Abstract
High cholesterol levels are associated with prostate cancer development. Androgens promote cholesterol accumulation by activating the sterol-regulatory element-binding protein isoform 2 (SREBP-2) transcription factor. However, SREBP-2 is in balance with the liver X receptor (LXR; NR1H2/NR1H3), a transcription factor that prevents cholesterol accumulation. Here, we show that LXR activity is down-regulated by the androgen receptor (AR; NR3C4). In turn, this reduces LXR target gene expression. This antagonism on LXR is also exerted by other steroid hormone receptors, including the estrogen, glucocorticoid, and progesterone receptors. This suggests a generalizable mechanism, but the AR does not affect LXR mRNA levels, protein degradation, or DNA binding. We also found that the AR does not require protein synthesis to influence LXR, suggesting a direct antagonism. However, the AR does not directly bind LXR. The AR N-terminal domain (involved in transactivation), but not its DNA-binding domain, is required to suppress LXR activity, suggesting coactivator competition. Overall, this androgen-mediated antagonism of LXR complements SREBP-2 activation, providing a more complete picture as to how androgens increase cellular cholesterol levels in a prostate cancer setting. Given the cross-talk between other steroid hormone receptors and LXR, hormonal regulation of cholesterol via LXR may occur in a variety of cellular contexts.
Keywords: Cholesterol Regulation, Gene Regulation, Hormone Receptors, Hormones, Transcription Regulation, ABCA1, ABCG1, Androgen Receptor, Liver X Receptor, Prostate Cancer
Introduction
Although cardiovascular disease has been the main focus for research into cholesterol regulation, there is now renewed interest in the relationship between cholesterol and prostate cancer (PCa).3 Epidemiological evidence associates PCa risk with high fat diets and lipid accumulation, whereas taking cholesterol-lowering drugs (e.g. statins) correlates with reduced PCa risk (1, 2). Studies at the cellular level, beginning with the 1942 study by Swyer (3), have led to the general observation that the aging prostate and PCa have elevated intracellular cholesterol levels (4). This could contribute to PCa development by providing a raw material for membrane synthesis, androgen production, and other signaling pathways (5).
Within the cell, cholesterol levels are regulated by uptake, synthesis, and efflux. A major homeostatic mechanism occurs at the transcriptional level, governed by the transcription factors sterol-regulatory element binding protein 2 (SREBP-2) and liver X receptor (LXR; NR1H2/NR1H3). SREBP-2 up-regulates a suite of genes involved in cholesterol uptake (e.g. low density lipoprotein receptor (LDLR)) and synthesis (e.g. HMG-CoA reductase (HMGCR)) (6, 7), promoting cholesterol accumulation. In contrast, as a heterodimer with the retinoid X receptor (RXR), LXR down-regulates genes involved in cholesterol synthesis (8) and increases those involved in efflux, such as the ATP-binding cassette transporter A1 (ABCA1) and G1 (ABCG1) (9). Combined with promoting the degradation of the LDLR (10), LXR thus prevents cholesterol accumulation. Consequently, it is possible that PCa cells have increased SREBP-2 activity and reduced LXR activity, thus leading to elevated cholesterol levels.
We previously investigated cholesterol homeostasis in two PCa laboratory cell lines, PC-3 and LNCaP, in the context of SREBP-2 (11). Although SREBP-2 responded to altered cholesterol status in both cell lines, PC-3 cells had higher basal SREBP-2 activity. Here, we began by further exploring how these cell lines vary and subsequently observed that a critical difference between these two cell lines, namely androgen receptor (AR; NR3C4) activity, influences LXR. This adds another level of complexity to cholesterol regulation in a PCa setting, and given the role of the AR in both PCa development and treatment (12), this has implications for PCa therapy.
EXPERIMENTAL PROCEDURES
Cell Culture
Cell lines used in this study were provided as gifts, including AR-positive LNCaP cells (Dr. Pamela Russell, Prince of Wales Hospital, Sydney, Australia), AR-negative PC-3 cells (Dr. Qihan Dong, University of Sydney), highly transfectable HeLa (HeLaT) cells (Dr. Noel Whitaker, University of New South Wales), and breast cancer MCF-7 cells (Dr. Ingrid Gelissen, University of New South Wales). LNCaP, PC-3, and HeLaT cells were maintained in Medium A (RPMI 1640, supplemented with 10% (v/v) fetal calf serum (FCS), 100 units/ml penicillin, and 100 μg/ml streptomycin), whereas MCF-7 cells were maintained in Medium B (high glucose DMEM, supplemented with 10% (v/v) FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin). All medium components were obtained from Invitrogen.
Hormones were removed from FCS with dextran-coated charcoal (Sigma-Aldrich), using a method similar to that described previously (13). Briefly, dextran-coated charcoal was added to FCS (1 g/100 ml of FCS) and stirred gently at room temperature for 1 h. This mixture was centrifuged (2,500 × g, 10 min), and the supernatant was similarly treated with dextran-coated charcoal. This was centrifuged again, and the supernatant was filter-sterilized (0.22-μm filter), generating charcoal-stripped FCS.
Prior to plating LNCaP and PC-3 cells, plates and dishes were treated with 25 μg/ml polyethyleneimine (Sigma-Aldrich) in 0.15 mm NaCl to enhance cellular adhesion (14). Treatment was performed in either Medium A or Medium C (RPMI 1640, supplemented with 10% (v/v) charcoal-stripped FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin).
Plasmid Constructs and Transfection
The sources of plasmids are listed in supplemental Table S1. For simplicity, the plasmid names have been condensed (supplemental Table S1). AR-ΔDBD (codons 555–612 deleted), AR-mDBD (codon 573 mutated from A to D, which disrupts the first zinc finger and subsequently all DNA binding (15)), and AR-ΔNTD (codons 2–536 deleted) expression constructs were generated from pcDNA-AR using PCR-based site-directed mutagenesis (16). To generate an AR expression construct driven by the viral thymidine kinase (TK) promoter (TK-AR), we replaced the squalene monooxygenase gene in TK-SM with the AR gene (from pcDNA-AR) using polymerase incomplete primer extension (PIPE) cloning (17). PIPE cloning was also used to generate the FLAG-LXRβ expression construct, by inserting a FLAG tag (DYKDDDDK) at the N terminus of the LXRβ gene (in pCMX-hLXRβ).
To generate Gluc fusion constructs, PIPE cloning was used to replace the GCN4 (zipper) gene in zipper-hGluc(1) with the LXRβ gene (from pCMX-hLXRβ) and insert an N-terminal FLAG tag, generating a FLAG-tagged LXRβ-hGluc(1) fusion construct. A similar protocol was used on zipper-hGluc(2) to generate FLAG-tagged RXRα-hGluc(2) and AR-hGluc(2) constructs, with genes sourced from pCMX-hRXRα and pcDNA-AR, respectively.
The primers used in these cloning and mutagenesis protocols are available upon request. Successful constructs were confirmed by sequencing and expression by Western blotting. Because our anti-AR antibody binds within the region deleted in AR-ΔNTD, we also generated MycHis-tagged AR and AR-ΔNTD expression constructs by (i) PIPE-cloning the AR gene (from pcDNA-AR) into pcDNA4-MycHisC, generating “AR-MycHis,” and then (ii) deleting codons 2–536 from AR-MycHis as was done for pcDNA-AR, generating “AR-ΔNTD-MycHis.” Expression was confirmed by probing against the Myc tag, and the MycHis-tagged constructs had similar effects on LXR activity as their respective untagged constructs (data not shown).
For transfection, cells were plated as described and transfected using TransIT-2020 reagent (MirusBio), according to the manufacturer's instructions. In addition, the media were refreshed prior to the addition of reagent-DNA complexes.
