Abstract
The lipocalin mouse 24p3 has been implicated in diverse physiological processes, including apoptosis, iron trafficking, development and innate immunity. Studies from our laboratory as well as others demonstrated the proapoptotic activity of 24p3 in a variety of cultured models. However, a general role for the lipocalin 24p3 in the hematopoietic system has not been tested in vivo. To study the role of 24p3, we derived 24p3 null mice and back-crossed them onto C57BL/6 and 129/SVE backgrounds. Homozygous 24p3−/− mice developed a progressive accumulation of lymphoid, myeloid, and erythroid cells, which was not due to enhanced hematopoiesis because competitive repopulation and recovery from myelosuppression were the same as for wild type. Instead, apoptotic defects were unique to many mature hematopoietic cell types, including neutrophils, cytokine-dependent mast cells, thymocytes, and erythroid cells. Thymocytes isolated from 24p3 null mice also displayed resistance to apoptosis-induced by dexamethasone. Bim response to various apoptotic stimuli was attenuated in 24p3−/− cells, thus explaining their resistance to the ensuing cell death. The results of these studies, in conjunction with those of previous studies, reveal 24p3 as a regulator of the hematopoietic compartment with important roles in normal physiology and disease progression. Interestingly, these functions are limited to relatively mature blood cell compartments.
Keywords: Apoptosis, Blood, Cell Death, Hematopoiesis, Transgenic, 24p3, Apoptosis, Hematopoietic Cells
Introduction
Lipocalins are a family of secreted proteins that can bind small molecular weight ligands (1, 2) and have diverse molecular recognition properties and functions (reviewed in Ref. 1). The lipocalin mouse 24p3 (also called lipocalin 2 (LCN2)) can mediate either apoptosis or iron uptake in cells expressing the 24p3 cell surface receptor, 24p3R (also called SLC22A17) (3). The ability of 24p3 to induce either apoptosis or iron uptake depends upon its iron status. Iron-loaded 24p3 binds to 24p3R, is internalized, and releases its bound iron, thereby increasing intracellular iron concentration without promoting apoptosis. By contrast, following association with an intracellular siderophore, iron-lacking 24p3 chelates iron and transfers it to the extracellular medium, thereby reducing intracellular iron concentration, which induces expression of the proapoptotic protein Bim, resulting in apoptosis (3). In addition, agents that reduce intracellular iron levels increase Bim expression. Most importantly, intracellular iron delivery blocks induction of Bim expression and apoptosis resulting from the loss of IL-3, the addition of 24p3, or an iron chelator. Therefore, induction of Bim in response to decreased intracellular iron levels is an important aspect of 24p3-induced cell death, and it is required for apoptosis resulting from the addition of 24p3 (3).
We have previously shown that the withdrawal of interleukin-3 (IL-3) induces expression of 24p3 in IL-3-dependent murine pro-B lymphocytic FL5.12 cells and that conditioned medium (CM) from IL-3-deprived FL5.12 cells contains 24p3 and induces apoptosis in naive FL5.12 cells (4). Moreover, 24p3 induces apoptosis in a variety of hematopoietic cells. Based upon these results, we have proposed that IL-3 withdrawal results in 24p3 transcription, leading to synthesis and secretion of 24p3, which induces apoptosis in cells expressing 24p3R through an autocrine/paracrine pathway.
To study the biological functions of 24p3 in a more physiological context, we have generated mutant mice bearing a homozygous deletion in the 24p3 gene. Our data demonstrate a critical role for 24p3 in diverse apoptotic pathways that are relevant to hematopoietic cells. Remarkably, 24p3 was uniquely required for maintenance of survival in committed hematopoietic cells and was not essential for early hematopoiesis.
EXPERIMENTAL PROCEDURES
Generation of 24p3−/− Mice
129/SVE embryonic stem cells were electroporated with a linearized targeting construct in which exons 1–5 of the 24p3 gene were replaced with a phosphoglycerol kinase-neomycin cassette; the construct contained 1.8 kb of 5′-flanking and 3.5 kb of 3′-flanking sequence homology. Following transfection, embryonic stem cells were selected with G418 (Invitrogen) and 1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil for 14 days. Recombinants were identified by PCR and confirmed by Southern blot analysis with BamHI-digested genomic DNA hybridized to an external probe. Positive clones were injected into C57BL/6 blastocysts. Germ line-transmitting male chimeric mice were intercrossed with female C57BL/6 mice to generate heterozygous F1 progeny, which were then intercrossed to derive 24p3−/− mice. The resulting hybrids were successively back-crossed for at least six generations to either 129/SVE or C57BL/6 wild type mice (Jackson Laboratories). Routine genotype analysis of offspring was carried out by multiplex PCR analysis of genomic DNA obtained from tail snips using primers P1 (5′-TGTACTGGCAATTACTTCATGGCTTCC-3′), P2 (5′-CAGGGATCAAGTTCTGAGTTGAGTCC-3′), and P3 (5′-TATGGCTTCTGAGGCGGAAAGAACC-3′). Immunoblot analysis of proteins extracted from concentrated urine samples of 24p3+/+, 24p3+/−, and 24p3−/− mice was performed using an anti-24p3 antibody (Santa Cruz Biotechnology, Inc.).
Hematological Profile Analysis
Bone marrow cells were flushed from femora and tibia of 8–12-week-old mice, and total cellularity was enumerated by hemocytometer. To assess cell composition, bone marrow cells were stained with fluorescently labeled antibodies against Gr-1, Mac-1, B220, or Ter-119 (BD Biosciences or eBioscience) or matching isotype control antibodies (BD Biosciences) in phosphate-buffered saline (PBS) containing 10% bovine serum albumin for 20 min on ice. Cells were then washed twice and fixed in PBS containing 1% formaldehyde (Sigma). Labeled cells were analyzed on a FACSCalibur or FACScan cytometer at the UMMS Flow Cytometry Core Laboratory or Case Flow Cytometry Core Facility and analyzed using CellQuest and FlowJo software.
For peripheral blood analysis, total leukocyte counts were performed using the Unopette system (BD Biosciences), and differential leukocyte counts were determined by staining with Diff-Quick (Dade-Behring). Hematological parameters were determined using a Hemavet 950FS apparatus (Drew Scientific).
Single cell suspensions of spleen were made by mincing the spleen tissue and straining the minced tissue through a 40-μm sieve (BD Biosciences). The spleen total cellularity was enumerated by hemocytometer. All animal experiments were performed in accordance with the Institutional Animal Care and Use Committee guidelines.
In some experiments, mice were treated with 200 mg/kg 5-FU to induce myelosuppression. On day 7 following treatment, blood counts were analyzed by a hematology analyzer, bone marrow total cellularity was counted by a hematocytometer, and the c-kit-positive, lineage-negative, and sca-1-positive (KLS) fraction of cells was determined by flow cytometry.
Analysis of Apoptosis in Primary and Cultured Bone Marrow Cells
Bone marrow cells were isolated as described above and cultured in the presence of a cytokine mixture comprising 20 ng/ml IL-3 and 50 ng/ml each IL-6, IL-7, and stem cell factor (all from Peprotech), and at various time points the total cell counts and cell subset composition were analyzed by flow cytometry as described above. For spontaneous apoptosis assays, cultured Gr-1+ bone marrow cells were stained with annexin V-FITC (BD Biosciences) and analyzed by FACS.
