Abstract
Increased levels of ADAM12 have been reported in a variety of human cancers. We have previously reported that urinary ADAM12 is predictive of disease status in breast cancer patients and that ADAM12 protein levels in urine increase with progression of disease. On the basis of these findings, the goal of this study was to elucidate the contribution of ADAM12 in breast tumor growth and progression. Overexpression of both the ADAM12-L (transmembrane) and ADAM12-S (secreted) isoforms in human breast tumor cells resulted in a significantly higher rate of tumor take and increased tumor size. Cells expressing the enzymatically inactive form of the secreted isoform, ADAM12-S, had tumor take rates and tumor volumes similar to those of wild-type cells, suggesting that the tumor-promoting activity of ADAM12-S was a function of its proteolytic activity. Of the two isoforms, only the secreted isoform, ADAM12-S, enhanced the ability of tumor cells to migrate and invade in vitro and resulted in a higher incidence of local and distant metastasis in vivo. This stimulatory effect of ADAM12-S on migration and invasion was dependent on its catalytic activity. Expression of both ADAM12 isoforms was found to be significantly elevated in human malignant breast tissue. Taken together, our results suggest that ADAM12 overexpression results in increased tumor take, tumor size, and metastasis in vivo. These findings suggest that ADAM12 may represent a potential therapeutic target in breast cancer.
Keywords: ADAM ADAMTS, Breast Cancer, Enzyme Catalysis, Tumor Metastases, Tumor Promoter, ADAM12, Cell Migration and Invasion
Introduction
Human ADAM12 (meltrin α, MCMP) is expressed as two alternatively spliced forms, a membrane-anchored long form (ADAM12-L) and a short secreted form (ADAM12-S) (1). ADAM12-L and -S share a high overall sequence homology, differing only in the transmembrane domain (that ADAM12-S lacks) and a C terminus that is distinct in each isoform. Discrete functions can be attributed to the individual ADAM12 domains. The catalytic domain of ADAM12 contains the consensus HEXGHXXGXXHD zinc-binding motif, and both isoforms are active proteases. ADAM12-L sheds several membrane-bound ligands, including heparin-binding EGF-like growth factor binding protein (HB-EGF) (2), EGF (3), betacellulin (3), Notch ligand delta-like 1 (4), and placental leucine aminopeptidase (5). ADAM12-S can cleave insulin-like growth factor binding protein (IGFBP)-3 and IGFBP-5 (6, 7) and degrade extracellular matrix substrates (8). ADAM12 mRNA and protein are highly expressed in a variety of malignant tumor tissues and tumor cell lines including breast, brain, bladder, gastric, colon, lung, laryngeal, and hepatocellular carcinomas (for a review, see Refs. 9–16). The role of the two isoforms of ADAM12 or mechanistic studies describing their functional role in cancer have been rare to date. In this report, we have utilized an in vivo orthotopic tumor model that reliably recapitulates human breast tumor growth to investigate the role of the two distinct isoforms of ADAM12 in the development of invasive breast cancer.
ADAM12 can be detected in the urine of breast (8) and bladder (11) cancer patients, and its levels have been shown to correlate with disease status, stage, and cancer risk (8, 17). The discovery of ADAM12 as a potential biomarker for breast cancer begged the question of its relevance in human breast tumorigenesis. We show that overexpression of both ADAM12 isoforms in breast tumor cells promotes tumor growth and that only ADAM12-S expression stimulates their migration and invasion in vitro and local and distant invasion in vivo as a function of its proteolytic activity. We further show that both ADAM12-L and ADAM12-S expression levels are significantly higher in human malignant breast tissue and metastatic lymph nodes compared with normal breast tissue.
EXPERIMENTAL PROCEDURES
Reagents, Antibodies and Cell Lines
ADAM12 antibody rb122 was a gift from Dr. Ulla Wewer (Copenhagen, Denmark). Other antibodies used in the study include ERα2 from Santa Cruz Biotechnology (Santa Cruz, CA), cytokeratin (Abcam, Cambridge, MA), GAPDH (Millipore, Temecula, CA), ADAM12 (Proteintech Group, Chicago, IL). HRP-conjugated anti-rabbit and anti-mouse antibodies were from Vector Biolabs (Burlingame, CA). MCF-7, MDA-MB-231, Hs578T, and T47-D cells were obtained from the ATCC and cultured according to ATCC protocols.
Transfection of Breast Cancer Cells with Human ADAM12-L and ADAM12-S and siRNA Studies
MCF-7 cells were stably transfected with the pcDNA3 plasmid encoding human full-length ADAM12-L, ADAM12-S, and ADAM12-Scatmut (with an E351Q point mutation) using an Amaxa nucleofector kit (Lonza, Walkersville, MD). Stable clones were selected based on neomycin-resistant growth (G418, 0.5 mg/ml; Invitrogen). For siRNA studies, the ADAM12 siRNA constructs and DharmaFECT1 transfection reagents (Thermo Scientific, Lafayette, CO) were used according to the manufacturer's instructions. Stable clone pools of ADAM12-expressing T47-D cells have been described previously by us (18).
Immunoblotting
Cell lysates were prepared using 1× lysis buffer (Cell Signaling Technology, Danvers, MA). For analysis of ADAM12-S, serum-free conditioned medium was concentrated using a 10-kDa cutoff filter (YM-3 Microcon, Millipore). Protein concentration of the lysates and conditioned medium was determined using the Bradford method (Bio-Rad). Immunoblotting was conducted as described before (8).
Semi-quantitative RT-PCR and Real-time RT-PCR Analysis
Total RNA was extracted from cells using the RNAeasy kit according to the manufacturer's protocol (Qiagen, Germantown, MD). cDNA was prepared by reverse transcription from 1 μg of total RNA using the Superscript III reverse transcriptase kit (Invitrogen). Forward and reverse primers are indicated in supplemental Table 1. Real time RT-PCR was performed using iQtm SYBR® Supermix (Bio-Rad). GAPDH expression was used for normalization. For ADAM12-L and ADAM12-S gene expression in normal breast and tumor tissue, TissueScantm tissue quantitative PCR array (Origene, Rockville, MD) was used according to the manufacturer's protocol. The panel consisted of cDNA derived from normal (n = 5), Stage I (n = 11), Stage II (n = 14), Stage III (n = 14), and Stage IV (n = 4) breast cancer tissue, respectively. β-Actin expression was used to normalize relative ADAM12-L and ADAM12-S expression in tissue samples.