Cholesterol Assay
Following treatment, cells were lysed with radioimmune precipitation buffer (1% (v/v) Nonidet P-40, 0.1% (w/v) SDS, 1 mm Na3VO4, 150 mm NaCl, 20 mm Tris-HCl (pH 7.4), 5 mm EDTA, 0.5% (w/v) sodium deoxycholate). Cholesterol content was analyzed using the Amplex Red cholesterol assay kit (Invitrogen) and normalized to protein content, which was determined using the Pierce BCA protein assay (Thermo Fisher Scientific).
Luciferase Assay
Cells were plated in 60-mm dishes for transfection. HeLaT cells were transfected with 3.6 μg of luciferase construct and 0.36 μg of expression construct, and LNCaP cells were transfected with 6 μg of luciferase construct. Following transfection, cells were trypsinized and seeded into 24-well plates in Medium C and allowed to adhere overnight. As described previously (11), the treatment was delivered in a small quantity of plating medium, added to the existing media in the wells. Following treatment, cells were washed twice with PBS and lysed with PPBT buffer (100 mm potassium phosphate (pH 7.8) with 0.2% (v/v) Triton X-100) (18). Firefly luciferase activity was determined using the luciferase assay system (Promega) and normalized to protein content, as determined using the Pierce BCA protein assay. This was then made relative to the vehicle condition to obtain “relative luciferase activity.” This approach controls for both transfection efficiency (seeding after transfection) and cell number (protein content).
Furthermore, because LXRE-luc is driven by both LXR response elements (LXREs) and the viral TK promoter (19), additional cells were transfected with the TK-luc construct instead of LXRE-luc in the same experiment, and relative LXRE-luc activity values were divided by those of TK-luc to determine LXRE-specific promoter activity. For simplicity, this has been depicted as “LXRE-luc activity” in the figures presented here.
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
Total RNA was harvested and reverse transcribed to cDNA, and mRNA levels were determined (from cDNA) by qRT-PCR, as described previously (11). Primer sequences used to amplify human porphobilinogen deaminase (PBGD) (20), prostate-specific antigen (PSA) (21), ABCA1 (20), ABCG1 (22), LDLR (22), and HMGCR (22) cDNA have been previously described. Primer sequences for human SREBP-1c and SREBP-2 were provided by Dr. Etienne Lefai (Faculté de Médecine Lyon Sud). Primer sequences for human LXRα and LXRβ were also obtained from Dr Lefai but slightly modified to increase stringency between LXRα/β isoforms and transcript variants, LXRα-F (5′-GTTATAACCGGGAAGACTTTGCCA-3′), LXRα-R (5′-GCCTCTCTACCTGGAGCTGGT-3′), LXRβ-F (5′-CGTGGACTTCGCTAAGCAAGTG-3′), and LXRβ-R (5′-GGTGGAAGTCGTCCTTGCTGTAGG-3′). Primer sequences used to amplify human fatty acid synthase (FASN) cDNA were FASN-F (5′-GCAGAAGCTGTACACACTGCAG-3′) and FASN-R (5′-CAGGATGGGCACCTGCTGCT-3′). Primer sequences used to amplify SREBP cleavage activation protein (SCAP) were hSCAP-F (5′-CAAGAGGCTGCGTGTTGTC-3′) and hSCAP-R (5′-CCAGGATGCCAATCCAGA-3′). Primer sequences used to amplify human v-myb myeloblastosis viral oncogene homolog (avian)-like 1 (MYBL1) cDNA were MYBL1-F (5′-AAAATGCGAGTGGGTCATTC-3′) and MYBL1-R (5′-CCAGGACATGTGTTGAAAAACT-3′), which detect both MYBL1 isoforms. All primers were tested for amplification efficiency. Amplification data were analyzed using Rotor-Gene Version 6.0 (Build 27) (Corbett Research). Melting curve analysis was performed to confirm the production of a single product in each reaction. The mRNA expression levels were normalized to that of PBGD and made relative to the vehicle condition using the ΔΔCt method.
Western Blotting
Cells were plated in 6-well plates and transfected and treated as described in the figure legends. Cells were harvested with SDS lysis buffer (1% (w/v) SDS, 10 mm Tris-HCl (pH 7.6), 100 mm NaCl), supplemented with 2% (v/v) protease inhibitor mixture (Sigma-Aldrich). Protein content was determined using the Pierce BCA protein assay. Protein aliquots (30 μg) were subjected to 7.5% (w/v) SDS-PAGE and transferred to Trans-Blot transfer medium (Bio-Rad), as described previously (11). Membranes were blocked in 5% (w/v) skim milk in PBST buffer (0.1% (v/v) Tween 20 in PBS) for 1 h at room temperature. This was followed by incubation in primary antibody for 1 h at room temperature, washing with PBST six times for 5 min, incubation in secondary antibody for 1 h at room temperature, and washing with PBST six times for 5 min.
Primary antibodies included anti-FLAG (mouse clone M2, Sigma-Aldrich), anti-Myc (mouse clone 9E10, Santa Cruz Biotechnology, Inc.), anti-α-tubulin (mouse clone B-5-1-2, Sigma-Aldrich), and anti-AR (rabbit clone, catalogue no. 3202, Cell Signaling Technology). Peroxidase-conjugated AffiniPure donkey anti-mouse and anti-rabbit secondary antibodies were obtained from Jackson ImmunoResearch Laboratories.
Antibodies were visualized on Hyperfilm (GE Healthcare) using the ECL detection system (Millipore). Between antibodies, membranes were treated with a stripping buffer (25 mm glycine (pH 2), 1.5% (w/v) SDS). Films were scanned using the HP Scanjet G3010 (Hewlett-Packard).
Electrophoretic Mobility Shift Assay (EMSA)
HeLaT cells were plated in 100-mm dishes (2 dishes/condition) and transfected with 5 μg of each expression construct per 100-mm dish, as described in the figure legends. Following transfection, cell nuclei were isolated using a modified version of the protocol described previously (23). Cells were harvested on ice. Cells were washed with cold PBS, scraped, and pelleted by centrifugation (1,000 × g, 5 min). Cell pellets were resuspended in 0.5 ml of resuspension buffer (10 mm HEPES-KOH (pH 7.4), 10 mm KCl, 1.5 mm MgCl2, 5 mm Na-EDTA, 5 mm Na-EGTA, 250 mm sucrose, 5 mm dithiothreitol, supplemented with 2% (v/v) protease inhibitor mixture). The cell suspension was passed through a 21-gauge needle 30 times and centrifuged (1,000 × g, 7 min, 4 °C). The pellet was resuspended in 50 μl of nuclear extract buffer (20 mm HEPES-KOH (pH 7.6), 25% (v/v) glycerol, 0.42 m NaCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, supplemented with 2% (v/v) protease inhibitor mixture) and rotated at 4 °C overnight, and supernatant was isolated after centrifugation (30,000 × g, 30 min, 4 °C). Protein content was determined using the Pierce BCA protein assay.
For the EMSA, nuclear extract was incubated with binding buffer (final concentrations: 10 mm Tris-HCl (pH 8), 40 mm KCl, 0.1% (v/v) Nonidet P-40, 6% (v/v) glycerol, 1 mm dithiothreitol, 0.25 mg/ml salmon sperm DNA in a 20-μl reaction, adapted from Ref. 24) for 5 min at 25 °C. 1 pmol of Cy5–5′-labeled LXRE probe (5′-AGCTTGAATGACCAGCAGTAACCTCAGC-3′, bound to its reverse complement, obtained from Ref. 25) was then added, and the mixture was incubated for 30 min at 15 °C. For the competition assay, 100 pmol of unlabeled LXRE probe or mutant LXRE probe (5′-AGCTTGAATGTTCAGCAGTATTCTCAGC-3′ (mutations underlined), obtained from Whitney et al. (25)) were added, and the mixture was incubated for 5 min at 25 °C before the addition of labeled probe. For the supershift assay, 5 μg of anti-FLAG antibody (mouse clone M2, Sigma-Aldrich) was added after incubation with the labeled probe, and the mixture was incubated for a further 30 min at 15 °C. The mixtures were then subjected to 6% (w/v) native PAGE at 75 V and 4 °C and visualized using the FLA-5100 fluorescence scanner (Fujifilm, Tokyo, Japan).
