Abstract
Editing of adenosine (A) to inosine (I) at the first anticodon position in tRNA is catalyzed by adenosine deaminases acting on tRNA (ADATs). This essential reaction in bacteria and eukarya permits a single tRNA to decode multiple codons. Bacterial ADATa is a homodimer with two bound essential Zn2+. The ADATa crystal structure revealed residues important for substrate binding and catalysis; however, such high resolution structural information is not available for eukaryotic tRNA deaminases. Despite significant sequence similarity among deaminases, we continue to uncover unexpected functional differences between Trypanosoma brucei ADAT2/3 (TbADAT2/3) and its bacterial counterpart. Previously, we demonstrated that TbADAT2/3 is unique in catalyzing two different deamination reactions. Here we show by kinetic analyses and inductively coupled plasma emission spectrometry that wild type TbADAT2/3 coordinates two Zn2+ per heterodimer, but unlike any other tRNA deaminase, mutation of one of the key Zn2+-coordinating cysteines in TbADAT2 yields a functional enzyme with a single-bound zinc. These data suggest that, at least, TbADAT3 may play a role in catalysis via direct coordination of the catalytic Zn2+. These observations raise the possibility of an unusual Zn2+ coordination interface with important implications for the function and evolution of editing deaminases.
Keywords: Enzyme Kinetics, RNA Editing, Transfer RNA (tRNA), Trypanosome, Zinc, ADAT2/3, Deaminase, tRNA Editing
Introduction
All genomes encode far less tRNA diversity than is required for all the codons used for translation (1). This apparent decoding conundrum is resolved by the inherent base pairing flexibility between the third codon position in mRNA and the first position of the anticodon in tRNA, as originally proposed in Crick's wobble rules (2). Wobbling permits some nucleotides to form sufficiently stable non-Watson and Crick base pairs providing critical flexibility, without sacrificing translational fidelity (3). Among the most extreme cases of base-pairing flexibility in protein synthesis is the use of the nucleotide inosine, a guanosine analog that can base pair with A, C, or U (3). Thus in the context of a codon-anticodon interaction, inosine at the first position of the anticodon (I34) effectively allows a single tRNA to decode three different codons for the same amino acid, obviating the need for additional tRNAs to be encoded in genomes. Given the role of I34 in decoding in bacteria and eukarya, it is not surprising that the enzymes responsible for inosine formation are essential for viability (4–6).
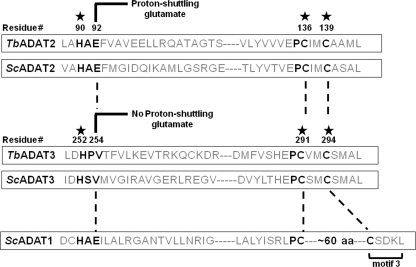
Inosine formation in tRNAs is not relegated to the anticodon and has been described at various positions in tRNAs from a number of organisms. For example, in eukaryotic tRNAs inosine can also occur at position 37 where it may affect anticodon loop structure (7, 8). Interestingly, the sequence of the enzyme responsible for synthesis of I37 resembles classic adenosine deaminases, whereas that for I34 has all the sequence signature motifs of cytidine deaminase, perhaps reflecting their different evolutionary paths (see Fig. 1).
FIGURE 1.
Evolutionarily conserved residues in the active sites of TbADAT2 and TbADAT3. The figure shows a schematic highlighting conserved amino acids involved in Zn2+ coordination in TbADAT2, ScADAT2, TbADAT3, and ScADAT3 (denoted by a star). The key glutamate required for catalysis is as indicated. This glutamate is involved in shuttling a proton from an activated water molecule into the leaving amino group, which is released as ammonia during the reaction. The glutamate has been naturally replaced during evolution by a valine in Sc and TbADAT3. The numbers indicate amino acid positions in each sequence relative to the first methionine in T. brucei. Key deaminase residues are highlighted with black letters. motif 3 refers to a motif characteristic of adenosine deaminase (ADAT1 for example), where the second zinc-coordinating cysteine is separated from the first by an ∼60-amino acid spacer. This motif is never found in cytidine deaminases.
Regardless of nucleotide position, all inosines in RNA are presumably formed by the zinc-dependent hydrolytic deamination of adenosine, catalyzed by adenosine deaminases acting on tRNAs (ADATs)4 or adenosine deaminases acting on RNA (ADARs) for all other RNAs. These enzymes can specifically target a variety of RNA substrates, for example tRNAs, mRNAs, miRNAs, and even rRNA, while excluding free nucleotides (9–12). Currently, the best studied example of inosine in tRNA is that occurring at the first position of the anticodon (I34) where the essential inosine has, as discussed above, a direct bearing on decoding. In bacteria, I34 is generated by bacterial homodimeric ADATa with each monomer coordinating one Zn2+ and containing all the additional amino acids needed for catalysis (13–15). Inosine has not been described in mitochondria, but a recent report showed that a bacteria-type ADATa enzyme plays key functions in inosine formation and thus translation in chloroplasts (16, 17). ADATa only targets a single tRNA (tRNAACGArg) and can efficiently catalyze deamination of an anticodon stem loop corresponding to a minimal version of tRNAACGArg (4). Importantly, RNA recognition was highlighted in the co-crystal structure of the Staphylococcus aureus ADATa with a bound anticodon stem loop, which suggested that all of the key contacts needed for catalysis were maintained even with this shortened substrate (18).
