Abstract
It was found recently that a diabetes-associated protein in insulin-sensitive tissue (DAPIT) is associated with mitochondrial ATP synthase. Here, we report that the suppressed expression of DAPIT in DAPIT-knockdown HeLa cells causes loss of the population of ATP synthase in mitochondria. Consequently, DAPIT-knockdown cells show smaller mitochondrial ATP synthesis activity, slower growth in normal medium, and poorer viability in glucose-free medium than the control cells. The mRNA levels of α- and β-subunits of ATP synthase remain unchanged by DAPIT knockdown. These results indicate a critical role of DAPIT in maintaining the ATP synthase population in mitochondria and raise an intriguing possibility of active role of DAPIT in cellular energy metabolism.
Keywords: ATP Synthase, ATPases, Cell Metabolism, Diabetes, Energy Metabolism, F1F0-ATPase, H+-ATPase, Insulin resistance, Mitochondria, Mitochondrial Metabolism
Introduction
ATP synthase is a ubiquitous enzyme found in plasma membranes of bacteria, inner membranes of mitochondria, and chloroplast thylakoid membranes. It is a rotary motor enzyme that synthesizes ATP by proton flow along the gradient of electrochemical potential of proton across membranes (1–10). ATP synthase consists of two major portions: a membrane-protruding globular F1 domain that acts as an ATPase when isolated, and a membrane-embedded Fo domain that acts as a proton channel when F1 is removed. In the typical bacterial ATP synthase, the subunit composition of F1 is α3β3γδϵ and that of Fo is ab2cn (n, different among species). The proton flow drives rotation of the oligomer ring of c-subunits (c-ring) that makes the γϵ central stalk rotate with it, generating torque and conformational changes in the catalytic α3β3 domain of F1 to synthesize ATP. To prevent the dragged rotation, the second stalk, made of δ and b2, connects the stators in F1 (α3β3) and F0 (a-subunit). In addition to these core subunits, mitochondrial ATP synthase contains several other subunits, that is, d, e, f, g, A6L, F6, and mitochondrial ϵ, which is different from bacterial ϵ (11). Further, it was found recently that two proteins with unknown function, DAPIT3 (diabetes-associated protein in insulin-sensitive tissue) and MLQ (6.8-kDa proteolipid), are associated with mammalian mitochondrial ATP synthase (12, 13).
In this report, we focused on DAPIT, a small (58-amino acid), basic (pI = 10.4) protein with a single putative transmembrane segment (14). DAPIT was first recognized as a gene product whose mRNA level increased during skeletal muscle growth in rats and decreased by treatment with streptozotocin, a drug that induces diabetes (15). DAPIT is associated with ATP synthase in a stoichiometric manner but is not found in ATP synthase purified in the absence of phospholipids (12). Also, DAPIT is prone to dissociation from ATP synthase in the presence of relatively strong detergents, but ATP synthase without DAPIT still retains the same ATP hydrolysis activity (13). Orthologs of DAPIT and MLQ are found in vertebrates and invertebrates but not in yeast and other fungi (12). From these observations, DAPIT has been assumed to have some minor roles that are dispensable for the core function of ATP synthase. However, here we report that knockdown of DAPIT resulted in a drastic decrease in the population of ATP synthase in human cells.
EXPERIMENTAL PROCEDURES
Cells and Materials
HeLa and Phoenix GP cells were purchased from Health Science Research Resources Bank (JCRB9004) and American Type Culture Collection (SD-3514), respectively. They were cultured at 37 °C in a 5% CO2 incubator in Dulbecco's modified Eagle's medium (DMEM) (Nissui Pharmaceutical) supplemented with 10% FBS (Invitrogen). Unless otherwise stated, DMEM containing 0.1% glucose was used. Oligomycin was purchased from Cell Signaling Technology. Antibodies against ATP5A (α-subunit), ATP5B (β-subunit), and voltage-dependent anion channel protein (VDAC) were purchased from Molecular Probes (A21350 and A21351) and Abcam (ab16816), respectively. HRP-linked anti-mouse and anti-rabbit antibodies were purchased from GE Healthcare (NA931 and NA934). Rabbit antibody against human DAPIT was obtained by using purified DAPIT with histidine tag at the C terminus expressed in Escherichia coli.
