Abstract
A plethora of peptides are generated intracellularly, and most peptide-human leukocyte antigen (HLA)-I interactions are of a transient, unproductive nature. Without a quality control mechanism, the HLA-I system would be stressed by futile attempts to present peptides not sufficient for the stable peptide-HLA-I complex formation required for long term presentation. Tapasin is thought to be central to this essential quality control, but the underlying mechanisms remain unknown. Here, we report that the N-terminal region of tapasin, Tpn1–87, assisted folding of peptide-HLA-A*02:01 complexes according to the identity of the peptide. The facilitation was also specific for the identity of the HLA-I heavy chain, where it correlated to established tapasin dependence hierarchies. Two large sets of HLA-A*02:01 binding peptides, one extracted from natural HLA-I ligands from the SYFPEITHI database and one consisting of medium to high affinity non-SYFPEITHI ligands, were studied in the context of HLA-A*02:01 binding and stability. We show that the SYFPEITHI peptides induced more stable HLA-A*02:01 molecules than the other ligands, although affinities were similar. Remarkably, Tpn1–87 could functionally discriminate the selected SYFPEITHI peptides from the other peptide binders with high sensitivity and specificity. We suggest that this HLA-I- and peptide-specific function, together with the functions exerted by the more C-terminal parts of tapasin, are major features of tapasin-mediated HLA-I quality control. These findings are important for understanding the biogenesis of HLA-I molecules, the selection of presented T-cell epitopes, and the identification of immunogenic targets in both basic research and vaccine design.
Keywords: Antigen Presentation, Antigen Processing, Chaperone Chaperonin, MHC, Peptides, Tapasin
Introduction
Mature HLA-I3 molecules are located on the surface of all nucleated cells where they present peptides to CD8+ T lymphocytes. Before the peptide-HLA-I complex is transported to the cell surface, maturation and assembly with peptides occur in the ER. During late stage maturation, HLA-I molecules are integrated in the peptide-loading complex (PLC), which at least consists of TAP1/2, tapasin, calreticulin, protein disulfide isomerase, ERp57, and HLA-I (1, 2). Integration of HLA-I into the PLC is mediated by tapasin, which structurally bridges HLA-I and TAP (3, 4). Tapasin is a multi-domain protein, which has been suggested to perform multiple functions. Tapasin has been shown to enhance peptide binding to TAP, facilitate peptide loading onto HLA-I, edit the HLA-I bound peptide repertoire, and retain and recycle suboptimally loaded peptide-HLA-I complexes (5–9). Consequently, in the absence of tapasin, cell surface-expressed HLA-I molecules are less stable and present a partly different peptide repertoire than HLA-I on wild-type cells (10–12). The effect of tapasin depends on the HLA-I allomorph where certain allomorphs, such as HLA-A*02:01, are dependent on tapasin for efficient presentation, whereas others are less influenced (10–12).
Recently, we showed that an N-terminal fragment of tapasin, Tpn1–87, assisted folding of peptide-HLA-A*02:01 complexes in the absence of other PLC proteins (13). Here, we set out to identify the peptide-HLA-I targets for the tapasin quality control hidden in Tpn1–87 and investigate the mechanisms for the Tpn1–87 facilitation of HLA-I. First, we asked whether the Tpn1–87 facilitation would depend on the identity of HLA-I heavy chain (HC) and whether the facilitation would correlate with the known tapasin dependence, i.e. the differentially facilitated cell surface expression of various HLA-I molecules by tapasin (10–12). Second, with focus on HLA-A*02:01 we asked whether and how peptide identity would influence the degree of Tpn1–87 facilitation. Two different large sets of HLA-A*02:01 binding peptides were used: 1) peptides that have been identified as constituents of the natural HLA-I presented peptide repertoire (eluted from HLA-I and present in the SYFPEITHI database of natural HLA-I binding peptides, whereof a major proportion also been identified as T-cell epitopes (14)) and 2) other HLA-I binders, identified in an unrelated biochemical epitope screening effort (15) but not up to present date qualified to be part of the SYFPEITHI database. We also analyzed the influence of specific amino acids on HLA-A*02:01 binding using positional scanning combinatorial peptide libraries (PSCPL), each with only one fixed amino acid at a certain position and the rest of the positions with random amino acids.
The findings in this paper show that the effect of Tpn1–87 varies with the identity of the HLA-I molecule in a manner that correlates perfectly with the described tapasin dependence of different HLA-I molecules (10–12). We also show that Tpn1–87 recognizes and facilitates the folding of HLA-A*02:01-peptide complexes of low intrinsic stability but does not change the peptide binding function in a way that affects the specificity. Finally, the data show that the degree of Tpn1–87 facilitation (i.e. increased HLA-I Bmax Tpn1–87/Ctrl) with high specificity and sensitivity separates HLA-A*02:01 SYFPEITHI ligands from other HLA-A*02:01 peptide binders. Based on these results, we would ascribe Tpn1–87 independency as a striking discriminator of natural ligands to other HLA-A*02:01 binding peptides of similar affinity. This provides a tool with high potential for improved predictions of immunogenicity of peptides. We propose that the chaperone function located in the N-terminal part of tapasin, together with a possible peptide editing function, involving tapasin Cys-95 and with retention/recycling of immature MHC-I molecules mediated by the C-terminal double lysine motif of tapasin, equip the multifunctional tapasin with a unique set of tools to assist in the MHC-I quality control and shape the resulting repertoire of presented CD8+ T-cell epitopes.
EXPERIMENTAL PROCEDURES
Peptide Synthesis
All of the peptides were purchased from Schafer-N (Copenhagen, Denmark) with a purity of 95% or higher. The HLA-I peptide panels were designed to be HLA-I binders. In each panel, some of the peptide sequences were derived from the SYFPEITHI database, which represent immunogenic peptides that bind stably to HLA-I and get presented on the cell surface (14). The non-SYFPEITHI peptide sequences did not exist in the SYFPEITHI database and have not been found to be immunogenic. The peptide sequences used in this work are listed in supplemental Table S1.
Positional Scanning Combinatorial Peptide Libraries
PSCPLs were purchased from Schafer-N (Copenhagen, Denmark). We used a previously reported method capable of determining the HLA-I peptide binding specificity using PSCPLs (17). In brief, peptides are synthesized with completely random amino acids at all positions in the peptide except at a chosen position. This position is then substituted with a fixed amino acid. Consequently, a complete PSCPL 9-mer peptide library would have 20 amino acid substitutions for each position and systematically address all nine positions of a 9-mer peptide resulting in 180 unique sublibraries. Relative binding (RB) values are defined as X9 = EC50/sublibrary EC50, the ratio between the affinity of the completely random library with no fixed amino acid residues and the affinity of the specific sublibrary. Amino acid substitutions leading to RB values below 0.5 are considered disfavored, whereas substitutions leading to RB values above 2.0 are considered favored. All of the experiments were done four times, and Student's t tests were used to determine significant differences between RB values in the absence and presence of Tpn1–87.