In contrast, the eukaryotic enzymes target seven or eight different tRNAs depending on the organism, raising questions about the evolutionary transition to this broader, multi-substrate specificity. The eukaryotic enzyme, as exemplified by the yeast system, functions as a heterodimer, comprised of two subunits, ADAT2 and ADAT3, that unlike the bacterial enzyme needs a full-length tRNA for activity (5, 19). Currently, however, because of the lack of a crystal structure or detailed kinetic characterization, little is known about how the zinc co-factor is coordinated or what the rules are for substrate recognition among eukaryotic tRNA anticodon deaminases (including yeast). Because of its similarity with other members of the cytidine deaminase superfamily, including bacterial ADATa, it has been suggested that this group of eukaryotic enzymes uses similar metal coordinating motifs, H(C)XE and PCXXC (where X represents any amino acid) (5). The conserved histidine and two cysteines coordinate the zinc ion, whereas the fourth ligand is an activated water molecule. A conserved glutamate in ADAT2 then acts as a proton shuttle between the activated water and the exocyclic nitrogen at C-6 of the purine ring. Interestingly, in the equivalent site of Trypanosoma brucei ADAT3, a valine is found in place of the conserved catalytic glutamate (see Fig. 1) (6). Similarly ADAT3 from yeast also has a valine at this position (Val218) (5), a trend that is seen in ∼60% of all ADAT3s. Isoleucine and threonine are also fairly well conserved in this position (combined total 25% of ADAT3s). Because ADAT3 still has all of the Zn2+-binding residues, it has been suggested that this subunit might only play a structural role, an untested premise (5, 6). In the absence of evidence to suggest otherwise, the eukaryotic A34-specific tRNA deaminases are thus presumed to bind two zinc ions (one catalytic and one structural), both essential for activity (5).
We exploited the intrinsic asymmetry of the heterodimeric ADAT2/3 to explore the general contribution of each subunit to activity, including the role of Zn2+-coordinating residues. We have previously shown that ADAT2/3 from T. brucei (TbADAT2/3) is unusual in that it performs both C to U and A to I deamination reactions (6). Using a combination of molecular and biochemical approaches, we have now examined the steady-state kinetic behavior of TbADAT2/3 and present the first complete kinetic characterization of a eukaryotic tRNA deaminase. We show that this enzyme is mechanistically distinct from other multimeric RNA deaminases in its ability to catalyze the deamination reaction with a single bound Zn2+. Taken together, the findings presented here highlight the possible tight interplay between subunits that generate a functional active site with implications for the specificity and evolution of multimeric editing deaminases.
EXPERIMENTAL PROCEDURES
Mutagenesis and Recombinant Expression of TbADAT2/3 Mutants in Escherichia coli
TbADAT2 and TbADAT3 single amino acid substitutions were generated by QuikChange site-directed mutagenesis (Stratagene) per manufacturer's protocol. E. coli (BL21 DE3) was transformed with expression vector pCDF-Duet, and recombinant protein expression was as described previously (20). The cells were harvested by centrifugation, suspended in binding buffer (20 mm Tris, pH 8.0, 500 mm NaCl, and 5 mm imidazole), and lysed by sonication with a Branson sonifier 450 (five times, 30-s intervals with 60 s rest between sonication). The resulting lysate was clarified by centrifugation at 15,000 × g for 15 min, followed by centrifugation of the resulting supernatant at 100,000 × g for 30 min (S100). The S100 was incubated with Ni2+-nitrilotriacetic acid-agarose beads, and purification was performed as per manufacturer instructions (Qiagen). Bound proteins were eluted with 500 mm imidazole, and the purified protein peak was dialyzed at 4 °C overnight in storage buffer (50 mm Hepes, pH 8.0, 0.1 mm EDTA, 0.5 mm MgCl2, and 2 mm 1,4-dithiothreitol). Glycerol (final concentration, 20%) was added to the dialyzed protein for storage in aliquots at −80 °C.
Adenosine Deaminase Assays
An A34 site-specifically radioactively labeled tRNAVal substrate was generated and refolded as described previously (20). Increasing concentrations of tRNA were incubated with wild type or mutant recombinant enzyme at 27 °C for 45 min (or longer as indicated). The sample was extracted with phenol and ethanol-precipitated. The resulting pellet was suspended in 9 μl of 1× nuclease P1 buffer (as supplied with enzyme; 30 mm NaOAc, pH 5.3) and 1 μl of nuclease P1 (MPBiomedicals) and incubated overnight at 37 °C. The samples were dried with heat under vacuum. Dried samples were suspended in 2.5 μl of H2O, and 1 μl was spotted on a TLC cellulose plate (Merck). The reaction products were separated by one-dimensional TLC in solvent C (0.1 m sodium phosphate, pH 6.8:ammonium sulfate:n-propanol (100:60:2, v/w/v)) and visualized using the Storm imaging system (GE Healthcare). The signals were quantified using ImageQuant software. Steady-state kinetic constants were calculated by nonlinear regression using SigmaPlot kinetic software (with the enzyme kinetic module) and Lineweaver-Burk plots (supplemental Fig. S1).
Electrophoretic Mobility Shift Assay
Gel-purified radiolabeled tRNAVal mimicking the product tRNA (T7-transcript containing G34 instead of A34) was incubated on ice in the presence of reaction buffer (50 mm Hepes, pH 8.0, 1 mm MgCl2, and 5 mm KCl) with increasing concentrations of protein for 15 min. Glycerol was added to the samples (final concentration, 10%), and the products were separated on a 6.5% nondenaturing polyacrylamide gel run in 1× NNB (67 mm Tris, pH 8.3, 22 mm boric acid, and 0.6 mm EDTA) at 100 volts for 1.5 h at 25 °C. The gel was dried and exposed to a PhosphorImager screen; the products were visualized and analyzed using the Storm imaging system and ImageQuant software (GE Healthcare), respectively. The binding data were fit to a single ligand-binding curve using SigmaPlot kinetic software.
Zinc Analysis
TbADAT2/3 wild type and mutant enzymes were analyzed for zinc content via inductively coupled plasma (ICP) emission spectrometry by the University of Georgia Chemical Analysis Laboratory. Wild type and mutant TbADAT2/3 were expressed as above but were extensively dialyzed in 50 mm Hepes, pH 8.0, 100 mm KCl, and 2 mm EDTA to remove loosely bound zinc and other metal ions. The buffer was further exchanged in metal-free Hepes, pH 8.0, and 50 mm KCl to remove excess salt and EDTA. Pure nitric acid was added to 2%. Protein samples and buffer controls were analyzed in 2% nitric acid. Zinc content was determined by ICP using buffer alone as the background control.