Gaussia Luciferase Complementation Assay
HeLaT cells were plated in 6-well plates and transfected with hGluc fusion constructs (1 μg for one construct alone and 0.5 μg/construct if two constructs per well), as specified in the figure legends. Following transfection, cells were harvested and assayed as described previously (26), with slight modifications. Cells were trypsinized and pelleted in cold PBS (4 °C, 2,000 × g, 5 min). The pellet was washed with cold PBS before resuspension in 500 μl of phenol red-free RPMI (supplemented with 2% (v/v) protease inhibitor mixture). Cells were flash-frozen at −80 °C for 10 min and then thawed in a 37 °C water bath for 10 min. This freeze-thaw cycle was repeated twice more before centrifugation (4 °C, 10,000 × g, 5 min). The supernatant (20 μl) was assayed by the addition of 50 μl of Gaussia luciferase assay reagent (Targeting Systems) with a 0.4 s delay before integrating signal intensity over 10 s using the Veritas luminometer (Promega).
RESULTS
Cholesterol Efflux Genes, ABCA1 and ABCG1, Are Negatively Regulated by Androgens
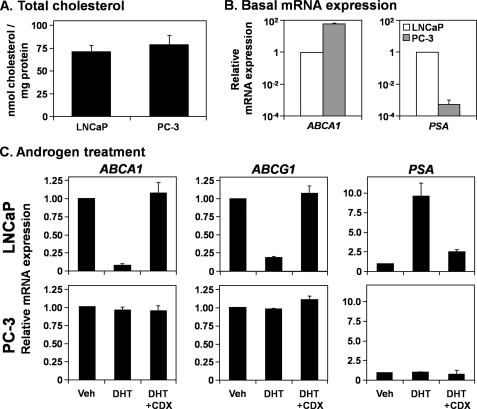
Following on from our previous work, in which we found that PC-3 cells had higher basal SREBP-2 activity than LNCaP cells (11), we predicted that PC-3 cells would have higher cholesterol levels. However, in the current study, we found that both cell lines had similar cholesterol content (Fig. 1A), with little esterified cholesterol (data not shown). Given the increased cholesterol uptake and synthesis associated with greater SREBP-2 activity, PC-3 cells may also have higher cholesterol efflux to maintain similar cholesterol levels to LNCaP cells. Two key efflux proteins are ABCA1 and ABCG1. We found that ABCA1 mRNA expression was ∼60-fold higher in PC-3 than LNCaP cells (Fig. 1B), which confirms previous findings (27); this study also showed that PC-3 cells have greater ABCG1 mRNA expression.
FIGURE 1.
Androgens influence cholesterol efflux gene expression in LNCaP, but not PC-3 cells. A, cells were grown in full serum (FCS, Medium A) for 24 h, after which cellular (total) cholesterol content was determined. B, cells were grown in Medium A for 24 h before ABCA1 and PSA mRNA expression levels were quantified by qRT-PCR. For each experiment, the ΔCt values, relative to the PBGD housekeeping gene, were normalized to that of the LNCaP cells in FCS to obtain the ΔΔCt values. These were converted into relative mRNA expression levels, whereby a one-unit decrease in ΔΔCt results in a 2-fold increase in mRNA expression. The scale on the y axis is logarithmic. C, cells were starved in androgen-deficient medium (Medium C) for 24 h, before treatment with DHT (1 nm), with or without casodex (CDX) (10 μm), in Medium C for another 24 h. Following treatment, ABCA1, ABCG1, and PSA mRNA expression levels were measured by qRT-PCR. Data are presented as mean ± S.E. (error bars), from three separate experiments, each performed with triplicate wells per condition. Veh, vehicle.
In contrast, mRNA expression of PSA, an androgen-regulated gene, was significantly (∼2000-fold) lower in PC-3 than LNCaP cells (Fig. 1B). Because PC-3 cells lack significant AR activity (28), whereas LNCaP cells are androgen-responsive (29), we hypothesized that the AR is down-regulating ABCA1 and ABCG1 mRNA expression in LNCaP cells. Treatment with dihydrotestosterone (DHT), a potent androgen, caused a significant decrease in mRNA expression of these genes in LNCaP cells (Fig. 1C). This effect was abolished by casodex, an AR antagonist. As a control, PSA mRNA expression increased with DHT treatment and was lowered to control levels with casodex cotreatment (Fig. 1C). In contrast, DHT and casodex had little effect in PC-3 cells (Fig. 1C). Thus, ABCA1 and ABCG1 transcription are negatively regulated by androgens in LNCaP cells.
The Androgen Receptor Antagonizes Liver X Receptor Activity
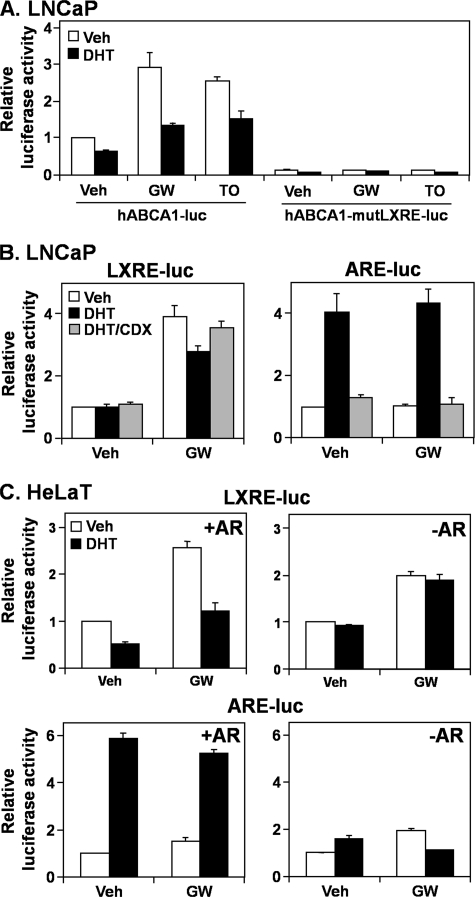
Because both ABCA1 and ABCG1 are LXR target genes, we hypothesized that AR antagonizes LXR. To explore this, we employed hABCA1-luc, a luciferase reporter construct driven by the ABCA1 promoter (−928/+10) (30). The LXR agonists, GW683965 and TO901317, increased promoter activity, which was reduced by DHT treatment in LNCaP cells (Fig. 2A). Mutating the LXRE abolished most of the ABCA1 promoter activity (Fig. 2A, hABCA1-mutLXRE-luc), including the DHT effect. Thus, it appears that DHT acts through the LXRE.
FIGURE 2.
The androgen receptor influences the liver X receptor activity at the promoter level. A, LNCaP cells were transfected with a luciferase construct containing the human ABCA1 promoter, either wild type (hABCA1-luc) or with the LXRE mutated (hABCA1-mutLXRE-luc). LNCaP (B) and HeLaT cells (C) were transfected with LXRE-luc or ARE-luc. HeLaT cells were also transfected with or without an AR expression vector. Following transfection, cells were treated for 24 h with DHT (1 nm), casodex (CDX) (10 μm), GW683965 (GW) (1 μm), or TO901317 (TO) (1 μm) in androgen-deficient medium (Medium C), after which relative firefly luciferase levels were assayed. Data are presented as mean ± S.E. (error bars) from at least three separate experiments, with each experiment performed with triplicate wells per condition. Veh, vehicle.