Retroviral Gene Transduction
For knocking down of DAPIT, we used four shRNAs: shDAPIT-1 (TACCAGTTCACTGGTATTA), shDAPIT-2 (CTGTGTACTGGCCACATAT), shDAPIT-3 (GTTAAGGTCCAAAAAAACT), and shDAPIT-4 (TCGTCTGTGCTGCCAATCGACTCGGCGTG) targeted to the coding sequence and 5′-UTR of the human DAPIT gene. They were cloned to shRNA-expressing retroviral vector of pSUPER.retro.puro (OligoEngine). A packaging cell Phoenix GP was transfected with a retroviral vector and pCI-VSVG (Addgene) to express vesicular stomatitis virus-G envelope protein. Each retrovirus was used to infect the target HeLa cells (16).
mRNA Quantification
mRNAs for ATP5A1 (α-subunit), ATP5B (β-subunit), and glyceraldehyde-3-phosphate dehydrogenase in the infected HeLa cells were quantified by PCR. The total RNA was isolated with an illustra RNAspin Mini RNA Isolation kit (GE Healthcare), and cDNA was synthesized with PrimeScript II reverse transcriptase using oligo(dT) primer (TAKARA). PCR was performed on the StepOnePlusTM instrument (Applied Biosystems) with a SYBR Green Master Mix (Applied Biosystems) using the following primers: ATP5A1 forward, 5′-CAAGCCTTGTTGGGCACTAT-3′ and reverse, 5′-AGCTTCAAATCCAGCCAAGA-3′; ATP5B forward, 5′-CCCTGAAGGAGACCATCAAA-3′ and reverse, 5′-TTGCCACAGCTTCTTCAATG-3′; glyceraldehyde-3-phosphate dehydrogenase forward, 5′-GCTCTCCAGAACATCATCCC-3′ and reverse, 5′-TTTCTAGACGGCAGGTCAGG-3′. The data were calculated according to the ΔΔCt method (17) using mRNA of glyceraldehyde-3-phosphate dehydrogenase as an internal calibration standard.
Cell Growth and Viability
Proliferation of cells was measured by Cell Count Reagent SF (Nacalai tesque), a method based on colorimetric quantification of NADH in the culture that is proportional to cell number (18). Briefly, cells were passaged at 5.0 × 102 cells in a 96-well plate and cultured overnight. Culture was continued another 2 days, and the amounts of NADH before and after the 2-day culture were compared with the estimated proliferation of cells. Viability of cells in the glucose-containing and glucose-free DMEM was tested as follows. After the cells were passaged at 1.0 × 105 cells in a 6-cm dish and incubated overnight, the culture medium was exchanged either with new DMEM, which contained 0.1% glucose, or new glucose-free DMEM, which contained 0.1% galactose instead of glucose. In this experiment, the dialyzed FBS was used for a supplement. After a 2-day culture, cells were trypsinized, collected, and treated with trypan blue, and the numbers of living and dead cells were counted by a CountessTM automated cell counter (Invitrogen).
Measurement of Membrane Potentials
HeLa cells (shMock, shDAPIT-2, shDAPIT-4) were grown in DMEM without phenol red on a 35-mm glass-bottom dish (IWAKI). The dish was set on a microscope (Olympus IX-81) and incubated with 50 nm tetramethylrhodamine ethylester (TMRE) (Molecular Probes) for 10 min at 37 °C in a CO2 chamber attached to the microscope. The fluorescent images of TMRE were observed with a filter unit (Olympus U-MWIG3; excitation 530–550 nm, emission ≥575 nm). Then, 10 μm (final concentration) carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added, and fluorescent images were observed after 10 min.