[35S]Met/Cys Labeling of Wild Type TbADAT2/3
Wild type enzyme was expressed and purified using the above protocol with the following modifications. First, BL21 DE3 RIL X, a methionine auxotroph cell line was used. At A600 0.6–0.8 cells were harvested (6000 rpm for 5 min) and suspended in M9 minimal medium supplemented with a trace metals mix, 2 mm MgSO4, 100 mm CaCl2, 0.2% glucose, and 100 mm “cold” methionine. The cells were then grown for an additional 30 min at 37 °C before induction. Protein expression was induced with isopropyl β-d-thiogalactopyranoside (final concentration, 0.5 mm) and 1 mCi of radiolabel [35S]Met/Cys. Following 15 h of induction (at 25 °C), the cells were harvested and processed as described above.
Modeling of T. brucei ADAT2/3 Heterodimer
Sequences of T. brucei ADAT2 and ADAT3 were submitted to the GeneSilico protein structure prediction MetaServer (21), which is a gateway for >200 primary methods for predicting various aspects of protein structure and in particular for the detection of remote homology to other proteins with known structure. Expectedly, the metaserver indicated homology between ADAT2 and ADAT3 and a large group of structures from the cytidine deaminase superfamily. As the modeling template, we selected the structure of tRNA adenosine deaminase ADATa (TadA) from S. aureus (Protein Data Bank code 2B3J), because it was one of the top scoring structures, and it has been solved as a complex with the RNA substrate. Three-dimensional models were generated for the heterodimer of TbADAT2/3 based on the ADATa homodimer using the FRankenstein's monster method (22). Subsequently, an alternative “swapped” model was built by exchanging parts of ADAT2 and ADAT3 containing the HXE and CXXC motifs to form hybrid active sites, followed by remodeling of loops connecting the “swapped” elements. Finally, the N-terminal domain of ADAT3 was added by de novo modeling with ROSETTA (23), in the same orientation with respect to the N terminus of ADAT3 in both initial models, i.e. the regular and the swapped one. The atomic details are only predictions; nonetheless, the models are expected to be accurate on the level of the inter-residue contacts within individual domains and hence can be used to aid in analyzing various types of data in the three-dimensional context and thereby help to interpret the effect of residue substitutions resulting from mutations. Sequence conservation has been calculated with ConSurf (24), based on multiple sequence alignments of TbADAT2 and TbADAT3 orthologs, respectively, using representative sequences from sequenced eukaryotic genomes, selected from the results of a PSI-BLAST search (25).
RESULTS
Mutation of a Key Zn2+-coordinating Cysteine in ADAT2 Still Yields an Active Enzyme
We previously showed that the heterodimeric T. brucei anticodon deaminase (TbADAT2/3) had sufficient catalytic flexibility to perform both adenosine and cytidine deamination reactions, albeit in two different substrates, tRNA and DNA, respectively (6). In light of these observations, we decided to probe further the general contribution of each subunit (TbADAT2 and TbADAT3) to enzymatic activity. Previous studies reported the identification and characterization of the S. cerevisiae ADAT2/3 (ScADAT2/3) enzyme (5). This enzyme, like in T. brucei, is heterodimeric, and whereas the ScADAT2 subunit harbors all of the key conserved residues required for deamination, the sequence of ScADAT3 is naturally missing the conserved active site glutamate essential for deaminase activity. ADAT2 was thus proposed to play a catalytic role, whereas ADAT3 plays solely a structural one (5). However, the general contribution of each subunit of ADAT2/3 to catalysis has not been formally established in any system. We systematically made a number of single-amino acid substitutions in each subunit of the TbADAT2/3, at residues conserved among most deaminases (Fig. 1). In most deaminases, Zn2+ coordination occurs via an amino acid triad composed of one histidine (or cysteine) separated by a spacer region (with a variable number of amino acids) followed by two additional cysteines (the so-called H…CXXC motif) (7, 26) (Fig. 1). In TbADAT2/3, alanine substitutions of either the conserved histidine (His252) or the cysteines (Cys291 and Cys294) of TbADAT3 produced an inactive enzyme (Table 1 and data not shown). This recombinant enzyme, however, could still form a heterodimer with ADAT2. Two inferences can be drawn. First, the lack of activity of this mutant is due to the inability of TbADAT3 to coordinate Zn2+. Second, Zn2+ coordination by TbADAT3 is not a prerequisite for heterodimerization. These observations are consistent with attributes reported for the yeast counterpart.
TABLE 1.
The effect of amino acid substitutions at evolutionarily conserved residues in TbADAT2 and TbADAT3
| Enzyme | Kma | kcata,b | kcat/Km |
|---|---|---|---|
| μm | min−1 | min−1/μm | |
| Wild type | 0.75 ± 0.11 | 0.19 ± 0.07 | 0.25 |
| ADAT3 H252A | — | — | — |
| ADAT3 C291A | — | — | — |
| ADAT3 C294A | — | — | — |
| ADAT3 V254E | — | — | — |
| ADAT3 V254L | 2.77 ± 0.58 | 0.05 ± 0.03 | 0.02 |
| ADAT3 V254T | 2.14 ± 0.79 | 0.11 ± 0.01 | 0.05 |
| ADAT2 E92A | — | — | — |
| ADAT2 H90A | — | — | — |
| ADAT2 C136A/ADAT3 C291A | — | — | — |
| ADAT2 C136A | 1.48 ± 0.40 | 0.05 ± 0.01 | 0.02 |
| ADAT2 C139A | — | — | — |
a The values are obtained from at least three independent trials. A dash denotes that no activity was detected.
b kcat values are calculated using experimental Vmax values and protein concentrations via Bradford assay.