We tested this hypothesis using the LXRE-luc construct, which has three LXREs in tandem upstream of a viral TK promoter (19). We utilized GW683965 alone because TO901317 antagonizes the AR (31), as observed with our own assays (data not shown). Cotreatment with DHT reduced the response to GW683965, but this was overcome by concurrent casodex treatment (Fig. 2B, left). Also, DHT and casodex had the reverse effect on AR-specific activity, assayed using ARE-luc (Fig. 2B, right), which is driven by five tandem AR response elements. Thus, the AR may mediate this androgen-induced antagonism of the LXRE.
To further demonstrate the role of the AR, we employed an AR-negative, highly transfectable cell line, HeLaT. Transfecting an AR expression construct made these cells androgen-responsive (Fig. 2C, bottom), which in turn allowed DHT to suppress GW683965-induced LXRE activity (Fig. 2C, top left). This effect was not observed without the AR expression construct (Fig. 2C, top right). Thus, using pharmacological (Fig. 2B) and genetic (Fig. 2C) approaches, we have shown that the AR antagonizes LXR target gene expression via the LXRE.
Other Steroid Hormone Receptors Antagonize LXR Activity at the Promoter Level
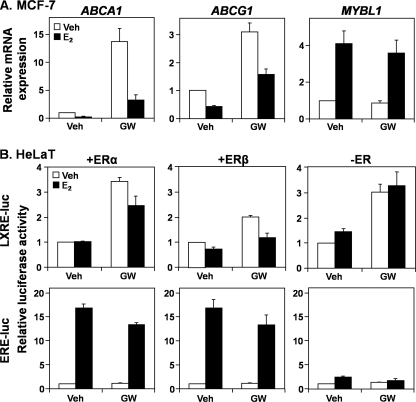
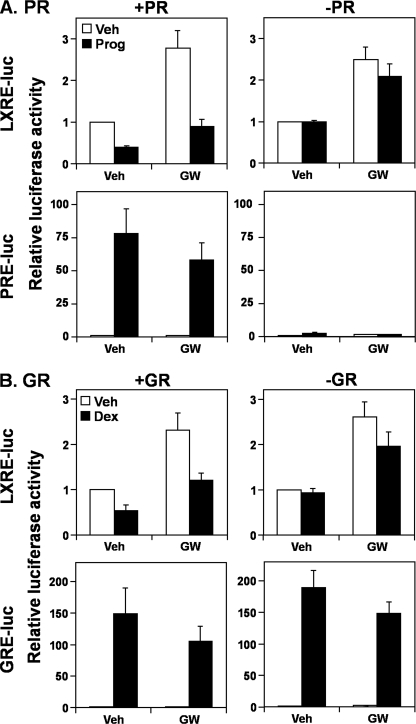
To gain insight into how AR antagonizes LXR, we examined whether this negative effect could be exerted by other steroid hormone receptors (type I nuclear receptors). Analogous to androgens in LNCaP cells, treatment with a potent estrogen, 17β-estradiol (E2), decreased ABCA1 and ABCG1 mRNA expression in estrogen-receptor (ER)-positive MCF-7 cells (Fig. 3A), whereas up-regulating a known estrogen-responsive gene, MYBL1 (32). Furthermore, similar to androgens in Fig. 2C, E2 reduced LXR activity in HeLaT cells only when the LXRE-luc construct was cotransfected with either an ERα or ERβ expression construct (Fig. 3B). We observed a similar phenomenon with exogenous progesterone receptor (PR) and glucocorticoid receptor (GR) as well as endogenous GR (p < 0.05, paired t test) (Fig. 4). Thus, it appears that other steroid hormone receptors also antagonize LXR activity, via the LXRE. In addition, LXR activation decreased ERα (Fig. 3B), PR and GR (Fig. 4) activity (p < 0.05, paired t test), and AR slightly (Fig. 2C, p ∼0.08, paired t test), suggesting that the antagonism is mutual.
FIGURE 3.
The estrogen receptor also reduces liver X receptor promoter activity. A, MCF-7 cells were starved in steroid hormone-deficient medium (Medium C) for 24 h before treatment with E2 (1 nm), with or without GW683965 (GW) (1 μm), in Medium C for another 24 h. Following treatment, ABCA1, ABCG1, and MYBL1 mRNA expression levels were measured by qRT-PCR. B, HeLaT cells were transfected with LXRE-luc or ERE-luc, with or without an ERα or ERβ expression vector. Following transfection, cells were treated for 24 h with E2 (1 nm) or GW683965 (1 μm), in androgen-deficient media (Medium C), after which relative firefly luciferase levels were assayed. Data are presented as mean ± S.E. (error bars) from three separate experiments, each performed with triplicate wells per condition. Veh, vehicle.
FIGURE 4.
Other steroid hormone receptors also influence the liver X receptor at the promoter level. A, HeLaT cells were transfected with LXRE-luc or PRE-luc, with or without a PR expression vector. Following transfection, cells were treated for 24 h with progesterone (Prog) (1 μm) or GW683965 (GW) (1 μm), in steroid hormone-deficient media (Medium C). B, HeLaT cells were transfected with LXRE-luc or GRE-luc, with or without a GR expression vector. Following transfection, cells were treated for 24 h with dexamethasone (Dex) (100 nm) or GW683965 (1 μm), in Medium C. Following treatment in A and B, relative firefly luciferase levels were assayed. Data are presented as mean ± S.E. (error bars) from three separate experiments, each performed with triplicate wells per condition. Veh, vehicle.
The AR Does Not Influence LXR Expression or LXR-DNA Binding or Bind LXR Directly
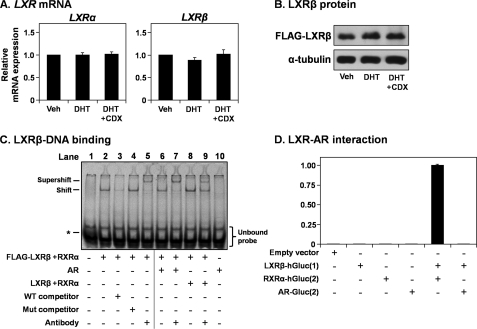
Our next set of experiments aimed to dissect the mechanism by which AR influences LXR activity. The AR did not influence the mRNA expression of either LXR isoform in LNCaP cells (Fig. 5A). In subsequent experiments, we focused on LXRβ, the ubiquitous isoform, which is also expressed at higher levels than LXRα in PCa cells (27). The AR did not affect the protein degradation of LXRβ (Fig. 5B).
FIGURE 5.
The androgen receptor does not influence LXR expression or LXR-DNA binding. A, RNA harvested in the experiments described in Fig. 1C were analyzed for LXRα and LXRβ mRNA expression levels by qRT-PCR. B, LNCaP cells were transfected with 2 μg of FLAG-LXRβ expression vector. Following transfection, cells were starved in Medium C for 24 h before treatment with DHT (1 nm), with or without casodex (CDX) (10 μm), in Medium C for another 24 h. Following treatment, protein was harvested and subjected to SDS-PAGE and immunoblotting. C, HeLaT cells were transfected with FLAG-LXRβ, LXRβ, RXRα, or AR expression constructs or empty vector (pcDNA3.1). After transfection, nuclear extract was harvested. For each condition, 5 μg of nuclear extract was used and made up to 10 μg using the nuclear extract of cells transfected with the empty vector. The EMSA was performed as described under “Experimental Procedures.” The band in the first lane (indicated by an asterisk) is bromophenol blue, used to track the progress of the electrophoresis. D, to perform the Gaussia luciferase complementation assay, HeLaT cells were transfected with the LXRβ-hGluc(1), RXRα-hGluc(2), AR-hGluc(2), or empty vector (pcDNA3.1). After transfection, cells were harvested, and Gaussia luciferase was assayed. Luciferase activity was made relative to the LXRβ-hGluc(1) + RXRα-hGluc(2) condition. Data in A are presented as mean ± S.E. (error bars) from three separate experiments, with each experiment performed with triplicate wells per condition. Data in B and C are representative of two independent experiments each. Data in D are presented as mean ± S.D. (error bars), representative of two separate experiments, with each experiment performed with triplicate wells per condition. Veh, vehicle.
