||
| 23 | 30.2% | Pos-pos | Neg-pos | 6 | 26% | |
| Pos-pos | Neg-neg | 4 | 17.4% | |||
| Pos-neg | Neg-neg | 2 | 8.7% | |||
| Neg-pos | Neg-pos | 6 | 26% | |||
| Neg-pos | Neg-neg | 2 | 8.7% | |||
| Neg-neg | Neg-neg | 3 | 13.2% |
a Genes modulated in Huh7 cells exposed to 1000 IU/ml IFNα for 2, 4, and 8 h were divided into three categories: activated, repressed, and not changed (see “Experimental Procedures” for more details).
b All combinations of STAT2/P-STAT2 occupancy at their ISRE sites are listed for activated and repressed ISGs. The frequency of each “ISRE occupancy group” is expressed both as absolute number of genes and as percentage within each category.
Scatter plots in supplemental Fig. S1D (S1D) show how activated (red spots) and repressed (green spots) genes divide according to their level of positivity for STAT2 or P-STAT2 enrichment before and after IFNα treatment. It is worth noting that both transcriptional activation and repression correlate with high levels of STAT2/P-STAT2 occupation. In addition, the scatter plot of P-STAT2 Cy5/Cy3 ratio in untreated Huh7 cells clearly shows that, differently from what observed with STAT2, there are no repressed genes having a basal P-STAT2 binding.
Altogether, these data suggest that many typical activated ISGs possess a basal STAT2 binding before IFNα stimulation, that is not always accompanied by a P-STAT2 binding either before or after IFNα and that, unexpectedly, IFNα repressed genes seem to require the presence of STAT2 before IFNα stimulation or the recruitment of both STAT2 and P-STAT2 after the treatment.
Chromatin Dynamics at STAT2/P-STAT2 Target Promoters
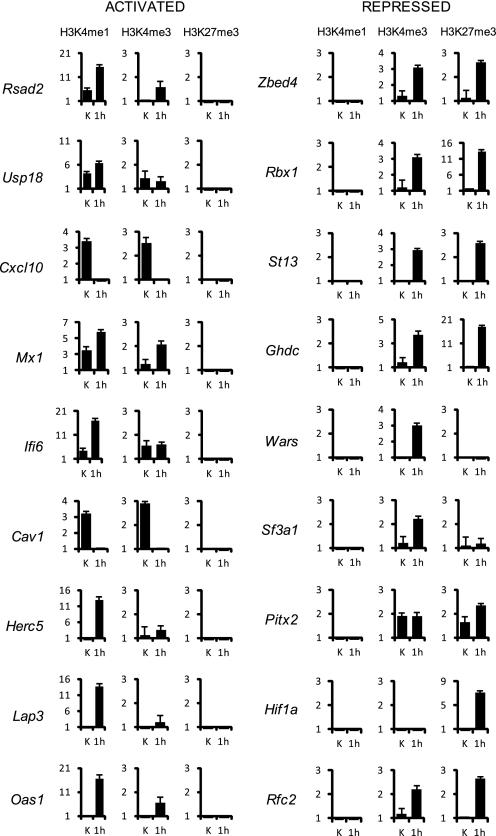
To investigate the chromatin context associated to IFNα stimulation and STAT2/P-STAT2 binding dynamics, we performed ChIP assays followed by real time PCR analysis on the same set of 18 promoters analyzed for STAT2/P-STAT2 occupation (see Fig. 3) with antibodies against, monomethyl- and trimethyl-Lys4 of histone H3 and trimethyl-Lys27 of histone H3 (Fig. 5). IFNα-activated genes show no association at all to the repressive mark H3K27me3. Conversely, the presence of H3K4me1 is restricted to activated promoters. Before IFNα stimulation, H3K4me1 can be found only on promoters that display a concomitant STAT2 occupation (RSAD2, USP18, CXCL10, MX1, IFI6 (in the STAT2 pos-pos group) and CAV1 (in the STAT2 pos-neg group)). Conversely, IFNα-repressed genes display a strong correlation with H3K27me3. In particular, H3K27me3 is associated to P-STAT2 presence/recruitment on target promoters (ZBED4, RBX1 (in the P-STAT2 pos-pos group), ST13, HIF1a and RFC2 (in the P-STAT2 neg-pos group)). SF3A1 and PITX2 represent an exception: both are STAT2 positive before IFNα and become STAT2 negative after the treatment; they display H3K27me3 but do not show any P-STAT2 binding either before or after IFN. The H3K4me3 modification is present both in activated and repressed genes. In repressed genes, it seems to mirror H3K27me3 and it is found also on those promoters that do not recruit P-STAT2 (GHDC and WARS).