Isolation, Purification, and Analysis of Neutrophils
24p3+/+ and 24p3−/− mice were injected intraperitoneally with 1 ml of 9% casein (Sigma), and 8 h later, mice were sacrificed, and the blood was drawn from the retro-orbital plexus into heparinized tubes. Peritoneal cells were harvested into 5 ml of PBS plus 1% BSA, washed, layered onto a three-step Percoll (Amersham Biosciences) gradient (1.07, 1.06, and 1.05 g/ml), and centrifuged at 400 rpm for 30 min at 4 °C (5). The neutrophil band was aspirated, the total number was determined by hemocytometer, and the neutrophil purity was determined by FACS analysis using a Gr-1 antibody. Cells were suspended at 106 cells/ml in PBS plus 1% BSA. Elicited polymorphonuclear leukocytes (PMNs)4 from mice (n = 5) were pooled for further analysis. Unelicited bone marrow PMNs were obtained by flushing cells from femora of 24p3+/+ and 24p3−/− mice (n = 5) and centrifugation through Percoll as described above. PMNs in peripheral blood were separated by lysing RBC with a lysis buffer (Gentra Systems). The percentage of PMNs in peripheral blood was determined by Gr-1 staining. To determine neutrophil apoptosis, elicited PMNs from 24p3+/+ and 24p3−/− mice were cultured in RPMI (Invitrogen) with or without granulocyte colony-stimulating factor (G-CSF) (25 ng/ml, Peprotech) for the indicated time periods. Apoptosis was determined by annexin V-FITC staining.
Myeloid Progenitor Colony Formation Assays
Bone marrow cells (4 × 105) were isolated as described above and cultured in medium containing 0.9% methylcellulose (Methocult M3234, Stem Cell Technology) in IMDM supplemented with 1% BSA, 100 mm 2-mercaptoethanol, 2 mm l-glutamine, 15% FBS, 10 mg/ml bovine pancreatic insulin, 200 mg/ml holotransferrin (all from Invitrogen), and 10 ng/ml recombinant murine IL-3 or 100 ng/ml G-CSF (Peprotech). Colonies were counted 7–10 days postculture.
Analysis of Apoptosis in IL-3-dependent Primary Bone Marrow Cells
IL-3-responsive primary cells were derived from bone marrow as described previously (6, 7). Briefly, bone marrow cells were aseptically flushed from femora of 8–12-week-old 24p3+/+ or 24p3−/− mice (n = 3) into RPMI medium containing 10% FBS. Red blood cells were lysed with a lysis solution (Gentra Systems), and unlysed cells were washed three times with RPMI plus 10% FBS. Cells were then suspended in RPMI supplemented with 10% FBS, 3 ng/ml IL-3 (Peprotech), 50 mm 2-mercaptoethanol, 25 mm HEPES, 1 mm sodium pyruvate, 4.5 g/liter d-glucose, 0.1 mg/ml streptomycin, 100 units/ml penicillin, and 2 mm glutamine (all from Invitrogen). Two days later, the non-adherent cells were collected and cultured for an additional 14 days in the above medium with periodic changes of medium. The culture was then enriched in IL-3-responsive mast cells as judged by toluidine blue staining (>90% positive) and surface staining with CD34, FcϵRI, and c-Kit antibodies coupled to fluorochromes (BD Biosciences). To determine apoptosis, cells were washed three times with RPMI and 10% FBS (lacking IL-3) and recultured in the wash medium. Cell death was then determined by annexin V-FITC staining (BD Biosciences). Stained cells were analyzed by flow cytometry in CD34-gated or ungated cells. Cells were supplemented with exogenous iron by the addition of 50 μm FeCl3 (Sigma). CM was prepared as described previously (4).
Analysis of Thymocyte Apoptosis
Freshly isolated thymocytes from 6–8-week-old 24p3+/+ or 24p3−/− mice were cultured in RPMI supplemented with 10% heat-inactivated FBS, 10 mm HEPES, 50 mm 2-mercaptoethanol, 0.1 mg/ml streptomycin, 100 units/ml penicillin, and 2 mm glutamine (all from Invitrogen) in the presence or absence of 1 μm dexamethasone (Sigma). The percentage of apoptosis was determined by annexin V-FITC (BD Biosciences) staining.
For in vivo experiments, 8–12-week-old 24p3+/+ or 24p3−/− mice were injected intraperitoneally with 125 mg of dexamethasone dissolved in normal saline containing 20 mg/ml propylene glycol (EMD Sciences) or with normal saline as control. Thymi, spleens, and bone marrow samples were harvested 20 h postinjection, and the total cellularity was enumerated by hemocytometer. Single cell suspensions from bone marrow, spleen, and thymus were surface-stained with CD4 or CD8 antibodies conjugated to FITC or PE (BD Biosciences) and analyzed by flow cytometry.
Bone Marrow Transplantation Experiments
Bone marrow was flushed from femora and tibia of 8–12-week-old male donor mice. Bone marrow cells were enumerated by hemocytometer, and 2 × 106 cells were injected via the tail vein into female recipient mice that had been lethally irradiated (1100 rads in two split doses). Recipient mice were analyzed at time points beginning 4 weeks and up to 12 months after transplantation. The success of transplantation was confirmed by PCR analysis for the presence of a Y chromosome-specific gene, Smcy, using primers SMC4-1 (5′-CTGAAGCCTTTGGCTTTGAGCAAGCTAC-3′) and SMCX-1 (5′-CAAAGAATTTGGCAGCGGTTTCCCT-3′) (8). For competitive repopulation studies, donor engraftment was measured using the CD4.1/CD45.2 ratio as described (9). Differential leukocyte and red blood cell counts were determined using a Hemavet 950FS counter (Drew Scientific).
Antibodies and Immunoblotting
A rabbit polyclonal anti-Bim antibody that recognizes all splice variants was purchased from Calbiochem. Anti-tubulin antibody was from Santa Cruz Biotechnology, Inc. Bim analysis in bone marrow-derived mast cells (BMMCs) and thymocytes was performed as described (3).
RESULTS
Targeted Disruption of the 24p3 Gene in Mice
To assess the proapoptotic role of 24p3 in vivo, we derived homozygous mice bearing a deletion in the 24p3 gene. The targeting vector was designed to replace exons 1–5 of the 24p3 coding sequences with a phosphoglycerate kinase-neomycin cassette (Fig. 1A). The targeting construct was transfected into murine embryonic stem cells, and neomycin-resistant colonies were analyzed by Southern blot hybridization and PCR to identify those that harbored the homologously targeted integrand. Three positive embryonic stem cell clones carrying the mutant 24p3 allele were injected into C57BL/6 blastocysts to generate chimeric mice. Heterozygous 24p3+/− mice originating from one of the clones were intercrossed to produce F2 progeny. Routine genotyping of the mice was carried out by PCR amplification of genomic tail DNA (Fig. 1B).
