Abstract
Mineralized tissues such as dentin and bone assemble extracellular matrices uniquely rich in a variety of acidic phosphoproteins. Although these proteins are presumed to play a role in the process of biomineralization, key questions regarding the nature of their contributions remain unanswered. First, it is not known whether highly phosphorylated proteins alone can induce matrix mineralization, or whether this activity requires the involvement of other bone/dentin non-collagenous proteins. Second, it remains to be established whether the protein kinases that phosphorylate these acidic proteins are unique to cells responsible for producing mineralized tissues. To begin to address these questions, we consider the case of phosphophoryn (PP), due to its high content of phosphate, high affinity for Ca2+, and its potential role in hydroxyapatite nucleation. We have created a model system of biomineralization in a cellular environment by expressing PP in NIH3T3 fibroblasts (which do not produce a mineralized matrix); as a positive control, PP was expressed in MC3T3-E1 osteoblastic cells, which normally mineralize their matrices. We show that expression of PP in NIH3T3 cells is sufficient for the induction of matrix mineralization. In addition, assessment of the phosphorylation status of PP in these cells reveals that the transfected NIH3T3 cells are able to phosphorylate PP. We suggest that the phosphorylation of PP is essential for mineral formation. The principle goal of this study is to enrich the current knowledge of mineralized tissue phosphorylation events by analyzing them in the context of a complete cellular environment.
Keywords: Bone, Calcium, Collagen, Extracellular matrix, Protein Phosphorylation, Biomineralization, Dentin
Introduction
In biologically induced mineralization, crystals are generally produced by a heterogeneous nucleation mechanism. The deposition of mineral crystals in bone, dentin, and cartilage is orchestrated by cells, and the growth of these crystals is facilitated through mineral-matrix interactions. Current data indicate an important role for non-collagenous extracellular matrix (ECM)2 proteins associated with collagen fibrils in the process of biomineralization. The high affinity of these non-collagenous proteins for divalent cations may facilitate control of the nucleation and growth of the initial mineral deposits. In addition, these proteins may subsequently regulate the size, composition, microstructure, morphology, and the orientation of the resulting crystals formed (crystal growth) (1).
Mineralized tissues are unique in their production of extracellular acidic phosphoproteins. These phosphoproteins may be of structural and/or regulatory significance to the formation of mineralized tissues, potentially acting as a key interface between the nucleating mineral crystal and surrounding collagen fibrils (2, 3). These phosphoproteins include: osteopontin (OPN) (4), bone sialoprotein (5, 6), dentin matrix protein 1 (DMP1) (7), phosphophoryn (PP) (8), osteonectin (9), bone acidic glycoprotein 75 kDa (BAG 75) (10), and MEPE (11), each accumulating in the extracellular matrices of bone and dentin to different extents. Establishing the presence of such a diversity of these phosphorylated proteins and the consequent potential array of phosphorylated polypeptides that could be derived by their post-translational modification may be significant to understanding their functional activity.
Although relatively little attention has been directed toward understanding the effects of phosphorylation of these ECM proteins, it is reasonable to speculate that the nature and extent of their phosphorylation could confer specificity in their function and specific behaviors between tissues. For example, the OPN gene is expressed and translated in many non-mineralizing tissues with very low levels of phosphorylation, compared with the extent of OPN phosphorylation in bone (12, 13). In contrast, the extent of OPN phosphorylation in bone (14) is less than that of OPN in milk (15). These differences suggest that the phosphorylation state can confer functional specificity to these proteins, and strongly implies the existence of tissue-specific kinases. One of our goals is to understand how post-translational phosphorylation of non-collagenous ECM proteins potentiates their contribution to the formation of mineral matrices. Second, we are interested in whether the protein kinases responsible for phosphorylating these proteins are unique to cells dedicated to producing mineralized tissues or whether they could be present in non-mineralizing tissues as well. Our proposed model system focuses on the highly phosphorylated ECM protein known as PP, due to its high content of phosphate and possession of one of the highest affinities for Ca2+ of the matrix proteins studied to date (16). In this study, PP is overexpressed in NIH3T3, a cell that does not normally mineralize its matrix, and in MC3T3-E1, an osteoblastic cell that is capable of forming a mineralized matrix. We further assess the phosphorylation status of secreted PP harvested from the extracellular matrix, and thereby speculate on its role in biomineralization in these distinct extracellular environments.
MATERIALS AND METHODS
Cloning of PP into a Bacterial Expression Vector
To clone the murine Pp gene into the glutathione S-transferase (GST) Gene Fusion System, the pGEX-4T-3 vector was used. Polymerase chain reaction (PCR) of exon 5, using the dentin sialophosphoprotein genomic DNA as a template, was performed. A PCR was carried out using gene-specific primers containing the DNA sequence for SalI and XbaI restriction enzymes added to the 5′ end of each primer. The forward primer with the SalI restriction site added to the 5′ end was: 5′-CTAATGTCGACATGGAGAGTGGCAGCCGTGGAGA-3′. The reverse primer with XbaI added to the 5′ end was 5′-GCATTCTAGATTAAAGCACCCGCCATTCAAATCG-3′. (Double underline signifies the gene-specific sequence, whereas the single underline represents the restriction enzyme and 5 random bases at the 5′ end.) The amplified 1.7-kb fragment of exon 5 was digested using SalI and XbaI restriction enzymes and subcloned into the GST vector. The Pp insert was confirmed using agarose gel electrophoresis and DNA sequencing. The newly generated clone was called pGEX-4T-3-PP.
Expression and Purification of the Recombinant Protein
The Amersham Biosciences/GE Healthcare GST Gene Fusion System is an integrated system for the expression, purification, and detection of fusion proteins produced in Escherichia coli. The construct pGEX-4T-3-PP was used to transform the bacterial host BL21. Ampicillin-resistant colonies containing the insert were grown in Luria broth (LB) containing ampicillin for 4 h at 30 °C. To induce expression of the fusion protein, isopropyl β-d-1-thiogalactopyranoside was then added at 1 mm, and incubation was carried out for another 3 h. The bacterial lysate was cleared of cellular debris by centrifugation and the cleared lysate was applied directly to glutathione-Sepharose 4B (Amersham Biosciences/GE Healthcare). After the fusion proteins were bound to the matrix, it was washed with 1× PBS to remove nonspecifically bound proteins. Bound GST fusion proteins were then released with thrombin. (Cleavage of the bound fusion protein eliminates the extra step of separating the released protein from GST, because the GST moiety remains bound to the matrix, whereas the cloned protein is eluted using wash buffer.) To remove thrombin from the eluates, p-aminobenzamidine immobilized on Sepharose 4 Fast Flow matrix was used (Amersham Biosciences/GE Healthcare). The purified protein was electrophoresed on polyacrylamide to verify the molecular mass, and an aliquot was used for amino acid composition and sequencing as previously reported (17, 18).