Other Procedures
ATP synthesis activities of cultured cells were measured by a newly developed assay that utilized streptolysin O to make plasma membrane-permeable (16). Mitochondria were fractionated from digitonin-solubilized cells as (19). To avoid proteolytic degradation, homogenization buffer (83 mm sucrose, 6.6 mm imidazole-HCl, pH 7.0) was supplemented with 1× Complete®, EDTA-free protease inhibitor mixture (Roche Applied Science). The protein content was measured by the BCA protein assay kit (Thermo Scientific), using BSA as a standard. The fractionated mitochondria were stored −80 °C in 20 mm imidazole-HCl, pH 7.0, containing 250 mm sucrose and 1× Complete®, EDTA-free protease inhibitor mixture. Clear Native PAGE was performed on a 3.5–18% polyacrylamide gradient gel as described (20). Mitochondrial fractions (5 μg) were suspended with 4.5 μl of solubilization buffer (50 mm NaCl, 50 mm imidazole-HCl, pH 7.0, 2 mm 6-aminohexanoic acid, 1 mm EDTA, and 15 μg of digitonin) on ice for 10 min. Samples were centrifuged at 100,000 × g for 15 min at 4 °C. The supernatant was mixed with 0.1 volume of a loading buffer (50% glycerol and 0.1% Ponceau S) and applied to electrophoresis at 4 °C. For Western blotting, gels were blotted onto PVDF membrane (Bio-Rad) by a wet blotter (Bio Craft) at 30 V for overnight with Towbin buffer supplemented with 20% ethanol and 0.037% SDS. Monoclonal antibodies against α-subunit (1:100,000), β-subunit (1:100,000), and VDAC (1:50,000), polyclonal antibody against DAPIT (1:20,000), and HRP-linked antibodies anti-rabbit (1:1,000,000), and anti-mouse (1:1,000,000) were used. The bands were then visualized by LumigenTM ECL Advance (GE Healthcare). The chemiluminescence was detected on an LAS-4000 system (Fujifilm). Staining intensities of the individual bands were quantified with ImageJ (National Institutes of Health).
RESULTS
DAPIT Production Is Suppressed in Knockdown Cells
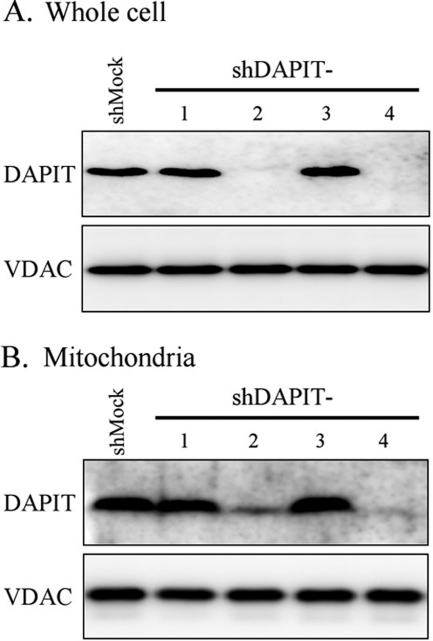
We have established four HeLa cell lines that constitutively produced the short hairpin RNAs (shRNAs), each targeting different coding sequence and 5′-UTR of the human DAPIT gene. Expression of DAPIT protein in the cell and localization in mitochondrial fraction were analyzed with SDS-PAGE followed by Western blotting. Among four shDAPIT cells, shDAPIT-2 cells and shDAPIT-4 cells showed a significantly suppressed expression of DAPIT protein (Fig. 1A, upper). Also, mitochondria in these two cells contained a decreased amount of DAPIT, that is, 45 and 2%, respectively, compared with mock-treated HeLa cells (shMock) (Fig. 1A, lower). Protein synthesis machineries (transcription and translation) were not impaired by DAPIT knockdown as seen in normal expression of a control mitochondrial protein VDAC (Fig. 1B).