Orthotopic Breast Tumor Xenografts in Nude Mice
WT MCF-7- and ADAM12-expressing stable clones were cultured as described until confluent. 4 × 106 cells of each type were suspended in 40 μl of cold Hanks' balanced salt solution and injected into the exposed fourth right inguinal mammary fat pad of 8- to 10-week old female BALB/c nude mice (Charles River Laboratories, Wilmington, MA) as described previously (19, 20). Slow-release 17-β-estradiol pellets (Innovative Research of America, Sarasota, FL) were implanted at the time of injection. Tumor volume was calculated based on the formula (length × width × width)/2. Animals were sacrificed when tumor sizes reached ∼1 cm in diameter or when mice were moribund. Primary tumor, axial and sentinel lymph nodes, and lung and liver tissues were collected for preparation of paraffin-embedded and fresh frozen sections for analysis. Lungs were harvested from all tumor-bearing animals, macerated, and cultured under sterile conditions to detect the presence of epithelial tumor cells of human origin.
Immunohistochemistry
Immunohistochemistry was conducted using paraffin-embedded tumor xenografts or human breast cancer tissue arrays (US Biomax, Rockville MD) as described before (19). The staining intensity of the tumor sections was evaluated by a pathologist and graded over a range, with 0 being negative and 3+ the most intense staining. ADAM12, ERα, and human cytokeratin were detected in lung tumor cell colonies using immunofluorescence. Cells were rinsed in PBS, fixed in 4% paraformaldehyde, and blocked with 5% sheep serum. After incubation with primary antibody, Alexa Fluor-labeled secondary anti-mouse or anti-rabbit was used to detect expression.
Migration and Invasion Assay
In vitro migration studies were conducted using a modified Boyden chamber assay as described previously (19). Briefly, cells (104 cells/well) were plated in the upper chamber (Costar transwell assay, Corning, Inc., Corning NY). 600 μl of 10% FBS containing medium was placed in the lower chamber. After 24 h, cells were fixed and stained with a Diff-Quik stain set (Siemens, Inc., Deerfield, IL). Cells adhering to the top of the filter were gently wiped away and those adhering to the lower surface of the filter were counted using a microscope at 400× magnification (24 fields). In invasion assays, cells migrated through a filter precoated with Matrigel (BD Falcontm HTS FluoroBloktm, BD Biosciences). After fixation, cells were fluorescently labeled (CellTrackerGreentm, Invitrogen) and counted as above.
Statistical Analysis
Results for migration, invasion, and proliferation assays are reported as mean ± S.D. of at least three independent experiments. Tumor growth studies were conducted twice. Differences between experimental groups were statistically analyzed using the Mann-Whitney test, and p ≤ 0.05 were considered to be statistically significant.
RESULTS
ADAM12 Expression Is Elevated in Human Malignant Breast Tissue and Metastatic Lymph Nodes
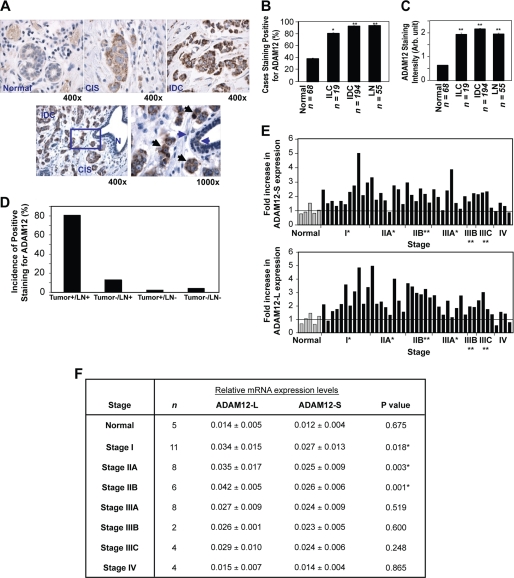
To assess the distinct roles of ADAM12 isoforms in breast tumor growth and progression, we conducted a comprehensive analysis of arrays of malignant human breast tissue and adjacent normal tissue for ADAM12 protein and mRNA expression by immunohistochemistry (IHC) and real-time RT-PCR, respectively. Although ADAM12 protein expression was absent or very low in normal tissue, higher levels were detected in carcinoma in situ (CIS) and invasive carcinoma tissues (IDC) (Fig. 1A, upper panel). The differential ADAM12 expression pattern could be observed in close proximity within the same tissue section. For example, normal breast duct displayed little or no ADAM12 staining; however, clusters of carcinoma in situ and invasive carcinoma tissue stained moderately and highly for this protein, respectively (Fig. 1A, lower panel). Positive staining for ADAM12 was detected with a higher frequency in locally invasive breast carcinoma (invasive lobular carcinoma, ILC, 80%), invasive carcinoma tissues (92%), and metastatic lymph nodes (LN) (95%, p < 0.0003) (Fig. 1B) as compared with normal breast epithelium (35%). Interestingly, staining in normal tissue was largely negative or at extremely low levels (average intensity, ∼ 0.60), whereas carcinoma samples (invasive lobular carcinoma and invasive carcinoma tissues; avg. intensity, ∼1.9 and ∼2.1, p < 0.0001) as well as metastatic LN (average intensity, ∼1.9, p < 0.0001) had significantly higher ADAM12 staining (Fig. 1C). Of the matched sets of primary tumor tissue and metastatic LN (n = 53) analyzed in this study, ∼80% had a positive expression of ADAM12 in both the primary tumor and matched LN, ∼13% were negative for ADAM12 in the primary tumor but had positive staining in the LN, and only ∼1% of the primary tumors were positive for ADAM12 expression, whereas their matched LN were negative (Fig. 1D), suggesting that ADAM12 expression may be essential for primary tumor growth as well as promoting and/or maintaining distant metastasis.
FIGURE 1.
ADAM12 expression is elevated in malignant human breast tissue. IHC analyses for ADAM12 in a human tissue array. Low or no ADAM12 was detected in normal breast tissue, whereas significantly higher ADAM12 protein was detected in carcinoma in situ (CIS) and invasive carcinoma (IDC) tissues (A). B, the frequency of positive ADAM12 expression was significantly higher in malignant tissue as compared with normal breast tissue. C, average ADAM12 staining intensity (range 0–3, where 0 = no staining and 3 = maximum staining intensity) was also significantly higher in malignant tissue as compared with normal breast tissue. These results are expressed as the mean ± S.D. Student's t test was performed. *, p = 0.001, **, p ≤ 0.0001. D, a majority (80%) of the matched sets of primary tumor and metastatic LN analyzed had positive expression of ADAM12 for both tissues. E and F, transcript levels of both ADAM12 isoforms are significantly higher in malignant breast tumor tissue. Relative ADAM12-L and ADAM12-S transcript levels were normalized to β-actin and calibrated to the mean mRNA level (arbitrary value of 1) in normal tissue (gray bars). The fold increase in gene expression relative to the mean value for each disease sample (ADAM12-S, *, p < 0.05; ADAM12-L, **, p < 0.005) is indicated (E). ADAM12-L is preferentially expressed in Stage I and II breast cancer, whereas both forms are expressed in the later stages of disease (F).