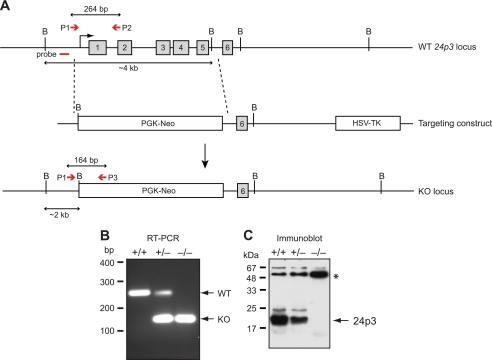
FIGURE 1.
Generation of 24p3−/− null mice. A, targeting strategy. Schematic diagram of the WT 24p3 gene (top), the targeting construct (middle), and the 24p3 KO allele (bottom). After homologous recombination, the targeting vector replaces exons 1–5 (indicated by gray boxes) with the phosphoglycerate kinase-neomycin resistance gene (PGK-neo) cassette. The external 5′-flanking probe is indicated, as are the positions of PCR primers for detecting WT (P1 and P2) and mutant KO (P1 and P3) alleles. The position of the external 5′-flanking probe used for Southern blot analysis is indicated, as are the BamHI sites (denoted by the letter B). B, multiplex PCR analysis for genotype identification of 24p3+/+, 24p3+/−, and 24p3−/− mice by multiplex PCR analysis for WT and mutant alleles. Genomic DNA was prepared from tail snips of 24p3+/+, 24p3+/−, and 24p3−/− mice and amplified with P1, P2, and P3 primer pairs (P1 and P2 or P1 and P3) as described in A (P1 and P2 are for the WT allele, and P1 and P3 are for the mutant allele). A PCR product of 264 bp represents the WT allele, whereas a product of 164 represents the targeted KO allele. C, immunoblot analysis of proteins extracted from monitoring the presence of 24p3 in concentrated urine samples of from 24p3+/+, 24p3+/−, and 24p3−/− mice. Immunoblot analysis was performed using specific antisera directed against 24p3 (Santa Cruz Biotechnology, Inc.). An immunoreactive band migrating at the size expected for 24p3 was detected in urine samples from 24p3+/+ and 24p3+/− mice but not in the urine sample from 24p3−/− mice. An asterisk indicates a contaminating band that serves as a loading control.
To confirm that the targeted knock-out successfully abolished 24p3 expression, we performed immunoblot analysis on concentrated urine samples from wild type (24p3+/+), heterozygous (24p3+/−), and homozygous (24p3−/−) mice using an anti-24p3 antibody. As expected, 24p3 was present in the urine of 24p3+/+ mice, reduced in 24p3+/− mice, and absent in 24p3−/− mice (Fig. 1C).
We derived congenic 24p3−/− mice on various genetic backgrounds by sequentially back-crossing F2 progeny to 129/SVE or C57BL/6 mice (see below). Mice deficient for 24p3 on 129/SVE, C57BL/6, or the mixed genetic background (129/SVE × C57BL/6) were analyzed in this report. In all three strains, 24p3−/− mice were normal in appearance and size, displayed no overt physical abnormalities, developed normally, exhibited normal growth, and were fertile.
24p3−/− Mice Have an Abnormal Hematological Profile
Based on the proapoptotic activity of 24p3 in a variety of hematopoietic cells (4), we hypothesized that 24p3−/− mice would display an accumulation of bone marrow cells. To test this idea, we first determined the total cellularity of the bone marrow of 24p3+/+ and 24p3−/− mice. The results, presented in Table 1, demonstrated that the total number of cells in the bone marrow of 24p3−/− mice was ∼1.6-fold higher than that of 24p3+/+ littermates.
TABLE 1.
Hematological profile of bone marrow, peripheral blood, and spleen from 24p3+/+ and 24p3−/− mice
Numbers represent the mean ± S.D. per mouse of n > 6 mice for bone marrow and spleen analysis and n = 9 mice for peripheral blood analysis. For peripheral blood, values are given per μl of blood. For bone marrow and spleen, the cell subset composition was expressed as the percentage relative to the total cell population. All mice were on the 129SVE genetic background.
| 24p3+/+ | 24p3−/− | p value | |
|---|---|---|---|
| Bone marrow | |||
| Bone marrow (× 107) | 4.05 ± 0.70 | 6.36 ± 1.20 | <0.01 |
| Gr-1+/Mac-1+ cells (%) | 46.10 ± 1.12 | 51.37 ± 1.23 | 0.0018 |
| B220+ cells (%) | 22.80 ± 2.00 | 27.22 ± 1.10 | 0.028 |
| Ter-119+ cells (%) | 17 ± 1.08 | 21 ± 2.48 | 0.017 |
| Peripheral blood | |||
| Leukocytes | |||
| White blood cells (× 103) | 4.75 ± 0.92 | 9.32 ± 0.65 | 0.006 |
| Lymphocytes (× 103) | 3.23 ± 0.64 | 5.12 ± 0.94 | 0.05 |
| Neutrophils (× 103) | 1.19 ± 0.47 | 3.56 ± 0.34 | 0.002 |
| Eosinophils (× 103) | 0.03 ± 0.025 | 0.19 ± 0.14 | 0.079 |
| Monocytes (× 103) | 0.28 ± 0.11 | 0.42 ± 0.1 | 0.176 |
| Basophils (× 103) | 0.007 ± 0.009 | 0.005 ± 0.01 | 0.788 |
| Erythrocytes | |||
| Red blood cells (106/μl) | 7.61 ± 2.64 | 8.94 ± 1.09 | 0.393 |
| Hematocrit (%) | 32.45 ± 1.72 | 39.06 ± 4.66 | 0.343 |
| Mean corpuscular hemoglobin (pg) | 12.17 ± 1.39 | 14.28 ± 0.28 | 0.073 |
| Platelets | 1733 ± 750 | 1092 ± 63.69 | 0.102 |
| Spleen | |||
| Splenocytes (× 107) | 12.00 ± 0.70 | 13.50 ± 3.20 | <0.01 |
| B220+ cells (%) | 46.00 ± 1.00 | 53.00 ± 2.00 | <0.05 |
| Ter-119+ cells (%) | 5.70 ± 1.82 | 16.42 ± 2.32 | 3.26 × 10−5 |
To assess the cell composition of the bone marrow, we performed flow cytometry with various lineage-specific markers, including Gr-1/Mac-1 (a myeloid marker), B220 (a B cell marker), and Ter-119 (an erythroid marker). The results revealed that the increase in bone marrow cell number in 24p3−/− mice was due to an abnormal accumulation of Gr-1+/Mac-1+ myeloid cells, as well as Ter-119+ and B220+ cells (Table 1).
Immature B cells that have passed checkpoints in the bone marrow emigrate to the spleen, where they undergo further transitional stages to become mature B cells. These transitional B cells express the surface marker B220 among others. Consistent with the findings of BM, the proportion of B220+ splenic cells in 24p3−/− mice was ∼1.5-fold higher when compared with control splenic samples (Table 1). The Ter119+ cells in the spleen of 24p3−/− mice was also higher (∼3-fold increase), and these results are consistent with the fact that 24p3 regulates mouse erythropoiesis (Table 1) (10).