In the TbADAT3 sequence, a valine is found in place of the key catalytic glutamate (HPV instead of HAE; Val254 in HPV; Fig. 1). Previous studies showed that in the ScADAT2/3 heterodimer replacement of the ADAT3 valine for glutamate (V218E) could not rescue the loss of activity seen in the active site mutant E56A, suggesting that the position of the glutamate on ScADAT2 was crucial for catalysis (5). However, ScADAT3 V218E supported full activity in the context of a wild type ScADAT2 (5). Analogous substitutions in TbADAT2/3 caused similar effects with one striking difference, TbADAT3 V254E in the context of a wild type ADAT2 completely inactivated the enzyme (Table 1). However, substitutions of TbADAT3 V254 by leucine, threonine, or serine were tolerated, albeit with a 3–7-fold increase in Km and smaller or insignificant changes in kcat (compared with the wild type). These data reinforce the view that Zn2+ coordination by the HPV region of TbADAT3 serves a structural and/or binding role but does not directly partake in catalysis. However, in light of the fact that changes of nearby residues do affect activity (Table 1, e.g. ADAT3 V254E), we conclude that minor perturbations of residues proximal to His252 of TbADAT3 can have drastic effects on enzyme function. Similarly, substitutions in the conserved HAE motif region in TbADAT2 showed that, like in all other deaminases, the key catalytic glutamate (Glu92) in TbADAT2 was essential for activity, but in contrast to the equivalent position in ADAT3, other amino acid substitutions at Glu92 were always deleterious (data not shown), highlighting the essential role of the conserved glutamate in catalysis.
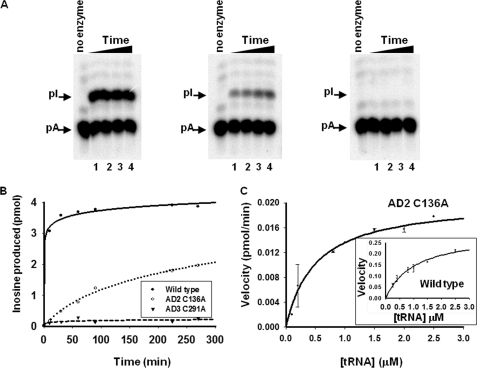
To further assess the specific contributions of TbADAT2 and TbADAT3 to catalysis, we mutated the other conserved active site domain, the PCXXC domain, in each subunit (Fig. 1). We made individual alanine substitutions in TbADAT2 and TbADAT3, so that one subunit was bearing the mutation in the context of the second subunit remaining wild type (Table 1). As expected, substitutions of cysteine 139 (the second cysteine in CXXC) to alanine in TbADAT2 abrogated enzymatic activity, indicating the essential role of this residue for activity, likely because of its ability to participate in Zn2+ coordination (data not shown). Unexpectedly, however, replacing the first cysteine in TbADAT2 (C136A) (Fig. 1) still generated an active enzyme (Fig. 2 and Table 1). This recombinant variant showed only a 4-fold reduction in kcat and a 12.5-fold reduction in kcat/Km compared with the wild type. The larger decrease in catalytic efficiency was partly due to a 2-fold increase in the Km, suggesting that the modest change in kcat was either partly structural or was due to small effects on substrate binding. This is in stark contrast with similar mutations in the cytidine deaminase from Escherichia coli, which led to either no detectable activity or a >100,000-fold decrease in specific activity (27). Our observation suggests that although both cysteines in the CXXC motif of TbADAT2 play a role in optimal enzyme function, there is a first among equals: Zn2+ coordination via the first cysteine is not essential for catalysis. Taken together, the results in Table 1 suggest marked differences in the functional significance of residues within conserved motifs between TbADAT2/3 and other nucleotide deaminases. These observations also reveal that all of the key Zn2+-coordinating residues of TbADAT3 are essential, but one of the three zinc-coordinating residues in TbADAT2 is dispensable for activity.
FIGURE 2.
Amino acid substitution of one of the key zinc-coordinating residue in TbADAT2 yields an active enzyme. A, a site-specifically 32P-labeled (at A34) tRNA substrate was incubated with constant concentrations of wild type and mutant enzyme for varying time. The reaction products were then digested to 5′-nucleotide monophosphates and separated by thin layer chromatography. The arrows indicate the positions of 5′-adenosine monophosphate (pA) and the product 5′-inosine monophosphate (pI). Nonradioactive nucleotides were used as markers (not shown). In all panels, the no enzyme lane refers to a negative control in which tRNA was incubated with buffer in the absence of enzyme; this also served as a control for background during quantitation. The samples were incubated with wild type or mutant enzymes for 60, 90, 120, and 180 min (lanes 1–4, respectively) (panels from left to right, wild type, AD C136A, and AD3 C291A). B, the left panel shows a graph of a time course of similar reactions as above but for extended periods of time. AD2 C136A denotes a substitution of one of the zinc-coordinating cysteines in TbADAT2 expressed in the context of a heterodimer with wild type TbADAT3. AD3 C291A denotes the analogous mutant but in TbADAT3 in the context of wild type TbADAT2. C, steady-state kinetic analysis of the TbADAT2 C136A mutant by incubation with increasing saturating concentrations of tRNA substrate. The reaction velocity (pmol of inosine formed per minute) was plotted against tRNA substrate concentration. This curve was fit to the Michaelis-Menten equation using SigmaPlot software. The inset shows a similar experiment but with the recombinant wild type enzyme. Each plot represents at least three independent experiments.
A Single Zn2+ Is Necessary and Sufficient for ADAT2/3 Activity
Two possibilities could account for the above results: 1) the TbADAT2 C136A mutant might somehow coordinate a loosely bound catalytic Zn2+ or 2) a single Zn2+ is sufficient for catalysis. To rule out the first possibility and test the idea that chelation of a potentially loose bound zinc ion would effectively inactivate the enzyme, we performed deamination reactions with the TbADAT2 C136A mutant in the presence of increasing concentrations of EDTA (Fig. 3A). However, no significant effect on the enzyme activity of the mutant was observed even in the presence of 1 mm EDTA, which corresponds to a >650-fold molar excess compared with the enzyme (1.5 μm in these reactions). Similar mutations in a number of deaminases can be partially rescued by increasing Zn2+ concentration (27). We found that exogenous Zn2+ addition failed to rescue any of the inactive mutants; importantly, the C136A mutant had comparable activity as in the absence of added Zn2+ (Fig. 3B). In fact, as in the case of wild type, this mutant was inhibited by ZnCl2 concentrations exceeding 50 μm. The lack of rescue by the addition of Zn2+ may be related to the observation that only after co-expression of both subunits in E. coli were we able to generate an active enzyme, and perhaps Zn2+ has already been incorporated into the enzyme during de novo assembly of the recombinant heterodimer.