Currently available ADAM12 antibodies do not distinguish between the two isoforms. Therefore, to determine which isoforms of ADAM12 are preferentially expressed in human malignant breast tissues, we measured the expression of each isoform in a cDNA array of normal and disease tissue (TissueScan qPCR array, Origene). The panel consists of cDNA derived from normal (n = 5), Stage I (n = 11), Stage II (n = 14), Stage III (n = 14), and Stage IV (n = 4) breast cancer tissue. Relative transcript levels of both the ADAM12-L (p = 0.007) and ADAM12-S (p = 0.008) isoforms were significantly higher (≥2-fold) for Stages I-III of breast cancer as compared with normal breast epithelium (Fig. 1E). Although levels of ADAM12 in Stage IV breast cancer samples did not appear significantly different from those for normal breast tissue, only four Stage IV cDNA samples were available for analysis. In fact, in our IHC analysis of breast cancer tissue microarrays we found that the ADAM12 staining of Stage IV metastatic breast tumors (n = 46; average staining intensity, ∼1.68) was significantly higher than that of normal breast epithelium (n = 68; average staining intensity, ∼ 0.63; p ≤ 0.0001). Comparison of individual isoform expression in breast cancer indicated that ADAM12-L mRNA levels appeared to be slightly but significantly higher in the early stage I, IIA, and IIB tumor tissues (Fig. 1F) as compared with ADAM12-S mRNA.
ADAM12 Expression Is Up-regulated in Aggressive and Metastatic Breast Cancer Cells
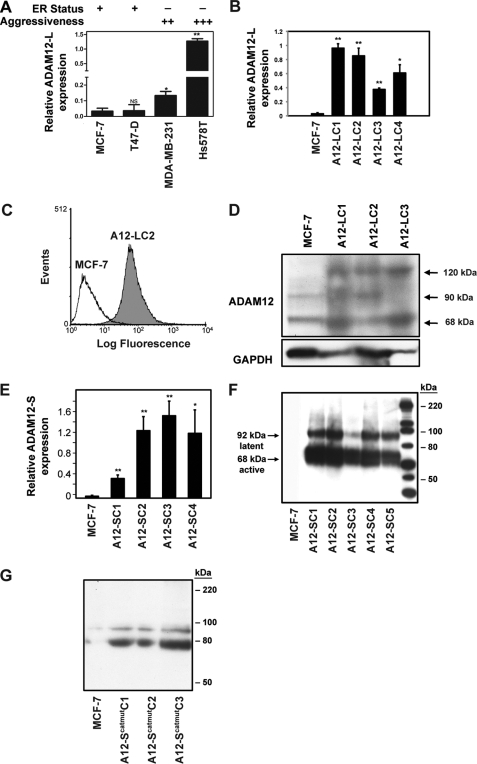
We analyzed ADAM12 levels in a panel of human breast cancer cells that differ in their aggressiveness, estrogen receptor (ER) status, and metastatic ability. Transcript levels of ADAM12 were lower in less aggressive, ER-positive cells such as MCF-7 and T47-D in contrast to highly aggressive, metastatic, and ER-negative cells such as MDA-MB-231 and Hs578T (Fig. 2A). These data raise the possibility that ADAM12 contributes to the more aggressive, metastatic breast cancer phenotype. To investigate the hypothesis that ADAM12 isoforms play a role in breast tumor growth and progression, we selected two model cell lines that express low levels of this metalloprotease, MCF-7 and T47-D, for overexpression studies.
FIGURE 2.
ADAM12 expression is elevated in aggressive breast cancer cells. A, real-time RT-PCR analysis indicated significantly higher expression of ADAM12-L in highly aggressive ER-negative cells as compared with ER-positive, less invasive breast cancer cells. *, p = 0.005; **, p ≤ 0.0001. B–G, stable expression of ADAM12 isoforms in MCF-7 cells. Individual ADAM12-l-expressing clones are indicated as LC1, LC2, LC3, and LC4. B, real-time RT-PCR analysis of ADAM12-L in WT MCF-7 and representative ADAM12-L-overexpressing clones. *, p = 0.001; **, p ≤ 0.0001. Cell-surface ADAM12-L expression was confirmed via FACS analysis. C, a representative ADAM12-L clone, LC2, had considerably higher cell surface staining as compared with the WT MCF-7. D, immunoblot analysis of cell lysates indicating increased expression of ADAM12-L in transfectants as compared with WT MCF-7 cells. Three distinct bands representing ADAM12-L were detected, including an ∼120-kDa latent form, an ∼90-kDa active form, and an ∼68-kDa truncated form, respectively. E, ADAM12-S stable clones displayed an ∼10- to 30-fold increase in mRNA expression as compared with WT MCF-7 cells. *, p ≤ 0.01; **, p ≤ 0.0001. Immunoblot of serum-free CM demonstrated expression of latent and the active species of ADAM12-S (F) and ADAM12-Scatmut (G) in the clones but not WT MCF-7 cells.
Stable Expression of ADAM12 Isoforms in MCF-7 Cells
MCF-7 cells were stably transfected with ADAM12-L, ADAM12-S, and ADAM12-Scatmut (with an E351Q point mutation) constructs. Individual clones were selected on the basis of ADAM12 mRNA and protein expression. A significantly higher ADAM12-L mRNA and protein expression was observed in the stable clones as compared with parental MCF-7 cells (Fig. 2, B and D). Cell-surface ADAM12-L expression was confirmed via FACS analysis (Fig. 2C). ADAM12-S-overexpressing stable clones displayed an approximate 10- to 30-fold increase in ADAM12-S mRNA expression (Fig. 2E) which was confirmed at the protein level by immunoblot analysis of serum-free conditioned media (F). Similarly, overexpression in ADAM12-Scatmut clones was confirmed via immunoblot analysis of serum-free conditioned media (Fig. 2G).