Next we compared the hematological profile of peripheral blood in 24p3−/− and 24p3+/+ mice (Table 1). Analysis of total leukocyte counts showed that the number of white blood cells was elevated ∼1.96-fold in 24p3−/− mice. Analysis of differential leukocyte counts revealed that, with the exception of basophils, all leukocyte cell types were increased in 24p3−/− mice. Furthermore, compared with 24p3+/+ mice, the peripheral blood of 24p3−/− mice contained an increased number of mature red blood cells, resulting in an elevated hematocrit. By contrast, the number of platelets in the peripheral blood of 24p3−/− and 24p3+/+ mice was similar.
To determine whether the hematological abnormalities were cell-intrinsic, bone marrow from 24p3+/+ or 24p3−/− male mice was transplanted into either 24p3+/+ or 24p3−/− female mice (Fig. 2A), and the hematological profile of recipient mice was analyzed 12 months after transplantation. PCR analysis using a Y chromosome-specific marker confirmed the success of transplantation (Fig. 2B). We found that 24p3+/+ or 24p3−/− mice transplanted with bone marrow from 24p3−/− mice developed severe leukocytosis with increased numbers of both neutrophils and lymphocytes, whereas mice transplanted with bone marrow from 24p3+/+ mice retained a relatively normal hematological profile (Table 2). Interestingly, we have also noticed a milder monocytosis in mice transplanted with bone marrow from 24p3−/− mice (∼2.5-fold increase in group 3 when compared with group 1). These results are consistent with the previous findings that 24p3 is a regulator of monocyte/macrophage growth (10, 11). Thus, the hematological abnormalities of 24p3−/− mice are cell-intrinsic.
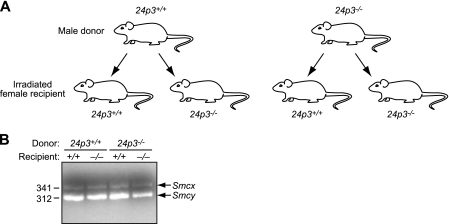
FIGURE 2.
Control experiments confirming the success of bone marrow transplantation. A, schematic representation of the experimental strategy for bone marrow transplantation experiments. B, PCR analysis confirming the success of transplantation. Bone marrow cells isolated from recipient mice were analyzed by PCR for the presence of a Y chromosome-specific gene, Smcy (represented by the 312 bp band). The primers also amplify a homologous gene on the X chromosome, Smcx, due to intronic differences, (represented by the 341-bp band; ref. 8). Both donor and recipient mice were on the 129/SVE genetic background.
TABLE 2.
Hematological profile of 24p3+/+ and 24p3−/− mice following bone marrow transplantation
Numbers represent the mean ± S.D. per mouse, and are given per μl of blood.
| Donor 24p3+/+ |
Donor 24p3−/− |
p value (Group 1 versus Group 3) | |||
|---|---|---|---|---|---|
| Recipient 24p3+/+ (Group 1) (n = 3) | Recipient 24p3−/− (Group 2) (n = 6) | Recipient 24p3+/+ (Group 3) (n = 6) | Recipient 24p3−/− (Group 4) (n = 4) | ||
| White blood cells (× 103) | 3.63 ± 0.35 | 3.70 ± 1.31 | 10.64 ± 0.7 | 11.52 ± 0.67 | 0.05 |
| Lymphocytes (× 103) | 2.46 ± 0.35 | 2.60 ± 0.43 | 4.64 ± 0.26 | 5.45 ± 0.53 | 0.0126 |
| Neutrophils (× 103) | 1.52 ± 0.19 | 0.82 ± 0.65 | 4.94 ± 0.65 | 5.39 ± 0.35 | 0.05 |
| Monocytes (× 103) | 0.29 ± 0.07 | 0.28 ± 0.02 | 0.73 ± 0.07 | 0.83 ± 0.17 | 0.04 |
| Red blood cells (× 106) | 7.78 ± 0.8 | 5.60 ± 0.50 | 7.92 ± 0.16 | 8.75 ± 0.70 | 0.807 |
24p3−/− Mice Develop Neutrophilia in Late Age That Is Not Due to Enhanced Myelopoiesis
Given the potential role of 24p3 in regulating hematopoietic system homeostasis, we aged 24p3−/− mice on the 129/SVE background up to 18 months and compared their hematological profiles with age-matched control 24p3+/+ mice. We did not observe any differences in the mortality rates of 24p3+/+ and 24p3−/− mice during this period (n = 30). After 18 months, there was pronounced leukocytosis in 24p3−/− mice (Table 3). In particular, the number of circulating neutrophils was ∼4.3-fold higher in 24p3−/− mice compared with 24p3+/+ mice. In addition, these mice also displayed a moderate increase in the number of circulating monocytes (∼1.75-fold increase in 24p3−/− mice), thus further confirming the two previous reports that 24p3 suppresses the growth of monocytes/macrophages (10, 11). These results further demonstrate that 24p3 regulates homeostasis of the myeloid compartment.
TABLE 3.
Hematological profile of aged 24p3+/+ and 24p3−/− mice
Mice were aged 18 months. Numbers represent the mean ± S.D. per mouse of n = 6 24p3+/+ mice and n = 8 24p3−/− mice. Values are given per μl of blood. All mice were on the 129SVE genetic background.
| 24p3+/+ | 24p3−/− | p value | |
|---|---|---|---|
| White blood cells (× 103) | 9.18 ± 1.01 | 23.6 ± 0.25 | 0.039 |
| Lymphocytes (× 103) | 5.32 ± 0.51 | 6.15 ± 0.51 | 0.45 |
| Neutrophils (× 103) | 3.8 ± 0.19 | 16.28 ± 1.13 | 0.04 |
| Monocytes (× 103) | 0.42 ± 0.03 | 0.24 ± 0.01 | 0.01 |
| Red blood cells (× 106) | 8.41 ± 0.07 | 5.93 ± 0.58 | 0.09 |
| Hematocrit (%) | 50.75 ± 4.31 | 45.5 ± 4.38 | 0.55 |
| Mean corpuscular hemoglobin (pg) | 16.35 ± 1.00 | 18.8 ± 4.1 | 0.58 |
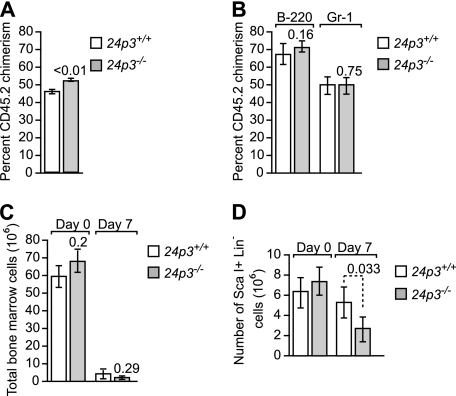
Because defects were observed in the myeloid compartment, we next examined whether the increase in neutrophils was due to increased hematopoiesis along the myeloid lineage. This was measured in two ways. First, bone marrow cells were isolated from 24p3+/+ or 24p3−/− mice (CD45.2) and were mixed at a 1:1 ratio with BoyJ competitor cells (CD45.1), followed by competitive repopulation in lethally irradiated recipient mice. Analysis of the transplanted mice 9 weeks later showed normal reconstitution of the total white blood cells, and multilineage analysis (performed 6 months post-transplantation) did not show any bias toward the myeloid lineage when comparing 24p3−/− with 24p3+/+ groups (Figs. 3, A and B). Similarly, 5-FU treatment of 24p3+/+ or 24p3−/− mice resulted in comparable declines in bone marrow cells (Fig. 3C). Although the impact of 5-FU was mild on the Sca-1+Lin− fraction (Fig. 3D), there was clearly no increased recovery in the 24p3−/− group; rather, these cells were modestly lower (p = 0.033). Therefore, 24p3 does not promote myelopoiesis through an affect on early hematopoietic progenitor cells.