Protein Production
The recombinant GrpE-FXa-Tpn1–87 protein, here termed Tpn1–87, was produced as described previously (13). Briefly, a gene encoding the Factor Xa cleavage site, FXa, and the first 87 amino acids of the mature human tapasin (NP_003181) was inserted into a GrpE/pET28a vector. Expression of Tpn1–87 was done in Escherichia coli BL21(DE3) cells as described previously (35). Urea-dissolved Tpn1–87 was purified by anion exchange and size exclusion chromatography (GE Healthcare, Äkta FPLC). HLA-I HCs and human β2-microglobulin (β2m) were produced as described previously (35). HLA-I HC and GrpE-FXa-Tpn1–87 was stored individually in 8 m urea at −20 °C until use.
HLA-I Folding Assay
The peptide-HLA-I folding assays were done as described previously (13, 36). Briefly, 2 nm of biotinylated recombinant HLA-I HC was diluted into a mixture of peptide and 30 nm recombinant human β2m in the absence or presence of 20 nm Tpn1–87 in PBS containing 50 mm Tris and maleic acid, pH 6.6. The reaction mixtures were incubated at 18 °C for 48 h to allow the peptide-HLA-I complex formation to reach steady state. Detection of folded peptide-HLA-I complexes was done by adding 15 μl of the folding reaction to 15 μl of PBS containing 10 μg/ml each of AlphaScreen donor beads (PerkinElmer Life Sciences, 6760002; conjugated with streptavidin) and Acceptor beads (PerkinElmer Life Sciences, 6762001; in-house conjugated with W6/32, a conformation-specific monoclonal anti-HLA-I antibody (37)). The peptide-HLA-I complex formation allows for a proximity-based signal transfer between the donor and acceptor beads. The plates were incubated at 18 °C overnight and then equilibrated for 1 h to reader temperature and read (EnVisionTM; PerkinElmer Life Sciences). The conversion of AlphaScreen signal to peptide-HLA-I complex concentration was done using a preformed peptide-HLA-A*02:01 complex as standard. All of the experiments were done in duplicate on the same day and repeated on separate days, and standard deviations for each folding reaction were calculated and visualized in the graphs. Tpn1–87-mediated folding facilitation was defined as the ratio between the maximum concentrations of folded peptide-HLA-I complexes obtained in the presence or absence of Tpn1–87 (Tpn1–87 Bmax/Ctrl Bmax). Student's t test was used to determine whether the means were significantly different.
HLA-I Stability Assay
The assay has been described elsewhere (38). Briefly, 50 nm biotinylated HLA-A*02:01 HC, 125I-labeled β2m (final specific activity of 250 cpm/μl), and 1 μm of a binding peptide was folded in the presence or absence of 500 nm Tpn1–87 in PBS containing 50 mm Tris and maleic acid, pH 6.6. The folding reactions were incubated in a streptavidin-coated FlashPlate (PerkinElmer Life Sciences, SMP103) at 18 °C for 24 h. Dissociation of the peptide-HLA-I complex was initiated by adding 1 μm unlabeled β2m and incubating the plate at 37 °C. HLA-I-bound 125I-labeled β2m was continuously read at 37 °C (TopCount NXT; PerkinElmer Life Sciences). All of the experiments were done at least two times with double setups. The half-lives were calculated from the dissociation curves using the exponential decay equation in Prism 5 (GraphPad).
Statistics
The AlphaScreen peptide dose-response experiments were done two times on separate days with fresh stock material. Each experiment was done in duplicate using the same stock material. Sigmoidal curve graphs were calculated from all of the data belonging to the same peptide-HLA-I experiment. The curves are calculated based on the mean values, and standard deviations are shown in both vertical directions. Only peptide-HLA-I experiments from which saturated sigmoidal curves could be calculated are included in this work. To determine the underlying mechanism of Tpn1–87 facilitation, the ability of the Bmax, EC50, peptide-HLA-I stability, and Tpn1–87 facilitation parameters to select between SYFPEITHI and non-SYFPEITHI peptides was measured using receiver operating characteristic (ROC) statistics. Here, a ROC curve was constructed from which the ability of the parameters to select with high sensitivity and specificity is demonstrated by the area under the curve (AUC). An excellent parameter demonstrates an AUC close to 1. To determine any significant differences between the AUCs, a jack knife-based ROC analysis was made in which the entire data set was partioned, and different partitions were included in different rounds of the ROC analysis, resulting in different AUCs for the same parameter. The AUCs were averaged, and a Student's t test was applied to determine statistical differences between the parameter AUCs.
RESULTS
Tpn1–87 Facilitates Folding According to Both HLA-I HC and Peptide Identity
Efficient antigen presentation of different HLA-I molecules differentially depends on tapasin, so that some HLA-I molecules are expressed at the cell surface at normal levels in the absence of tapasin, whereas the expression of other HLA-I molecules is dramatically decreased. In addition, not only the quantity but also the quality of these peptide-HLA-I complexes is altered (11). The reason behind these differences in tapasin dependence is not known.
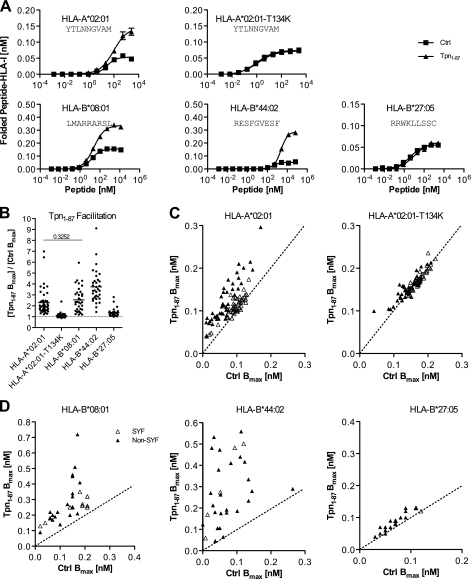
We recently observed that Tpn1–87 facilitates folding of HLA-A*02:01, but not the tapasin-independent HLA-A*02:01-T134K (13), suggesting that Tpn1–87 facilitation may be coupled to the tapasin dependence for various HLA-I molecules. We therefore produced recombinant versions of HLA-B*27:05, HLA-A*02:01, HLA-A*02:01-T134K, HLA-B*08:01, and HLA-B*44:02 and studied their folding in the presence and absence of Tpn1–87. These HLA-I molecules cover a wide spectrum of tapasin dependence as defined by decreased cell surface expression in the absence of tapasin (10, 12). Studies took place under equilibrium conditions, and titrated amounts of peptides were offered to the folding HLA-I complexes in the presence or absence of Tpn1–87. The resulting formation of peptide-HLA-I complexes and the maximum obtainable concentration of folded peptide-HLA-I complexes (Bmax) were initially determined for a few peptide-HLA-I combinations (Fig. 1A and supplemental Fig. S1) and then extended to larger allomorph-specific peptide sets (Fig. 1B). The analysis showed that the folding of the peptide-HLA-I complexes was differentially facilitated by Tpn1–87 according to the identity of the HLA-I HC (Fig. 1B). Underlining the relevance of our in vitro system, the results were in perfect accordance with previous studies of tapasin in cellular models: the facilitation was very pronounced for HLA-B*44:02, intermediate for HLA-B*08:01 and HLA-A*02:01, and very low or absent for the HLA-B*27:05 and HLA-A*02:01-T134K molecules (10, 12).