ADAM12-S Overexpression Significantly Increases Breast Tumor Cell Migration and Invasion
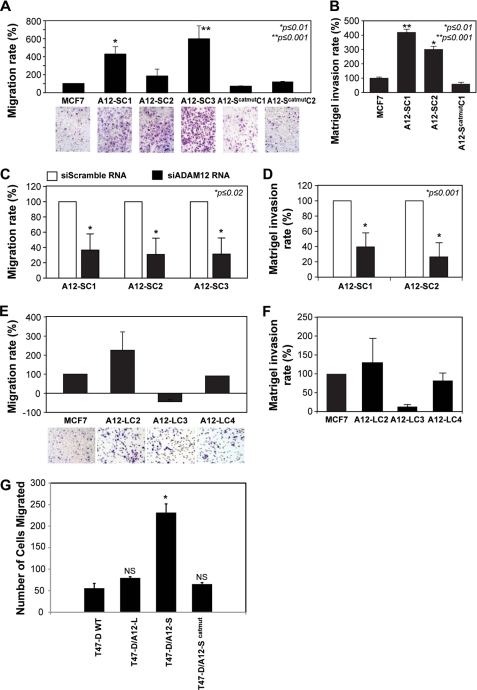
ADAM12-S expression induced significantly higher rates of migration (Fig. 3A) in contrast to WT MCF-7. The increased migratory capacity of the stable clones appeared to be directly proportional to the levels of ADAM12 expression in the individual clones (data not shown). Stable clones expressing the enzymatically inactive form of ADAM12-S (ADAM12-Scatmut), on the other hand, displayed migration rates similar to WT MCF-7 (Fig. 3A). In a standard Matrigel invasion assay, minimal invasion was observed for WT MCF-7 (Fig. 3B), whereas ADAM12-S expression induced significantly more invasion of the Matrigel layer. As with migration, expression of ADAM12-Scatmut resulted in very little or no invasion across the Matrigel layer (Fig. 3B). Silencing of ADAM12-S expression in the stable clones confirmed that this stimulatory effect on migration was specific. Down-regulation of ADAM12-S mRNA expression (supplemental Fig. 1, 60–75% decrease) resulted in a significant reduction in migration (70–80% reduction, Fig. 3C) and invasion (60–80% reduction, D) of the clones as compared with the controls. These data suggest that the catalytic activity of ADAM12-S is crucial for stimulating the migratory and invasive potential of breast cancer cells. Interestingly, overexpression of ADAM12-L did not result in a similar stimulatory effect on migration or invasion. Although a single ADAM12-L clone did display an approximately 2-fold increased migration, this property was not replicated in the other clones, where migration or invasion rates were equal to or slightly (but not significantly) lower than WT MCF-7 (Fig. 3, E and F). The differential effects on migration may partly be explained in terms of the cellular localization and/or substrate specificity of the two forms.
FIGURE 3.
The secreted isoform, ADAM12-S, promotes migration and invasion in breast cancer cells. Migration and Matrigel invasion analysis of ADAM12-S-expressing breast tumor cells. In vitro migration (A) and Matrigel invasion (B) analyses of representative ADAM12-S and ADAM12-Scatmut clones and WT MCF-7 cells. ADAM12-S-expressing cells displayed significantly (1.5- to 5-fold) higher migration and invasion rates as compared with WT MCF-7 or cells expressing enzymatically inactive ADAM12-Scatmut. C and D, the stimulation of migration and invasion was reversed when ADAM12-S expression was down-regulated using siRNA. Expression of the transmembrane form, ADAM12-L, did not stimulate migration or Matrigel invasion. E and F, migration rates of ADAM12-L clones were equal to or slightly (but not significantly) lower than WT MCF-7. G, ADAM12-S-expressing T47-D cells displayed enhanced migration rates compared with mock-transfected T47-D cells, cells expressing ADAM12-Scatmut, or ADAM12-L. *, p < 0.01. NS, not significant).
We next asked whether the effect of ADAM12-S on migration and invasion could be generalized to other breast cancer cell lines. Wild-type T47-D, which, like MCF-7 cells, are an ER-positive, low metastatic line and stable clone pools of ADAM12-expressing T47-D cells (18) were analyzed. ADAM12-S-expressing T47-D breast tumor cells (Fig. 3G) migrated at a faster rate than the empty vector-transfected cells, in contrast to which, ADAM12-L-overexpressing T47-D cells did not display enhanced migration (G), thereby recapitulating the effect observed in ADAM12-expressing MCF-7 cells.
ADAM12 Expression Results in Increased Tumorigenicity in an Orthotopic Breast Tumor Model
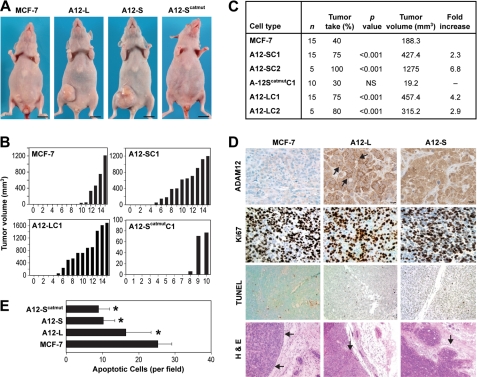
We next examined the effect of ADAM12 expression on tumor progression in vivo using an orthotopic mammary fat pad xenograft model in immunodeficient mice. Multiple MCF-7 clones expressing ADAM12-L, ADAM12-S, ADAM12-Scatmut, and WT MCF-7 cells, respectively, were injected into the mammary fat pad of 8-week old athymic mice in the presence of subcutaneously implanted slow-release 17β-estradiol pellets. The tumor injection site was monitored weekly, and tumors were measured when they became palpable starting at ∼3 weeks after injection of cells. Mice bearing representative tumors from each group and tumor volumes of individual animals from each experimental group are summarized in Fig. 4. Overexpression of both isoforms of ADAM12 resulted in a significantly higher tumor take and increased tumor growth. Tumors arising from individual ADAM12-S-expressing clones had significantly higher tumor take rates (ADAM12-Sclone1, 11 of 15, 75%, p < 0.001; ADAM12-Sclone2, 5 of 5, 100%, p < 0.001) as compared with WT MCF-7 (6 of 15, 40%), whereas expression of ADAM12-Scatmut resulted in a tumor take similar to the WT cells (3 of 10, 30%) (Fig. 4, A and B). By week 12, mean tumor volumes were significantly greater in ADAM12-S-overexpressing tumors (∼2.5- to 6-fold increase, p < 0.001) than WT MCF-7 tumors (Fig. 4C), and these differences remained significant until the animals were sacrificed at week 16. Interestingly, whereas the tumor take rates for the tumors expressing the ADAM12-Scatmut were comparable with the WT MCF-7 tumors, the mean tumor volumes were significantly lower (10-fold decrease, p < 0.01) in the former (Fig. 4C). Similarly, individual ADAM12-L-expressing MCF-7 clones also had significantly higher tumor take rates (75–80%, p < 0.001) than WT MCF-7 tumors (40%), respectively (Fig. 4B). Further, overexpression of the ADAM12-L isoform also resulted in significantly higher tumor volumes (∼2- to 4-fold increase, p ≤ 0.002) as compared with the WT MCF-7 tumors (Fig. 4C).