FIGURE 3.
Role of the 24p3/24p3R pathway in stress hematopoiesis. A, bone marrow cells were harvested from both hind limbs of either 24p3−/− or control WT mice (both on C57BL/6 background CD45.2 +) and mixed with competitor Ly-5.1 BoyJ bone marrow cells (CD45.2+) at a ratio of 1:1. The mixed bone marrow cells were injected into lethally irradiated (11 Gy (1100 rads)) Ly-5.1 BoyJ recipient mice at a ratio of 1 donor equivalent to five recipients. The percentage of donor-derived CD45.2 chimerism in the peripheral blood of each recipient mouse was determined by flow cytometry. The donor engraftment was analyzed 9 weeks or later after the transplantation. Results shown are mean ± S.D. (error bars) from two independent transplantations with a total of 10 mice in each group. B, percentage of donor-derived chimerism in Gr-1- and B220-positive populations in the peripheral blood of each recipient mouse. The donor engraftment was analyzed 6 months post-transplantation. Results shown are mean ± S.D. from 24p3+/+ (n = 7) and 24p3−/− (n = 8). C and D, 5-FU was injected intraperitoneally (200 mg/kg), and at 0 days (before injection) and 7 days (after injection), mice were euthanized, and bone marrow from hind limbs (femur and tibia) was collected. The bone marrow counts are shown for total white blood cells (C) and for the Sca-1+Lin− fraction (D).
The 24p3/24p3R Pathway Controls Neutrophil Apoptosis
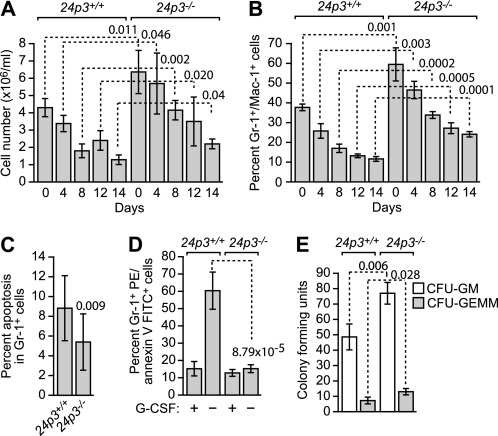
To determine whether the abnormal hematological profiles observed in 24p3−/− mice resulted from an apoptotic defect, we performed a series of in vitro experiments. Bone marrow cells from 24p3−/− and 24p3+/+ mice were cultured in the presence of a cytokine mixture comprising IL-3, IL-6, IL-7, and stem cell factor, and at various time points, the total cell counts and cell subset composition were analyzed by flow cytometry. Consistent with the in vivo results (Table 1), we observed an increase in the total number of cells (Fig. 4A) and the percentage of Gr-1+/Mac-1+ cells (Fig. 4B) in bone marrow cultures derived from 24p3−/− mice.
FIGURE 4.
24p3/24p3R proapoptotic pathway regulates neutrophil apoptosis. A, total number of cells in bone marrow cultures derived from 24p3+/+ and 24p3−/− mice. B, percentage of Gr-1+/Mac-1+ cells in bone marrow cultures derived from 24p3+/+ and 24p3−/− mice. C, apoptosis assays. Cultured Gr-1+ bone marrow cells from 24p3+/+ and 24p3−/− mice were analyzed by annexin V-FITC staining. D, cultured Gr-1+ bone marrow cells from 24p3+/+ and 24p3−/− mice were analyzed for apoptosis by annexin V-FITC staining following the removal of G-CSF. E, colony formation assay in bone marrow cell cultures from 24p3+/+ and 24p3−/− mice. CFU-GM, colony-forming units, granulocyte and macrophage; CFU-GEMM, colony-forming units, granulocyte, erythroid, macrophage, and megakaryocyte. Error bars, S.D.
To determine whether the increased number of Gr-1+ cells was due to resistance of 24p3−/− cells to apoptosis, we first analyzed spontaneous apoptosis of cultured Gr-1+ bone marrow cells by annexin V-FITC staining. The results of Fig. 4C show that Gr-1+ 24p3−/− cells had a reduced rate of spontaneous apoptosis compared with Gr-1+ 24p3+/+ cells.
We also analyzed apoptosis induction following cytokine deprivation in peritoneal neutrophils. Fig. 4D shows, as expected, that removal of G-CSF resulted in a substantial induction of apoptosis in 24p3+/+ Gr-1+ cells. By contrast, 24p3−/− Gr-1+ cells were largely resistant to apoptosis resulting from G-CSF deprivation.
Finally, we measured granulocyte precursor frequencies by colony formation in bone marrow cell cultures supported by cytokines. The results of Fig. 4E indicate that the number of colony-forming units from bone marrow cells from 24p3−/− mice was substantially higher than that from 24p3+/+ mice. In addition, the size of the G-CSF-dependent colony was much larger in 24p3−/− mice than that from 24p3+/+ mice.
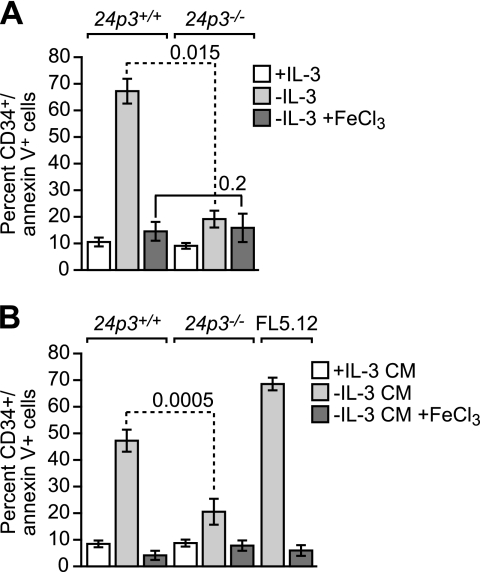
The 24p3/24p3R Pathway Modulates Apoptosis Induction in IL-3-dependent Mast Cells
Prolonged culture of bone marrow cells in IL-3-containing medium results in a uniform population of bone mucosal mast cells, which are commonly referred to as BMMCs (6, 12, 13). When IL-3 is removed from the culture medium, BMMCs undergo apoptosis. Therefore, these cells serve as model to study IL-3 deprivation-induced apoptosis in primary cells (7, 14).