FIGURE 1.
Tpn1–87 facilitates folding of peptide-HLA-I complexes according to peptide and HLA-I identity. A, folding of peptide-HLA-I complexes in single peptide dose-response experiments. Fixed concentrations of β2m and HLA-I HCs were mixed with titrated concentrations of peptide in the presence (▴) or absence (■) of Tpn1–87. The mixtures were incubated at 18 °C for 48 h, and folded peptide-HLA-I complexes were detected by the HLA-I conformation-specific W6/32 monoclonal antibody in a homogenous assay (36). B, a study of Tpn1–87 facilitation based on the maximum amount of folded peptide-HLA-I complexes, Bmax. Peptide dose-response curves were made by offering each peptide in different concentrations to the folding reaction. The saturation plateaus were calculated as Bmax from the curves. Binding curves were made in the presence (Tpn1–87 Bmax) and absence (Ctrl Bmax) of Tpn1–87 with SYFPEITHI (△) and non-SYFPEITHI (▴) peptides. B, the degree of Tpn1–87 facilitation for each of the tested HLA-I molecules is shown. C, the Bmax values with and without Tpn1–87 for the binding of each of the tested peptides to HLA-A*02:01 and HLA-A*02:01-T134K are plotted. D, the Bmax values for the binding of peptides specific for HLA-B*44:02, -B*08:01, and -B*27:05 with and without Tpn1–87 are plotted. All of the experiments were done in quadruplicate, and standard deviations for each folding reaction were calculated and visualized in the graphs. A Student's t test was applied to determine whether the means were significantly different. All of the means were significantly different (p < 0.001), except for the one with the p value shown in the graph.
These experiments also showed that for the tapasin-dependent allomorphs, there was a large variation of tapasin facilitation that seemed to depend on the different peptides. For a more extensive study of the relation of Tpn1–87 facilitation to peptide identity, we chose to study HLA-A*02:01 in detail and used a panel of 88 HLA-A*02:01-specific peptides. In contrast to many other HLA allele families, HLA-A2 is frequent in all studied ethnic groups, making it a strong candidate for development of peptide-based vaccines. HLA-A*02:01 is both the most prominent member of the HLA-A2 family and one of the most examined HLA-I molecules; hence many of the known HLA-A*02:01-restricted peptides are registered in the SYFPEITHI database of naturally occurring ligands. We have also previously identified a large body of HLA-A*02:01 binding peptides that are not in the SYFPEITHI database (termed non-SYFPEITHI peptides) and have not yet been observed to be presented on the cell surface as T-cell epitopes (15, 16). We examined folding with 44 SYFPEITHI and 44 non-SYFPEITHI peptides. Tpn1–87 facilitated folding of HLA-A*02:01 with SYFPEITHI peptides to a strikingly lesser degree than with non-SYFPEITHI peptides (Fig. 1C). As expected, the folding of the mutant HLA-A*02:01-T134K, which cannot interact with tapasin, was not facilitated with either the studied SYFPEITHI or non-SYFPEITHI peptides. Together, these results suggested that both the identity of the HLA-I HC and the peptide determines the degree of Tpn1–87 facilitation.
To clarify whether the difference between SYFPEITHI and non-SYFPEITHI peptides in terms of Tpn1–87 facilitation could also be observed for other HLA-I molecules, we analyzed the binding of SYFPEITHI and non-SYFPEITHI peptides restricted to HLA-B*08:01, HLA-B*27:05, or HLA-B*44:02 (Fig. 1D). The folding of HLA-B*08:01 resembled folding of HLA-A*02:01 because folding with SYFPEITHI peptides was less facilitated than folding with non-SYFPEITHI peptides. In contrast, the tapasin-independent HLA-B*27:05 resembled HLA-A*02:01-T134K and was not or was only slightly facilitated irrespective of peptide identity. HLA-B*44:02 is by far the most tapasin-dependent allomorph and showed a large Tpn1–87 facilitation of folding with both SYFPEITHI and non-SYFPEITHI peptides.
Tpn1–87 Increases the Binding Affinity of Natural Ligands for HLA-A02:01
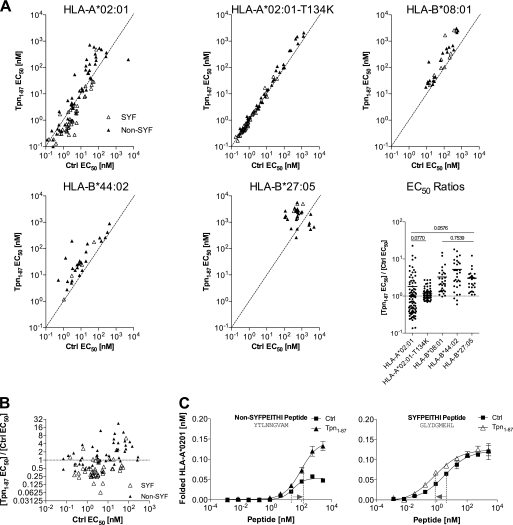
In an attempt to explain the facilitation mechanism, we tested whether Tpn1–87 would alter the peptide binding affinity to HLA-I. Therefore, we calculated the affinities from the folding of multiple peptide-HLA-I combinations in peptide dose-response experiments in the absence or presence of Tpn1–87. For a majority of peptide-HLA-I complexes containing HLA-B*08:01, HLA-B*44:02, and HLA-B*27:05 molecules, Tpn1–87 decreased the affinity (increased EC50) of the peptide-HLA-I interaction (Fig. 2A). Although there seemed to be a small average decrease also in HLA-A*02:01-peptide affinity, the situation was more complex per se, because Tpn1–87 also increased the affinity of a large number of peptide-HLA-A*02:01 complexes (Fig. 2A). A closer analysis showed that the peptides with increased affinity in the presence of Tpn1–87 to a large extent were contributed from the SYFPEITHI group, whereas the peptides with decreased affinities were predominantly non-SYFPEITHI peptides (Fig. 2, B and C). The affinities of the same HLA-A*02:01 binding peptides on the HLA-A*02:01-T134K mutant were not influenced by Tpn1–87 (Fig. 2A), correlating this effect on affinity to tapasin dependence (18).
FIGURE 2.
Tpn1–87 influences the peptide binding affinity to HLA-I. A, Tpn1–87 influences the peptide affinity to HLA-I. Multiple binding curves were made, and the peptide concentration resulting in half-saturation was calculated as EC50. Binding curves were made in the presence (Tpn1–87 EC50) and absence (Ctrl EC50) of Tpn1–87. 44 SYFPEITHI (△) and 44 non-SYFPEITHI (▴) peptides were tested on HLA-A*02:01. The same peptides were tested on HLA-A*02:01-T134K. Other peptide panels were tested on HLA-B*08:01, HLA-B*44:02, and HLA-B*27:05. The EC50 ratios (Tpn1–87 EC50/Ctrl EC50) are shown in the EC50 ratios graph. A Student's t test was used to determine whether the means were significantly different (p < 0.05) between the HLA-I molecules. The p values are shown in cases where no significant differences were found (for a complete list see supplemental Table S2). B, the EC50 ratios on HLA-A*02:01 were plotted against Ctrl EC50 and grouped in non-SYFPEITHI and SYFPEITHI peptide groups. C, peptide dose-response curves representing the non-SYFPEITHI and SYFPEITHI peptide-HLA-A*02:01 complexes were analyzed for shifts in EC50. For the non-SYFPEITHI peptide, the EC50 values increased in the presence of Tpn1–87 corresponding to a decrease in affinity. For the SYFPEITHI peptide, the EC50 values decreased in the presence of Tpn1–87, corresponding to an increase in affinity.