FIGURE 3.
A single tightly bound zinc is sufficient for enzyme activity. A, deamination reactions as described above (Fig. 2) were performed using recombinant mutant TbADAT2 (in the context of wild type TbADAT3) in the presence of increasing concentrations of EDTA. After thin layer chromatography, 5′-adenosine and 5′-inosine monophosphate signals were quantitated using ImageQuant software. The amount of inosine produced was plotted over time for each concentration of EDTA. “No EDTA” (filled circle) is a control reaction in the absence of EDTA. 0.1 mm EDTA (open circle), 0.5 mm EDTA (filled triangle), and 1.0 mm EDTA (filled triangle) represent reactions in which EDTA was added in 60, 300, and >650 molar excess compared with the enzyme concentration. B, wild type and mutant recombinant enzymes where incubated as above (Fig. 2) for 1 h in the presence of increasing concentration of zinc (ZnCl2). Picomoles of inosine produced were plotted versus zinc concentration (in μm). In all cases the mutant protein is expressed in the context of its wild type partner. C, picomoles of Zn2+ per pmol of TbADAT2/3 heterodimer in the recombinant wild type and mutant enzyme were calculated via ICP. TbADAT2 denotes recombinant TbADAT2 expressed and purified as a homodimer in the absence of ADAT3.
We then explored the second possibility and determined the Zn2+ content of the wild type and C136A mutant TbADAT2/3 heterodimers by ICP emission spectrometry (28). We purified recombinant wild type and/or mutant enzyme by Ni2+-chelate chromatography followed by extensive dialysis in the presence of EDTA to remove any loosely bound metals and to also rule out the possibility of metals binding via the His tag. These samples were then subjected to ICP, which revealed that, as expected, the wild type enzyme contained an average of 1.99 ± 0.12 zinc ions/heterodimer. Interestingly, the TbADAT2 mutant C136A contained an average of 0.78 ± 0.22 pmol of zinc bound per pmol of heterodimeric enzyme (Fig. 3C). It is possible that a different metal is still coordinated by residues other than the conserved HCC triad and that this second metal is the one catalyzing the deamination reaction in the C136A mutant. Replacement of zinc by cobalt in other deaminases can still yield an active enzyme, albeit with a reduced catalytic rate (29). To test this possibility, we also screened the mutant and wild type for the presence of 19 other metals including cobalt, magnesium, and iron. We found no measurable levels of any other metal in our preparations (data not shown). The lack of binding of any other metals strongly suggests that the His6 tag does not contribute to the measured zinc content in our studies. Therefore, the observation that mutant C136A retained activity, in the context of a heterodimer, strongly supports the view that only one zinc ion is necessary and sufficient for catalysis, although two zinc ions are tightly associated with the active wild type enzyme (Figs. 2 and 3).
TbADAT2 Forms Inactive Homodimers in Solution Despite coordinating Zn2+
To further assess the role of Zn2+ coordination in enzymatic activity, we also measured the Zn2+ content of TbADAT2 expressed in the absence of TbADAT3. We show that TbADAT2 could still bind 2 mol of Zn2+/mol of homodimer (Fig. 3C). This is similar to the Zn2+ content of TbADAT2/3 and likewise suggested a multimeric state for TbADAT2 expressed alone. To explore this idea, we subjected TbADAT2 to size exclusion chromatography. We found that TbADAT2 has an elution volume corresponding to that of a 54-kDa protein, consistent with a stable homodimer; however, no enzymatic activity was observed even after prolonged incubation (>24 h) with substrate (data not shown). Therefore, although binding the amount of Zn2+ typically found in dimeric deaminases (e.g. bacterial ADATa), TbADAT2 by itself could not catalyze adenosine deamination of its natural substrates (tRNAThr, tRNAVal, etc.), perhaps hinting at the acute functional interdependence of the two subunits in the heterodimer.
TbADAT2/3 Binds One tRNA per Heterodimer
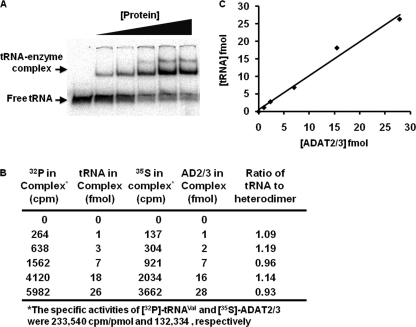
Other deaminases, notably ADATa, bind two tRNAs per homodimer, one per subunit, as shown in the co-crystal of the S. aureus enzyme bound to a tRNAArg stem loop (18). Our model for activity based on a single catalytic Zn2+, as seen in the ADAT2 C136A mutant, suggests that there is only one active site, despite the heterodimeric wild type enzyme coordinating two zinc ions. We therefore postulated that the heterodimer, differing from other deaminases, might bind only a single substrate per catalytic cycle. To determine the number of tRNAs bound to TbADAT2/3, 32P-labeled tRNAVal was incubated with increasing concentrations of [35S]Met/Cys-labeled TbADAT2/3; this double labeling permitted determination of an accurate stoichiometry in the RNP complex. Electrophoresis mobility shift assays showed that ∼1 pmol of TbADAT2/3 binds 1.06 pmol of tRNA (Fig. 4). Similar numbers were obtained when the protein concentration was kept constant, and the tRNA concentration was varied. This indicates that the heterodimer binds a single tRNA per catalytic cycle. We arrived at the same stoichiometry whether we used numbers obtained from one or both bands seen in the EMSA. This suggests that the two bands seen in our binding studies at higher protein concentrations likely represent either alternative multimeric states of the tRNA-protein complex or alternative conformations of the same complex. Currently, however, we are unable to distinguish between these possibilities.
FIGURE 4.