Previous studies have suggested that the 24p3/24p3R pathway has a role in inducing apoptosis in IL-3-dependent cell lines and primary cells following cytokine deprivation (4). Fig. 5A shows that following cytokine deprivation, BMMCs from 24p3+/+ mice underwent efficient apoptosis, as expected. Significantly, in 24p3−/− BMMCs, apoptosis following cytokine deprivation was substantially reduced.
FIGURE 5.
The 24p3/24p3R proapoptotic pathway is critical for apoptosis induction in IL-3-dependent mast cells. A, apoptosis assays. BMMCs were cultured in the presence or absence of IL-3 or in the absence of IL-3 with the addition of FeCl3 and analyzed for apoptosis by annexin V-FITC staining. B, naive BMMCs were treated with CM from BMMCs cultured in the presence or absence of IL-3, or in the absence of IL-3 with the addition of FeCl3. As a control, BMMCs were treated with CM from IL-3-deprived FL5.12 cells. Error bars, S.D.
We have previously shown that 24p3 induces apoptosis through a pathway culminating in decreased intracellular iron levels (3). If 24p3 is responsible for apoptosis induction in cytokine-deprived 24p3+/+ BMMCs, then supplementation with exogenous iron should prevent cell death. In support of this possibility, Fig. 5A shows that, following IL-3 deprivation, the addition of FeCl3 substantially reduced apoptosis in 24p3+/+ BMMCs.
24p3 is a secreted protein, and CM from cytokine-deprived IL-3-dependent cell lines, such as FL5.12 cells, induces apoptosis following the addition to naive cells (4). Fig. 5B shows that the CM from IL-3-deprived 24p3+/+ BMMCs or, as a control, FL5.12 cells efficiently induced apoptosis when added to naive BMMCs. By contrast, the CM from cytokine-deprived 24p3−/− BMMCs was substantially less effective in inducing apoptosis. As expected, apoptosis induction was suppressed by the addition of FeCl3.
The 24p3/24p3R Pathway Mediates Thymocyte Apoptosis and Glucocorticoid-mediated Apoptosis
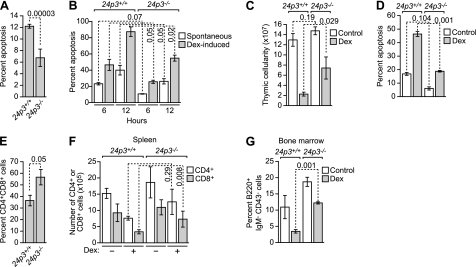
Glucocorticoids (GCs), produced by cortical cells of the thymus, are physiological inducers of thymocyte apoptosis. The 24p3 promoter contains a glucocorticoid-response element, and accordingly, 24p3 is transcriptionally activated by glucocorticoids (15). We have previously shown that 24p3 can induce apoptosis in primary thymocytes and that 24p3 expression correlates with the levels of spontaneous or GC-induced apoptosis (4). These considerations prompted us to analyze the role of 24p3 in thymocyte apoptosis.
To assess the role of 24p3 in spontaneous thymocyte apoptosis, we first analyzed freshly isolated thymocytes from 24p3+/+ or 24p3−/− mice by annexin V-FITC staining. As shown in Fig. 6A, apoptosis in primary thymocytes from 24p3−/− mice was markedly reduced compared with that of 24p3+/+ thymocytes. Thymocytes undergo spontaneous apoptosis when cultured in vitro 16, 17). Fig. 6B shows that the level of apoptosis in cultured 24p3+/+ thymocytes was higher than that in 24p3−/− thymocytes.
FIGURE 6.
The 24p3/24p3R proapoptotic pathway is required for glucocorticoid-mediated apoptosis in thymocytes. A, analysis of spontaneous apoptosis in primary thymocytes from 24p3+/+ and 24p3−/− mice. B, analysis of spontaneous and Dex-induced apoptosis in cultured thymocytes from 24p3+/+ and 24p3−/− mice. C, total thymic cellularity in 24p3+/+ and 24p3−/− mice following systemic administration of Dex or, as a control, saline. D, analysis of apoptosis in thymocytes in 24p3+/+ and 24p3−/− mice following systemic administration of Dex. E, FACS analysis monitoring the percentage of CD4+/CD8+ thymocytes in 24p3+/+ and 24p3−/− mice. F, analysis of splenic lymphocyte number in 24p3+/+ and 24p3−/− mice following systemic administration of Dex. G, analysis of B220+ bone marrow-derived pre-B cells in 24p3+/+ and 24p3−/− mice following systemic administration of Dex. Error bars, S.D.
We next asked whether 24p3+/+ and 24p3−/− thymocytes are also differentially sensitive to GC-induced apoptosis. Fig. 6B shows that treatment with the synthetic GC dexamethasone (Dex) resulted in a level of apoptosis in 24p3+/+ thymocytes that was higher than that in 24p3−/− thymocytes.
To evaluate the contribution of 24p3 to GC-induced apoptosis in vivo, 24p3+/+ or 24p3−/− mice were injected intraperitoneally with Dex. After 20 h, mice were sacrificed, and thymi, spleens, and bone marrow samples were collected and analyzed for cellularity and cell subset compositions. Fig. 6C shows that, following Dex treatment, total thymic cellularity was decreased in 24p3+/+ compared with 24p3−/− mice. As expected, the percentage of apoptotic cells in thymi from Dex-treated 24p3+/+ mice was substantially higher than that from Dex-treated 24p3−/− mice (Fig. 6D). CD4+/CD8+ immature thymocytes are very sensitive to the effects of GCs (18). FACS analysis demonstrated that, following Dex treatment, there was a substantial increase in the number of CD4+/CD8+ thymocytes in 24p3−/− mice compared with 24p3+/+ mice (Fig. 6E). Mature lymphocytes are also sensitive to GCs, albeit to a lesser extent than CD4+/CD8+ thymocytes. Therefore, we also evaluated the effect of Dex on the survival of mature splenic lymphocytes. Fig. 6F shows that the number of splenic lymphocytes (either CD4+ or CD8+) was higher in Dex-treated 24p3−/− mice than in Dex-treated 24p3+/+ mice. Likewise, following Dex treatment, the number of B220+ bone marrow-derived pre-B cells was significantly higher in 24p3−/− mice than their 24p3+/+ counterparts (Fig. 6G). Collectively, these results indicate that 24p3 is an important target gene involved in GC-mediated apoptosis of lymphocytes and other hematopoietic cell types.