FIGURE 5.
Histone marks at ISRE sites and transcriptional modulation of selected array genes. Chromatin from untreated (K) and IFNα-treated (1000 UI/ml for 1 h) Huh7 cells was immunoprecipitated with α-H3K4me1, α-H3K4me3 or an anti-H3K27me3 antibodies and analyzed by real-time PCR with primers amplifying the regions corresponding to the oligonucleotide specifically enriched according to the ChIP-chip arrays data analysis (see supplemental Tables S1 and S4). Results are expressed as specific enrichment (% of input values on selected regions divided for % of input values on the control (CTL) region) (see “Experimental Procedures” for more details). The figure shows mean values derived from three independent ChIPs experiments. Bars indicate standard deviation.
DISCUSSION
In this work we investigated by ChIP-chip the binding dynamics of STAT2 and its phosphorylated form P-STAT2 to the ISRE sequences present on a large repertoire of IFNα/β specific target genes before and after IFNα induction in hepatocellular carcinoma Huh7 cells and in primary human hepatocytes (PHH). The STAT2 and P-STAT2 occupancy and the chromatin context around the STAT2-bound ISREs were correlated with activation and repression of genes expression. We found that the 62% of the investigated promoters show STAT2 binding prior to IFNα treatment. The large majority of these genes belongs to the Immune Modulation/Host Defense pathway and include most of the “typical” ISGs. Although the presence of STAT2 on target promoters before its activation by the IFNα signaling cascade might be unexpected, it has to be underlined that recent work indicates that p53, known to be also “signal” activated, binds to its target sequences prior to “activation” by DNA damaging agents or other stimuli (30, 51–54). TCF4, the nuclear effector of Wnt signaling, has been shown to bind to its DNA target sequences on Wnt-responsive genes in a stimulus-independent manner and TCF4 chromatin recruitment seems to correlate with a specific epigenetic structure (55). A note of caution in the interpretation of chromatin occupancy by supposedly “latent” and “signal activated” transcription factors came from the observation that this phenomenon, in the case of p53 (Shaked et al., 54), might be restricted to transformed cells and not observed in non transformed cells. Our results do not support this view, and, indeed, we found a good correlation between STAT2 basal occupancy of target ISREs in Huh7 cells and in primary hepatocytes. Moreover, STAT2 is able to shuttle constitutively in and out of the nucleus without being phosphorylated (9). We found that only 30% of STAT2 basally bound promoters also show a P-STAT2 positivity and the percentage raises to 74% after IFNα stimulation. Although we cannot formally exclude that false-negatives results for P-STAT2 occupancy may have occurred for one or few genes, the combined analysis of the STAT2/P-STAT2 binding and target genes expression, strongly support the notion that STAT2 plays a role in regulating ISGs expression in a phoshorylation-independent manner. Similarly, un-phosphorylated STAT1 has been recently found to drive the constitutive expression of some target genes and to prolong the expression of specific ISGs without phosphorylation (56). Finally, Lou et al. (57) have shown that STAT2-IRF9 heterodimers can drive the activation of RIG-G, a “typical” ISG, without requiring STAT2 phosphorylation. Thus, different STATs or P-STATs complexes could alternate each other on specific targets and exert both distinct and overlapping functions.
Our data also show that P-STAT2 can be directly involved in repressing some interferon target genes. Indeed, we have recently reported that the recruitment of a P-STAT2 containing ISGF3 complex is required for transcriptional repression of the DNp73 promoter in response to IFNα (58). 26% of our repressed genes recruit both STAT2 and P-STAT2 after IFNα treatment, thus behaving as the DNp73 promoter. Moreover, 6 out of 12 genes repressed by IFNα that display STAT2 binding before interferon recruit P-STAT2 after treatment. These observations might be consistent with a role of basal STAT2 in the maintenance of a pre-existing “chromatin repressive mark,” whereas the “de novo” recruitment of a repressive complex would require P-STAT2. Notably, by the analysis of multiple ISRE sequence alignment, we could not find any significant differences in nucleotide composition or conservation between activated and repressed genes that might explain the recruitment of transcriptionally repressive versus active ISGF3 complexes(supplemental Fig. S2). Interestingly, in activated target genes, ISREs are mostly located between 0 and 200 from the tss, while in repressed genes the ISREs are far upstream or downstream.