Wild type TbADAT2/3 binds one tRNA per heterodimer. A, 35S-labeled recombinant TbADAT2/3 was incubated with increasing concentrations of 32P-labeled tRNAVal; protein-tRNA complexes were separated from free tRNA via polyacrylamide gel electrophoresis. The first lane is a control reaction in which the RNA was incubated with buffer only. The arrows indicate where free tRNA versus TbADAT2/3-tRNA complex migrates in the gel. B, each band from A was individually excised from the dried gel, added to scintillation liquid, and counted for both 35S and 32P signal (expressed as cpm). Control lanes with tRNA or protein alone were counted and used to calculate the specific activity of each reactant (not shown). Counts per minute of 35S-TbADAT2/3 and 32P-tRNAVal per band are shown. Counts per minute values were converted to fentomoles using the calculated specific activity of the tRNA or protein as labeled. The last column displays the ratio of tRNA per enzyme (heterodimer). C, the data from B was used to plot the concentration of tRNA versus the protein concentration in each protein-tRNA complex. This plot yields a slope of 1 for the best fitting straight line consistent with one tRNA bound per heterodimer.
We also tested recombinant TbADAT2 alone in similar binding studies. This protein, however, showed severe defects in substrate binding (supplemental Fig. S2) partly explaining its lack of enzymatic activity.
DISCUSSION
Common to all polynucleotide deaminases is the use of a hydrolytic mechanism mediated by zinc found in their active sites (30). This co-factor is coordinated by the evolutionarily conserved motif (H/C)…CXXC, which constitutes a key signature sequence for members of the Cytidine Deaminase (CDA) superfamily. In addition, a majority of nucleotide and editing deaminases exist in cells as functional multimers, generally homodimers or homotetramers (30, 31), formed by identical subunits. Regardless of their multimeric state, one Zn2+ ion is bound per subunit, and it has been suggested that in many cases each active site acts independently. However, given that each subunit is identical in the multimeric state, it has been difficult to assess what the general contribution of each active site is to enzyme activity and specificity. This question is especially important with polynucleotide deaminases in that the system must ensure that only the intended nucleotide target is deaminated; otherwise rampant deamination may lead to the creation of defective products that could have serious consequences to the health and viability of cells.
Unique among polynucleotide deaminases are the eukaryotic ADAT2/3 enzymes composed of two different subunits that function as heterodimers. In these enzymes, each subunit still coordinates a Zn2+ but as described earlier, the larger subunit (ADAT3) is missing the key catalytic glutamate and contains numerous additional amino acid substitutions, making it distinct from the second subunit (ADAT2) in size and sequence. We took advantage of this naturally occurring asymmetry to assess the relative contribution of each subunit to catalysis. We found that a cysteine to alanine substitution (C136A) in one of the key zinc-coordinating residues of TbADAT2 still maintained a substantial amount of activity. This is surprising given that ADAT2, which contains the full set of residues needed for catalysis, was proposed to be the catalytic subunit of the enzyme. Two different possibilities may explain these results: 1) when Cys136 is replaced by alanine, TbADAT2 can still coordinate a Zn2+ via vicinal cysteines or histidines, used as an alternative to the highly conserved zinc-coordinating His90…136CXXC139 triad; this then yields an enzyme that remains active by virtue of still having Zn2+ ions bound in its active site; or 2) a single Zn2+ is necessary and sufficient for activity. The second possibility is strongly supported by the observation, obtained from ICP, that the catalytically active C136A mutant only has one bound Zn2+/heterodimer. Similar experiments performed with nucleotide deaminases led to inactive or impaired enzyme activity, but their activity could be rescued by Zn2+ addition, indicating that despite losing Zn2+ because of the substitution(s), the enzymes maintain a preformed active site (27). The addition of increasing concentrations of ZnCl2 to the TbADAT2/3 C136A mutant not only failed to rescue activity but also at higher concentrations became inhibitory to both the mutant and the wild type enzyme (used as control). Significantly, alanine substitution of the equivalent cysteine (Cys291) in TbADAT3 generated an inactive enzyme that again could not be rescued by zinc addition.
Studies with ScADAT2/3 showed that ADAT2 by itself is inactive (5); the same is true of TbADAT2. In addition we show that TbADAT2 can form stable homodimers in solution and that the TbADAT2 homodimer binds 2 mol of Zn2+/mol of dimer (or 1 Zn2+/subunit) analogous to the ADATa enzyme. This suggests that for TbADAT2 to be active, it still requires the presence of TbADAT3 and that activity is not a simple matter of whether or not the protein can bind Zn2+. Interestingly, the TbADAT2 homodimer could bind tRNA, although poorly, with a dissociation constant (Kd) in the micromolar range, suggesting that its inability to catalyze the reaction is partly due to a substrate binding defect, but beyond that it is not clear whether it is not also due to improper Zn2+ alignment in its core, whose geometry can be of crucial importance for deaminase activity (27). These observations indicate that the active site local structure is very sensitive to amino acid substitutions, but importantly the fact that a single Zn2+ is sufficient for activity supports a model by which TbADAT3 contributes to catalysis, and it is not simply a passive player in the reaction. A corollary of these observations is the fact that TbADAT2/3 contains a single active site and therefore must bind one substrate per heterodimer. Indeed, we show in our binding studies that the stoichiometry of enzyme to substrate was near 1:1, clearly supporting our proposal.