Tpn1–87 Discriminates Affinity-paired Natural and Non-natural Peptides
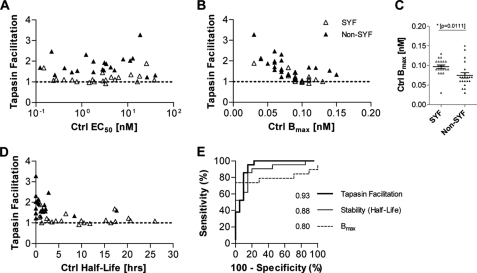
Although Tpn1–87 increased the binding affinity of SYFPEITHI peptides to HLA-A*02:01, the number of the HLA-A*02:01 complexes folded with SYFPEITHI peptides were not at all or only slightly increased by Tpn1–87 (i.e. Tpn1–87 facilitation), whereas those with non-SYFPEITHI peptides in general were much more facilitated (Fig. 1B). We next set out to determine whether the effects of Tpn1–87 on SYFPEITHI versus non-SYFPEITHI peptide-HLA-I complexes reflected differences in peptide-HLA-I binding affinities or not. To this end, we analyzed the Tpn1–87 facilitation using a selection of SYFPEITHI and non-SYFPEITHI peptides, which had been paired according to affinity to HLA-A*02:01, thus eliminating peptide affinity differences between SYFPEITHI and non-SYFPEITHI peptides. Strikingly, the Tpn1–87 facilitation was much higher for non-SYFPEITHI peptides than for SYFPEITHI peptides, even though there was no difference in affinity, and the two peptide groups were clearly defined based on the Tpn1–87 facilitation (Fig. 3A). Hence, the degree of Tpn1–87 facilitation did not depend on the peptide affinity to HLA-A*02:01, at least not for the high affinity binding interactions studied here. These results also suggested that Tpn1–87 is able to distinguish natural ligands from other binders in a manner not dependent on peptide affinity.
FIGURE 3.
Tpn1–87 facilitates folding and discriminates immunogenic peptides independent of peptide affinity to HLA-A*02:01. 21 SYFPEITHI and 21 non-SYFPEITHI peptides were paired, based upon affinity to HLA-A*02:01. Fixed concentrations of β2m and HLA-A*02:01 HC were mixed with various concentrations of peptide in the presence or absence of Tpn1–87. A, the peptide affinities (EC50) to the HLA-I molecules were calculated as the peptide concentration required to reach the half-saturation point on the sigmoidal dose-response curve. The Tpn1–87 facilitation was plotted against EC50. B, the Tpn1–87 facilitation was plotted against the saturation plateau, Bmax. C, the Bmax values for the SYFPEITHI and non-SYFPEITHI peptides in the absence of Tpn1–87 were plotted in a vertical scatter diagram. D, the Tpn1–87 facilitation was plotted against measured stabilities of the peptide-HLA-A*02:01. E, ROC analysis was performed for the ability of each parameter (Bmax, stability, and Tpn1–87 facilitation) to discriminate between SYFPEITHI peptides and non-SYFPEITHI peptides. The AUC values are shown. To determine whether significant differences exist between the areas under the ROC curves, a jack knife analysis was performed on the ROC areas. A Student's t test was used to determine statistically significant differences (p < 0.05) between the parameters tested. All of the AUCs differed significantly in the t test. ***, p < 0.0001.
Tpn1–87 Does Not Facilitate Folding of HLA-I Complexes with SYFPEITHI Peptides
Because peptide-HLA-I affinity could not explain the Tpn1–87-based discrimination of SYFPEITHI from non-SYFPEITHI peptides, some other mechanisms must exist. We observed that each peptide-HLA-I combination is unique regarding the maximum concentration of folded peptide-HLA-I complex obtainable (Ctrl Bmax) (Fig. 1B). Consequently, we used the affinity-paired peptide panel to analyze whether the Tpn1–87 facilitation correlated with Ctrl Bmax. The resulting Tpn1–87 facilitation inversely correlated with the maximum concentration of folded peptide-HLA-I complexes, suggesting that Tpn1–87 in general facilitated the folding of HLA-A*02:01 with peptides otherwise unable to efficiently support folding (low Ctrl Bmax; Fig. 3B) and that SYFPEITHI peptides could be characterized as being able to support HLA-I folding (exhibiting a high Ctrl Bmax) with little or no direct need for Tpn1–87 (Fig. 3C). For the mutant HLA-A*02:01-T134K, which is known to be unable to interact with tapasin, the SYFPEITHI peptides gave a slightly higher Bmax than the non-SYFPEITHI peptides (supplemental Fig. S5A), but folding of T134K was not assisted by Tpn1–87, and there was naturally no correlation between Tpn1–87 facilitation with neither affinity nor Bmax (supplemental Fig. S5, B and C).
Another parameter that might characterize peptide HLA-I complexes involving SYFPEITHI peptides is peptide-HLA-I stability, which has been suggested to be a better indicator of peptide immunogenicity than affinity (19, 20). Thus, we speculated whether Tpn1–87 would affect the stability of peptide-HLA-I complexes and whether the Tpn1–87 facilitation would depend upon the peptide-HLA-I stability. To answer these questions, we first measured the stability of peptide-HLA-A*02:01 complexes folded with the affinity-paired peptides in the presence or absence of Tpn1–87 (supplemental Fig. S3). In general, we observed no differences in the stability of the peptide-HLA-A*02:01 complexes in the absence or presence of Tpn1–87, suggesting that Tpn1–87 does not affect the stability of already folded HLA-A*02:01 complexes, at least not for the high affinity interactions studied here. To discover whether differences in stability still could explain Tpn1–87 dependence, we measured the stability of the affinity-paired peptide-HLA-A*02:01 complexes. The results showed that the stability varied remarkably across the different peptide-HLA-A*02:01 complexes, where complexes folded with SYFPEITHI peptides were more stable than those folded with non-SYFPEITHI peptides (Fig. 3D). For the studied peptides here, Tpn1–87 facilitation inversely correlated with the intrinsic stability of the peptide-HLA-A*02:01 complexes (Fig. 3D). Thus, stable peptide-HLA-A*02:01 complexes (exemplified by those folded with SYFPEITHI peptides) were only slightly or not at all facilitated by Tpn1–87. These results suggest that the intrinsic stability of peptide-HLA-A*02:01 complexes is of importance for the degree of Tpn1–87 facilitation.