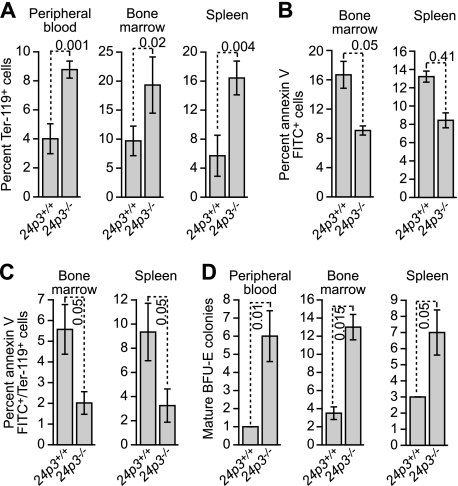
The 24p3/24p3R Pathway Controls Apoptosis Induction in Erythroid Cells
The hematological profiles described above (see Table 1) raised the possibility that 24p3−/− mice would have abnormalities in erythropoiesis. To investigate this issue more carefully, we quantified Ter-119+ cells in 24p3+/+ and 24p3−/− mice. The results of Fig. 7A show that there was a greater number of Ter-119+ cells in peripheral blood, bone marrow, and spleen of 24p3−/− mice compared with 24p3+/+ littermates. Likewise, there was a greater number of annexin V-positive cells (Fig. 7B), and annexin V+/Ter-119+ cells (Fig. 7C) in bone marrow and spleen of 24p3+/+ mice compared with 24p3−/− mice. Finally, we analyzed erythroid precursor frequencies using an in vitro colony formation assay. The results of Fig. 7D indicate that peripheral blood, bone marrow, and spleen from 24p3−/− mice yielded more burst forming unit-erythroid colonies (the earliest known erythroid precursor cells) than the corresponding samples from 24p3+/+ mice.
FIGURE 7.
The 24p3/24p3R proapoptotic pathway regulates apoptosis in erythroid cells. A, analysis of Ter-119+ cells in peripheral blood (left), bone marrow (middle), and spleen (right) from 24p3+/+ and 24p3−/− mice. B, apoptosis assays in bone marrow and spleen from 24p3+/+ and 24p3−/− mice. C, analysis of apoptosis in Ter-119+ cells in bone marrow and spleen from 24p3+/+ and 24p3−/− mice. D, in vitro colony formation assay monitoring the number of BFU-E colonies from peripheral blood, bone marrow, and spleen from 24p3+/+ and 24p3−/− mice. Error bars, S.D.
Influence of Genetic Background on the Role of 24p3
In several previous studies (e.g. see Refs. 19 and 20), 24p3−/− mice were derived and analyzed, but a significant hematologic effect was not reported. In these previous studies, all of the experiments were performed in a mixed genetic background, whereas our studies were performed in back-crossed mice (129/SVE and C57BL/6 genetic backgrounds). To analyze the role of genetic background, we compared apoptosis in cytokine-deprived BMMCs derived from 24p3+/+ and 24p3−/− mice on three genetic backgrounds: 129/SVE (back-crossed at least 10 generations), C57BL/6 (back-crossed 9 generations), and the mixed genetic background (129/SVE × C57BL/6).
As expected, IL-3 deprivation efficiently induced apoptosis in BMMCs derived from 24p3+/+ mice on all three genetic backgrounds (supplemental Fig. 1A). Moreover, on all three genetic backgrounds, the level of apoptosis in 24p3−/− BMMCs was lower than that in 24p3+/+ BMMCs. However, this difference was the greatest on the 129/SVE background and least on the mixed 129/SVE × C57BL/6 background. Collectively, these results indicate that the genetic background of mice can modulate the effects of 24p3 deficiency.
We have previously shown that the 24p3 proapoptotic pathway is regulated through intracellular iron concentration, and an important aspect of this apoptotic mechanism is induction of the proapoptotic Bim protein in response to decreased intracellular iron (3). Bim is required for apoptosis resulting from IL-3 deprivation or the addition of 24p3 (3, 21) (reviewed in Ref. 22). We hypothesize that the resistance to IL-3 deprivation-induced cell death in 24p3−/− BMMCs may be related to variation in Bim induction. To test this prediction, we analyzed Bim levels in BMMCs by immunoblotting. Results presented in supplemental Fig. 1B show that levels of Bim (Bim is spliced alternatively into two predominant isoforms: Bim extra long (Bim-EL) and Bim long (Bim-L)) remain unchanged despite the loss of IL-3 in 24p3−/− BMMCs from mice on 129/SVE or C57BL/6 backgrounds, and in contrast, a modest increase in Bim levels was noticed in cells obtained from mice on the mixed 129/SVE × C57BL/6 genetic background. As expected, Bim expression levels were robustly induced following the loss of IL-3 in BMMCs of 24p3+/+ mice on all three genetic backgrounds (supplemental Fig. 1B).
Bim also plays an important role in GC-induced cell death (23, 24). We next analyzed Bim levels in thymocytes derived from 24p3+/+ and 24p3−/− mice. As expected, GC treatment induced Bim (both isoforms) in thymocytes obtained from 24p3+/+ mice on all genetic backgrounds. Residual Bim-L expression in untreated cells perhaps reflected spontaneous apoptosis (see Refs. 4 and 16). Interestingly, only Bim-EL was moderately induced in GC-treated thymocytes obtained from 24p3−/− mice on 129/SVE or C57BL/6 backgrounds. In contrast, induced Bim levels in thymocytes from 24p3−/− mice on the mixed 129/SVE × C57BL/6 genetic background were comparable with those observed in 24p3+/+ mice (supplemental Fig. 1C). Collectively, these results suggest that 24p3 is required for efficient induction of Bim.
Based on these findings, we propose a model (supplemental Fig. 1D) in which IL-3 loss or GC treatment leads to the synthesis and secretion of 24p3, which then induces cell death in an autocrine/paracrine pathway by lowering intracellular iron levels (3, 4). An important aspect of this process is the induction of the proapoptotic protein, Bim (3). Abrogation of Bim by RNAi suppresses cell death resulting from either the addition of 24p3 or the loss of IL-3 (3, 21). Consistent with these findings, here we show that the induction of Bim following the loss of IL-3 or GC treatment in cells obtained from 24p3−/− mice is severely reduced. Therefore, variations in Bim induction upon IL-3 withdrawal or GC treatment in 24p3−/− cells may explain the resistance to the ensuing apoptosis.
DISCUSSION
Several previous studies have reported that 24p3 can induce apoptosis in cultured cell lines (4, 10). Here for the first time we show that 24p3 is an important regulator of multiple hematopoietic lineages in vivo. The progressive accumulation of lymphocytes, monocytes, and erythrocytes in 24p3−/− mice demonstrates the importance of 24p3 in the regulation of homeostasis of hematopoietic cells in adults. Strikingly, regulation of hematopoietic cell homeostasis was related to the ability of 24p3 to induce apoptosis in mature hematopoietic cells. For example, hematopoietic cells prepared from 24p3−/− mice display increased resistance to multiple death stimuli, including cytokine deprivation and GC treatment. We have previously shown that induction of apoptosis by 24p3 is due, at least in part, to up-regulation of Bim, a proapoptotic BCL-2 family member (3). In this regard, it is notable that, like 24p3−/− mice, bim null mice are also resistant to steroid-induced cell death (23).
The genetic background onto which a targeted allele is placed can result in a significant variation in the phenotype due to incomplete penetrance and variable expressivity (reviewed in Ref. 25). Strain-dependent modifiers are known to influence the phenotype of mice bearing deletions in genes that regulate cell death (reviewed in Ref. 22). For instance, similar to what we have found for 24p3, the effect of Bim, Bcl-2, or caspase-3 deficiency is highly dependent upon the genetic background (reviewed in Refs. 22 and 26).