FIGURE 1.
Expression of DAPIT protein in whole cells (A) and the isolated mitochondria of the mock-treated (shMock) and DAPIT-knockdown (shDAPIT-1, 2, 3, 4) HeLa cells (B). Whole cell (20 μg of protein) and mitochondria (5 μg of protein) were analyzed with SDS-PAGE, and DAPIT was stained with Western blotting. VDAC was also stained as an internal standard of mitochondrial protein. Experiments were carried out at least four times, and representative results are shown.
DAPIT-knockdown Cells Lose Population of ATP Synthase in Mitochondria
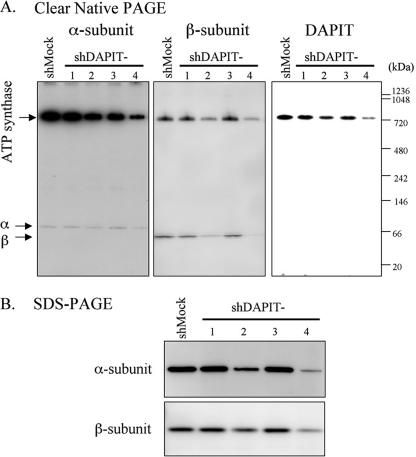
Next, mitochondrial fractions of shDAPIT cells were analyzed with Clear Native PAGE, and α-subunit, β-subunit, and DAPIT were visualized by Western blotting (Fig. 2A). In mitochondria from all cells, DAPIT, as well as α- and β-subunits, existed as components of ATP synthase, confirming the previous reports that DAPIT is one of the subunits of mammalian mitochondrial ATP synthase (12, 13). Judging from the staining intensities of the bands of ATP synthase-integrated α-, β-subunit, and DAPIT, the population of ATP synthase in mitochondria in shDAPIT-2 and shDAPIT-4 cells was significantly decreased, ∼50 and ∼25% of that of the shMock cell, respectively. Thus, it appears that the amount of available DAPIT limits the population of ATP synthase in mitochondria. Free monomers of α- and β-subunits were also seen faintly and clearly, respectively. For each subunit, the amount of free subunit appeared to correlate with the amount of ATP synthase, suggesting that the free subunits are products of disassembly of ATP synthase. Because free monomers were found even in shMock cells, disassembly was not characteristic of DAPIT knockdown but occurred naturally in all cells. The reason why the amount of free α-subunit was much smaller than that of free β-subunit is not known. SDS-PAGE analysis showed that total amounts of α- and β-subunits in mitochondria of shDAPIT-2 and shDAPIT-4 cells matched their amounts in Clear Native PAGE (Fig. 2B). These results show that DAPIT knockdown results in loss of population of mature ATP synthase in mitochondria.
FIGURE 2.
ATP synthase in the mitochondria in shMock and DAPIT-knockdown cells. Mitochondria (5 μg of protein) were analyzed with Clear Native PAGE (A) or SDS-PAGE (B). The protein bands of α-subunit, β-subunit, and DAPIT were stained with Western blotting. α-Subunit was stained strongly to show the presence of free subunit. The positions of ATP synthase, free α-subunit, and free β-subunit in A are indicated. Experiments were carried out at least five times, and representative results are shown.
Transcription and Mitochondrial Transport of α- and β-Subunits Are Normal in DAPIT-knockdown Cells
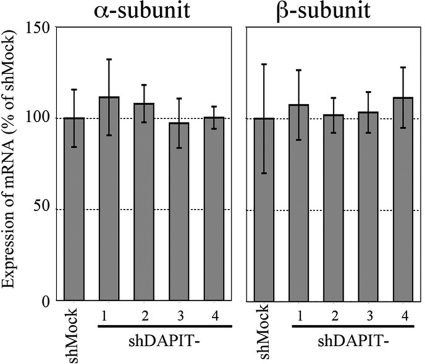
To examine a possibility that shRNA directly suppressed transcription of genes of ATP synthase, levels of mRNAs of α- and β-subunits were measured by real-time PCR (Fig. 3). As seen, there was no significant difference in mRNA levels of α- and β-subunits between the DAPIT-knockdown cells and the shMock cell. Given that translation of mRNA by ribosomes was not impaired in the knockdown cells as shown for normal VDAC expression (Fig. 1), this result indicated that α- and β-subunits, and most likely other subunits as well, should be synthesized normally in the DAPIT-knockdown cells.