Tpn1–87 Facilitation Accurately Identifies Immunogenic Peptides
Improved prediction of the immunogenicity of epitopes is highly desirable for a variety of purposes including selection of peptide-based vaccine candidates. To increase the proportion of correctly identified immunogenic HLA-I presented peptides, not only the HLA-I allomorph specific peptide binding motif must be considered but also other parameters such as the influence of the antigen processing machinery components and CD8+ T-cell receptor features. The studies of SYFPEITHI and non-SYFPEITHI peptides above showed that the maximum concentration of folded peptide-HLA-I complex (Ctrl Bmax), peptide-HLA-A*02:01 stability, and/or Tpn1–87 facilitation could be used to separate the two groups of peptides. The majority of peptides present in the SYFPEITHI database has been demonstrated to activate CD8+ T-cells and is considered immunogenic. To statistically evaluate the accuracy of the three parameters in correctly identifying SYFPEITHI from non-SYFPEITHI peptides, a ROC analysis was performed on the affinity-paired peptides binding to HLA-A*02:01. Using a sliding threshold, the y axis depicts the sensitivity (the ability to find SYFPEITHI peptides), and the x axis depicts 1 − the specificity (equivalent to the risk of including non-SYFPEITHI peptides) (Fig. 3E). A random nondiscrimination would follow the y = x diagonal, whereas useful discriminations would be shifted up and to the left. The AUC is a performance measurement of the discrimination parameter, and the higher the AUC, the better the parameter performs. The ROC analysis of the paired peptides showed that all three parameters performed well, but that Tpn1–87 facilitation was the best parameter to discriminate the SYFPEITHI peptides (Fig. 3E). The intrinsic stability performed second best, again suggesting it to be a vital part of the underlying mechanism of Tpn1–87 facilitation. We also measured affinity, stability, and Tpn1–87 facilitation on the entire peptide panel tested on HLA-A*02:01 (supplemental Fig. S4). Even though the peptides were not affinity-paired and the affinity range included high-to-medium affinity peptides, the ROC analysis showed that Tpn1–87 facilitation and intrinsic stability were the best parameters in discriminating SYFPEITHI from non-SYFPEITHI peptides, whereas Bmax and affinity were inferior.
Tpn1–87 Alters the Peptide Binding Specificity to Only a Minor Extent for HLA-A*02:01
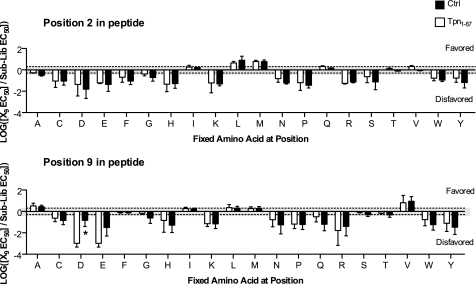
Tpn1–87 has a minor effect on HLA-A*02:01 peptide binding specificity. Only certain peptides bind to any given HLA-I molecule, and these peptides share common amino acid features. The peptide binding specificity of a given HLA-I is determined by favoring or disfavoring of certain amino acids in certain positions of the peptide. An unbiased method has previously been reported capable of determining the peptide binding specificities of HLA-I molecules using PSCPLs (17). To investigate whether Tpn1–87 alters or shows a preference for facilitation based on the peptide binding specificity of HLA-A*02:01, i.e. the peptide-HLA-I complex formation based on occupancy of certain amino acids at specified positions of the peptide, we here set out to use PSCPL in the presence or absence of Tpn1–87. The peptide binding specificity was analyzed for the HLA-A*02:01 anchor positions 2 and 9 (Fig. 4) and showed that the peptide specificity was largely unaltered when Tpn1–87 was present during the peptide-HLA-I folding. Expanding the PSCPL analysis to the nonanchor positions showed that Tpn1–87 only to a minor extent altered the amino acid preferences at these positions (supplemental Fig. S2), suggesting that more complex features of peptides and HLA-I molecules are the major denominators of the degree of tapasin facilitation. Analysis of the SYFPEITHI and non-SYFPEITHI peptides used in our folding assay showed a larger fraction of suboptimal anchors, i.e. in position 2 (p value 0.0004) and the C terminus (p value 0.0103) in the non-SYFPEITHI peptides (supplemental Table S3), suggesting this to be one property that separates the SYFPEITHI- from non-SYFPEITHI-HLA-A*02:01 complexes.
FIGURE 4.
Tpn1–87 alters the peptide binding specificity of HLA-A*02:01 to a minor extent. The peptide binding specificity of HLA-A*02:01 was tested using PSCPLs and shown for substitution positions 2 and 9 in the peptide. Log values of RB values (X9 EC50/sublibrary EC50) are plotted on the y axis, and the amino acid substitutions are shown on the x axis. Amino acid substitutions leading to RB values above 0.3 (log(2)) are considered favored, and values below −0.3 (log(0.5)) are considered unfavored (these boundaries are indicated by the gray shading). Significant differences (p < 0.05) are marked with an asterisk.
DISCUSSION
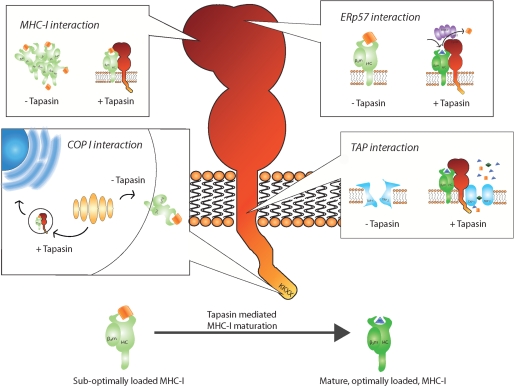
The regulation of MHC-I maturation is complex and controversial. In particular, the interactions of the PLC proteins and the mechanisms and functions of tapasin are debated. Because of the multi-functionality of tapasin, it seems plausible that different domains or regions of the protein contribute with different functions. Indeed, it has been shown that an ER retention/recycling motif and TAP interaction interface are located at the C-terminal part of tapasin (21, 22). The cysteine in position 95 in the ER luminal part of tapasin forms a disulfide conjugate with ERp57, and this conjugation has been proposed to be required for tapasin peptide editing (23). Furthermore, the ER luminal part of tapasin associates with MHC-I but not with TAP (1, 24). We recently demonstrated that the N-terminal region of tapasin, Tpn1–87, which is contained in the ER luminal domain, facilitates folding of HLA-A*02:01 in the absence of other ER proteins (13) (Fig. 5).
FIGURE 5.
Different sites and regions of tapasin work in concert to quality control MHC-I. Top left box, entire regions and single residues in tapasin, from the cytoplasmic tail to the most N-terminal region, have been suggested to be involved in MHC-I binding. Tapasin incorporates MHC-I into the PLC and brings it into close proximity of the TAP transported peptides. Major MHC-I binding sites are located in the ER luminal part of tapasin. In the first 87 amino acids of tapasin, a chaperone function is located that is suggested to keep the MHC-I in a peptide-receptive state and prevent MHC-I aggregation and degradation. Top right box, Cys-95 in tapasin forms a disulfide conjugate with Cys-57 in ERp57, which was suggested to allow tapasin to function as a MHC-I peptide editor. Bottom right box, sites in the cytosolic and transmembrane region of tapasin are important for binding to TAP1 and TAP2. Tapasin both stabilizes TAP and promotes binding of peptides to TAP before the ATP-dependent peptide translocation across the ER membrane. Bottom left box, a double lysine motif is located in the C-terminal of tapasin and mediates interaction with coat protein type I (COP I) vesicles. Coat protein type I vesicles have been proposed to recycle immature/peptide-receptive MHC-I molecules from the Golgi back to the ER. Binding of optimal peptide releases MHC-I from tapasin, allowing efficient antigen presentation on the cell surface.