We also investigated the chromatin context associated with STAT2/P-STAT2 binding by analyzing selected histone marks (i.e. H3K4me1, H3K4me3, and H3K27me3) around the ISREs involved in STAT2-mediated modulation of ISGs. It is well established that all monomethylated lysines are associated with active transcription (59). H3K4me3 is also widely associated to active transcription, but the results are more controversial (59). On the other hand, H3K27me3 appears to be a dominant mark and invariably mediates transcriptional repression (60). We found H3K4me1 only on activated promoters that display a concomitant STAT2 occupation before IFNα stimulation. Moreover, H3K4me1 is also found together with H3K4me3, confirming the frequent association of these two modifications at transcription start sites (61). These observations could suggest that basally bound STAT2 and H3K4me1/H3K4me3 may contribute together to “prime” chromatin loci that will be actively transcribed after IFNα stimulation. This would be also consistent with the finding that for the large majority of STAT1 binding sites after IFNγ stimulation, dominant H3K4me1/H3K4me3 combinations were already established before activation (62). Conversely, repressed genes display a strong correlation with the presence of H3K27me3, that is also associated to P-STAT2 presence/recruitment on target promoters. In repressed genes, H3K4me3 mirrors H3K27me3 pattern. The co-occurrence of the “positive” H3K4me3 and the “negative” H3K27me3 marks on the same locus has been already reported but it is not proven that both modifications occur on the same nucleosome.
Acetylation has been reported to play a dual role in the activation of ISGs and IFNα signaling, as it is required for a correct ISGF3 assembly (63, 64) but, after transcriptional complexes are recruited on the chromatin, it assumes a repressive role and HDACs activity is needed (65). When Huh7 cells were treated with both IFNα and TSA we could fully reproduce these findings. On the other hand, TSA treatment has no impact on IFNα-induced repression,4 indicating that acetylation does not play a pivotal role in IFN-repressive activity and probably histone methylation assumes a dominant role in driving IFNα-mediated ISGs repression.
In conclusion, we provide extensive new mechanistic knowledge on the transcriptional regulation at/around the ISRE regulatory sites of a large subset of ISGs and we define the molecular basis for IFNα-induced repression. Altogether, our observations further challenge the classical paradigm of STAT2 as a latent transcription factor activated only in response to cytokine stimulation. In this regard, we clearly show that STAT2 may regulate ISGs expression independently from its phosphorylation and that, on the other hand, P-Stat2 is involved in ISGs repression. We also found that histone methylation, rather than histones deacetylation, seems to have a dominant role in driving IFNα-mediated ISGs repression.
Our results may also translate into new insights for the clinical use of class I interferons. IFNα is the backbone drug for chronic hepatitis C patient (CHC) and an important option for chronic hepatitis B patients (CHB) (66, 67). Current IFNα-based therapies cure 50% of CHC patients but the molecular mechanisms that differentiate IFN responder and non-responder CHC patients are still unclear. Genome wide expression profiling studies have shown that the up-regulation of a specific set of IFNα responsive genes, including a number of “non classical” ISGs, prior to IFNα treatment in chronic HCV hepatitis patients predicts a subsequent non response to exogenous IFNα therapy (68, 69). A number of these “predictive” ISGs (namely USP18, CXCL10, MX1, IFI6, RSAD2, GHDC, LAP3, and HERC5) are included in this study and share some regulatory features and, in particular, the ability to be repressed by HDACs inhibitors. It will be interesting to expand the characterization of these “predictive” genes and to test whether the detailed knowledge of their chromatin dynamics might help in establishing meaningful ways to modulate their expression in CHC patients.
Supplementary Material
Acknowledgment
We thank Frauke Goeman for help in spotting the arrays.
This work was supported in part by grants from AIRC, European Community (LSHC-CT-2004-503576), MIUR-Cofin, and Progetti di Ateneo, Sapienza University of Rome and Schering-Plough (to M. L.). This work was also supported in part by fellowships from the AICF (American Italian Cancer Foundation) and from the Fondazione Adriano Buzzati-Traverso (to B. T.) and fellowships from the Fondazione A. Cesalpino and Rome Oncogenomic Center at the Regina Elena Cancer Institute (to F. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1–S5.