24p3 is a direct target gene of STAT5-mediated repression (27) and its down-regulation might be expected to confer improved survival. Reciprocally, 24p3 is induced by Foxo3a downstream of the PI3K pathway (28). Therefore, it is regulated by pathways that are critical in early hematopoietic development. In these studies, we determined that regulation of blood cell counts was not due to improved hematopoiesis along multiple lineages. This was tested by both competitive repopulation and 5-FU-induced myelosuppression, both of which were normal in mice lacking 24p3. Therefore, the increase in lymphocytes, monocytes, and erythrocytes was the result of an alternative mechanism that is active in relatively mature committed progenitors and precursor cells.
Studies from our laboratory and others involving cultured cells, mouse models, and patient samples have shown that misregulation of 24p3 expression can contribute to certain hematopoietic diseases, in particular chronic myelogenous leukemia (3, 29, 30). However, those studies did not examine the stage of hematopoiesis in which 24p3 was active. The results presented here, in conjunction with those of previous studies, reveal 24p3 as a regulator of hematopoietic cell survival and suggest that the impact of survival via this pathway may be unique to the differentiated progeny. The implications of this finding for diseases such as CML will require further study.
Supplementary Material
Acknowledgments
We thank the University of Massachusetts Medical School Transgenic Animal Modeling Core, the University of Massachusetts Flow Cytometry Core, and the Case Comprehensive Cancer Center Flow Cytometry Core; Jay Myers, Alex Huang, Alieta Ciocea, Victor Boyartchuk, and Mike Sramkowski for suggestions and help with animal experimentation; and Sara Evans for editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DK059380 (to K. D. B.), K01CA113838 and R01DK081395 (to L. R. D.), and R01CA115817 (to M. R. G.). This work was also supported by Case Western Reserve University start up funds (to L. R. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- PMN
- polymorphonuclear leukocyte
- G-CSF
- granulocyte colony-stimulating factor
- BMMC
- bone marrow-derived mast cell
- GC
- glucocorticoid
- Dex
- dexamethasone
- 5-FU
- 5-fluorouracil.
REFERENCES
- 1. Flower D. R. (1996) Biochem. J. 318, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akerstrom B., Flower D. R., Salier J. P. (2000) Biochim. Biophys. Acta 1482, 1–8 [DOI] [PubMed] [Google Scholar]
- 3. Devireddy L. R., Gazin C., Zhu X., Green M. R. (2005) Cell 123, 1293–1305 [DOI] [PubMed] [Google Scholar]
- 4. Devireddy L. R., Teodoro J. G., Richard F. A., Green M. R. (2001) Science 293, 829–834 [DOI] [PubMed] [Google Scholar]
- 5. Dong X., Mo Z., Bokoch G., Guo C., Li Z., Wu D. (2005) Curr. Biol. 15, 1874–1879 [DOI] [PubMed] [Google Scholar]
- 6. Rodriguez-Tarduchy G., Collins M., López-Rivas A. (1990) EMBO J. 9, 2997–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mekori Y. A., Oh C. K., Metcalfe D. D. (1993) J. Immunol. 151, 3775–3784 [PubMed] [Google Scholar]
- 8. Agulnik A. I., Longepied G., Ty M. T., Bishop C. E., Mitchell M. (1999) Mamm. Genome 10, 926–929 [DOI] [PubMed] [Google Scholar]
- 9. Bunting K. D., Bradley H. L., Hawley T. S., Moriggl R., Sorrentino B. P., Ihle J. N. (2002) Blood 99, 479–487 [DOI] [PubMed] [Google Scholar]
- 10. Miharada K., Hiroyama T., Sudo K., Danjo I., Nagasawa T., Nakamura Y. (2008) J. Cell. Physiol. 215, 526–537 [DOI] [PubMed] [Google Scholar]
- 11. Lee S., Lee J., Kim S., Park J. Y., Lee W. H., Mori K., Kim S. H., Kim I. K., Suk K. (2007) J. Immunol. 179, 3231–3241 [DOI] [PubMed] [Google Scholar]
- 12. Tsuji K., Zsebo K. M., Ogawa M. (1991) J. Cell. Physiol. 148, 362–369 [DOI] [PubMed] [Google Scholar]
- 13. Rodriguez-Tarduchy G., Collins M. K., García I., López-Rivas A. (1992) J. Immunol. 149, 535–540 [PubMed] [Google Scholar]
- 14. Metcalfe D. D., Mekori J. A., Rottem M. (1995) Exp. Dermatol. 4, 227–230 [DOI] [PubMed] [Google Scholar]
- 15. Garay-Rojas E., Harper M., Hraba-Renevey S., Kress M. (1996) Gene 170, 173–180 [DOI] [PubMed] [Google Scholar]
- 16. Tiso M., Gangemi R., Bargellesi Severi A., Pizzolitto S., Fabbi M., Risso A. (1995) Am. J. Pathol. 147, 434–444 [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang J., Mikecz K., Finnegan A., Glant T. T. (2000) J. Immunol. 165, 2970–2974 [DOI] [PubMed] [Google Scholar]
- 18. Sentman C. L., Shutter J. R., Hockenbery D., Kanagawa O., Korsmeyer S. J. (1991) Cell 67, 879–888 [DOI] [PubMed] [Google Scholar]
- 19. Flo T. H., Smith K. D., Sato S., Rodriguez D. J., Holmes M. A., Strong R. K., Akira S., Aderem A. (2004) Nature 432, 917–921 [DOI] [PubMed] [Google Scholar]
- 20. Berger T., Togawa A., Duncan G. S., Elia A. J., You-Ten A., Wakeham A., Fong H. E., Cheung C. C., Mak T. W. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1834–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harada H., Quearry B., Ruiz-Vela A., Korsmeyer S. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15313–15317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 23. Bouillet P., Purton J. F., Godfrey D. I., Zhang L. C., Coultas L., Puthalakath H., Pellegrini M., Cory S., Adams J. M., Strasser A. (2002) Nature 415, 922–926 [DOI] [PubMed] [Google Scholar]
- 24. Molitoris J. K., McColl K. S., Distelhorst C. W. (2011) Mol. Endocrinol. 25, 409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rivera J., Tessarollo L. (2008) Immunity 28, 1–4 [DOI] [PubMed] [Google Scholar]
- 26. Leonard J. R., Klocke B. J., D'Sa C., Flavell R. A., Roth K. A. (2002) J. Neuropathol. Exp. Neurol. 61, 673–677 [DOI] [PubMed] [Google Scholar]
- 27. Sheng Z., Wang S. Z., Green M. R. (2009) EMBO J. 28, 866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park S., Guo J., Kim D., Cheng J. Q. (2009) J. Biol. Chem. 284, 2187–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin H., Monaco G., Sun T., Ling X., Stephens C., Xie S., Belmont J., Arlinghaus R. (2005) Oncogene 24, 3246–3256 [DOI] [PubMed] [Google Scholar]
- 30. Leng X., Lin H., Ding T., Wang Y., Wu Y., Klumpp S., Sun T., Zhou Y., Monaco P., Belmont J., Aderem A., Akira S., Strong R., Arlinghaus R. (2008) Oncogene 27, 6110–6119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.