Here, we demonstrate that the Tpn1–87 assisted folding of peptide-HLA-I complexes varies according to both the HLA-I molecule and the peptide identity. It is noteworthy that the Tpn1–87 facilitation was in agreement with the established tapasin dependence hierarchy of HLA-I molecules in cellular contexts (10, 12). The reason for the HLA-I molecules being differentially affected by tapasin is to date not well understood, but one possibility is that the primary sequence of the HLA-I HC affects the HLA-I stability or directly modulates the three-dimensional conformation of the tapasin interaction site on HLA-I. The tapasin interaction site has been suggested to be located near the HLA-I HC α2-helix, close to the C-terminal part of the peptide when situated in the peptide-binding groove (18). This explanation is supported by the observation that mutation of the threonine to lysine in position 134 (T134K) on an exterior loop near the α2-helix renders HLA-A*02:01 tapasin-independent (18, 25, 26). Consistently, we observed here that the folding of HLA-A*02:01-T134K could not be facilitated by Tpn1–87. All of the wild-type HLA-A*02:01, HLA-B*44:02, HLA-B*08:01, or HLA-B*27:05 molecules have a threonine at position 134, but there are other structural differences between these HLA-I molecules, some of which might directly affect the tapasin binding. Single amino acid mutations in positions 114 and 116 in the HLA-I HC have been reported to influence tapasin dependence (12, 27). We here observed that the peptide identity was also a determinant for the Tpn1–87 facilitation for the HLA-A*02:01, HLA-B*44:02, and HLA-B*08:01 molecules (Fig. 1). Tpn1–87 did not facilitate the folding of the tapasin-independent HLA-B*27:05 and HLA-A*02:01-T134K molecules regardless of peptide offered. Hence, the effect of Tpn1–87 on peptide binding specificity differences could not be studied with these HLA-I molecules. PSCPLs were tested on HLA-A*02:01 to determine whether Tpn1–87 would alter the HLA-A*02:01 amino acid preferences at any of the nine positions of the binding peptide. No major differences were detected, and there were only two significant differences (p = 0.05), showing that at position 7 histidine was more disfavored in the peptides in the absence of Tpn1–87, and at position 9 aspartic acid was more disfavored in the peptides in the presence of Tpn1–87 (Fig. 4 and supplemental Fig. S2). Therefore, the Tpn1–87 facilitation in this in vitro setting cannot be said to allow a greater number of different peptides identities to bind to the HLA-A*02:01 molecule because of an alteration of the HLA-A*02:01 peptide binding specificity. Importantly, the PSCPL analyses are based on average affinities of a large (if not infinite) population of peptides varying at all positions, except in the position analyzed. Analyses of singular peptides with defined primary sequences differ, in that the affinities are not “average” but “exact.” Differences between all of the singular peptides (an infinite high number) in the PSCPL pool (and subpool) could pull the observed average affinity of a sublibrary in different directions, and the average direction may not be changed by the presence of Tpn1–87.
To explain the affinity alterations in peptide-HLA-I binding, we thoroughly examined the binding at different peptide concentrations. The decreased peptide affinities could be explained by the absence of Tpn1–87 facilitation at lower peptide concentrations and Tpn1–87 facilitation at higher peptide concentrations, whereas increased peptide affinities could be explained by an increased Tpn1–87 facilitation at lower peptide concentrations, but not necessarily a simultaneous increase in Bmax (Fig. 2C). For the majority of the peptide-HLA-I complexes tested, Tpn1–87 decreased the affinity (increased EC50 value) of the peptide to the HLA-I molecules, but for HLA-A*02:01, Tpn1–87 increased and decreased the affinity in a peptide-dependent manner separating the SYFPEITHI and non-SYFPEITHI peptides (Fig. 2A). This could indicate that although Tpn1–87 has no major effect on the number of formed complexes, it catalyzes the induction of a locked, stable conformation of HLA-A*02:01 with SYFPEITHI peptides.
Intact, stable peptide-MHC-I complexes at the cell surface are of major importance both to allow for proper signaling in case of infection and to prevent false signaling by uninfected cells whose surface MHC-I molecules could inadvertently pick up peptides in the surrounding if a MHC-I molecule would become reactivated (i.e. peptide-receptive) upon peptide dissociation. HLA-I molecules are known to be very unstable in the absence of HLA-I binding peptides. Here, we suggest that the intrinsic stability of the peptide-HLA-I complex is of importance for determination of the tapasin dependence, because the intrinsic stability of peptide complexes formed with HLA-A*02:01 inversely correlated with the Tpn1–87 facilitation (Fig. 3D and supplemental Fig. S4). Moreover, the significantly larger fraction of suboptimal amino acids in the anchor positions in the non-SYFPEITHI peptides (supplemental Table S3) could indicate an increased need for chaperoning by tapasin, which is also suggested by the lower stability of these peptide-MHC-I complexes (Fig. 3 and supplemental Fig. S4). We suggest that a complex combinatorial effect of amino acids on several positions, including the anchor positions, dictates facilitation by Tpn1–87.
Having analyzed the peptide repertoire presented by MHC-I on the cell surface, a previous study suggested that tapasin controls the peptide repertoire, resulting in the presentation of more stable peptide-MHC-I complexes (28). At first glance, our finding that the Tpn1–87 facilitation was high for unstable peptide-HLA-A*02:01 complexes seems to contradict the suggested role of tapasin as a mediator of cell surface expression of stable peptide-HLA-I complexes. However, findings from several different experimental systems suggest that tapasin selectively associates with peptide-receptive MHC-I molecules: 1) a direct interaction between recombinant tapasin and peptide-empty HLA-A*02:01 was demonstrated, and this interaction was sensitive to and could be disrupted by MHC-I binding peptide (29); 2) when using purified microsomes, the addition of high affinity peptides efficiently released MHC-I from tapasin (30); 3) in the absence of suitable peptides, i.e. in the TAP-deficient T2 cells, MHC-I molecules accumulate bound to tapasin for over 40 min before dissociation (6, 30); 4) another study using recombinant tapasin and HLA-B*08:01 showed that tapasin acts directly on HLA-B*08:01 as a chaperone increasing the number of peptide-receptive MHC-I molecules (31); and 5) finally, tapasin was demonstrated to increase the average affinity of the peptides to MHC-I presented at the cell surface (32) and to increase the stability of peptide-MHC-I complexes (20). Hence, inside the PLC, tapasin is thought to retain and keep MHC-I molecules in a peptide-receptive state until trimming of suboptimal peptides or replacement with optimal peptides allows the release of stable peptide-MHC-I complexes from the PLC (33). We propose that by using both the ER retention and a chaperone function, tapasin would be able to exert key quality control of HLA-I by promoting the presence in the ER of a high number of suboptimally loaded peptide-HLA-I complexes (Fig. 5).