FIGURE 4.
ADAM12 expression promotes orthotopic tumor growth. A, mice injected with ADAM12-L- and ADAM12-S-expressing clones formed larger tumors (center panels) than WT MCF-7 cells (left panel) or ADAM12-Scatmut (right panel). B and C, histograms and table indicate mean tumor volumes. B and C, ADAM12-S and ADAM12-L-expressing tumors displayed a significantly higher (≥ 75%) tumor take and tumor volumes (2- to 6-fold) as compared with the WT MCF-7 or ADAM12-Scatmut tumors. D, increased staining of ADAM12 in tumors expressing ADAM12-L and ADAM12-S, respectively, compared with WT MCF-7 tumors. Proliferation (Ki67-positive nuclei) for WT MCF-7and ADAM12-expressing cells remained the same (D), whereas apoptosis rates (TUNEL-positive nuclei) were significantly lower in the ADAM12-L and ADAM12-S clones, respectively (D and E). H&E staining of representative tumor sections show neatly demarcated boundaries for WT MCF-7 tumors (D), whereas tumor-invasive fronts in ADAM12-L-expressing tumors were slightly more uneven. ADAM12-S tumors had much wider and more irregular tumor invasive boundaries.
IHC analysis confirmed ADAM12 overexpression in vivo (Fig. 4D). As expected, localization of the two isoforms was different. ADAM12-L staining was positive on the membrane as well as in the cytoplasm, whereas ADAM12-S-expressing tumors had more diffuse cytoplasmic staining, with some staining in the extracellular space. Because the ADAM12-expressing tumors had significantly higher growth rates, we analyzed proliferation in these tumors. The number of Ki67-positive nuclei were slightly but not significantly higher in the ADAM12-expressing tumors compared with the WT MCF-7 tumors (Fig. 4D); however, TUNEL analysis indicated that the number of apoptotic nuclei were significantly lower in ADAM12-L tumors (∼1.5-fold decrease, p < 0.00001) as well as in ADAM12-S tumors (∼2.5-fold decrease, p < 0.00001) (Fig. 4E). The proliferation index of the tumors (rate of proliferation/apoptosis) was higher in both ADAM12-L (1.5-fold) and ADAM12-S (2.2-fold) as compared with WT MCF-7 tumors (data not shown), suggesting that the increased tumor size may be a consequence of lower apoptotic rates observed in the ADAM12-expressing tumors. However, the reduced apoptotic rates were also observed in tumors expressing ADAM12-Scatmut, suggesting that this effect is not dependent on ADAM12 catalytic function (Fig. 4E). Microvascular density of WT and ADAM12 tumors were similar (data not shown).
ADAM12-S Expression Promotes Local Tumor Invasion and Distant Metastasis
To assess the effect of ADAM12 expression on local tumor invasion, microscopic examination of H&E-stained tumor sections was conducted. WT MCF-7 tumors appeared encapsulated with well demarcated boundaries that were distinct from the surrounding mammary fat pad (Fig. 4D), whereas tumor invasive fronts in a majority of the ADAM12-L-expressing tumors were more uneven, for e.g. the tumor cells appeared to invade into the mammary fat pad and to engulf some of the ducts in the surrounding area (Fig. 4D). Remarkably, the phenotype of the ADAM12-S tumors was more aggressive, with much wider and more irregular tumor invasive boundaries that were sometimes indistinguishable from the surrounding stroma, with clusters of breast tumor cells that appear to have migrated away from the primary tumor to grow in nests in the surrounding subcutaneous zone and mammary fat pad (Fig. 4D). In some cases, the primary tumor encroached into the muscle layer of the abdomen (supplemental Fig. 2A). When the tumor sections were stained for CD34, a marker specific for the vasculature, we detected incidences of tumor cell extravasation into the intratumoral blood vessels and formation of metastatic foci (supplemental Fig. 2B). The incidences of blood vessel extravasation were sporadic but limited to the ADAM12-S-expressing tumors and were not detected in ADAM12-L or WT MCF-7 tumors.
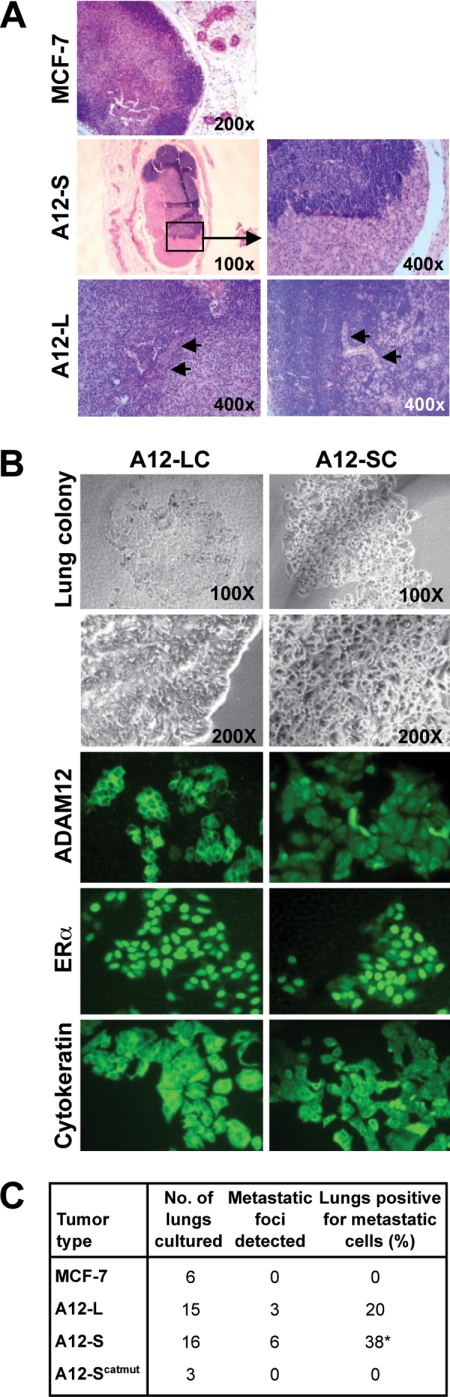
To assess distant metastases, sentinel and inguinal LN were immunostained for H&E (Fig. 5A). A majority of the LN from ADAM12 tumor-bearing mice appeared to be enlarged and, in some cases, hematogenous. Metastatic cell clusters were present with a significantly higher frequency in LN from ADAM12-S (7 of 18, 40%, p = 0.047) as compared with ADAM12-L (3 of 14; 21%, p = 0.24) and WT MCF-7 tumor-bearing animals (0 of 6, 0%) of tumor-bearing mice, respectively. Gross and microscopic examination of lung tissue from all tumor-bearing mice revealed no detectable micrometastases (data not shown). Because MCF-7 is a relatively non-metastatic cell line, we cultured harvested lung tissue from the sacrificed mice to assess the possible presence of solitary tumor cells in the lungs, which would be indicative of distant metastasis. Lungs were harvested from tumor-bearing animals at week 16, macerated, and cultured under sterile conditions routinely used for MCF-7 cells (21). Within two to 3 weeks, colonies of tumor cells were observed that appeared to be morphologically indistinguishable from the breast tumor cells grown in culture (Fig. 5B). A significantly higher number of tumor cell colonies were detected from lungs of animals bearing ADAM12-S tumors (6 of 16, 38%, p < 0.05), whereas no colonies were detected for the animals bearing ADAM12-Scatmut (0%) or the WT MCF-7 tumors (0%) and 20% of animals bearing ADAM12-L tumors (3 of 15, p = 0.13), respectively (Fig. 5C). To confirm that these colonies of epithelial cells had indeed originated from the primary orthotopic tumors and were of human origin, we analyzed expression of ADAM12, ERα, and cytokeratin in these cells. Lung cultures from both ADAM12-L- and ADAM12-S-expressing tumors stained positive for ADAM12, with the distinctive cell surface pattern of localization for the former and the diffuse cytoplasmic staining observed for the latter (Fig. 5B). Further, both cell types were positive for ERα nuclear and human cytokeratin staining (Fig. 5B), confirming that the lung colonies of epithelial cells had indeed originated from the primary orthotopic tumors and were of human origin. RT-PCR analysis of murine GAPDH and pulmonary surfactant protein SP-C and SP-A further ascertained that these isolated lung cultures were not mouse lung epithelial cells but rather of human origin (supplemental Fig. 2C). Therefore, consistent with the in vivo LN metastasis data, a significantly higher number of mice bearing ADAM12-S-expressing tumors displayed lung metastasis as compared with ADAM12-L or WT MCF-7 tumors.