Cloning of PP into a Mammalian Expression Vector (pShooter Vector)
A phosphophoryn gene fragment (exon 5) was subcloned into the pShooter/ER vector (Invitrogen) using PCR. The primers used were the same as described above to amplify the expected 1.7-kb fragment of exon 5. The fragment was digested using SalI and XbaI restriction enzymes and subcloned into the pShooter/ER vector. The pShooter/ER vector employs an N-terminal signal peptide targeting sequence that results in the recombinant protein product being directed to the endoplasmic reticulum in mammalian cells.
Transfection of pShooter-ER-PP Construct into MC3T3-E1 and NIH3T3 Cell Lines: Selection of Stable Transfectants and Detection of Transcripts
The pShooter-ER-PP construct was transfected into MC3T3-E1 and NIH3T3 cell lines using a FuGENE 6 transfection kit (Roche) according to the manufacturer's recommendations. Following DNA transfection, neomycin (G418) was used to select and maintain stable eukaryotic cell lines stably expressing the p-Shooter-ER-PP construct. A range of 800 μg/ml to 1 mg/ml of G418 was used for selection of neomycin resistance (a control of non-transfected cells was also used). The transfected cells were plated in 150-cm2 flasks at a high dilution. Sterile glass rings were used to select surviving colonies. The isolated colonies were trypsinized and transferred to a new flask where the cells were allowed to expand and then screened for gene and protein expression. Over 6 weeks, we isolated 10 and 12 colonies of NIH3T3 and MC3T3-E1, respectively, stably transfected with the Pp gene.
Transcript Detection in Stably Transfected Cells
Transfected NIH3T3 and MC3T3-E1 mRNA was extracted using the QuickPrep Micro mRNA Purification Kit (GE Healthcare). Extraction was carried out according to the protocol with addition of RNase-free DNase (Qiagen) treatment for 15 min, followed by a low salt buffer wash, just prior to mRNA elution from a MicroSpin column (GE Healthcare). The freshly isolated mRNA was used to carry out RT-PCR using the Access RT-PCR System (Promega). The primers used for the PCR were: forward 5′-GAG TGA GGA CAA GGA CGA ATC T-3′ and reverse 5′-ATC TAA TCA TCA CTG GTT GAG TG-3′. These primers should yield a 1.3-kb PCR fragment.
Isolation of PP Protein from the Stably Transfected Cells
NIH3T3-PP cells were cultured for 7 and 10 days in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin (Invitrogen), 50 mm l-ascorbic acid, and 4 mm inorganic phosphate (Sigma). Medium and cells/matrix were collected separately at days 7 and 10. The proteins were precipitated from the medium using trichloroacetic acid (TCA). The remaining cells (with associated extracellular matrix) were lysed in a RIPA buffer containing 0.1% (w/v) SDS. The RIPA buffer consists of 10 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm EDTA, 250 μm phenylmethylsulfonyl fluoride (PMSF), 1 mm N-ethylmaleimide, 2 mm 1-methionine, 2 mm 1-cysteine, 0.3% (v/v) Nonidet P-40, 0.05% (v/v) Triton X-100, 0.3% (w/v) sodium deoxycholate, 0.1% (w/v) bovine serum albumin. The precipitated proteins from the medium and cell/matrix were either further purified by column chromatography, or blotted on nitrocellulose paper using dot blot or Western blot techniques using both the anti-PP antibody (generated in our laboratory) and the anti-dentin matrix protein 2 (DMP2) antibody (a generous gift from Dr. Arthur Veis, Northwestern University). This antibody has been previously shown to have cross-reactivity with phosphophoryn due to the common content of (DSS)n repeats (19, 20). As a positive control, we used our recombinant PP as well as recombinant DMP2, provided by Dr. Veis. BSA was used as a negative control.
von Kossa Staining of the NIH3T3-PP Cell Cultures
Cells were grown for 7 and 10 days in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin (Invitrogen), 50 mm l-ascorbic acid, and 4 mm inorganic phosphate (Sigma). This phosphate concentration does not cause dystrophic calcification as shown in other culture studies (21, 22). At days 7 and 10, von Kossa staining for phosphate deposition was performed as follows: cell culture layers were washed twice with PBS, fixed in phosphate-buffered formalin for 10 min, and serially dehydrated in 70, 95, and 100% ethanol, two times each, and air dried. The plates were then rehydrated from 100 to 95 to 80% ethanol to H2O. The water was removed, a 2% silver nitrate solution was added, and the plate was exposed to sunlight for 20 min, after which the plate was rinsed with water. Sodium thiosulfate (5%) was added for 3 min, the plates were then rinsed in water, and the cells were counterstained with nuclear fast red for 5 min. The plates were washed with H2O, then twice with 95% ethanol and twice with 100% ethanol, and dried for image analysis.
Alizarin Red Staining of NIH3T3-PP Cells
To estimate the amount of calcium accumulated in the cultures, cells were cultured as described before. Cell layers were fixed in ice-cold 70% ethanol for 1 h and rinsed with double distilled H2O. Cells were stained with 40 mm alizarin red-S, pH 4.2, for 10 min with gentle agitation. Cell layers were rinsed five times with ddH2O and then rinsed for 15 min with 1× PBS and gentle agitation. Alizarin red was extracted from fixed cell layers by treatment with 500 μl of 10% cetylpyridinium chloride for 20 min with gentle agitation. Absorbance of extracted alizarin red in cetylpyridinium chloride solution was measured at 570 nm. The amount of calcium (in μg) was determined according to an alizarin red-Ca2+ standard curve and normalized to total protein of the cell lysate.
Phosphoprotein SDS-PAGE Stain
The relative phosphate content of the phosphoproteins was determined using Pro-Q® Diamond (Invitrogen) phosphoprotein gel stain, which selectively stains phosphoproteins in polyacrylamide gels. This proprietary fluorescent stain allows direct, in-gel detection of phosphate groups attached to tyrosine, serine, or threonine residues. The SDS-PAGE gels were stained according to the manufacturer's recommendation. In summary, the gels were fixed and stained with phosphoprotein gel stain equivalent to 10 times the volume of the gel with gentle agitation in the dark for 60–90 min. The gel was then destained in 80–100 ml of destaining solution with gentle agitation for 30 min at room temperature. The gel was then washed twice with ultrapure water at room temperature for 5 min per wash and imaged.
Measurement of Calcium and Phosphate
The specimens from tissue culture were washed three times with pH 8 ammoniated water, air dried, and then lyophilized.