Next, we tested whether AR influences LXR-DNA binding using EMSA. Proteins were overexpressed in HeLaT cells, and nuclear extracts were mixed prior to determining binding to a Cy5-labeled LXRE probe (Fig. 5C). Nuclear extract from cells transfected with FLAG-LXRβ and RXRα generated a shift (lane 2), which was diminished by competition with wild type (lane 3) but not mutated (lane 4) unlabeled probe. Furthermore, incubation with an anti-FLAG antibody generated a supershift (lane 5). Together, these controls confirm that the EMSA can detect LXR-DNA binding. Mixing the FLAG-LXRβ/RXRα lysate with AR lysate affected neither the shift nor supershift (lanes 6 and 7). In contrast, the addition of (untagged) LXRβ/RXRα lysate diminished the supershift band (lane 9). Furthermore, AR lysate alone did not generate a shift (lane 10). Collectively, this indicates that AR does not affect LXR-DNA binding, interacting with neither LXR nor the LXRE.
To further test a direct interaction between AR and LXR, we employed a Gaussia luciferase complementation assay (26). The two putative interaction partners are fused to different halves of Gaussia luciferase (either the N-terminal hGluc(1) or C-terminal hGluc(2)). These fusion constructs have no luciferase activity alone (Fig. 5D). However, when co-expressed, if there is an interaction between the putative partners, the hGluc halves are brought within close proximity, enabling luciferase activity. This can be seen with LXR and RXR but not between LXR and AR (Fig. 5D), confirming that there is no direct interaction between LXR and AR.
The AR Requires the N-terminal Domain to Antagonize LXR Activity
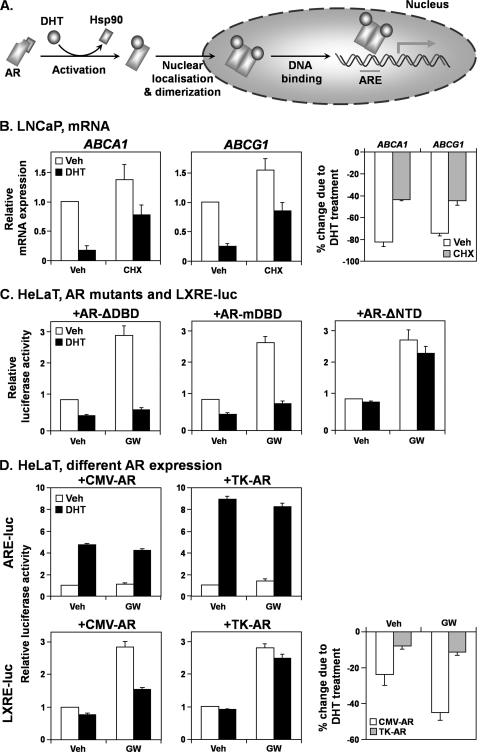
We next shifted our focus toward the AR; since both AR and ligand (DHT) are needed for antagonizing LXR activity (Fig. 2), what aspects of AR activation are required to influence LXR? The stages of AR activation are summarized in Fig. 6A. Briefly, a ligand (e.g. DHT) binds to AR, displacing a bound heat-shock protein (e.g. Hsp90) chaperone. This allows AR to migrate into the nucleus and dimerize, subsequently binding to DNA and up-regulating target gene expression (12).
FIGURE 6.
The androgen receptor influences the liver X receptor by a direct mechanism involving the N-terminal domain. A, schematic depicting different stages of AR activation. Details are provided under “Results.” The requirement of different stages in LXR antagonism are tested. B, LNCaP cells were starved in androgen-deficient media (Medium C) for 24 h, before treatment with DHT (1 nm), with or without cycloheximide (CHX) (10 μg/ml), in Medium C for another 24 h. Following treatment, ABCA1 and ABCG1 mRNA expression levels were measured by qRT-PCR (left and middle, respectively). The percentage decreases in ABCA1 and ABCG1 mRNA expression due to DHT are depicted on the right. C, HeLaT cells were transfected with LXRE-luc and either AR-ΔDBD, AR-mDBD, or AR-ΔNTD expression constructs. Following transfection, cells were treated for 24 h with DHT (1 nm) or GW683965 (GW) (1 μm), in androgen-deficient media (Medium C), after which relative firefly luciferase activity was assayed. As a control, the AR expression construct was included in these experiments, yielding results similar to those shown in Fig. 2C. D, HeLaT cells were transfected with either ARE-luc or LXRE-luc and either CMV-AR or TK-AR expression constructs. Following transfection, cells were treated and relative firefly luciferase activity was assayed as in C. The percentage decreases in LXRE-luc luciferase activity due to DHT treatment are depicted at the bottom right. Data are presented as mean ± S.E. (error bars) from three separate experiments, with each experiment performed with triplicate wells per condition. Veh, vehicle.
To test if AR target expression is required, LNCaP cells were treated with cycloheximide to inhibit protein translation. Even in the presence of cycloheximide, DHT reduced ABCA1 and ABCG1 expression (Fig. 6B), suggesting that AR does not antagonize LXR as a secondary effect.
We tested different aspects of AR activity by determining if mutating the AR could protect LXR activity, as measured by LXRE-luc in HeLaT cells (Fig. 2C). The DNA-binding domain (DBD) was mutated by direct deletion (AR-ΔDBD) or the point mutation A573D (AR-mDBD), which has been shown to ablate the DNA-binding capacity of the first zinc finger of the AR (15). Neither DBD mutation ablated the LXR antagonism (Fig. 6C, left and middle), suggesting that AR-DNA binding is not required, thus confirming that 1) AR does not bind to the LXRE (Fig. 5C), 2) AR does not influence gene expression of LXR (Fig. 5A) or an intermediate, and 3) AR does not require microRNA-associated mechanisms.
However, deleting the N-terminal domain (NTD) of AR (AR-ΔNTD) abolished the AR-LXR interaction (Fig. 6C, right). Because a transactivation domain, which interacts with transcriptional coregulators, is the major feature found within the NTD (33), this suggests that coregulator interaction is crucial for this antagonism.
This would suggest that differences in AR expression would influence LXR activity. To explore this, we compared the cytomegalovirus (CMV)-driven AR expression construct (CMV-AR, used previously in Figs. 2C and 6C) with one that is TK-driven (TK-AR). The CMV promoter is stronger than the TK promoter, resulting in higher AR expression with CMV-AR (not shown). Although the relative response to androgens was higher for TK-AR (Fig. 6D, top), TK-AR antagonized LXR activity less than CMV-driven AR (Fig. 6D, bottom). Thus, AR expression influences the LXR antagonism, which does not directly correlate with relative androgen responsiveness.
Androgens Influence SREBP-2 and SREBP-1c Inversely to LXR
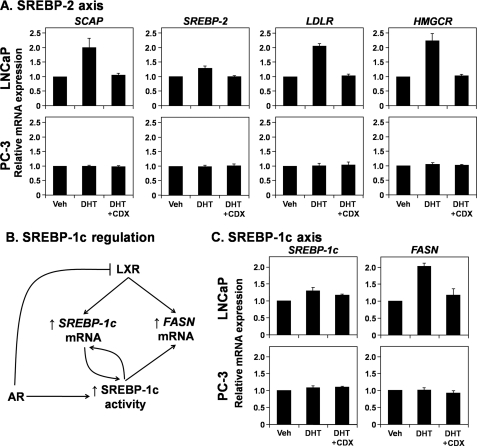
We next examined the impact that androgens have on the other lipogenic transcription factors, the SREBPs. All SREBP isoforms are activated when an escort protein, SCAP, facilitates migration to the Golgi apparatus (34). It is known that androgens up-regulate SCAP mRNA expression, thus promoting SREBP activation (35). This is confirmed in Fig. 7A, whereby androgens increase mRNA levels of SCAP and the SREBP-2-target genes, LDLR and HMGCR, while having little effect on SREBP-2 expression itself (Fig. 7A).