B. Testoni and M. Levrero, unpublished observations.
- IFNα
- interferon α
- ISRE
- interferon-stimulated responsive element
- ISG
- interferon-stimulated gene
- STAT
- signal transducer and activator of transcription
- PHH
- primary human hepatocytes.
REFERENCES
- 1. Stark G. R., Kerr I. M., Williams B. R., Silverman R. H., Schreiber R. D. (1998) Annu. Rev. Biochem. 67, 227–264 [DOI] [PubMed] [Google Scholar]
- 2. Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T. E., Kuestner R., Garrigues U., Birks C., Roraback J., Ostrander C., Dong D., Shin J., Presnell S., Fox B., Haldeman B., Cooper E., Taft D., Gilbert T., Grant F. J., Tackett M., Krivan W., McKnight G., Clegg C., Foster D., Klucher K. M. (2003) Nat. Immunol. 4, 63–68 [DOI] [PubMed] [Google Scholar]
- 3. Kotenko S. V., Gallagher G., Baurin V. V., Lewis-Antes A., Shen M., Shah N. K., Langer J. A., Sheikh F., Dickensheets H., Donnelly R. P. (2003) Nat. Immunol. 4, 69–77 [DOI] [PubMed] [Google Scholar]
- 4. Brierley M. M., Fish E. N. (2002) J. Interferon. Cytokine. Res. 22, 835–845 [DOI] [PubMed] [Google Scholar]
- 5. Uddin S., Platanias L. C. (2004) J. Biochem. Mol. Biol. 37, 635–641 [DOI] [PubMed] [Google Scholar]
- 6. Levy D. E., Darnell J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3, 651–662 [DOI] [PubMed] [Google Scholar]
- 7. Bowman T., Garcia R., Turkson J., Jove R. (2000) Oncogene 19, 2474–2488 [DOI] [PubMed] [Google Scholar]
- 8. Fu X. Y., Schindler C., Improta T., Aebersold R., Darnell J. E., Jr. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 7840–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Banninger G., Reich N. C. (2004) J. Biol. Chem. 279, 39199–39206 [DOI] [PubMed] [Google Scholar]
- 10. Fu X. Y., Kessler D. S., Veals S. A., Levy D. E., Darnell J. E., Jr. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 8555–8559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gupta S., Jiang M., Pernis A. B. (1999) J. Immunol. 163, 3834–3841 [PubMed] [Google Scholar]
- 12. Kisseleva T., Bhattacharya S., Braunstein J., Schindler C. W. (2002) Gene 285, 1–24 [DOI] [PubMed] [Google Scholar]
- 13. Wesoly J., Szweykowska-Kulinska Z., Bluyssen H. A. (2007) Acta Biochim. Pol. 54, 27–38 [PubMed] [Google Scholar]
- 14. Bluyssen H. A., Levy D. E. (1997) J. Biol. Chem. 272, 4600–4605 [DOI] [PubMed] [Google Scholar]
- 15. Leung S., Qureshi S. A., Kerr I. M., Darnell J. E., Jr., Stark G. R. (1995) Mol. Cell. Biol. 15, 1312–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kraus T. A., Lau J. F., Parisien J. P., Horvath C. M. (2003) J. Biol. Chem. 278, 13033–13038 [DOI] [PubMed] [Google Scholar]
- 17. Der S. D., Zhou A., Williams B. R., Silverman R. H. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geiss G. K., Carter V. S., He Y., Kwieciszewski B. K., Holzman T., Korth M. J., Lazaro C. A., Fausto N., Bumgarner R. E., Katze M. G. (2003) J. Virol. 77, 6367–6375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu W. Z., Sun H. C., Wang L., Chen J., Liu K. D., Tang Z. Y. (2005) World J. Gastroenterol. 11, 6613–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ward S. V., Samuel C. E. (2003) Virology 313, 553–566 [DOI] [PubMed] [Google Scholar]
- 21. Hartman S. E., Bertone P., Nath A. K., Royce T. E., Gerstein M., Weissman S., Snyder M. (2005) Genes Dev. 19, 2953–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wormald S., Hilton D. J., Smyth G. K., Speed T. P. (2006) BMC. Genomics. 7, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bhinge A. A., Kim J., Euskirchen G. M., Snyder M., Iyer V. R. (2007) Genome Res. 17, 910–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robertson G., Hirst M., Bainbridge M., Bilenky M., Zhao Y., Zeng T., Euskirchen G., Bernier B., Varhol R., Delaney A., Thiessen N., Griffith O. L., He A., Marra M., Snyder M., Jones S. (2007) Nat. Methods. 4, 651–657 [DOI] [PubMed] [Google Scholar]