Aliquots (∼1.0 mg) were hydrolyzed in 1 n HCl and diluted 1:1 to 1:10 depending on the initial calcium concentration observed. Calcium concentration was determined by atomic absorption spectrophotometry as described by Willis et al. (23). An equivalent volume was assayed for phosphate concentration (24). Calcium/phosphate ion molar ratios were calculated and compared with a hydroxyapatite standard included in all assays.
Affinity Column Purification
The HiTrap NHS-activated HP column (Amersham Biosciences/GE Healthcare) was used to couple the anti-PP antibody according to the manufacturer's recommendation. In summary, the antibody was first coupled to the column using a coupling buffer that consists of 0.2 m NaHCO3, 0.5 m NaCl, pH 8, washed to remove any uncoupled antibodies, and then deactivated to remove any excess active groups. PP purification was carried out using an affinity column pre-equilibrated with tissue culture medium. The isolated medium containing PP was loaded and washed. PP was eluted in a 0.1 m glycine, pH 3, solution.
Mineral Characterization in Tissue Culture by X-ray Diffraction
MC3T3-E1 and NIH3T3 cells were seeded on glass slides that were subsequently placed in 150 × 25-mm polystyrene tissue culture dishes. The cells were grown in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin and kept in a humidified incubator (37 °C and 5% CO2) until they reached ∼80% confluence. Once the cells reached 80% confluence, a 4 mm inorganic phosphate (prepared from 0.5 m NaH2PO4·2(H2O) stock solution was used) solution was created in DMEM containing 10% FBS, 1% penicillin/streptomycin, 2.5% Hepes. Day 1 was defined as the day of inorganic phosphate supplementation to the cell culture. At day 15, the medium was aspirated and culture dishes were washed with ammonia water (1000:1 deionized water to 5% ammonium hydroxide solution) to remove any unincorporated salts that might be present. The culture dishes were then air dried. The glass slides were removed from the culture dishes for analysis. Samples were analyzed using the Philips x-ray diffractometer (Philips X'pert Pro diffractometer with X'Celerator) employing CuKα radiation (λ = 1.5418 Å) from the 2θ angle of 21 to 80 degrees with time per step being 100 s, and step size (2θ) being 0.033 degrees. Some of the mineralized samples were also scanned from 2θ angles of 2 to 90 degrees to detect the presence of inorganic calcium phosphates, particularly, octacalcium phosphates that could also likely form under the conditions used for this study.
In Vitro Mineral Characterization
Recombinant PP (rPP) at concentrations from 2.5 to 100 ng/ml was assessed for its ability to initiate the formation and growth of the mineral phase, hydroxyapatite (HA). De novo HA formation and growth were monitored in the dynamic collagen gel HA growth system (25). In brief, in this double diffusion system, Ca2+ and HPO42- (Pi) ions circulate at room temperature diffusing into opposite ends of a 6-cm long 10% gelatin (Bloom 275, Fisher Chemical) gel. At a site 3.4 cm from the P entrance point, the gel includes a 100-μl band containing the proteins to be tested. At 4.5 days, the CaxPi mm product in the absence of protein at this site is 5.5 mm2. For de novo formation and growth studies, calcium and phosphate ions accumulation in the precipitant band containing 0.15 m Tris buffer (control) was compared with experimental gels with 0.01–100 μg/ml of recombinant protein in Tris buffer. For all experiments, identical “single diffusion” gels, into which only calcium and phosphate ions diffused, served as additional controls to correct for ion accretion due only to diffusion or to binding of the ion to the matrix protein. Control, non-phosphorylated recombinant double and single diffusion gels were run parallel to each recombinant experiment; comparisons were also made to these controls to avoid errors inherent in experiments that might have undergone variations in temperature, time span, solution flow, or reservoir concentrations.
At 4.5 days, the gels were removed from the apparatus and either the precipitant band was cut out for mineral analysis (x-ray diffraction or Fourier transform infrared spectroscopy (FTIR)) or the entire gel was cut into slices (0.30 ml volume), and the calcium (23) and phosphate ion (24) content of each slice was then determined by atomic absorption and spectrophotometric analyses, respectively. The calcium/phosphate ion analyses were performed following hydrolysis (18 h, 110 °C) of the gel in 4 n HCl. The calcium and phosphate ion contents of the precipitant band were calculated by subtracting the content of the comparable slice in the single diffusion gels from that of the precipitant band. The calcium and phosphate ion contents of adjacent bands were also compared, and used to verify that the gels had been sliced equivalently. All data were compared with the mineral content of the protein-free gels using the Student's t test. Each data point represents 3–12 gel analyses.
RESULTS
rPP Expression in Bacteria
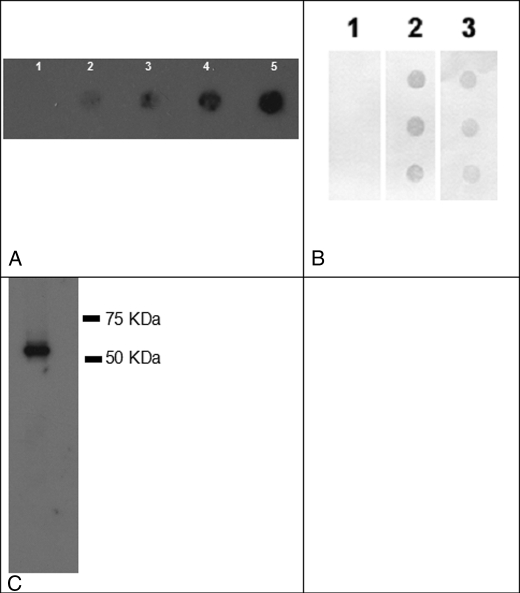
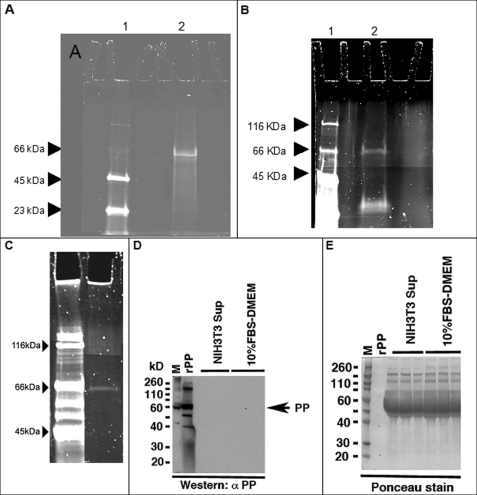
To compare the same protein made without post-translational modification (bacterially), or with post-translational modification (expressed in mammalian cells), we cloned the PP cDNA insert into both pGEX-4T-3 and pShooter expression vectors. The new constructs expressing PP were called: pGEX-4T-3-PP and pShooter-ER-PP. The construct pGEX-4T-3-PP was used to transform the bacterial host BL21, which in turn was induced to express PP. The purified protein, following removal of GST, was electrophoresed on polyacrylamide gels to verify a molecular mass of ∼55 kDa (17, 18), and stained with both Coomassie Blue and Stains-All. As expected, the acidic nature of the protein resulted in staining with Stains-All (28) but not with Coomassie Blue (data not shown). Dot blot analysis with an anti-PP antibody (Fig. 1A), verified that the recombinant protein is indeed rPP. These data were strengthened through a second dot blot using a polyclonal anti-DMP2 antibody (Fig. 1B). This antibody (a gift from Dr. A. Veis) has been previously shown to have cross-reactivity with phosphophoryn due to the common content of (DSS)n repeats (19, 20). We confirmed the correct size and migration of the recombinant PP through Western blot analysis (Fig. 1C). Furthermore, amino acid composition analysis of an aliquot of the preparation showed a protein rich in aspartic acid and serine, with the N-terminal end sequence corresponding to phosphophoryn (data not shown).