FIGURE 7.
The SREBP-2 and SREBP-1c axes are stimulated by androgens. A and C, RNA harvested in the experiments described in Fig. 1C were analyzed for SCAP, SREBP-2, LDLR, and HMGCR (A) and SREBP-1c and FASN (C) mRNA expression. B, schematic showing how AR and LXR influence SREBP-1c and FASN expression. Details are provided under “Results.” Data presented in A and C are mean ± S.E. (error bars) from three separate experiments, with each experiment performed with triplicate wells per condition. Veh, vehicle.
Increased SCAP expression also influences SREBP-1c, which targets genes in fatty acid metabolism, including FASN and SREBP-1c itself. However, in contrast to the SREBP-2, both SREBP-1c and FASN are also LXR target genes. Thus, it is interesting to consider the net effect of AR on SREBP-1c activity (Fig. 7B), because AR up-regulates SCAP mRNA (Fig. 7A) yet down-regulates LXR activity (Figs. 1 and 2). We found that altering androgen status had little effect on SREBP-1c mRNA expression, whereas DHT slightly increased FASN expression (Fig. 7C).
The AR-LXR Interaction Influences Cellular Cholesterol Levels
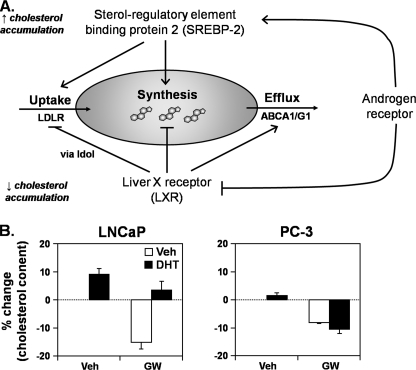
Returning our focus to cholesterol regulation, our results show that the AR down-regulates LXR activity (Figs. 1 and 2) and up-regulates SREBP-2 activity (Fig. 7A), which together would lead to increased cholesterol accumulation (Fig. 8A). Our investigation began with a comparison of cellular cholesterol levels (Fig. 1A), thus we last examined whether AR and LXR have opposing effects on cholesterol levels. Although treatment with LXR agonist (GW683965) reduced cholesterol levels in both LNCaP and PC-3 cells, androgen (DHT) treatment only reversed this effect in LNCaP cells and not in androgen-insensitive PC-3 cells (Fig. 8B). These results demonstrate that the AR-LXR interaction has a significant impact on cholesterol homeostasis.
FIGURE 8.
The cross-talk between the androgen receptor and liver X receptor influences cellular cholesterol homeostasis. A, androgens stimulate cholesterol accumulation by acting on both SREBP and LXR transcription factors. Details are provided under “Results.” B, LNCaP and PC-3 cells were treated for 48 h with DHT (1 nm) or GW683965 (GW) (1 μm) in Medium A. Every condition was cotreated with 5 μm compactin (mevastatin), to inhibit cholesterol synthesis and thus ensure that cholesterol levels fluctuate by only cholesterol import/export. Cells were harvested and assayed for total cholesterol content. The percentage change from the vehicle (Veh) (first) condition is depicted. Data are presented as mean ± S.E. (error bars) from three separate experiments, with each experiment performed with triplicate wells per condition.
DISCUSSION
In this study, we investigated the AR, a key hormone receptor in PCa, and its role in lipid metabolism. Androgens down-regulate ABCA1 mRNA expression (Fig. 1); although this has been observed previously at the transcriptional (36–38) and promoter (36) levels, our study sheds light on the mechanism. We demonstrate that this antagonism requires the AR and occurs via LXR (Fig. 2).
Other steroid hormone receptors down-regulate LXR activity (Figs. 3 and 4), suggesting a generalizable mechanism. It has been shown that ER down-regulates LXRα mRNA in mouse macrophages (39–40), through an ER response element in the LXRα promoter (39), but we found that E2 treatment did not influence LXRα or LXRβ mRNA expression in MCF-7 cells (data not shown). Likewise, GR down-regulates LXRα in mouse macrophages, through multiple GR response elements in the LXRα promoter, but GR agonists did not influence LXRα or LXRβ expression in human liver cell lines (41). To the best of our knowledge, little is known about the cross-talk between PR and LXR. In contrast to these steroid hormone receptors, the AR influences LXR directly (Figs. 2 and 6) but does not affect LXR mRNA levels, protein degradation, or LXR-DNA binding (Fig. 5). Also, there is no direct interaction between AR and LXR (Fig. 5D). Thus, we suggest that LXR and AR compete for coactivators, based on several lines of evidence.
First, mutating the AR DBD did not abolish the antagonism, whereas deleting the NTD did (Fig. 6C). The NTD is responsible for coactivator recruitment (33) and dimerization by interaction with the C terminus (42). Second, casodex treatment overcame the androgen effect on ABCA1/G1 mRNA levels (Fig. 1) and LXRE-specific activity (Fig. 2). Although the casodex mode of action remains controversial, one group has shown that casodex allows translocation of AR into the nucleus and DNA binding in LNCaP cells (43) but prevents NH2/COOH-terminal interaction and coactivator recruitment (43, 44). This casodex antagonism occurred independently of the corepressors NCoR and SMRT (44), suggesting competition for coactivators rather than corepressors. Third, weaker AR expression results in a higher androgen response but weaker LXR antagonism (Fig. 6D), suggesting that the antagonism is dependent upon the stoichiometry of AR in relation to other cellular components rather than relative levels of AR activity alone. Last, a subtle mutual antagonism was seen between the LXR and AR (Fig. 2C), more strongly observed with ERα (Fig. 3B), PR, and GR (Fig. 4), suggesting that this is a global mechanism.
Furthermore, if the AR is competing with LXR for coactivators, then there may be similar cross-talk between AR and other type II nuclear receptors (e.g. PPARγ, FXR, PXR, and CAR). For instance, AR competes with PPARγ for ARA70 (45), whereas the interaction may instead involve direct binding to the nuclear receptor, such as between AR and PXR (46) or RXR (47), the latter of which may influence LXR. Thus, the precise mechanism by which androgens antagonize LXR remains to be fully elucidated.
Nevertheless, this antagonism explains the increased LXR target gene expression in (AR-negative) PC-3 cells (48) and why androgens down-regulate ABCA1/G1 expression in vitro (36–38) (this study). This correlates with in vivo findings, including reduced ABCA1 expression when hypogonodal (androgen-deficient) mice were treated with testosterone (49), increased ABCA1/G1 expression in PCa xenografts when mice were treated with the androgen synthesis inhibitor dutasteride (50), and increased ABCA1 mRNA levels in PCa tissues from patients receiving androgen deprivation therapy (38). Thus, the cross-talk between AR and LXR is observed in vivo and hence has biological relevance.
In turn, the AR reverses LXR-mediated cholesterol loss in (AR-positive) LNCaP cells (Fig. 8B). This adds another layer of complexity to cholesterol homeostasis in a PCa setting. Whereas a suite of hormone receptors was shown to antagonize LXR activity (Figs. 3 and 4), the AR has also been shown to up-regulate SREBP-2 activity (35) (Fig. 7A). Thus, the AR acts through multiple avenues to promote cholesterol accumulation in PCa (Fig. 8A), which in turn provides raw material for PCa cell growth and signaling (5).