It is debated whether tapasin functions as a chaperone for MHC-I or a peptide editor in the sense of removing nonoptimal peptides during peptide loading of MHC-I. We believe that both functions may co-exist in the sense that tapasin may act as a chaperone keeping MHC-I in a peptide-receptive state. Using a large set of peptides, we observed Tpn1–87 to have chaperone activity and no peptide editing capabilities in terms of direct removal of unstably bound peptide. Rather, this most N-terminal part of tapasin may have an indirect peptide editing function in maintaining a peptide-receptive conformation of empty HLA-I molecules, a state that the HLA-I molecule would have to assume at least briefly during peptide exchange, in line with the model of MHC-I encounter complexes, as recently suggested (34). Our data on the effect of Tpn1–87, however, do certainly not exclude the possibility that wild-type tapasin in its natural environment of the PLC in the ER has a peptide-editing function, although our data would argue that additional requirements to such functionality would reside outside the Tpn1–87 region.
Identifying peptides that are suitable as targets in basic research or clinical applications is a critical question, and it is not always possible to determine what peptides will be presented, when such analysis is based only on the affinities between peptide and the MHC-I molecules or the stability of the resulting complex. We have here introduced a completely novel tool, Tpn1–87, with high potential for prediction of immunogenicity, and we have used large sets of HLA-A*02:01 binding peptides allowing comparisons and analysis of groups with statistically significant numbers of peptides. The relevance of discrimination between SYFPEITHI and non-SYFPETHI peptides is corroborated by the large proportion of the SYFPEITHI peptides that have been demonstrated to activate T-cells (in our here used affinity-paired data set 14:21). However, the proportion of immunogenic peptides from a SYFPEITHI set is supposed to be even higher, because all SYFPEITHI peptides have not been studied in T-cell activation assays and were consequently not identified as immunogenic or not, but all studied SYFPEITHI peptides are eluted from HLA-I molecules expressed and purified from cells. Hence, we here present a both specific and sensitive tool of high relevance for identification of T-cell epitopes. Moreover, the results presented here allow interpretation in more generalized terms than previous studies based on small numbers of peptides. In addition to being able to study large numbers of peptides, our in vitro model has several advantages, i.e. the effect of defined molecules or even specific parts of molecules (i.e. Tpn1–87), and only these, are studied, the assay is run in a format allowing large numbers of peptides and quadruplicate to be run in each experiment, and the assay is standardized and highly reproducible. Crucially, the functionality of the in vitro system developed perfectly reflects in vivo observations of the tapasin dependence of the studied HLA-I molecules. A previous study showed that mild cleavage of tapasin suggested an N-terminal proximal domain within the first 85 amino acids (39). However, with the more recent publication of the tapasin structure published by Dong et al. (40), it is clear that the Tpn1–87 is only the most N-terminal part of the larger Tpn1–270 domain. Our results in this paper demonstrate that some, if not all, HLA-I folding facilitation activity is preserved in the Tpn1–87 part of the domain. Importantly, perfectly in line with our results for Tpn1–87, the E. coli expressed Tpn1–271 fragment not only facilitates peptide-HLA-I complex formation but also shows similar ability for discrimination of SYFPEITHI versus non-SYFPEITHI peptides (data not shown).
For HLA-B*08:01, the facilitation effect of Tpn1–87 was indicated to discriminate SYFPEITHI from non-SYFPEITHI peptides, similarly to HLA-A*02:01 (Fig. 1 and supplemental Fig. S1). However, for proper conclusions and statements to be made, the number of peptides studied for binding to HLA-B*08:01 is too low, and the data sets need to be significantly extended. For HLA-A*02:01-T134K and HLA-B*27:05, Tpn1–87 does not facilitate neither SYFPEITHI nor non-SYFPEITHI complexes, which is in agreement with the tapasin independence of these allomorphs (Fig. 1 and supplemental Fig. S1). Because HLA-B*44:02 is the most tapasin-dependent allomorph studied so far, we included it in this study, but we could not detect any difference in Tpn1–87 facilitation of complexes formed with SYFPEITHI or non-SYFPEITHI peptides (Fig. 1 and supplemental Fig. S1). Unfortunately, in the assays used in this work, HLA-B*44:02 is difficult to work with because it does not fold or generate signal above background level, with the majority of peptides tested by us. We speculate that the reason why we do not see a difference in the facilitation of Tpn1–87 on SYFPEITHI versus non-SYFPEITHI peptide-HLA-B*44:02 complex formation is due to HLA-B*44:02 being intrinsically less stable than many other HLA-I allomorphs, and even SYFPEITHI peptides may not, at least not with the conditions used in our in vitro assays, induce a locked conformation that renders the peptide-HLA-I complex independent of Tpn1–87.
Finally and very importantly, we demonstrate that the N-terminal region of tapasin in the absence of other PLC proteins selectively with high specificity and sensitivity facilitates folding of, but does not dissociate, peptide-HLA-I complexes and thereby discriminates between natural and non-natural HLA-A*02:01 binding peptides (Figs. 1 and 3). This opens up possibilities to further disseminate features of and responses to peptides presented and not presented on the surface of human cells in cancer, autoimmune diseases and during positive and negative selection in the thymus. Our results also present a novel tool relevant for the development of predictors of peptide immunogenicity and of candidates for peptide-based vaccines. Based on these findings, we propose that tapasin prevents suboptimally loaded HLA-I molecules from aggregating, keep them peptide-receptive, and that both peptide and HLA-I HC identity are important parameters for the tapasin quality control.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance of Lotte Nielsen, Tasja Ebersole, Mette Olsen, and Sara Pedersen. We thank Camilla Thuring for help with the illustrations and David Liberg for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant HHSN266200400025C. This work was also supported by the Novo Nordisk Foundation, the Lundbeck Foundation, the Wedell-Wedellsborgs Foundation, the Benzon Foundation, the Copenhagen Cluster of Immunology, Swedish Medical Research Council Grant 2006-6500, the Crafoord Foundation, the Royal Physiographic Society in Lund, and the Thelma Zoegas, Magnus Bergvalls, Groshinskys, and Greta and Johan Kocks Foundations.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S5.
- HLA
- human leukocyte antigen
- MHC
- major histocompatibility complex
- PLC
- peptide loading complex
- TAP
- transporter associated with antigen processing
- HC
- heavy chain
- β2m
- β2-microglobulin
- ER
- endoplasmic reticulum
- PSCPL
- positional scanning combinatorial peptide libraries
- RB
- relative binding
- ROC
- receiver operating curve
- AUC
- area under the curve
- Ctrl
- control.