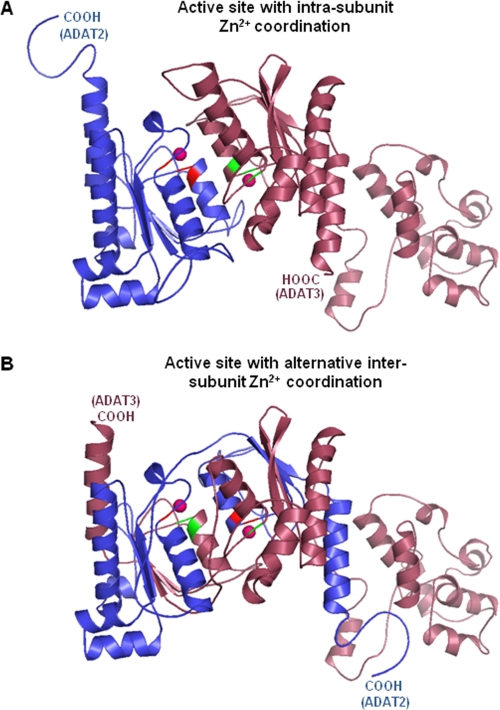
How then could the Zn2+ be coordinated in the C136A mutant? Again there are two viable possibilities. One possibility would be that Zn2+ coordination is still standard, as in the case of bacterial ADATa (13–15, 18), where each subunit binds one Zn2+ (Fig. 5A and supplemental Fig. S3A). In this model, because of the lack of the key catalytic glutamate in TbADAT3, TbADAT2 is the only catalytic subunit. A pitfall of this model is the fact that it does not explain how mutations that impair zinc coordination by TbADAT2 can still produce an active enzyme. We, however, suggest an alternative intersubunit zinc coordination model by TbADAT2/3 (Fig. 5B and supplemental Fig. S3B). This model is strongly supported by our observation that a single Zn2+ ion is necessary and sufficient for activity while still in the context of a heterodimer. Three-dimensional structure modeling provides us with an atomic model for how this intersubunit zinc coordination is possible. Two different three-dimensional models were generated for the TbADAT2/3 heterodimer catalytic domains based on the ADATa (TadA) homodimer using the FRankenstein's monster method (22) (Fig. 5A and supplemental Fig. S3A). An alternative “swapped” model, built by exchanging parts of ADAT2 and ADAT3 containing the HXE and CXXC motifs to form hybrid active sites, supports, at the very least, the possibility that ADAT3 contributes to catalysis via active site zinc coordination (Fig. 5B and supplemental Fig. S3B). To highlight the similarity (likelihood of the swapped model), the two models in Fig. 5 were superposed (supplemental Fig. S3C), showing negligible differences in the two predicted structures and nearly identical active sites (supplemental Fig. S3D).
FIGURE 5.
Structural model supports the possibility of alternative intersubunit zinc coordination in TbADAT2/3. Using the ADATa (TadA) homodimer (co-crystallized with tRNA anticodon loop) from S. aureus (Protein Data Bank code 2B3J) as a modeling template, two ADAT2/3 heterodimer models were built using the FRankenstein's monster method (22). A, this model represents the heterodimer most like the ADATa homodimer in that each subunit individually coordinates Zn2+ (intrasubunit coordination). B, this model represents an alternative “swapped” model, which was built by exchanging parts of ADAT2 and ADAT3 containing the HXE and CXXC motifs; this model supports the possibility of intersubunit zinc coordination. The models are colored by subunits: ADAT2 in dark blue and ADAT3 in dark pink. Active site residues (His, Cys, and Cys) of ADAT2 and ADAT3 are red and green, respectively. In both models, the C-terminal ends are as labeled, and the magenta spheres represent zinc ions.
Although both models are in line with the observed 1:1 stoichiometry for the wild type enzyme, the second model suggests that both subunits partake in catalysis and that ADAT3 is not simply a structural component as previously speculated. Clearly, the single Zn2+ that remains bound to the C136A mutant adopts the proper geometry and permits proper substrate positioning in the active site. This is in contrast to TbADAT2 alone, which remains inactive despite forming a homodimer and binding two Zn2+ ions. Admittedly, it is quite puzzling that an alanine substitution of the other TbADAT2 Zn2+ coordinating cysteine (C139A) yields an inactive enzyme. It is possible that this mutation, more so than C136A, affects not only zinc coordination but also local protein structure and perhaps substrate binding. Consistent with this view, alanine substitutions in either of the two cysteines (Cys136 or Cys139) of TbADAT2 leads to defects in tRNA binding. Analogous changes in TbADAT3 had negligible effects on substrate binding but deleterious effects on enzyme activity (35).
Taken together, the observations presented here have implications for the evolution of editing deaminases. ADAT2/3s from various organisms resemble cytidine deaminases in their primary sequence, despite catalyzing A to I editing reactions. Although it remains feasible that all polynucleotide deaminases evolved from an ancestral cytidine deaminase, it may not be necessarily true that all polynucleotide deaminases are derived via gene duplication of ancestral tRNA enzymes as has been proposed (5, 32). For example, it has been suggested that ADAT1 (adenosine deaminase for position 37 of eukaryotic tRNAs) evolved from acquisition of an additional motif (motif 3) (Fig. 1) by ADAT2. However, the observed sensitivity of TbADAT2 and TbADAT3 to seemingly modest amino acid substitutions may suggest otherwise. Acquisition of motif 3 would have required addition of an average of 60 amino acids between the two Zn2+-coordinating cysteines of ADAT2, which may pose a significant structural constraint to this motif acquisition yielding an active enzyme, if placed in the context of ADAT2. The structure of ADAR2 also provides a clue to the distinct evolutionary path of these enzymes. ADAR2 and ADAT1 share the common feature of requiring binding of inositol hexakisphospate for activity (33). This essential co-factor is bound to these enzymes by evolutionarily conserved residues, none of which are obvious in other deaminases, including ADAT2. Thus for ADAT1 to evolve from ADAT2, it would require an additional domain acquisition, making their evolutionary affinity even more distant. In fact, phylogenetic comparative analysis of nucleotide and polynucleotide deaminases from a variety of organisms, including single-cell protists (like trypanosomes), does not support an evolutionary model by which mRNA adenosine deaminases arise from ancestral tRNA anticodon deaminases (34). Given the data presented here, the active site structure of TbADAT2/3 is predictably different from other deaminases in the way it may coordinate its catalytic Zn2+; whether or not this is true of all ADAT2/3 deaminases will remain an open question.
In terms of specificity, what could the eukaryotic enzymes possibly gain by incorporating TbADAT3 and adopting a heterodimeric structure? Although a homodimer may confer certain functional advantages including cooperativity in substrate recognition, a heterodimer affords a rapid route for functional diversification. Because the heterodimeric eukaryotic deaminases display broader substrate specificity compared with their homodimeric bacterial homologs, it is conceivable that versatility in substrate recognition is the result of gene duplication and divergence of an ancestral ADAT to encode the two subunits in the heterodimeric ADAT2/3. Although it is premature to delineate the exact functional interdependence between these two subunits, it is evident that fully realizing and preserving the functional gains afforded by each subunit will require obligatory cross-talk between the two. The gradual acquisition of new functions (as in ADAT2/3) through modular additions is not unexpected. However, the likely repositioning of a catalytic metal ion, from independently binding each primordial module, to now bridge the two modules is an elegant solution to generate an efficient and high fidelity enzyme. This intersubunit metal coordination arrangement may best exploit the substrate binding and catalytic properties of the two modules while still enabling the evolution of multi-substrate specificity.