FIGURE 5.
ADAM12 expression promotes distant metastasis. A, H&E staining of lymph node sections indicated that metastatic cell clusters were present in LN from ADAM12-S (7 of 18, 40%, p = 0.047) and ADAM12-L-tumor bearing mice (3 of 14, 21%, p = 0.24), respectively, and completely absent in WT MCF-7 tumor-bearing animals. B and C, incidences of tumor cell lung colonization were significantly higher in ADAM12-S tumor-bearing mice (6 of 16, 38%, p < 0.05), whereas no colonies were detected for the animals bearing ADAM12-Scatmut (0%) or the WT MCF-7 tumors (0%) and 20% of animals bearing ADAM12-L tumors (3 of 15, p = 0.13), respectively. B, lung colonies of tumor cells were positive for ADAM12, ERα, and human cytokeratin immunostaining, confirming human and epithelial origin of these cells.
DISCUSSION
We have previously reported that urinary ADAM12 is predictive of disease status in breast cancer patients and that its levels in urine increase with progression of the disease. Increased levels of ADAM12 mRNA and protein have previously been reported in a number of cancers and tumor cell lines, suggesting an active role for ADAM12 in tumor progression. However, these studies did not identify either the causal role or the distinct contributions of either ADAM12 isoform in tumorigenesis. This study is the only one to date to describe a role for ADAM12 overexpression in human cancer cells using an orthotopic breast tumor model.
IHC of a panel human breast tumors indicated that ADAM12 expression is up-regulated in primary tumors at early stages and is maintained at later stages of tumorigenesis and in metastatic LN. Of the matched sets of primary tumor tissue and metastatic LN analyzed in this study, a majority had positive expression of ADAM12 in both the primary tumor and matched LN, suggesting that ADAM12 expression may be essential for primary tumor growth as well as promoting and/or maintaining distant metastasis. Interestingly, within this cohort, ∼13% of the primary tumors did not stain positive for ADAM12, whereas their matched LNs did (Fig. 1D), suggesting that metastatic cells originating from primary tumors expressing low levels or no ADAM12 may up-regulate ADAM12 expression during the process of invasion and that ADAM12 function may be critical for the establishment of distant metastasis.
This is the first report to analyze expression of both the transmembrane and secreted isoforms of ADAM12 in human breast cancer. Our data indicate that there is increased expression of both isoforms of ADAM12 in human breast cancer tissue. Interestingly, expression of the transmembrane isoform, ADAM12-L, was found to be slightly but significantly higher in early stages of breast tumorigenesis, whereas both forms are expressed in late-stage disease. The tumor stage-dependent differences in the expression levels of ADAM12 isoforms may result in their distinct contributions to the tumor growth and metastasis process. Increased expression of the transmembrane isoform, ADAM12-L, in Stage I and II breast cancer may reflect its role in promoting tumor growth, perhaps via its sheddase and growth factor receptor activation function. The high levels of ADAM12-S in late-stage human breast tumors support its potential role in tumor invasion and metastasis.
ADAM12 expression and catalytic function has not previously been correlated with enhanced tumor cell proliferation, although its effect on the proliferation of normal cells has been reported. ADAM12-L was reported to regulate bronchial epithelial cell proliferation and apoptosis in an HB-EGF- and EGFR-dependent manner (22). In addition, ADAM12-L has been reported to enhance chondrocyte proliferation in osteoarthritis via the degradation of IGFBP-5 and increased bioavailability of insulin-like growth factor (23). Therefore, an increased expression of ADAM12-L in early-stage breast cancer, coupled with its unique sheddase function, may suggest a role for this protease in breast tumor growth in vivo. The secreted isoform, ADAM12-S, which is rarely expressed in normal adult tissue (placenta is an exception) and which we found to promote tumor cell migration and invasion in vitro and local and distant metastasis in vivo, was expressed in late stage human breast tumors, indicating a potential role in tumor invasion and metastasis. Surprisingly, ADAM12 expression in human breast tumors appeared to be primarily epithelial rather than stromal. This is in contrast to mouse models of breast and prostate tumors, where ADAM12 staining was detected almost exclusively in the stroma (24). This difference suggests that the ADAM12-overexpressing orthotopic breast tumor model, used in this study, may reliably recapitulate human breast tumor growth compared with other models.