FIGURE 3.
Expression of mRNAs of α-subunit and β-subunit in DAPIT-knockdown cells relative to that in shMock cells. The amount of mRNA was measured by real-time quantitative PCR. Details are described under “Experimental Procedures.”
Further, transport of α- and β-subunits into mitochondria in the DAPIT-knockdown cells was not impaired because accumulation of precursor α- and β-subunits with a mitochondria-targeting presequence was not found in SDS-PAGE of whole cell extracts of all tested cells (data not shown). Therefore, it appears that α- and β-subunits are synthesized and transported into mitochondria normally.
ATP Synthesis, Cell Growth, and Viability Are Impaired by DAPIT Knockdown
Mitochondrial ATP synthesis activity of the DAPIT-knockdown cells was measured (16). The proton motive force necessary for ATP synthesis was generated by succinate oxidation. As expected from the decreased population of ATP synthase, ATP synthesis activity of shDAPIT-2 cells and that of shDAPIT-4 cells were ∼90 and ∼60% of that of the shMock cells, respectively (Fig. 4A). The respiratory electron transfer chains in these cells were functioning to produce proton motive force as demonstrated by accumulation of a membrane-potential probing dye, TMRE, in mitochondria that was dissipated by CCCP (Fig. 4B). ATP synthesis was completely inhibited by oligomycin in all cases, ensuring that we observed ATP synthesis by mitochondrial ATP synthase but not by adenylate kinase. The measured mitochondrial ATP synthesis activity did not agree with the population of ATP synthase in mitochondria; in the case of shDAPIT-4, ∼25% of ATP synthase was responsible for ∼60% ATP synthesis. This apparent disagreement can be explained by the presence of 2-fold more population of ATP synthase in mitochondria than needed for the maximum ATP synthesis activity as seen in β-subunit knockdown cells (16).
FIGURE 4.
Mitochondrial activities of ATP synthesis (A) and membrane potential generation (B) of shMock and DAPIT-knockdown cells. A, ATP synthesis activities were measured with luciferase (16). Filled symbols represent the data in the presence of oligomycin (10 μg/ml) of the corresponding open symbols. B, −CCCP, fluorescent images of cells incubated for 10 min with TMRE that accumulates in mitochondria in response to establishment of membrane potential. +CCCP, fluorescent images of the same field after addition of an uncoupler CCCP that dissipates membrane potential and proton gradient. Details are described under “Experimental Procedures.”
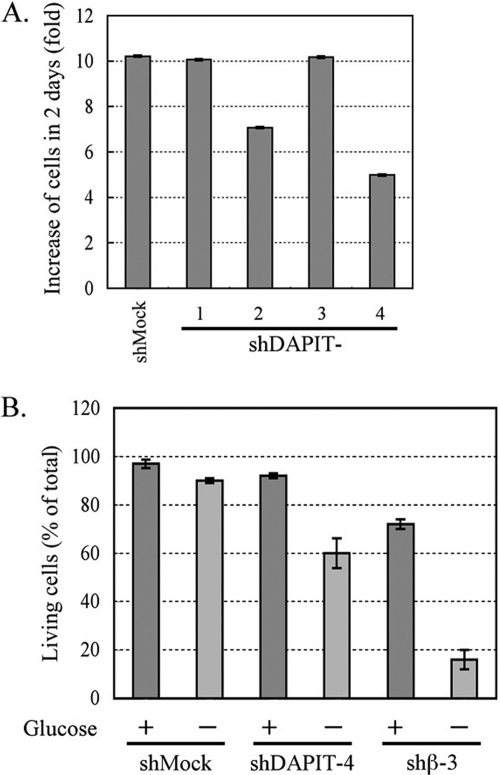
Cell proliferation of shDAPIT-2 and shDAPIT-4 after a 2-day culture was 70 and 50% of that of shMock cells (Fig. 5A). As the population of ATP synthase in the cell decreases, ATP production should become more dependent on glycolytic pathway, and viability of cells should be more susceptible to deprivation of glucose. Indeed, this was observed for the β-subunit knockdown cells (shβ-3), in which population of ATP synthase was <5% of the mock cell (16). When glucose in the culture medium of the shβ-3 cells was substituted with galactose that was unavailable for glycolysis, >80% of the cells died in 2 days (Fig. 5B). Under the same conditions, 40% of shDAPIT-4 cells died in galactose whereas only 10% of the shMock cells died in galactose (Fig. 5B).