There is also great interest in the regulation of fatty acid metabolism in PCa (51–53) and in cancers in general (54, 55). LXR differentially influences cholesterol and fatty acid homeostasis; while LXR promotes cholesterol depletion, it promotes fatty acid synthesis by directly up-regulating FASN and SREBP-1c expression. Thus, the AR-LXR antagonism should negatively influence SREBP-1c activity, but the AR also promotes SREBP-1c activation by increasing SCAP mRNA levels (35). We found that androgen treatment increased FASN expression and had little influence on SREBP-1c expression (Fig. 7C); this resembles the SREBP-2 axis (Fig. 7A), suggesting that increased SCAP expression outweighs LXR antagonism by the AR. Overall, it appears that LXR antagonism by AR impacts differently upon the cholesterol (ABCA1/ABCG1) and fatty acid (SREBP-1c/FASN) axes, probably due to the interaction between AR and other transcription factors.
Given that the AR plays a major role in prostate cell biology and PCa development, its antagonism of LXR suggests that activating LXR has therapeutic potential. Indeed, it has been shown that LXR reduces growth of PCa cells and xenografts (56, 57), delaying the progression to androgen independence in a xenograft model (58). This has been attributed to cholesterol depletion, which disrupts cholesterol-rich lipid rafts and thus down-regulates Akt activity (57). Cholesterol-independent mechanisms have also been proposed (59), such as influencing G1-S transition via Skp (56). The antiproliferative effect of LXR has also been described outside the cancerous setting, in the regenerating liver, whereby LXR lowers cholesterol required for hepatocyte proliferation (60).
In view of AR-LXR cross-talk, whereas we have shown here that AR antagonizes LXR, the relationship is bidirectional; LXR inhibited androgen-dependent PCa cell growth in vitro and prostate regeneration in mice by up-regulating sulfotransferases (e.g. sult2a1) and down-regulating steroid sulfatase (sts), the net effect being deactivated androgens (61). An analogous LXR effect has been found with estrogens (62, 63), albeit liver-specific, suggesting that this cross-talk extends beyond different androgens in a PCa setting. LXR reduces the growth of breast cancer cells (56, 62, 64), particularly ERα-positive cells (64), partially through an ER-dependent mechanism. Collectively, this supports the idea of two-way cross-talk between LXR and steroid hormone receptors, between lipid metabolism and hormonal signaling, with implications not only for PCa therapy but for a variety of cellular contexts.
Acknowledgments
We thank the members of the Brown Research Laboratory for feedback throughout the investigation and Dr. Ingrid Gelissen for introducing us to the estrogen angle of this investigation. We are also grateful to Dr. Fahri Saatcioglu and Dr. Jin Yang for assistance in transfecting LNCaP cells and to numerous colleagues for providing the cell lines, plasmids, and qRT-PCR primer designs used in this study. We thank Dr. David Handelsman for providing dihydrotestosterone.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- PCa
- prostate cancer
- SREBP
- sterol-regulatory element-binding protein
- LXR
- liver X receptor
- RXR
- retinoid X receptor
- LDLR
- low density lipoprotein receptor
- HMGCR
- HMG-CoA reductase
- AR
- androgen receptor
- HeLaT
- highly transfectable HeLa
- TK
- thymidine kinase
- LXRE
- LXR response element
- qRT-PCR
- quantitative reverse transcription-PCR
- PBGD
- porphobilinogen deaminase
- PSA
- prostate-specific antigen
- FASN
- fatty acid synthase
- SCAP
- SREBP cleavage activation protein
- DHT
- dihydrotestosterone
- ER
- estrogen receptor
- E2
- 17β-estradiol
- GR
- glucocorticoid receptor
- PR
- progesterone receptor
- DBD
- DNA-binding domain
- NTD
- N-terminal domain
- PIPE
- polymerase incomplete primer extension.
REFERENCES
- 1. Brown A. J. (2007) Clin. Exp. Pharmacol. Physiol. 34, 135–14117250629 [Google Scholar]
- 2. Solomon K. R., Freeman M. R. (2008) Trends Endocrinol. Metab. 19, 113–121 [DOI] [PubMed] [Google Scholar]
- 3. Swyer G. I. (1942) Cancer Res. 2, 372–375 [Google Scholar]
- 4. Schaffner C. P. (1981) Prog. Clin. Biol. Res. 75A, 279–324 [PubMed] [Google Scholar]
- 5. Krycer J. R., Sharpe L. J., Luu W., Brown A. J. (2010) Trends Endocrinol. Metab. 21, 268–276 [DOI] [PubMed] [Google Scholar]
- 6. Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., Goldstein J. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amemiya-Kudo M., Shimano H., Hasty A. H., Yahagi N., Yoshikawa T., Matsuzaka T., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., Osuga J., Harada K., Gotoda T., Sato R., Kimura S., Ishibashi S., Yamada N. (2002) J. Lipid Res. 43, 1220–1235 [PubMed] [Google Scholar]
- 8. Wang Y., Rogers P. M., Su C., Varga G., Stayrook K. R., Burris T. P. (2008) J. Biol. Chem. 283, 26332–26339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tall A. R., Costet P., Wang N. (2002) J. Clin. Invest. 110, 899–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zelcer N., Hong C., Boyadjian R., Tontonoz P. (2009) Science 325, 100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krycer J. R., Kristiana I., Brown A. J. (2009) PLoS One 4, e8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feldman B. J., Feldman D. (2001) Nat. Rev. Cancer 1, 34–45 [DOI] [PubMed] [Google Scholar]
- 13. Murillo H., Huang H., Schmidt L. J., Smith D. I., Tindall D. J. (2001) Endocrinology 142, 4795–4805 [DOI] [PubMed] [Google Scholar]
- 14. Vancha A. R., Govindaraju S., Parsa K. V., Jasti M., González-García M., Ballestero R. P. (2004) BMC Biotechnol. 4, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brüggenwirth H. T., Boehmer A. L., Lobaccaro J. M., Chiche L., Sultan C., Trapman J., Brinkmann A. O. (1998) Endocrinology 139, 103–110 [DOI] [PubMed] [Google Scholar]
- 16. Sanchis J., Fernández L., Carballeira J. D., Drone J., Gumulya Y., Höbenreich H., Kahakeaw D., Kille S., Lohmer R., Peyralans J. J., Podtetenieff J., Prasad S., Soni P., Taglieber A., Wu S., Zilly F. E., Reetz M. T. (2008) Appl. Microbiol. Biotechnol. 81, 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klock H. E., Lesley S. A. (2009) Methods Mol. Biol. 498, 91–103 [DOI] [PubMed] [Google Scholar]
- 18. Hampf M., Gossen M. (2006) Anal. Biochem. 356, 94–99 [DOI] [PubMed] [Google Scholar]
- 19. Wong J., Quinn C. M., Brown A. J. (2007) Lipids Health Dis. 6, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kielar D., Dietmaier W., Langmann T., Aslanidis C., Probst M., Naruszewicz M., Schmitz G. (2001) Clin. Chem. 47, 2089–2097 [PubMed] [Google Scholar]