Two recent studies have begun to investigate a role for ADAM12 in tumorigenesis using transgenic mouse models. One report used the mouse mammary tumor virus-polyomavirus middle T oncogene (MMTV-PyMT) mouse breast tumor model to show that ADAM12 transcripts were strongly up-regulated in the stromal cell subpopulation adjacent to epithelial tumor cells, which were largely negative for ADAM12 expression (24). In a gain of function study, transgenic overexpression of human ADAM12 in MMTV-PyMT mice was reported to accelerate breast tumor progression and increase tumor burden and aggressiveness (25). This report attributed the effect of Adam 12 on tumor growth to its opposing effects on apoptosis (i.e. ADAM12 expression increased sensitivity of stromal cells to apoptosis but rendered tumor cells resistant to apoptosis). However, this observed effect of ADAM12 on apoptosis was independent of its proteolytic function (25). Although we observe a similar decrease in apoptotic rates in ADAM12-expressing breast tumor cells, our results are in contrast to the previous study in that our data indicate that catalytic activity of the secreted isoform, ADAM12-S, is essential for its protumorigenic and prometastatic effects in vitro as well as in vivo. In the case of the transmembrane isoform, we cannot rule out the possibility that the cysteine-rich, disintegrin or cytoplasmic domains of ADAM12-L may independently influence its tumorigenic potential. It is widely appreciated that use of the MMTV-PyMT model results in rapid development of multifocal mammary adenocarcinomas as well as local and distant metastasis, regardless of the expression of the gene/protein of interest, making it difficult to rigorously and fairly relegate any effects observed specifically to ADAM12. To address this, we have tested the effects of ADAM12 overexpression in the breast tumor epithelium in a less aggressive tumor model, which provides the opportunity to more clearly assess the potential role of this protease in the initial establishment of the human breast tumors (tumor take), growth, and metastasis. Using human cancer cells in an in vivo orthotopic breast tumor model, we demonstrate that ADAM12 expression significantly increased tumor take and tumor growth.
We have recently reported that ADAM12 expression in breast tumor cells results in up-regulation of alternative growth pathways (18). Expression of ADAM12-L resulted in increased amphiregulin shedding and a concomitant up-regulation of EGFR protein expression and activation, ultimately resulting in estrogen-independent breast tumor cell proliferation. Similarly, increased ADAM12-S expression resulted in up-regulation of pIGF-1R and pMAPK levels in breast tumor cells (18). Therefore, one might speculate that ADAM12-induced higher tumor growth rates observed in this study may be associated with its effect on the up-regulation of alternative growth pathways. In the case of the secreted isoform, ADAM12-S, the tumor-promoting activity was a function of its catalytic function because cells expressing the enzymatically inactive form of the protease had tumor take rates and tumor volumes similar to parental MCF-7 cells.
Comparative analysis of ADAM12 mRNA in breast cancer cell lines indicated that expression is significantly elevated in highly invasive, estrogen-receptor-negative lines such as MDA-MB-231 and Hs578T as compared with low metastatic, estrogen-responsive lines such as MCF-7 and T47-D. To begin to understand the contribution of the distinct isoforms of ADAM12 in promoting breast tumorigenesis, we established stable clones that overexpress ADAM12-L, ADAM12-S, and ADAM12-Scatmut, respectively, in MCF-7 cells. Expression of ADAM12-S resulted in increased migration and invasion of MCF-7 and T47-D breast tumor cells. This effect was specific for ADAM12-S, as silencing of expression led to a reversal of increased migration. Therefore, the metalloprotease function of ADAM12-S is crucial for the stimulation of migration because MCF-7 cells expressing the catalytically inactive form of the protein do not display increased migration in vitro. Interestingly, ADAM12-L-expressing cells did not display a similar increase in migratory capacity. This effect was recapitulated in the in vivo tumor studies. ADAM12-S tumors had increased local invasion of the surrounding area, a higher incidence of vascular invasion, a significantly higher incidence of LN metastasis (40%), as well as lung colonization of individual tumor cells (38%) as compared with WT MCF-7 tumors that displayed no LN or lung colonization. The ADAM12-l-expressing tumors were moderately invasive (LN metastasis, 21% and lung colonies, 20%). However, these rates were not found to be significantly higher than WT MCF-7-tumor-bearing animals. Our data suggests that expression of the secreted isoform, ADAM12-S, but not the transmembrane isoform, ADAM12-L, selectively increased local tumor invasion and incidence of vascular invasion and promoted significantly higher incidence of LN metastasis as well as lung colonization as compared with WT MCF-7 tumors. Interestingly, ADAM12 appears to exert distinct and opposing effects on tumor versus normal cell migration. For example, ADAM12 was reported to inhibit Chinese hamster ovary cell migration, although this activity was independent of its catalytic function (26). Similarly, ADAM12-deficient keratinocytes migrated significantly more than wild-type keratinocytes in a wound-healing mouse model (27). Importantly, both of these studies were conducted with normal cells, and neither of these studies explored isoform-specific effects on cell migration. Our results suggest that the secreted isoform, ADAM12-S, enhances the migratory and invasive capacity of breast tumor cells, thereby allowing malignant cells to locally invade as well as to form distant metastasis. Therefore, in normal tissue, ADAM12 may inhibit cell migration by mediating cell adhesion via integrin interactions, whereas, in tumor cells, higher levels of ADAM12-S proteolytic function stimulate cell migration and invasion. Although we cannot rule out the possibility that the effect of the secreted isoform, ADAM12-S, on tumor cell migration and invasion as well as tumor metastasis is mediated via integrin interactions, it has been previously reported that several ADAM family members can cleave extracellular matrix proteins (28). Therefore, given that the effect of ADAM12-S on tumor cell migration and invasion is dependent on catalytic function, it is probable that ADAM12-S stimulates migration and invasion via ECM cleavage and remodeling. Experiments exploring the mechanism by which ADAM12 may exert its effects of tumor proliferation and invasion are currently being conducted.
Our study establishes distinct roles for ADAM12 isoforms in breast tumor progression and in local and distant tumor metastasis. These findings reflect the increased expression of ADAM12 in human breast cancer and metastatic lymph nodes and highlight the therapeutic potential of down-regulating or targeting ADAM12 in breast cancer, perhaps through the development of ADAM12-specific inhibitors (29).
Supplementary Material
Acknowledgments
We thank Dr. Ulla M. Wewer (University of Copenhagen, Copenhagen, Denmark) for providing the expression plasmids for ADAM12 isoforms and Kristin Johnson for assistance with the figures.
This work was supported, in whole or in part, by National Institutes of Health Grants PO1 CA45548 and R01 CA118764. This work was also supported by The Breast Cancer Research Foundation, The JoAnn Webb Fund for Angiogenesis Research, The Ellison Foundation, and The Fortin Foundation.
This work is dedicated to the memory of our mentor, the late M. Judah Folkman, M.D. We remain extremely grateful for his enthusiastic interest in this work and for many helpful discussions and suggestions.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S2 and Table 1.
- ER
- estrogen receptor
- IHC
- immunohistochemistry
- LN
- lymph node(s).
