Abstract
Plant pathogenic fungi use a wide range of different strategies to gain access to the carbon sources of their host plants. The hemibiotrophic maize pathogen Colletotrichum graminicola (teleomorph Glomerella graminicola) colonizes its host plants, and, after a short biotrophic phase, switches to destructive, necrotrophic development. Here we present the identification of five hexose transporter genes from C. graminicola, CgHXT1 to CgHXT5, the functional characterization of the encoded proteins, and detailed expression studies for these genes during vegetative and pathogenic development. Whereas CgHXT4 is expressed under all conditions analyzed, transcript abundances of CgHXT1 and CgHXT3 are transiently up-regulated during the biotrophic phase, and CgHXT2 and CgHXT5 are expressed exclusively during necrotrophic development. Analyses of the encoded proteins characterized CgHXT5 as a low-affinity/high-capacity hexose transporter with a narrow substrate specificity for glucose and mannose. In contrast, CgHXT1 to CgHXT3 are high affinity/low capacity transporters that also accept other substrates, including fructose, galactose, or xylose. CgHXT4, the largest of the identified proteins, has only little transport activity and may function as a sugar sensor. Phylogenetic studies revealed hexose transporters closely related to the five CgHXT proteins also in other pathogenic fungi suggesting conserved functions of these proteins during fungal pathogenesis.
Keywords: Fungi, Glucose Transport, Membrane Proteins, Plasma Membrane, Sugar Transport, Colletotrichum graminicola, hemibiotrophic Fungus
Introduction
The ascomycete Colletotrichum graminicola (Cesati) Wilson [teleomorph Glomerella graminicola (Politis)] is the causal agent of the worldwide occurring stem rot and leaf anthracnose of maize (Zea mays). Its hemibiotrophic lifestyle is characterized by an enduring destructive (necrotrophic) development that follows a short non-destructive (biotrophic) period of primary invasion and colonization (1, 2). The primary hyphae formed during this initial biotrophic phase invaginate but do not breach the plasma membrane of the host, as C. graminicola still depends on living host cells for an efficient transfer of nutrients (3). During the necrotrophic phase, however, which is initiated 48 to 72 h postinoculation, fast growing thin secondary hyphae breach the plant plasma membrane, kill the cell, and start to ramify within the host tissue. At the same time, secreted enzymes (4) degrade plant extracellular polysaccharides releasing a variety of mono-, di-, and oligosaccharides that are immediately accessibly for the fungus. Obviously, the fungus will need to respond to the changing carbon environment during these infection stages, to establish different transport systems in its plasma membrane, and to adjust the uptake of organic carbon sources to the changing concentrations and composition of its environment.
So far, nothing is known about the carbon sources used by C. graminicola or other hemibiotrophic fungi at these different infection stages or about the transport proteins used for carbon acquisition in infected plant material. However, over the last years plasma membrane-localized carbohydrate transporters were identified from the biotrophic basidiomycetes Uromyces fabae (5) and Ustilago maydis (6), the symbiotic, mycorrhiza-forming glomeromycete Geosiphon pyriformis (7), and from the root-colonizing ascomycete Metarhizium robertsii that is beneficial for plant health as it antagonizes pathogens and herbivores (8).
Whereas the transporters used in these four fungi belong to the major facilitator superfamily (9) and within this superfamily to the sugar transporter family, the nature of the transported substrates differed. The proteins identified in U. fabae (UfHXT1) and G. pyriformis (GpMST1) were characterized as classical monosaccharide transporters that reconstituted the growth defect of hexose uptake-deficient baker's yeast strains on glucose and mannose (7) and/or reconstituted the lost transport activities in baker's yeast (5, 7). In contrast, the U. maydis transporter UmSRT1 is absolutely specific for sucrose. The UmSRT1 gene is expressed only in fungal hyphae growing in planta. Cells growing in axenic cultures do never express UmSRT1 irrespective of the available carbon source. Most importantly, U. maydis mutants with a deletion in the UmSRT1 gene (Δsrt1) showed strongly reduced virulence on maize plants (6). Interestingly, virulence was restored by expressing the cDNA of the Arabidopsis thaliana SUC9 sucrose transporter (10) from the SRT1 promoter.