FIGURE 1.
Use of two distinct polyclonal antibodies to confirm that the newly synthesized recombinant protein is PP. Panel A, dot blot with anti-PP, lane 1, BSA control; lanes 2-5, increasing concentrations of PP, 3, 10, 15, and 30 ng. We show the expected reactivity with anti-PP. Panel B, dot blot with anti-DMP2, known to cross-react with PP. Lane 1, BSA control; lane 2, rDMP2 provided by Dr. Arthur Veis (+ ctrl); lane 3, rPP. As expected, we show cross-reactivity with anti-DMP2 antibody. Panel C, Western blot analysis of rPP using anti-PP antibody.
Stable Transfection of PP into NIH3T3 and MC3T3-E1
The pShooter-ER-PP construct was transfected into MC3T3-E1 and NIH3T3 cells. To determine that the NIH3T3 and MC3T3-E1 cells were stably transfected, we performed PCR using freshly isolated mRNA and forward and reverse primers specific to PP, which yielded a 1.3-kb PCR product (expected size using the specified primers).
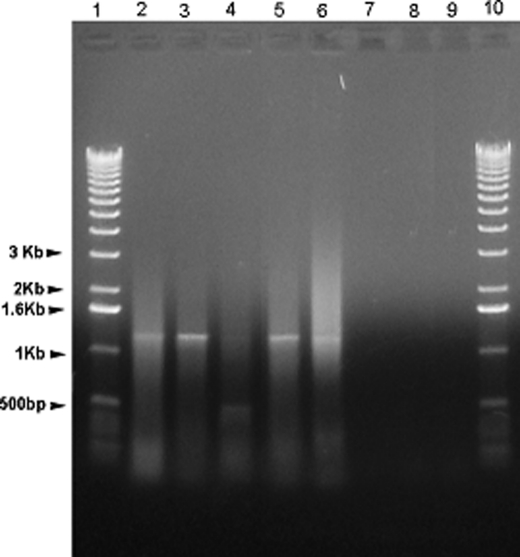
As shown in Fig. 2, most of the NIH3T3 isolated G418-resistant cell colonies produced the PP transcript as demonstrated by the presence of the 1.3-kb PCR band, whereas the control (non-transfected NIH3T3 cells) did not show the presence of the PP transcript. We also obtained similar results from MC3T3-E1 cells (data not shown).
FIGURE 2.
RT-PCR detection of the PP transcript from stably transfected NIH3T3 clones. mRNA was isolated from mouse NIH3T3 cells stably transfected with pShooter-ER-PP. RT-PCR was then performed to screen for presence of the Pp gene transcript. A 1.3-kb band is expected, and observed in clones 1, 2, 4, and 5. Lane 1, 1-kb ladder; lanes 2–6, 5 individual clones; lane 7, non-transfected NIH3T3 control; lane 8, negative control, no RNA added; lane 9, negative control, no reverse transcriptase added.
PP Secretion and Its Effect on Matrix Mineralization in NIH3T3 and MC3T3-E1 Cells
To establish that the PP produced in our stable transfectants was actually finding its way to the extracellular matrix, we examined whether PP was detectable in the media of transfected cells. The pShooter-ER-PP construct relies on a peptide at the N-terminal end of PP to direct newly translated protein to the ER and Golgi where its post-translational modification takes place (29, 30). As shown in Fig. 3, A–C, the stably transfected NIH3T3 and MC3T3-E1 cells secreted PP; the protein was readily precipitated from the medium of NIH3T3-PP and MC3T3-E1-PP and detectable by dot blot (Fig. 3A) and Western blot (Fig. 3, B–D) using anti-PP antibody. No phosphophoryn expression was detected in non-transfected cells (control). The stably transfected cells secreted ∼0.5 μg of phosphophoryn/ml of medium.
FIGURE 3.
Blot analysis of rPP precipitated from stably transfected cells. Panel A shows protein isolated from NIH3T3 stably transfected with PP cDNA (NIH3T3-PP) analyzed by dot blot using anti-DMP2 antibody. Lane 1, positive control, rDMP2; lane 2, PP isolated from NIH3T3-PP medium; lane 3, PP isolated from NIH3T3-PP cells/matrix; lane 4, BSA negative control. Panel B shows PP isolated PP from NIH3T3-PP medium and analyzed by Western blot using anti-PP antibody. Panel C, PP isolated from MC3T3-E1-PP medium, analyzed using anti-PP antibody. Panel D, precipitated NIH3T3 medium analyzed using anti-PP antibody.