FIGURE 5.
Cell proliferation in the glucose-containing medium (A) and viability in the glucose-free medium (B) of shMock and DAPIT-knockdown cells. A, proliferation of cells in 0.1% glucose is shown as an increase of cell numbers after a 2-day culture (-fold). B, glucose in the medium was replaced with 0.1% galactose and the culture continued another 2 days. Then, numbers of living cells and dead cells were counted. Viability was expressed as percent of living cells per total cells. Details are described under “Experimental Procedures.”
DISCUSSION
DAPIT is not essential for function and structure of mature ATP synthase. It is easily lost from ATP synthase during purification (12) or by exposure to relatively strong detergent (13) without losing the structural integrity and the core function of ATP synthase (13). This work, however, reveals that DAPIT is essential to maintain the population of ATP synthase in mitochondria in living cells. It appears that the transcription and translation are not impaired in DAPIT-knockdown cells (Fig. 1, 3), and therefore, assembly or degradation of ATP synthase should be affected by DAPIT. Given that DAPIT-less ATP synthase is structurally stable (13), it is likely that DAPIT plays a role in assembly of ATP synthase even if a potential role in protection from degradation is not excluded. The absence of structurally indispensable subunit should cause the failure of the assembly of ATP synthase as observed for knockdown cells of ϵ-subunit (21) and β-subunit (16). However, because DAPIT is dispensable for mature ATP synthase, it might play a chaperone-like role in the assembly of ATP synthase. The assembly of mitochondrial ATP synthase proceeds through several candidate intermediate subcomplexes such as F1, c-ring-F1, c-ring-a, and peripheral stalk (22) and, different from prokaryotic ATP synthase, the assembly depends on many factors and chaperones (23, 24). DAPIT has a putative transmembrane segment and should be associated with the F0 portion in ATP synthase. It is, therefore, tempting to assume that DAPIT assists subunit assembly of the F0 portion. Without assembled F0 portion, F1 or even its subcomplex cannot exist stably as demonstrated in Clear Native PAGE (Fig. 2A), where no protein band other than free and the ATP synthase-integrated subunit was seen.
Our finding provides a plausible explanation for physiological consequence caused from failure of the controlled DAPIT production. It was reported that the mRNA level of DAPIT in skeletal muscle in rat was elevated after active stretching of muscles, apparently to meet the increasing demand for ATP by increasing population of ATP synthase. The DAPIT gene was located within the locus whose deletion caused diabetes-associated phenotype of a diabetic rat (25) and mRNA level of DAPIT was lowered when diabetes was induced by streptozotocin (15). The poor DAPIT production should result in scarce population of ATP synthase and low level of ATP production, which in turn suppresses mitochondrial respiration first and glucose consumption next. Low level of cellular glucose consumption suppresses glucose uptake from blood and weakens the effect of insulin. The factor controlling DAPIT expression and actual roles of DAPIT in mitochondrial ATP production and in glucose metabolism are worth studying especially in the insulin-sensitive cells.