The gene identified in M. robertsii encodes an oligosaccharide transporter (MRT; 8). The MRT gene is expressed exclusively by M. robertsii germlings in the vicinity of grass roots, and disruption of the gene resulted in reduced growth on sucrose and galactosides such as raffinose, stachyose, or verbascose. Disrupting the gene did not alter the virulence to insects suggesting that MRT is involved only in the uptake of plant-derived carbohydrates. In fact, compared with the MRT wild-type strain, a Δmrt mutant of M. robertsii showed reduced growth in the rhizosphere of grasses (8).
The observed differences in the substrate specificities of the identified transporters reflect alternative strategies of symbiotic fungi (8) that may either hydrolyze extracellular plant-derived sucrose and feed on the resulting monosaccharides (Glomeromycota) or use plant-derived oligosaccharides without extracellular hydrolysis (Ascomycota). Direct uptake of sucrose by the UmSRT1 protein of the biotrophic pathogen U. maydis was interpreted as a mechanism to avoid apoplastic signals (glucose) potentially recognized by the host (6). In fact, plants use individual molecular patterns to recognize and discriminate pathogens, leading to activation of different defense strategies against biotrophic and necrotrophic fungi (11, 12). For biotrophic fungi direct uptake of sucrose may help to avoid elicitation of plant responses. After infection of maize leaves with C. graminicola (13), however, transcript abundance of incw1, which encodes a cell wall-localized invertase of maize, was significantly increased suggesting that composition and concentration of apoplastic sugars vary between different pathosystems and during different phases of the infection process.
Here we present the analyses of five transporters of the hemibiotrophic fungus C. graminicola and discuss their potential roles in the biotrophic or necrotrophic growth phase of this fungus. We determined the expression levels of the corresponding genes during these two growth phases, studied potential regulatory effects of glucose and leaf extracts, and characterized the proteins after expression of the corresponding cDNAs in baker's yeast. The presented data show that one of the transporter gens, CgHXT4, shows comparable expression levels under all growth conditions and may act as sugar sensor or regulator. Expression of two genes, CgHXT1 and CgHXT3, is transiently up-regulated during the biotrophic phase, and CgHXT2 and CgHXT5 are expressed exclusively during the necrotrophic development of the fungus. Analyses of pH optima, sensitivities to uncouplers, Km values and substrate specificities were performed and allow a detailed characterization of the different proteins. Interestingly, one of the genes, CgHXT1, yields alternatively spliced mRNAs that encode two functionally active transporters.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
Escherichia coli strain DH5α was used for cloning purposes (14). The C. graminicola strain M2 was used in this study. Plant infection assays with C. graminicola were performed as described (15). Briefly, 2-week-old maize plants were inoculated with fungal spore suspensions (106 spores per plant) and incubated at appropriate conditions in the climate chamber. Axenic cultures of C. graminicola were grown in minimal medium containing either 1.5% (w/v) glucose or 1.5% glucose plus 1.5% (w/v) maize cell walls plus 1.5% (v/v) soluble maize leave extract, or 1.5% (w/v) maize cell walls plus 1.5% (v/v) soluble maize leave extract (4). The S. cerevisiae strains used for expression analyses of CgHXT cDNAs was EBY.VW4000 (16). Cells were grown in minimal medium (0.67% yeast nitrogen base w/o amino acids plus required amino acids) containing 2% of the indicated sugar or sugar alcohol at 29 °C.
Identification of the 5 CgHXT Genes
For the identification of the 5 CgHXT genes analyzed in this study, extensive BLAST analyses were performed with predicted hexose transporter sequences from several fungi, including Ustilago maydis, Neurospora crassa, Magnaporthe grisea, Gibberella zeae, Neosartorya fischeri, Chaetomium globosum, and others against “Colletotrichum graminicola - WGS” sequences provided via the “trace archives” of this WEB-site. Identified shotgun sequences (typical size: about 300 base pairs) were assembled to yield contiguous genomic sequences (contigs).
cDNA Cloning and Expression in Yeast
CgHXT cDNA sequences were amplified from total RNA of C. graminicola using the primers (Table 1) predicted to be specific for the 5′-ends or 3′-ends of the identified CgHXT genes. Obtained fragments were sequenced and cloned into the yeast/E. coli shuttle vector NEV-E (17), and the resulting plasmid was used for yeast transformation (18).