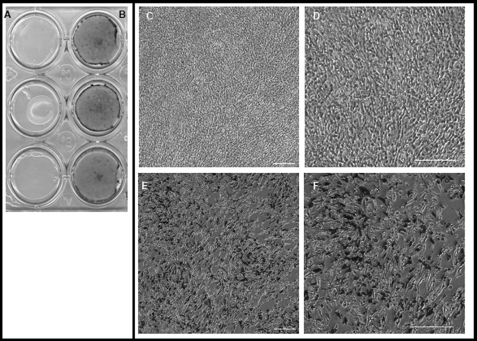
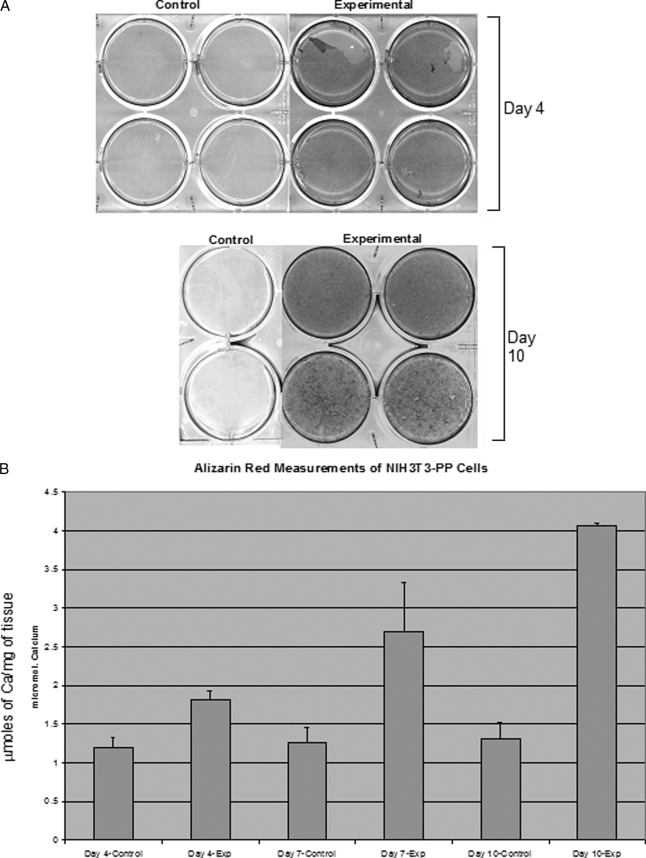
Of particular interest in this study was to see whether expressing phosphophoryn in cells that do not normally produce a mineralized matrix would alone induce these cells to form a mineralized matrix. In other words, would PP expression be sufficient to establish the necessary framework for establishing a mineralized ECM? Cells grown for 4, 7, and 10 days showed the presence of mineral when PP was present (Figs. 4 and 5). Phosphate (von Kossa) staining at days 7 and 10 showed negligible staining of the mineral deposits in control NIH3T3 cells (non-transfected) (Fig. 4A), whereas NIH3T3 cells expressing PP showed extensive mineral deposition (Fig. 4B). Higher magnification images (Fig. 4, E and F) verified these findings. Fig. 5A shows the presence of calcium (positive alizarin red stain) at days 4 and 10 (day 7 not shown), whereas the control cultures show only background alizarin red staining. Spectrophotometric quantification of alizarin red staining (Fig. 5B) demonstrated a 2-fold increase at day 4 and a 4-fold increase at day 10 in the staining levels, whereas the control cells remained at background levels. The differences between experimental and control were statistically significant (p < 0.001) based on analysis of variance and Fisher's Least Significant Difference post hoc test (Protected t test).
FIGURE 4.
Visualization of the mineralization process in NIH 3T3-PP cells. Panel A shows a von Kossa stain of control NIH 3T3 cells (day 10); negligible staining indicates a lack of mineralization. Panel B shows a positive von Kossa stain at day 10 of transfected NIH 3T3-PP cells indicating the presence of mineralization. Panels C–F, cells stained at day 10 with von Kossa and counterstained with fuchsin red. Panels C and D, non-transfected cells (control) shown at ×10 and 20 magnification, respectively (scale bar = 200 μm). Panels E and F show transfected cells with multiple mineral deposit foci at ×10 and 20 magnification, respectively.
FIGURE 5.
Assessment of calcium deposition by cells expressing PP. Panel A, alizarin red staining of control and transfected NIH3T3 at days 4 and 10. Panel B, alizarin red measurements of NIH3T3-PP cells conducted to determine calcium deposition at different time points. Bars represent mean ± S.D. of 4 replicates. Micromoles of the calcium ion/total protein concentration are shown.
Because alizarin red staining is rather a qualitative measure of calcium deposition, we performed quantitative analysis of the calcium concentration by atomic absorption spectrophotometry and phosphate concentration. From these calcium and phosphate concentrations we have calculated the calcium/phosphate ion molar ratios compared with a hydroxyapatite standard included in all assays. As shown in Table 1, the NIH3T3 cells expressing PP in the presence of inorganic phosphate had the highest concentration of calcium and a calcium/phosphate ion molar ratio of approximately 1.5 implying the presence of hydroxyapatite. When cells were grown in the absence of inorganic phosphate or PP expression, calcium/phosphate ion ratios were not indicative of hydroxyapatite.
TABLE 1.
Calcium content and calcium (Ca)/phosphate (P) molar ratios of matrices produced by NIH3T3 and NIH3T3-PP, with and without inorganic phosphate (+Pi)
| Ca/mg average | StDv | Ca/P molar ratios | |
|---|---|---|---|
| mm | |||
| HA | 0.05 | 1.61 | |
| NIH3T3-PP + Pi | 0.57 | 0.01 | 1.46 |
| NIH3T3-PP + Pi | 1.04 | 0.07 | 1.64 |
| NIH3T3-PP + Pi | 1.03 | 0.07 | 1.41 |
| NIH3T3-PP + Pi | 0.64 | 0.00 | 1.49 |
| NIH3T3-PP + Pi | 0.63 | 0.01 | 1.53 |
| NIH3T3 + Pi | 0.16 | 0.00 | 1.21 |
| NIH3T3 + Pi | 0.19 | 0.00 | 1.30 |
| NIH3T3 + Pi | 0.19 | 0.00 | 1.23 |
| NIH3T3 + Pi | 0.12 | 0.00 | 1.15 |
| NIH3T3 + Pi | 0.20 | 0.00 | 1.19 |
| NIH3T3-PP | 0.24 | 0.01 | 0.95 |
| NIH3T3-PP | 0.20 | 0.01 | 0.95 |
| NIH3T3-PP | 0.17 | 0.00 | 0.72 |
| NIH3T3-PP | 0.20 | 0.00 | 0.94 |
| NIH3T3-PP | 0.19 | 0.00 | 0.95 |
| NIH3T3-PP | 0.18 | 0.01 | 0.83 |
| NIH3T3 | 0.15 | 0.00 | 0.94 |
| NIH3T3 | 0.16 | 0.00 | 0.80 |
| NIH3T3 | 0.17 | 0.01 | 0.86 |
Phosphorylation Status of PP Purified from NIH3T3
The next key question we sought to answer was whether cells that do not normally have the ability to mineralize their extracellular matrix (such as NIH3T3), could post-translationally modify phosphophoryn, or if this could only be achieved by cells that natively produce mineralizing matrices such as osteoblasts or odontoblasts. Secreted PP was purified from cell medium and visualized on SDS-PAGE with ProQ Diamond stain, which is specific for phosphoproteins. Fig. 6A shows the result of TCA precipitation from NIH3T3-PP medium, whereas Fig. 6, B and C, show antibody-affinity purified PP from NIH3T3-PP and MC3T3-E1-PP, respectively. In all three cases, the purified PP was phosphorylated PP, indicating that both the NIH3T3 and the MC3T3-E1 cells are capable of this post-translational modification.