Footnotes
- DAPIT
- diabetes-associated protein in insulin-sensitive tissue
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- TMRE
- tetramethylrhodamine ethylester
- VDAC
- voltage-dependent anion channel.
REFERENCES
- 1. Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. (1994) Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 2. Noji H., Yasuda R., Yoshida M., Kinosita K., Jr. (1997) Nature 386, 299–302 [DOI] [PubMed] [Google Scholar]
- 3. Boyer P. D. (1997) Annu. Rev. Biochem. 66, 717–749 [DOI] [PubMed] [Google Scholar]
- 4. Yoshida M., Muneyuki E., Hisabori T. (2001) Nat. Rev. Mol. Cell Biol. 2, 669–677 [DOI] [PubMed] [Google Scholar]
- 5. Weber J., Senior A. E. (2003) FEBS Lett. 545, 61–70 [DOI] [PubMed] [Google Scholar]
- 6. Nakamoto R. K., Baylis Scanlon J. A., Al-Shawi M. K. (2008) Arch. Biochem. Biophys. 476, 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Junge W., Sielaff H., Engelbrecht S. (2009) Nature 459, 364–370 [DOI] [PubMed] [Google Scholar]
- 8. von Ballmoos C., Wiedenmann A., Dimroth P. (2009) Annu. Rev. Biochem. 78, 649–672 [DOI] [PubMed] [Google Scholar]
- 9. Kagawa Y. (2010) Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 86, 667–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Düser M. G., Zarrabi N., Cipriano D. J., Ernst S., Glick G. D., Dunn S. D., Börsch M. (2009) EMBO J. 28, 2689–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walker J. E., Dickson V. K. (2006) Biochim. Biophys. Acta 1757, 286–296 [DOI] [PubMed] [Google Scholar]
- 12. Chen R., Runswick M. J., Carroll J., Fearnley I. M., Walker J. E. (2007) FEBS Lett. 581, 3145–3148 [DOI] [PubMed] [Google Scholar]
- 13. Meyer B., Wittig I., Trifilieff E., Karas M., Schägger H. (2007) Mol. Cell. Proteomics 6, 1690–1699 [DOI] [PubMed] [Google Scholar]
- 14. Carroll J., Fearnley I. M., Walker J. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16170–16175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Päivärinne H., Kainulainen H. (2001) Acta Diabetol. 38, 83–86 [DOI] [PubMed] [Google Scholar]
- 16. Fujikawa M., Yoshida M. (2010) Biochem. Biophys. Res. Commun. 401, 538–543 [DOI] [PubMed] [Google Scholar]
- 17. Bookout A. L., Mangelsdorf D. J. (2003) Nucl. Recept. Signal. 1, e012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ishiyama M., Miyazono Y., Sasamoto K., Ohkura Y., Ueno K. (1997) Talanta 44, 1299–12305 [DOI] [PubMed] [Google Scholar]
- 19. Pallotti F., Lenaz G. (2007) Methods Cell Biol. 80, 3–44 [DOI] [PubMed] [Google Scholar]
- 20. Wittig I., Karas M., Schägger H. (2007) Mol. Cell. Proteomics 6, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 21. Havlícková V., Kaplanová V., Nùsková H., Drahota Z., Houstek J. (2010) Biochim. Biophys. Acta 1797, 1124–1129 [DOI] [PubMed] [Google Scholar]
- 22. Wittig I., Schägger H. (2008) Biochim. Biophys. Acta. 1777, 592–598 [DOI] [PubMed] [Google Scholar]
- 23. Rak M., Zeng X., Brière J. J., Tzagoloff A. (2009) Biochim. Biophys. Acta 1793, 108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ludlam A., Brunzelle J., Pribyl T., Xu X., Gatti D. L., Ackerman S. H. (2009) J. Biol. Chem. 284, 17138–17146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Granhall C., Park H. B., Fakhrai-Rad H., Luthman H. (2006) Genetics 174, 1565–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]