TABLE 1.
Primers used for the amplification of CgHXT1 to CgHXT5 cDNAs or for the quantification of mRNA abundance by semiquantitative or quantitative RT-PCR
| Gene | Primer name | Sequence |
|---|---|---|
| cDNA amplification: | ||
| CgHXT1 | CgHXT1(#13)fw | 5′-CTCTCTGAATTCCCCCTTCGTCATCATGGGTA-3′ |
| CgHXT1(#13)rev | 5′-GAGAGAGAATTCCACACGCAATAAACAG | |
| CAGACA-3′ | ||
| CgHXT2 | CgHXT2(#29)fw1 | 5′-CTCTCTGAATTCATGGGGTTGGGCAAACTGTA-3′ |
| CgHXT2(#29)rev | 5′-GAGAGAGAATTCCCCTGTCTCTTATACAA | |
| CATTCATACG-3′ | ||
| CgHXT3 | CgHXT3(#27)fw | 5′-CTCTCTGAATTCAAACCACAACAGCCCTCCAC-3′ |
| CgHXT3(#27)rev | 5′-GAGAGAGAATTCCGAGACGTCGGCTTAAACAT-3′ | |
| CgHXT4 | CgHXT4(III)fw | 5′-CTCTCTGAATTCCCCACAGTCCCATTCACCAT-3′ |
| CgHXT4(III)rev | 5′-GAGAGAGAATTCAGTGGCAATGGTAAC | |
| CAGCA-3′ | ||
| CgHXT4-pEX-GFPr | 5′-TCTCTGAATTCCATAGCTGAAGTCCACATTGCC-3′ | |
| CgHXT5 | CgHXT5(IV)fw | 5′-CTCTCTGAATTCGATACCCCCGTCAACAGC-3′ |
| CgHXT5rev#3 | 5′-GAGAGAGAATTCTTAGACGGTGTTTTCA | |
| GCCTTC-3′ | ||
| Quantification by RT-PCR: | ||
| CgACT | CgACT-qRT.F1 | 5′-TTCTACGAGCTTCCTGACGG-3′ |
| CgACT-qRT.R1 | 5′-CCGCTCTCAAGACCAAGGAC-3′ | |
| CgHXT1 | CgHXT1(#13)fw | 5′-ATCGCCTTCGTCTACTTCTTCAT-3′ |
| CgHXT1(#13)rev | 5′-CTAGGCAGAGGCGGTCTTCT-3′ | |
| CgHXT2 | UL13(fw) | 5′-TTGTCTCTGCCTGCATGTTC-3′ |
| UL14(rev) | 5′-AGTTGCGTAGGGAGTGAGGA-3′ | |
| CgHXT3 | UL19(fw) | 5′-ATTGTCGCTGGTCGTCTCAT-3′ |
| UL20(rev) | 5′-CAATGGTGATGCAGAACTGG-3′ | |
| CgHXT4 | UL1(fw) | 5′-ATTCTTCTTCGCAGCATC-3′ |
| UL2(rev) | 5′-AAGACCATGCACCACACAAA-3′ | |
| CgHXT5 | UL3(fw) | 5′-GACACCAGCATCTGGGAAAT-3′ |
| UL4(rev) | 5′-GGTGACGTAGCTGTTGCTGA-3′ | |
Transport Studies with Radiolabeled Substrates
Yeast cells were grown to an A600 nm of 1.0, harvested, washed twice with water and re-suspended in buffer to an A600 nm of 20.0. If not otherwise indicated, uptake experiments were performed in 50 mm sodium phosphate buffer pH 5.0 with an initial glucose concentration of 1 mm. Cells were shaken in a rotary shaker at 29 °C and transport tests were started by adding labeled substrate. Samples were withdrawn at given intervals, filtered on glass microfiber filters (1.5 μm pore size) and washed with an excess of distilled H2O. Incorporation of radioactivity was determined by scintillation counting. Competition analyses were performed with 0.1 mm (CgHXT1, CgHXT3, CgHXT5) or 1 mm [14C]glucose (CgHXT2) in the presence of 10 mm competitor (100-fold excess; CgHXT1, CgHXT2, CgHXT5) or 5 mm (50-fold excess; CgHXT3). For inhibitor analyses carbonylcyanide m-chlorophenylhydrazone (CCCP)2 was used at a final concentration of 50 μm.