Supplementary Material
Acknowledgments
We thank all members of the Alfonzo laboratory for helpful comments and discussions. We also thank Drs. Jane Jackman, Venkat Gopalan, Silvo Conticello, and Harold Smith for helpful suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM084065 (to J. D. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- ADAT
- adenosine deaminases acting on tRNA
- ADAR
- adenosine deaminases acting on RNA
- ICP
- inductively coupled plasma.
REFERENCES
- 1. Agris P. F., Vendeix F. A., Graham W. D. (2007) J. Mol. Biol. 366, 1–13 [DOI] [PubMed] [Google Scholar]
- 2. Crick F. H. (1966) J. Mol. Biol. 19, 548–555 [DOI] [PubMed] [Google Scholar]
- 3. Murphy F. V., 4th, Ramakrishnan V. (2004) Nat. Struct. Mol. Biol. 11, 1251–1252 [DOI] [PubMed] [Google Scholar]
- 4. Wolf J., Gerber A. P., Keller W. (2002) EMBO J. 21, 3841–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gerber A. P., Keller W. (1999) Science 286, 1146–1149 [DOI] [PubMed] [Google Scholar]
- 6. Rubio M. A., Pastar I., Gaston K. W., Ragone F. L., Janzen C. J., Cross G. A., Papavasiliou F. N., Alfonzo J. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7821–7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gerber A., Grosjean H., Melcher T., Keller W. (1998) EMBO J. 17, 4780–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grosjean H., Auxilien S., Constantinesco F., Simon C., Corda Y., Becker H. F., Foiret D., Morin A., Jin Y. X., Fournier M., Fourrey J. L. (1996) Biochimie 78, 488–501 [DOI] [PubMed] [Google Scholar]
- 9. Habig J. W., Dale T., Bass B. L. (2007) Mol. Cell 25, 792–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A. G., Nishikura K. (2007) Science 315, 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hurst S. R., Hough R. F., Aruscavage P. J., Bass B. L. (1995) RNA 1, 1051–1060 [PMC free article] [PubMed] [Google Scholar]
- 12. Gray M. W. (1976) Nucleic Acids Res. 3, 977–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elias Y., Huang R. H. (2005) Biochemistry 44, 12057–12065 [DOI] [PubMed] [Google Scholar]
- 14. Kim J., Malashkevich V., Roday S., Lisbin M., Schramm V. L., Almo S. C. (2006) Biochemistry 45, 6407–6416 [DOI] [PubMed] [Google Scholar]
- 15. Kuratani M., Ishii R., Bessho Y., Fukunaga R., Sengoku T., Shirouzu M., Sekine S., Yokoyama S. (2005) J. Biol. Chem. 280, 16002–16008 [DOI] [PubMed] [Google Scholar]
- 16. Karcher D., Bock R. (2009) RNA 15, 1251–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Delannoy E., Le Ret M., Faivre-Nitschke E., Estavillo G. M., Bergdoll M., Taylor N. L., Pogson B. J., Small I., Imbault P., Gualberto J. M. (2009) Plant Cell 21, 2058–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Losey H. C., Ruthenburg A. J., Verdine G. L. (2006) Nat. Struct. Mol. Biol. 13, 153–159 [DOI] [PubMed] [Google Scholar]
- 19. Auxilien S., Crain P. F., Trewyn R. W., Grosjean H. (1996) J. Mol. Biol. 262, 437–458 [DOI] [PubMed] [Google Scholar]
- 20. Moore M. J., Sharp P. A. (1992) Science 256, 992–997 [DOI] [PubMed] [Google Scholar]
- 21. Kurowski M. A., Bujnicki J. M. (2003) Nucleic Acids Res. 31, 3305–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kosinski J., Cymerman I. A., Feder M., Kurowski M. A., Sasin J. M., Bujnicki J. M. (2003) Proteins 53, (Suppl. 6) 369–379 [DOI] [PubMed] [Google Scholar]
- 23. Simons K. T., Kooperberg C., Huang E., Baker D. (1997) J. Mol. Biol. 268, 209–225 [DOI] [PubMed] [Google Scholar]
- 24. Armon A., Graur D., Ben-Tal N. (2001) J. Mol. Biol. 307, 447–463 [DOI] [PubMed] [Google Scholar]
- 25. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xie K., Sowden M. P., Dance G. S., Torelli A. T., Smith H. C., Wedekind J. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8114–8119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith A. A., Carlow D. C., Wolfenden R., Short S. A. (1994) Biochemistry 33, 6468–6474 [DOI] [PubMed] [Google Scholar]
- 28. Fassel V. A. (1978) Science 202, 183–191 [DOI] [PubMed] [Google Scholar]
- 29. Betts L., Xiang S., Short S. A., Wolfenden R., Carter C. W., Jr. (1994) J. Mol. Biol. 235, 635–656 [DOI] [PubMed] [Google Scholar]
- 30. MacElrevey C., Wedekind J. E. (2008) in RNA and DNA Editing (Smith H. C. ed) pp. 369–419, Wiley and Sons, Hoboken, NJ [Google Scholar]
- 31. Salter J. D., Krucinska J., Raina J., Smith H. C., Wedekind J. E. (2009) Biochemistry 48, 10685–10687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schaub M., Keller W. (2002) Biochimie 84, 791–803 [DOI] [PubMed] [Google Scholar]
- 33. Macbeth M. R., Schubert H. L., Vandemark A. P., Lingam A. T., Hill C. P., Bass B. L. (2005) Science 309, 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Conticello S. G., Thomas C. J., Petersen-Mahrt S. K., Neuberger M. S. (2005) Mol. Biol. Evol. 22, 367–377 [DOI] [PubMed] [Google Scholar]
- 35. Ragone F. L., Spears J. L., Alfonzo J. D. (2011) RNA, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.