- 25. Xu D., Erickson S., Szeps M., Gruber A., Sangfelt O., Einhorn S., Pisa P., Grandér D. (2000) Blood 96, 4313–4318 [PubMed] [Google Scholar]
- 26. Radaeva S., Jaruga B., Hong F., Kim W. H., Fan S., Cai H., Strom S., Liu Y., El-Assal O., Gao B. (2002) Gastroenterology 122, 1020–1034 [DOI] [PubMed] [Google Scholar]
- 27. Inagaki Y., Nemoto T., Kushida M., Sheng Y., Higashi K., Ikeda K., Kawada N., Shirasaki F., Takehara K., Sugiyama K., Fujii M., Yamauchi H., Nakao A., de Crombrugghe B., Watanabe T., Okazaki I. (2003) Hepatology 38, 890–899 [DOI] [PubMed] [Google Scholar]
- 28. O'Geen H., Nicolet C. M., Blahnik K., Green R., Farnham P. J. (2006) BioTechniques 41, 577–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Testoni B., Mantovani R. (2006) Nucleic Acids Res. 34, 928–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ceribelli M., Alcalay M., Viganò M. A., Mantovani R. (2006) Cell Cycle. 5, 1102–1110 [DOI] [PubMed] [Google Scholar]
- 31. Jen K. Y., Cheung V. G. (2005) Cancer Res. 65, 7666–7673 [DOI] [PubMed] [Google Scholar]
- 32. Pichard-Garcia L., Gerbal-Chaloin S., Ferrini J. B., Fabre J. M., Maurel P. (2002) Methods Enzymol. 357, 311–321 [DOI] [PubMed] [Google Scholar]
- 33. Malakhov M. P., Malakhova O. A., Kim K. I., Ritchie K. J., Zhang D. E. (2002) J. Biol. Chem. 277, 9976–9981 [DOI] [PubMed] [Google Scholar]
- 34. Sarasin-Filipowicz M., Wang X., Yan M., Duong F. H., Poli V., Hilton D. J., Zhang D. E., Heim M. H. (2009) Mol. Cell. Biol. 29, 4841–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haller O., Stertz S., Kochs G. (2007) Microbes. Infect. 9, 1636–1643 [DOI] [PubMed] [Google Scholar]
- 36. Zhu H., Zhao H., Collins C. D., Eckenrode S. E., Run Q., McIndoe R. A., Crawford J. M., Nelson D. R., She J. X., Liu C. (2003) Hepatology 37, 1180–1188 [DOI] [PubMed] [Google Scholar]
- 37. Helbig K. J., Lau D. T., Semendric L., Harley H. A., Beard M. R. (2005) Hepatology 42, 702–710 [DOI] [PubMed] [Google Scholar]
- 38. Barry M., Früh K. (2006) Sci. STKE. 2006, pe21. [DOI] [PubMed] [Google Scholar]
- 39. Shi Z. Z., Zhang J. W., Zheng S. (2007) J. Zhejiang. Univ Sci. B. 8, 170–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kisselev L., Frolova L., Haenni A. L. (1993) Trends Biochem. Sci. 18, 263–267 [DOI] [PubMed] [Google Scholar]
- 41. D'Acunto C. W., Carratù A., Rodriquez M., Taddei M., Parente L., Petrella A. (2010) Anticancer. Res. 30, 887–894 [PubMed] [Google Scholar]
- 42. Noskov V. N., Araki H., Sugino A. (1998) Mol. Cell. Biol. 18, 4914–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gerber S. A., Pober J. S. (2008) J. Immunol. 181, 1052–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hong S. W., Yoo J. W., Kang H. S., Kim S., Lee D. K. (2009) Mol. Cells. 27, 243–250 [DOI] [PubMed] [Google Scholar]
- 45. Wong J. J., Pung Y. F., Sze N. S., Chin K. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10735–10740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao X., Liu Y., Ma Q., Wang X., Jin H., Mehrpour M., Chen Q. (2009) Biochem. Biophys. Res. Commun. 378, 21–26 [DOI] [PubMed] [Google Scholar]
- 47. Kioussi C., Briata P., Baek S. H., Rose D. W., Hamblet N. S., Herman T., Ohgi K. A., Lin C., Gleiberman A., Wang J., Brault V., Ruiz-Lozano P., Nguyen H. D., Kemler R., Glass C. K., Wynshaw-Boris A., Rosenfeld M. G. (2002) Cell 111, 673–685 [DOI] [PubMed] [Google Scholar]