- 21. Bièche I., Parfait B., Tozlu S., Lidereau R., Vidaud M. (2001) Carcinogenesis 22, 1521–1526 [DOI] [PubMed] [Google Scholar]
- 22. Wong J., Quinn C. M., Brown A. J. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 2365–2371 [DOI] [PubMed] [Google Scholar]
- 23. Sever N., Yang T., Brown M. S., Goldstein J. L., DeBose-Boyd R. A. (2003) Mol. Cell 11, 25–33 [DOI] [PubMed] [Google Scholar]
- 24. Lehmann J. M., Kliewer S. A., Moore L. B., Smith-Oliver T. A., Oliver B. B., Su J. L., Sundseth S. S., Winegar D. A., Blanchard D. E., Spencer T. A., Willson T. M. (1997) J. Biol. Chem. 272, 3137–3140 [DOI] [PubMed] [Google Scholar]
- 25. Whitney K. D., Watson M. A., Goodwin B., Galardi C. M., Maglich J. M., Wilson J. G., Willson T. M., Collins J. L., Kliewer S. A. (2001) J. Biol. Chem. 276, 43509–43515 [DOI] [PubMed] [Google Scholar]
- 26. Remy I., Michnick S. W. (2006) Nat. Methods 3, 977–979 [DOI] [PubMed] [Google Scholar]
- 27. Trasino S. E., Kim Y. S., Wang T. T. (2009) Mol. Cancer Ther. 8, 1934–1945 [DOI] [PubMed] [Google Scholar]
- 28. Kaighn M. E., Narayan K. S., Ohnuki Y., Lechner J. F., Jones L. W. (1979) Invest. Urol. 17, 16–23 [PubMed] [Google Scholar]
- 29. Horoszewicz J. S., Leong S. S., Kawinski E., Karr J. P., Rosenthal H., Chu T. M., Mirand E. A., Murphy G. P. (1983) Cancer Res. 43, 1809–1818 [PubMed] [Google Scholar]
- 30. Costet P., Luo Y., Wang N., Tall A. R. (2000) J. Biol. Chem. 275, 28240–28245 [DOI] [PubMed] [Google Scholar]
- 31. Chuu C. P., Chen R. Y., Hiipakka R. A., Kokontis J. M., Warner K. V., Xiang J., Liao S. (2007) Biochem. Biophys. Res. Commun. 357, 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bourdeau V., Deschênes J., Laperrière D., Aid M., White J. H., Mader S. (2008) Nucleic Acids Res. 36, 76–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jenster G., van der Korput H. A., Trapman J., Brinkmann A. O. (1995) J. Biol. Chem. 270, 7341–7346 [DOI] [PubMed] [Google Scholar]
- 34. Brown M. S., Goldstein J. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11041–11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heemers H. V., Verhoeven G., Swinnen J. V. (2006) Mol. Endocrinol. 20, 2265–2277 [DOI] [PubMed] [Google Scholar]
- 36. Fukuchi J., Hiipakka R. A., Kokontis J. M., Hsu S., Ko A. L., Fitzgerald M. L., Liao S. (2004) Cancer Res. 64, 7682–7685 [DOI] [PubMed] [Google Scholar]
- 37. Wang J. H., Tuohimaa P. (2008) Gene Regul. Syst. Bio. 2, 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sekine Y., Demosky S. J., Stonik J. A., Furuya Y., Koike H., Suzuki K., Remaley A. T. (2010) Mol. Cancer Res. 8, 1284–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lundholm L., Movérare S., Steffensen K. R., Nilsson M., Otsuki M., Ohlsson C., Gustafsson J. A., Dahlman-Wright K. (2004) J. Mol. Endocrinol. 32, 879–892 [DOI] [PubMed] [Google Scholar]
- 40. Kramer P. R., Wray S. (2002) J. Steroid Biochem. Mol. Biol. 81, 203–216 [DOI] [PubMed] [Google Scholar]
- 41. Steffensen K. R., Holter E., Alikhani N., Eskild W., Gustafsson J. A. (2003) Biochem. Biophys. Res. Commun. 312, 716–724 [DOI] [PubMed] [Google Scholar]
- 42. Schaufele F., Carbonell X., Guerbadot M., Borngraeber S., Chapman M. S., Ma A. A., Miner J. N., Diamond M. I. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9802–9807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Masiello D., Cheng S., Bubley G. J., Lu M. L., Balk S. P. (2002) J. Biol. Chem. 277, 26321–26326 [DOI] [PubMed] [Google Scholar]
- 44. Hodgson M. C., Astapova I., Hollenberg A. N., Balk S. P. (2007) Cancer Res. 67, 8388–8395 [DOI] [PubMed] [Google Scholar]
- 45. Heinlein C. A., Ting H. J., Yeh S., Chang C. (1999) J. Biol. Chem. 274, 16147–16152 [DOI] [PubMed] [Google Scholar]
- 46. Chaturvedi N. K., Kumar S., Negi S., Tyagi R. K. (2010) Mol. Cell Biochem. 345, 291–308 [DOI] [PubMed] [Google Scholar]
- 47. Chuang K. H., Lee Y. F., Lin W. J., Chu C. Y., Altuwaijri S., Wan Y. J., Chang C. (2005) Mol. Endocrinol. 19, 1200–1212 [DOI] [PubMed] [Google Scholar]
- 48. Dozmorov M. G., Hurst R. E., Culkin D. J., Kropp B. P., Frank M. B., Osban J., Penning T. M., Lin H. K. (2009) Prostate 69, 1077–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Singh J., Manickam P., Shmoish M., Natik S., Denyer G., Handelsman D., Gong D. W., Dong Q. (2006) Cancer Lett. 237, 298–304 [DOI] [PubMed] [Google Scholar]
- 50. Schmidt L. J., Regan K. M., Anderson S. K., Sun Z., Ballman K. V., Tindall D. J. (2009) Prostate 69, 1730–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Swinnen J. V., Roskams T., Joniau S., Van Poppel H., Oyen R., Baert L., Heyns W., Verhoeven G. (2002) Int. J. Cancer 98, 19–22 [DOI] [PubMed] [Google Scholar]
- 52. Rossi S., Graner E., Febbo P., Weinstein L., Bhattacharya N., Onody T., Bubley G., Balk S., Loda M. (2003) Mol. Cancer Res. 1, 707–715 [PubMed] [Google Scholar]
- 53. Pizer E. S., Pflug B. R., Bova G. S., Han W. F., Udan M. S., Nelson J. B. (2001) Prostate 47, 102–110 [DOI] [PubMed] [Google Scholar]
- 54. Swinnen J. V., Brusselmans K., Verhoeven G. (2006) Curr. Opin Clin. Nutr. Metab. Care 9, 358–365 [DOI] [PubMed] [Google Scholar]
- 55. Menendez J. A., Lupu R. (2007) Nat. Rev. Cancer 7, 763–777 [DOI] [PubMed] [Google Scholar]
- 56. Fukuchi J., Kokontis J. M., Hiipakka R. A., Chuu C. P., Liao S. (2004) Cancer Res. 64, 7686–7689 [DOI] [PubMed] [Google Scholar]
- 57. Pommier A. J., Alves G., Viennois E., Bernard S., Communal Y., Sion B., Marceau G., Damon C., Mouzat K., Caira F., Baron S., Lobaccaro J. M. (2010) Oncogene 29, 2712–2723 [DOI] [PubMed] [Google Scholar]
- 58. Chuu C. P., Hiipakka R. A., Kokontis J. M., Fukuchi J., Chen R. Y., Liao S. (2006) Cancer Res. 66, 6482–6486 [DOI] [PubMed] [Google Scholar]
- 59. Chuu C. P., Kokontis J. M., Hiipakka R. A., Liao S. (2007) J. Biomed. Sci. 14, 543–553 [DOI] [PubMed] [Google Scholar]
- 60. Lo Sasso G., Celli N., Caboni M., Murzilli S., Salvatore L., Morgano A., Vacca M., Pagliani T., Parini P., Moschetta A. (2010) Hepatology 51, 1334–1344 [DOI] [PubMed] [Google Scholar]
- 61. Lee J. H., Gong H., Khadem S., Lu Y., Gao X., Li S., Zhang J., Xie W. (2008) Endocrinology 149, 3778–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gong H., Guo P., Zhai Y., Zhou J., Uppal H., Jarzynka M. J., Song W. C., Cheng S. Y., Xie W. (2007) Mol. Endocrinol. 21, 1781–1790 [DOI] [PubMed] [Google Scholar]
- 63. Khor V. K., Tong M. H., Qian Y., Song W. C. (2008) Endocrinology 149, 5440–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vedin L. L., Lewandowski S. A., Parini P., Gustafsson J. A., Steffensen K. R. (2009) Carcinogenesis 30, 575–579 [DOI] [PubMed] [Google Scholar]