FIGURE 6.
Assessment of the phosphorylation state of secreted recombinant PP. NIH3T3-PP and MC3T3-E1-PP cells were cultured for 3 days, medium was collected, and TCA precipitation was performed; the precipitated proteins were further purified by an affinity column and analyzed by SDS-PAGE. The gels were stained with ProQ Diamond stain, specific for phosphoproteins. Lane 1 in panels A–C shows the ProQ phosphoprotein standard. Panel A shows the TCA precipitation from NIH3T3-PP medium, stained with ProQ; lane 2, major phosphoprotein band at ∼55 kDa following TCA precipitation. Panels B and C, lane 2, shows PP purified using an affinity column, from NIH3T3-PP and MC3T3-E1 medium, respectively. The 55-kDa band stains positive with ProQ, indicating that the PP secreted by NIH3T3 and MC3T3-E1 is indeed phosphorylated. Panels D and E verify that the antibody is specific for PP, and does not cross-react with any proteins derived from the medium itself.
Mineral Characterization
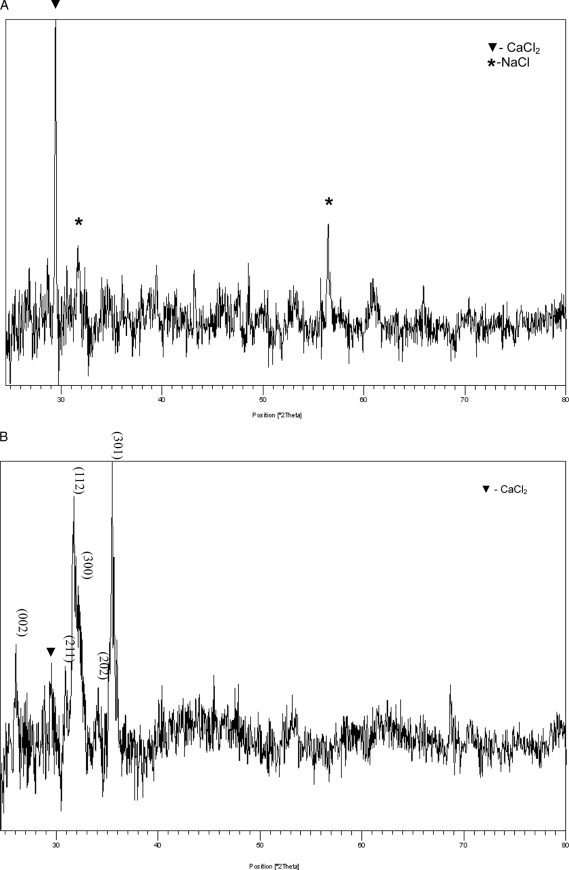
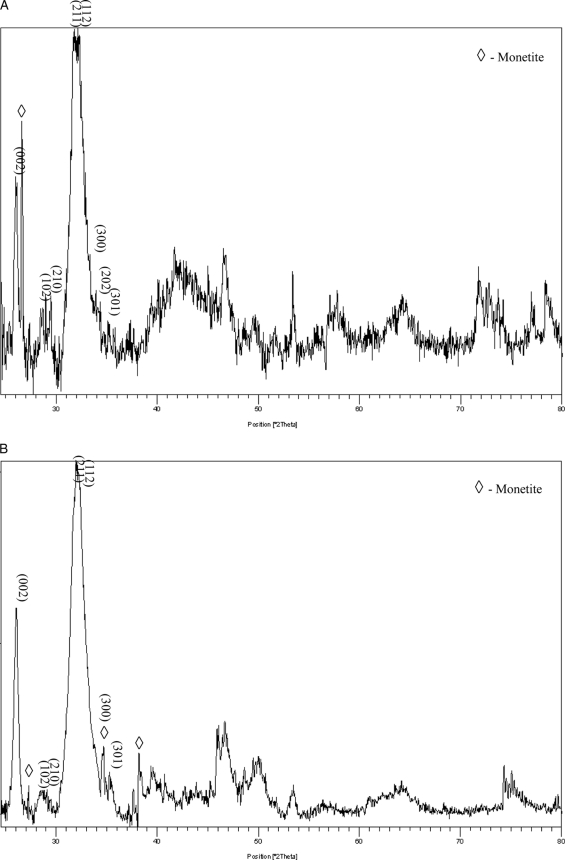
As we have demonstrated above through staining techniques, the expression of PP in NIH3T3 and MC3T3 cells alone was sufficient to induce the extracellular matrix to mineralize. To characterize the mineral nature of this newly formed matrix, the mineral in the collected matrices was examined by high resolution powder x-ray diffraction analysis. Fig. 7B shows the x-ray diffraction data of NIH3T3-PP cell cultures grown for 15 days. The most prominent peaks indexed in the pattern confirm the presence of hydroxyapatite. In contrast, in the NIH3T3-control cell cultures (Fig. 7A) there was no detectable mineral phase present, indicating the likelihood of a low fraction of mineralized volume or the existence of a low volume fraction of an amorphous phase or phases. More detailed systematic studies on the nucleation and growth kinetics of the mineral phase are essential to the elucidation of the mineral structure, and these studies are currently underway. However, in osteoblast-like MC3T3-E1 cells, the MC3T3-E1-control and MC3T3-E1-PP both produced a matrix that facilitated the formation of hydroxyapatite as expected; this is evidenced from the presence of the most prominent indexed hydroxyapatite peaks, shown in Fig. 8. Minor peaks of monetite can also be seen in Fig. 7, A and B, for both the MC3T3-E1 control and MC3T3-E1-PP cells, which could have formed as a result of a localized variation in pH. In regard to MC3T3-E1 mineralization, there is no difference in the x-ray diffraction pattern collected for the control and experimentally obtained mineralized matrix from MC3T3-E1-PP cells. Based on the indexed peaks in the XRD patterns, it appears that hydroxyapatite is readily apparent. However, this does not exclude differences in the process of mineralization, and the investigation of this possibility is ongoing. Considering that in the case of the cells that do not possess the ability to mineralize their extracellular matrix (such as NIH3T3), overexpression of PP clearly results in the formation of mineralized hydroxyapatite. It is possible that in the case of the cells that have the ability to secrete a matrix that will mineralize, the expression of PP could affect the mineralization process to further induce mineralization. This is particularly relevant in the MC3T3-E1 cells where we observed floating mineral particles, which were absent in both the MC3T3-E1 control as well as the NIH3T3-PP cells. These aspects would require a detailed quantitative assessment of the volume fraction of the crystallized phase; this work is currently in progress and will be part of a subsequent manuscript.3
FIGURE 7.