Subcellular Localization of CgHXT4 in Baker's Yeast
A CgHXT4-GFP fusion was generated by amplification of the CgHXT4 open reading frame (ORF) with the primers CgHXT4(III)fw and CgHXT4-pEX-GFPr (Table 1) and by insertion of the resulting fragment upstream of the GFP ORF into the pEX-Tag plasmid (19). Subcellular localization of the CgHXT4-GFP fusion in transformed yeast cells (Gietz et al., 18) was determined by confocal microscopy (Leica TCS SPII; Leica Microsystems, Bensheim, Germany) as described (6).
RNA Isolation, Semiquantitative, and Quantitative Real-time PCR Analysis
From infected and uninfected maize plants 3rd leaves were harvested at 2, 3, and 4 dpi and immediately shock frozen in liquid nitrogen. Total RNA was isolated with the isolation kit peqGOLD Plant RNA from Peqlab (Erlangen, Germany) according to the manufacturer's instructions. Mycelium from axenic cultures was harvested and shock frozen in liquid nitrogen 1, 2, and 3 days after cultivation in minimal medium supplemented with different carbon sources as described above. Subsequent RNA isolation procedure was as described for infected maize plants.
For cDNA synthesis, 1 μg of total RNA was reverse transcribed with the QuantiTect Reverse Transcription kit from Qiagen (Hilden, Germany). Semiquantitative RT-PCR was performed with cDNA amounts adjusted to yield equal amounts of actin mRNA (CgACTIN). Oligonucleotide sequences used for semiquantitative or quantitative RT-PCRs are shown in Table 1. Quantitative real-time RT-PCRs were performed on a RotorGene 2000 (Corbett Research, Sydney, Australia) with QuantiTect_SYBR_Green PCR Master Mix from Qiagen. Samples were normalized to CgACTIN mRNA levels.
Phylogenetic Analysis
For the phylogenetic analysis of the C. graminicola transporters and of other transporters and sensors, protein sequences were aligned (ClustalW2, 20) using the following settings: Protein weight matrix, Gonnet; gap open, 10; gap extension, 0.2; gap distances, 5; clustering, neighbor joining; format, PHYLIP. The PhyML 3.0 software package (21) was used for tree calculation with the JTT model for amino acid substitutions and 100 bootstrap samplings. TREEVIEW X 0.4.1 for Mac (22) was used for tree viewing.
RESULTS
Basic Characterization of the Five CgHXT Sequences
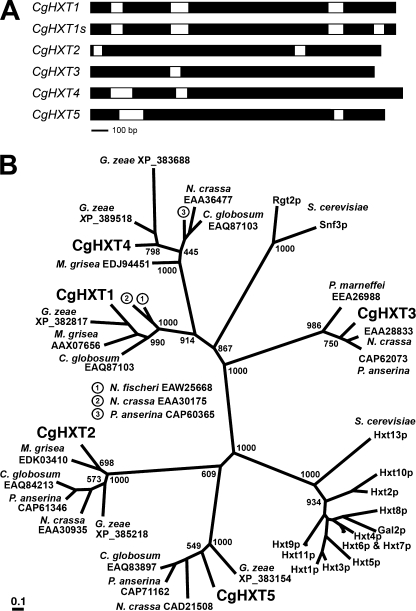
Blast searches with HXT and HXT-like cDNA sequences from several fungi (Gibberella zeae, Magnaporthe grisea, Neurospora crassa, Saccharomyces cerevisiae) in the NCBI trace archives (Colletotrichum, WGS) identified 5 genes encoding putative HXT proteins. The genes are interrupted by 1 to 4 introns (Fig. 1A) and were named CgHXT1 to CgHXT5. To confirm the predicted ORFs and the intron/exon organization, cDNAs were amplified with primers specific for the predicted 5′- and 3′-ends of the coding region and sequenced (EMBL accession numbers: FN433101 to FN433105). The protein sequences deduced from these 5 ORFs had 12 predicted transmembrane helices and belong to the major facilitator superfamily (9). The proteins shared between 36.9% (CgHXT1 and CgHXT5) and 49.8% (CgHXT1 and CgHXT4) identical amino acids. The residue numbers, deduced molecular masses and isoelectric points of the proteins are shown in Table 2.