- 48. Park B. L., Kim Y. J., Cheong H. S., Lee S. O., Han C. S., Yoon J. H., Park J. H., Chang H. S., Park C. S., Lee H. S., Shin H. D. (2007) Biochem. Biophys. Res. Commun. 363, 776–781 [DOI] [PubMed] [Google Scholar]
- 49. Cui K., Tailor P., Liu H., Chen X., Ozato K., Zhao K. (2004) Mol. Cell. Biol. 24, 4476–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rosati A., Ammirante M., Gentilella A., Basile A., Festa M., Pascale M., Marzullo L., Belisario M. A., Tosco A., Franceschelli S., Moltedo O., Pagliuca G., Lerose R., Turco M. C. (2007) Int. J. Biochem. Cell Biol. 39, 1337–1342 [DOI] [PubMed] [Google Scholar]
- 51. Kaeser M. D., Iggo R. D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Espinosa J. M., Verdun R. E., Emerson B. M. (2003) Mol. Cell. 12, 1015–1027 [DOI] [PubMed] [Google Scholar]
- 53. Imbriano C., Gurtner A., Cocchiarella F., Di Agostino S., Basile V., Gostissa M., Dobbelstein M., Del Sal G., Piaggio G., Mantovani R. (2005) Mol. Cell. Biol. 25, 3737–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shaked H., Shiff I., Kott-Gutkowski M., Siegfried Z., Haupt Y., Simon I. (2008) Cancer Res. 68, 9671–9677 [DOI] [PubMed] [Google Scholar]
- 55. Wöhrle S., Wallmen B., Hecht A. (2007) Mol. Cell. Biol. 27, 8164–8177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cheon H., Stark G. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9373–9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lou Y. J., Pan X. R., Jia P. M., Li D., Xiao S., Zhang Z. L., Chen S. J., Chen Z., Tong J. H. (2009) Cancer Res. 69, 3673–3680 [DOI] [PubMed] [Google Scholar]
- 58. Testoni B., Schinzari V., Guerrieri F., Gerbal-Chaloin S., Blandino G., Levrero M. (2011) Oncogene, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang Z., Schones D. E., Zhao K. (2009) Curr. Opin. Genet. Dev. 19, 127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang Z., Zang C., Rosenfeld J. A., Schones D. E., Barski A., Cuddapah S., Cui K., Roh T. Y., Peng W., Zhang M. Q., Zhao K. (2008) Nat. Genet. 40, 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 62. Robertson A. G., Bilenky M., Tam A., Zhao Y., Zeng T., Thiessen N., Cezard T., Fejes A. P., Wederell E. D., Cullum R., Euskirchen G., Krzywinski M., Birol I., Snyder M., Hoodless P. A., Hirst M., Marra M. A., Jones S. J. (2008) Genome Res. 18, 1906–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tang X., Gao J. S., Guan Y. J., McLane K. E., Yuan Z. L., Ramratnam B., Chin Y. E. (2007) Cell 131, 93–105 [DOI] [PubMed] [Google Scholar]
- 64. Krämer O. H., Heinzel T. (2010) Mol. Cell. Endocrinol. 315, 40–48 [DOI] [PubMed] [Google Scholar]
- 65. Chang H. M., Paulson M., Holko M., Rice C. M., Williams B. R., Marié I., Levy D. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9578–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ghany M. G., Strader D. B., Thomas D. L., Seeff L. B. (2009) Hepatology 49, 1335–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. (2009) J. Hepatol. 50, 227–242 [DOI] [PubMed] [Google Scholar]
- 68. Chen L., Borozan I., Feld J., Sun J., Tannis L. L., Coltescu C., Heathcote J., Edwards A. M., McGilvray I. D. (2005) Gastroenterology 128, 1437–1444 [DOI] [PubMed] [Google Scholar]
- 69. Sarasin-Filipowicz M., Oakeley E. J., Duong F. H., Christen V., Terracciano L., Filipowicz W., Heim M. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7034–7039 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.