REFERENCES
- 1. Gilpin B. J., Loechel F., Mattei M. G., Engvall E., Albrechtsen R., Wewer U. M. (1998) J. Biol. Chem. 273, 157–166 [DOI] [PubMed] [Google Scholar]
- 2. Asakura M., Kitakaze M., Takashima S., Liao Y., Ishikura F., Yoshinaka T., Ohmoto H., Node K., Yoshino K., Ishiguro H., Asanuma H., Sanada S., Matsumura Y., Takeda H., Beppu S., Tada M., Hori M., Higashiyama S. (2002) Nat. Med. 8, 35–40 [DOI] [PubMed] [Google Scholar]
- 3. Horiuchi K., Zhou H. M., Kelly K., Manova K., Blobel C. P. (2005) Dev. Biol. 283, 459–471 [DOI] [PubMed] [Google Scholar]
- 4. Dyczynska E., Sun D., Yi H., Sehara-Fujisawa A., Blobel C. P., Zolkiewska A. (2007) J. Biol. Chem. 282, 436–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ito N., Nomura S., Iwase A., Ito T., Kikkawa F., Tsujimoto M., Ishiura S., Mizutani S. (2004) Biochem. Biophys. Res. Commun. 314, 1008–1013 [DOI] [PubMed] [Google Scholar]
- 6. Loechel F., Gilpin B. J., Engvall E., Albrechtsen R., Wewer U. M. (1998) J. Biol. Chem. 273, 16993–16997 [DOI] [PubMed] [Google Scholar]
- 7. Loechel F., Fox J. W., Murphy G., Albrechtsen R., Wewer U. M. (2000) Biochem. Biophys. Res. Commun. 278, 511–515 [DOI] [PubMed] [Google Scholar]
- 8. Roy R., Wewer U. M., Zurakowski D., Pories S. E., Moses M. A. (2004) J. Biol. Chem. 279, 51323–51330 [DOI] [PubMed] [Google Scholar]
- 9. Kveiborg M., Albrechtsen R., Couchman J. R., Wewer U. M. (2008) Int. J. Biochem. Cell Biol. 40, 1685–1702 [DOI] [PubMed] [Google Scholar]
- 10. Lendeckel U., Kohl J., Arndt M., Carl-McGrath S., Donat H., Röcken C. (2005) J. Cancer Res. Clin. Oncol. 131, 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fröhlich C., Albrechtsen R., Dyrskjøt L., Rudkjaer L., Ørntoft T. F., Wewer U. M. (2006) Clin. Cancer Res. 12, 7359–7368 [DOI] [PubMed] [Google Scholar]
- 12. Kodama T., Ikeda E., Okada A., Ohtsuka T., Shimoda M., Shiomi T., Yoshida K., Nakada M., Ohuchi E., Okada Y. (2004) Am. J. Pathol. 165, 1743–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carl-McGrath S., Lendeckel U., Ebert M., Roessner A., Röcken C. (2005) Int. J. Oncol. 26, 17–24 [PubMed] [Google Scholar]
- 14. Markowski J., Oczko-Wojciechowska M., Gierek T., Jarzab M., Paluch J., Kowalska M., Wygoda Z., Pfeifer A., Tyszkiewicz T., Jarzab B., Niedzielska I., Borgiel-Marek H. (2009) J. Physiol. Pharmacol. 60 Suppl 1: 57–63 [PubMed] [Google Scholar]
- 15. Mino N., Miyahara R., Nakayama E., Takahashi T., Takahashi A., Iwakiri S., Sonobe M., Okubo K., Hirata T., Sehara A., Date H. (2009) J. Surg. Oncol. 100, 267–272 [DOI] [PubMed] [Google Scholar]
- 16. Le Pabic H., Bonnier D., Wewer U. M., Coutand A., Musso O., Baffet G., Clément B., Théret N. (2003) Hepatology 37, 1056–1066 [DOI] [PubMed] [Google Scholar]
- 17. Pories S. E., Zurakowski D., Roy R., Lamb C. C., Raza S., Exarhopoulos A., Scheib R. G., Schumer S., Lenahan C., Borges V., Louis G. W., Anand A., Isakovich N., Hirshfield-Bartek J., Wewer U., Lotz M. M., Moses M. A. (2008) Cancer Epidemiol. Biomark. Prev. 17, 1034–1042 [DOI] [PubMed] [Google Scholar]
- 18. Roy R., Moses M. A. (2011) Breast Cancer Res. Treat., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang J., Bielenberg D. R., Rodig S. J., Doiron R., Clifton M. C., Kung A. L., Strong R. K., Zurakowski D., Moses M. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3913–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fernández C. A., Yan L., Louis G., Yang J., Kutok J. L., Moses M. A. (2005) Clin. Cancer Res. 11, 5390–5395 [DOI] [PubMed] [Google Scholar]
- 21. Perrotte P., Bielenberg D. R., Eve B. Y., Dinney C. P. N. (1997) Mol. Urol. 1, 299–307 [Google Scholar]
- 22. Rocks N., Estrella C., Paulissen G., Quesada-Calvo F., Gilles C., Guéders M. M., Crahay C., Foidart J. M., Gosset P., Noel A., Cataldo D. D. (2008) Cell Prolif 41, 988–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Okada A., Mochizuki S., Yatabe T., Kimura T., Shiomi T., Fujita Y., Matsumoto H., Sehara-Fujisawa A., Iwamoto Y., Okada Y. (2008) Arthritis Rheum. 58, 778–789 [DOI] [PubMed] [Google Scholar]
- 24. Peduto L., Reuter V. E., Sehara-Fujisawa A., Shaffer D. R., Scher H. I., Blobel C. P. (2006) Oncogene 25, 5462–5466 [DOI] [PubMed] [Google Scholar]
- 25. Kveiborg M., Fröhlich C., Albrechtsen R., Tischler V., Dietrich N., Holck P., Kronqvist P., Rank F., Mercurio A. M., Wewer U. M. (2005) Cancer Res. 65, 4754–4761 [DOI] [PubMed] [Google Scholar]
- 26. Huang J., Bridges L. C., White J. M. (2005) Mol. Biol. Cell 16, 4982–4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harsha A., Stojadinovic O., Brem H., Sehara-Fujisawa A., Wewer U., Loomis C. A., Blobel C. P., Tomic-Canic M. (2008) J. Mol. Med. 86, 961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roy R., Zhang B., Moses M. A. (2006) Exp. Cell Res. 312, 608–622 [DOI] [PubMed] [Google Scholar]
- 29. Kveiborg M., Jacobsen J., Lee M. H., Nagase H., Wewer U. M., Murphy G. (2010) Biochem. J. 430, 79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.