REFERENCES
- 1. Cresswell P., Bangia N., Dick T., Diedrich G. (1999) Immunol. Rev. 172, 21–28 [DOI] [PubMed] [Google Scholar]
- 2. Park B., Lee S., Kim E., Cho K., Riddell S. R., Cho S., Ahn K. (2006) Cell 127, 369–382 [DOI] [PubMed] [Google Scholar]
- 3. Li S., Sjögren H. O., Hellman U., Pettersson R. F., Wang P. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8708–8713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ortmann B., Copeman J., Lehner P. J., Sadasivan B., Herberg J. A., Grandea A. G., Riddell S. R., Tampé R., Spies T., Trowsdale J., Cresswell P. (1997) Science 277, 1306–1309 [DOI] [PubMed] [Google Scholar]
- 5. Grandea A. G., 3rd, Lehner P. J., Cresswell P., Spies T. (1997) Immunogenetics 46, 477–483 [DOI] [PubMed] [Google Scholar]
- 6. Paulsson K. M., Kleijmeer M. J., Griffith J., Jevon M., Chen S., Anderson P. O., Sjogren H. O., Li S., Wang P. (2002) J. Biol. Chem. 277, 18266–18271 [DOI] [PubMed] [Google Scholar]
- 7. Zarling A. L., Luckey C. J., Marto J. A., White F. M., Brame C. J., Evans A. M., Lehner P. J., Cresswell P., Shabanowitz J., Hunt D. F., Engelhard V. H. (2003) J. Immunol. 171, 5287–5295 [DOI] [PubMed] [Google Scholar]
- 8. Paulsson K. M., Wang P. (2004) FASEB J. 18, 31–38 [DOI] [PubMed] [Google Scholar]
- 9. Paulsson K. M., Jevon M., Wang J. W., Li S., Wang P. (2006) J. Immunol. 176, 7482–7488 [DOI] [PubMed] [Google Scholar]
- 10. Peh C. A., Burrows S. R., Barnden M., Khanna R., Cresswell P., Moss D. J., McCluskey J. (1998) Immunity 8, 531–542 [DOI] [PubMed] [Google Scholar]
- 11. Purcell A. W., Gorman J. J., Garcia-Peydró M., Paradela A., Burrows S. R., Talbo G. H., Laham N., Peh C. A., Reynolds E. C., López De Castro J. A., McCluskey J. (2001) J. Immunol. 166, 1016–1027 [DOI] [PubMed] [Google Scholar]
- 12. Park B., Lee S., Kim E., Ahn K. (2003) J. Immunol. 170, 961–968 [DOI] [PubMed] [Google Scholar]
- 13. Roder G., Geironson L., Darabi A., Harndahl M., Schafer-Nielsen C., Skjødt K., Buus S., Paulsson K. (2009) Eur. J. Immunol. 39, 2682–2694 [DOI] [PubMed] [Google Scholar]
- 14. Rammensee H., Bachmann J., Emmerich N. P., Bachor O. A., Stevanović S. (1999) Immunogenetics 50, 213–219 [DOI] [PubMed] [Google Scholar]
- 15. Vita R., Zarebski L., Greenbaum J. A., Emami H., Hoof I., Salimi N., Damle R., Sette A., Peters B. (2010) Nucleic Acids Res. 38, D854–D862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yewdell J. W., Bennink J. R. (1999) Annu. Rev. Immunol. 17, 51–88 [DOI] [PubMed] [Google Scholar]
- 17. Stryhn A., Pedersen L. O., Romme T., Holm C. B., Holm A., Buus S. (1996) Eur. J. Immunol. 26, 1911–1918 [DOI] [PubMed] [Google Scholar]
- 18. Lewis J. W., Neisig A., Neefjes J., Elliott T. (1996) Curr. Biol. 6, 873–883 [DOI] [PubMed] [Google Scholar]
- 19. van der Burg S. H., Visseren M. J., Brandt R. M., Kast W. M., Melief C. J. (1996) J. Immunol. 156, 3308–3314 [PubMed] [Google Scholar]
- 20. Thirdborough S. M., Roddick J. S., Radcliffe J. N., Howarth M., Stevenson F. K., Elliott T. (2008) Eur. J. Immunol. 38, 364–369 [DOI] [PubMed] [Google Scholar]
- 21. Petersen J. L., Hickman-Miller H. D., McIlhaney M. M., Vargas S. E., Purcell A. W., Hildebrand W. H., Solheim J. C. (2005) J. Immunol. 174, 962–969 [DOI] [PubMed] [Google Scholar]
- 22. Papadopoulos M., Momburg F. (2007) J. Biol. Chem. 282, 9401–9410 [DOI] [PubMed] [Google Scholar]
- 23. Wearsch P. A., Cresswell P. (2007) Nat. Immunol. 8, 873–881 [DOI] [PubMed] [Google Scholar]
- 24. Lehner P. J., Surman M. J., Cresswell P. (1998) Immunity 8, 221–231 [DOI] [PubMed] [Google Scholar]
- 25. Peace-Brewer A. L., Tussey L. G., Matsui M., Li G., Quinn D. G., Frelinger J. A. (1996) Immunity 4, 505–514 [DOI] [PubMed] [Google Scholar]
- 26. Yu Y. Y., Turnquist H. R., Myers N. B., Balendiran G. K., Hansen T. H., Solheim J. C. (1999) J. Immunol. 163, 4427–4433 [PubMed] [Google Scholar]
- 27. Turnquist H. R., Thomas H. J., Prilliman K. R., Lutz C. T., Hildebrand W. H., Solheim J. C. (2000) Eur. J. Immunol. 30, 3021–3028 [DOI] [PubMed] [Google Scholar]
- 28. Howarth M., Williams A., Tolstrup A. B., Elliott T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11737–11742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rizvi S. M., Raghavan M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18220–18225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paulsson K. M., Anderson P. O., Chen S., Sjögren H. O., Ljunggren H. G., Wang P., Li S. (2001) Int. Immunol. 13, 23–29 [DOI] [PubMed] [Google Scholar]
- 31. Chen M., Bouvier M. (2007) EMBO J. 26, 1681–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Williams A. P., Peh C. A., Purcell A. W., McCluskey J., Elliott T. (2002) Immunity 16, 509–520 [DOI] [PubMed] [Google Scholar]
- 33. Purcell A. W., Elliott T. (2008) Curr. Opin. Immunol. 20, 75–81 [DOI] [PubMed] [Google Scholar]
- 34. Praveen P. V., Yaneva R., Kalbacher H., Springer S. (2010) Eur. J. Immunol. 40, 214–224 [DOI] [PubMed] [Google Scholar]
- 35. Ferré H., Ruffet E., Blicher T., Sylvester-Hvid C., Nielsen L. L., Hobley T. J., Thomas O. R., Buus S. (2003) Protein Sci. 12, 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harndahl M., Justesen S., Lamberth K., Røder G., Nielsen M., Buus S. (2009) J. Biomol. Screen 14, 173–180 [DOI] [PubMed] [Google Scholar]
- 37. Parham P., Barnstable C. J., Bodmer W. F. (1979) J. Immunol. 123, 342–349 [PubMed] [Google Scholar]
- 38. Harndahl M., Rasmussen M., Roder G., Buus S. (2010) J. Immunol. Methods, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen M., Stafford W. F., Diedrich G., Khan A., Bouvier M. (2002) Biochemistry 49, 14539–14545 [DOI] [PubMed] [Google Scholar]
- 40. Dong G., Wearsch P. A., Peaper D. R., Cresswell P., Reinisch K. M. (2009) Immunity 1, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.