X-ray diffraction analysis of matrix from PP expressing NIH3T3 cells. Panel A, pattern of the mineralized matrix extracted from NIH3T3-control cells grown for 15 days. Panel B, NIH3T3-PP cells grown for 15 days showing the most prominent indexed peaks of the hydroxyapatite phase.
FIGURE 8.
X-ray diffraction analysis of matrix from PP expressing MC3T3 cells. Panel A shows the diffraction spectrum from MC3T3-E1-control cells grown for 15 days. In panel B, diffraction analysis of MC3T3-E1-PP cells grown for 15 days. Both MC3T3-E1-PP and MC3T3-E1-control cells exhibit the presence of hydroxyapatite peaks. The most prominent peaks are also indexed to confirm the presence of the hydroxyapatite phase.
Assessment of the Ability of rPP to Induce Mineral Formation
Finally, we were interested in whether non-phosphorylated PP could induce mineral formation, or whether phosphorylation is a requirement for this particular function of the protein. In a dynamic gelatin gel system, rPP harvested from bacteria (2.5–100 μg/ml) did not induce any mineral formation (data not shown). In each result, the calcium/phosphate ion ratio of the accumulated mineral was typical of HA, and representative x-ray diffraction and FTIR demonstrated HA formation, but the mineral yield in systems with non-phosphorylated PP was never significantly different from that in the control protein-free system. Thus, in the same system and at concentrations similar to those at which PP from rats was able to induce HA formation (31), non-phosphorylated rPP exhibited no induction of mineral formation. We suggest that in the case of PP, phosphorylation is a clear requirement for potentiation of mineral formation.
DISCUSSION
The process of biomineralization is complex, and the creation of a template for mineral deposition requires a high degree of organization among the involved proteins (for recent reviews, see Boskey et al. (1) and Veis (32)). Current approaches to studying the role of different proteins in biomineralization involve isolation through chromatography or preparation by recombinant protein technology. The purified protein is then assessed for its ability to induce or inhibit mineralization in a variety of in vitro systems (reviewed by Silverman et al. (33)). Although such systems are indisputably valuable and we have used them ourselves, they are limited by the reductive nature of an acellular environment. This limitation has driven us to consider a tissue culture system where aspects of biomineralization can be studied within the context of a normally complex cellular milieu. Indeed, to date very few studies have examined mineralization through protein modification in cell culture systems (34–37). The system described in this study allows for the manipulation of cells to achieve either expression or inhibition of proteins of interest to the mineralization process.
In this paper, we establish a biomineralization model using mammalian cells. Specifically, we focus on a unique extracellular matrix protein, phosphophoryn, which undergoes a high degree of phosphorylation during its synthesis. As described, our vector design directs the recombinant PP first to the endoplasmic reticulum (ER), subsequently channeling the protein through the secretory pathway (ER, Golgi, and secretory vesicles) where it undergoes post-translational modification (26, 27, 29, 30).
The investigative goal of the study was aimed at two particular questions. 1) Are the protein kinases responsible for the phosphorylation of such a highly phosphorylated protein (PP) restricted only to cells that natively assemble a mineralized matrix? 2) Can the expression of PP induce matrix mineralization in an environment not normally known to mineralize?
Our experimental approach is to take PP out of the context of a natively mineralizing environment, and to instead express it in the background of NIH3T3 cells. This setup allows for a clean comparison of mineralization potential with and without PP. Our results indicate that the expression of PP is sufficient for the induction of a matrix mineralizing phenotype in these cells.
Furthermore, this system allows us to ask whether these non-mineralizing fibroblast cells express the necessary kinases required for the phosphorylation of PP. Our data clearly show that PP can be phosphorylated by NIH3T3, thus indicating that the machinery necessary for the post-translational modification of PP is not restricted to cells that produce a mineralized matrix. It will be of interest in further studies to determine the extent of phosphorylation by the NIH3T3s and compare it to the degree of PP phosphorylation by MC3T3-E1 (osteoblastic cells). These quantitative data will provide us a better understanding of whether the degree and/or nature of post-translational modification is indeed different between cells that produce a matrix that is capable of mineralizing and those that do not. We also suggest that phosphorylation of PP is required for assembly of the mineralized matrix, based on the fact that our recombinant (bacterially expressed) PP did not induce mineral formation in the dynamic gel system.
Our current data indicate that PP can induce mineralization in an ectopic environment without the exogenous addition of other non-collagenous proteins known to be involved in the mineralization process. Whether this is exclusively through a physicochemical or templating effect or a signaling effect is yet to be determined, data from our laboratory and others indicate that through mitogen-activated protein kinase (MAPK) and SMAD pathways, PP can up-regulate the expression of other bone/dentin non-collagenous proteins (17, 18) involved in the biomineralization process.
Analysis of the mineral phase extracted from both matrices of NIH3T3-PP and MC3T3-E1-PP by XRD show the presence of hydroxyapatite (Figs. 7 and 8). These data indicate that PP plays a role in matrix mineralization, both in cells that natively mineralize their matrix, and in those that do not.
In conclusion, PP can be phosphorylated by cells that do not normally produce a mineralized matrix, such as NIH3T3 fibroblasts. We infer from this that the kinases required for PP modification are not restricted to mineralizing cells. Second, PP can efficiently induce matrix mineralization when expressed in NIH3T3 cells, and it appears to also play a role in MC3T3-E1 mineralization. We thus conclude that PP expression alone in a cell that also produces a collagenous matrix is sufficient to facilitate the initiation of mineralization. Finally, the tissue culture system described in this article will support further study of biomineralization events by allowing key players to be taken out of their native context, yet still expressed within the complexity of a cellular environment. We suggest that logical follow-on studies shall involve engineering cells to express other biomineralization proteins and their mutants. Future work will leverage proteomic methods and mineral characterization techniques toward the finer elucidation of mechanisms in biomineralization biology.
Acknowledgment
We acknowledge the editorial and scientific contribution of Dr. Leslie Bannon.
This work was supported, in whole or in part, by National Institutes of Health Grants DE016123 (to C. S.) and DE04141 (to A. L. B.).
C. Sfeir, D. Lee, J. Li, X. Zhang, A. L. Boskey, and P. N. Kumta, unpublished data.
- ECM
- extracellular matrix
- OPN
- osteopontin
- PP
- phosphophoryn
- rPP
- recombinant PP
- DMP
- dentin matrix protein
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Boskey A. L. (2003) Connect. Tissue Res. 44, Suppl. 1, 5–9 [PubMed] [Google Scholar]
- 2. Traub W., Jodaikin A., Arad T., Veis A., Sabsay B. (1992) Matrix 12, 197–201 [DOI] [PubMed] [Google Scholar]
- 3. Veis A., Sharkey M., Dickson I. (1977) in Calcium Binding Proteins and Calcium Function (Wasserman R. H., Corradino R. A., Carafoli E., Kretsinger R. H., MacLennan D. H., Siegel F. L. eds) pp. 409–415, Elsvier-North Holland, Amsterdam [Google Scholar]
- 4. Prince C. W., Oosawa T., Butler W. T., Tomana M., Bhown A. S., Bhown M., Schrohenloher R. E. (1987) J. Biol. Chem. 262, 2900–2907 [PubMed] [Google Scholar]
- 5. Fisher L. W., Whitson S. W., Avioli L. V., Termine J. D. (1983) J. Biol. Chem. 258, 12723–12727 [PubMed] [Google Scholar]
- 6. Stubbs J. T., 3rd, Mintz K. P., Eanes E. D., Torchia D. A., Fisher L. W. (1997) J. Bone Miner. Res. 12, 1210–1222 [DOI] [PubMed] [Google Scholar]
- 7. George A., Sabsay B., Simonian P. A., Veis A. (1993) J. Biol. Chem. 268, 12624–12630 [PubMed] [Google Scholar]
- 8. Dimuzio M. T., Veis A. (1978) Calcified Tissue Res. 25, 169–178 [DOI] [PubMed] [Google Scholar]
- 9. Termine J. D., Kleinman H. K., Whitson S. W., Conn K. M., McGarvey M. L., Martin G. R. (1981) Cell 26, 99–105 [DOI] [PubMed] [Google Scholar]
- 10. Gorski J. P. (1998) Crit. Rev. Oral Biol. Med. 9, 201–223 [DOI] [PubMed] [Google Scholar]
- 11. Fisher L. W., Torchia D. A., Fohr B., Young M. F., Fedarko N. S. (2001) Biochem. Biophys. Res. Commun. 280, 460–465 [DOI] [PubMed] [Google Scholar]
- 12. Nomura S., Wills A. J., Edwards D. R., Heath J. K., Hogan B. L. (1988) J. Cell Biol. 106, 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mark M. P., Prince C. W., Gay S., Austin R. L., Butler W. T. (1988) Cell Tissue Res. 251, 23–30 [DOI] [PubMed] [Google Scholar]
- 14. Salih E., Zhou H. Y., Glimcher M. J. (1996) J. Biol. Chem. 271, 16897–16905 [DOI] [PubMed] [Google Scholar]
- 15. Lasa M., Chang P. L., Prince C. W., Pinna L. A. (1997) Biochem. Biophys. Res. Commun. 240, 602–605 [DOI] [PubMed] [Google Scholar]
- 16. Stetler-Stevenson W. G., Veis A. (1987) Calcif. Tissue Int. 40, 97–102 [DOI] [PubMed] [Google Scholar]
- 17. Jadlowiec J., Koch H., Zhang X., Campbell P. G., Seyedain M., Sfeir C. (2004) J. Biol. Chem. 279, 53323–53330 [DOI] [PubMed] [Google Scholar]
- 18. Jadlowiec J. A., Zhang X., Li J., Campbell P. G., Sfeir C. (2006) J. Biol. Chem. 281, 5341–5347 [DOI] [PubMed] [Google Scholar]
- 19. Veis A., Wei K., Sfeir C., George A., Malone J. (1998) Eur. J. Oral Sci. 106, Suppl. 1, 234–238 [DOI] [PubMed] [Google Scholar]
- 20. He G., Ramachandran A., Dahl T., George S., Schultz D., Cookson D., Veis A., George A. (2005) J. Biol. Chem. 280, 33109–33114 [DOI] [PubMed] [Google Scholar]
- 21. Magne D., Bluteau G., Lopez-Cazaux S., Weiss P., Pilet P., Ritchie H. H., Daculsi G., Guicheux J. (2004) Connect. Tissue Res. 45, 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pourmand E. P., Binderman I., Doty S. B., Kudryashov V., Boskey A. L. (2007) J. Cell. Biochem. 100, 43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Willis J. B. (1960) Spectrochim. Acta 16, 259–272 [Google Scholar]
- 24. Heinonen J. K., Lahti R. J. (1981) Anal. Biochem. 113, 313–317 [DOI] [PubMed] [Google Scholar]
- 25. Boskey A. L. (1989) J. Phys. Chem. 91, 1628–1633 [Google Scholar]
- 26. Wu C. B., Pelech S. L., Veis A. (1992) J. Biol. Chem. 267, 16588–16594 [PubMed] [Google Scholar]
- 27. Wu C. B., Shimizu Y., Ng A., Pan Y. M. (1996) Connect. Tissue Res. 34, 23–32 [DOI] [PubMed] [Google Scholar]
- 28. Campbell K. P., MacLennan D. H., Jorgensen A. O. (1983) J. Biol. Chem. 258, 11267–11273 [PubMed] [Google Scholar]
- 29. Sfeir C., Veis A. (1996) Connect. Tissue Res. 35, 215–222 [DOI] [PubMed] [Google Scholar]
- 30. Sfeir C., Veis A. (1995) J. Bone Miner. Res. 10, 607–615 [DOI] [PubMed] [Google Scholar]
- 31. Boskey A. L., Maresca M., Doty S., Sabsay B., Veis A. (1990) Bone Miner. 11, 55–65 [DOI] [PubMed] [Google Scholar]
- 32. Veis A. (2005) Science 307, 1419–1420 [DOI] [PubMed] [Google Scholar]
- 33. Silverman L., Boskey A. L. (2004) Calcif. Tissue Int. 75, 494–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ueno A., Miwa Y., Miyoshi K., Horiguchi T., Inoue H., Ruspita I., Abe K., Yamashita K., Hayashi E., Noma T. (2006) J. Cell. Physiol. 209, 322–332 [DOI] [PubMed] [Google Scholar]
- 35. Morais S., Sousa J. P., Fernandes M. H., Carvalho G. S. (1998) Biomaterials 19, 13–21 [DOI] [PubMed] [Google Scholar]
- 36. Niemann A., von Bohlen A., Klockenkämper R., Keck E. (1990) Biochem. Biophys. Res. Commun. 170, 1216–1222 [DOI] [PubMed] [Google Scholar]
- 37. Harris N. L., Rattray K. R., Tye C. E., Underhill T. M., Somerman M. J., D'Errico J. A., Chambers A. F., Hunter G. K., Goldberg H. A. (2000) Bone 27, 795–802 [DOI] [PubMed] [Google Scholar]