FIGURE 1.
Phylogenetic analyses and genomic organization. A, intron/exon organization in the CgHXT1 to CgHXT5 genes. The two alternative genomic organizations shown for CgHXT1 and CgHXT1s indicate two alternative splice variants. B, phylogenetic tree (maximum likelihood) of CgHXT1 to CgHXT5 protein sequences and of characterized and predicted hexose transporters and sensors from other fungi. Abbreviations of fungus names and GenBankTM accessions or protein names are given. Bootstrap values of 1000 samplings are given at central branches.
TABLE 2.
Gene names, number of amino acids, calculated molecular masses, and isoelectric points (IP) of the proteins analyzed in the present study
| Gene | Amino acids | Mol. mass | IP |
|---|---|---|---|
| CgHXT1 | 531 | 57.93 | 7.96 |
| CgHXT1s | 508 | 55.40 | 7.67 |
| CgHXT2 | 551 | 60.32 | 7.01 |
| CgHXT3 | 559 | 61.05 | 7.25 |
| CgHXT4 | 569 | 61.63 | 5.35 |
| CgHXT5 | 532 | 57.83 | 6.83 |
Reproducibly, but with much lower frequency, a second ORF was obtained for the CgHXT1 gene. In contrast to the regular ORF (CgHXT1; EMBL accession number: FN433101; encodes a protein of 531 amino acids), this second ORF encodes a shorter protein of only 508 amino acids and was named CgHXT1s (s for short; EMBL accession number: FN433106). The size difference is due to a 23-amino acid deletion corresponding to residues 488–510 in the cytoplasmic C terminus of the longer protein. This deletion results from an additional intron that is removed during the formation of the CgHXT1s but not of the CgHXT1 mRNA (Fig. 1A). This additional intron is 69 bp long, and its removal does not alter the reading frame of the downstream sequence.
Phylogenetic calculations (maximum likelihood; 1000 bootstrap replications) were performed with CgHXT1 to CgHXT5 (CgHXT1s was not treated as an extra protein in these calculations), with 23 functionally uncharacterized proteins from other phytopathogenic (Gibberella zeae, 23; Magnaporthe grisea, 24), endophytic (Chaetomium globosum; 25), animal-pathogenic (Penicillium marneffei; 26) or saprophytic (Neurospora crassa, 27; Neosartorya fischeri, 28; Podospora anserina, 29) fungi, with 13 functionally characterized HXT-type transporters (Hxt1p to Hxt11p, Hxt13p and Gal2p; 30) from Saccharomyces cerevisiae (baker's yeast), and with the functionally characterized Snf3p and Rgt2p glucose sensors from baker's yeast (31, 32). Fig. 1B demonstrates that the 5 CgHXT proteins form separate clusters together with HXT-type proteins from other phytopathogenic, animal pathogenic, and saprophytic fungi. Interestingly, closely related homologs for CgHXT1, CgHXT2, CgHXT4 and CgHXT5 were found in G. zeae, for CgHXT1, CgHXT2 and CgHXT4 in M. grisea, and for CgHXT2 to CgHXT5 in P. anserina. Moreover, Fig. 1B shows that the 5 CgHXT clusters do not include any of the well-characterized transporters or sensors from baker's yeast.
Growth of CgHXT-expressing Baker's Yeast on Different Carbon Sources and Functional Analyses of CgHXT1, CgHXT2, CgHXT3, and CgHXT5 in Transgenic Yeast Cells
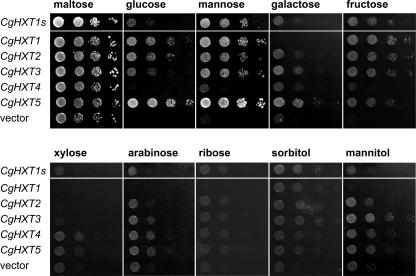
For functional analyses we cloned the CgHXT1 to CgHXT5 ORFs and the ORF of the CgHXT1s splice variant into the unique EcoRI cloning site of the yeast expression vector NEV-E (17). The resulting constructs NEV-CgHXT1 to NEV-CgHXT5 and NEV-CgHXT1s were used to transform yeast strain EBY.VW4000, which lacks 20 genes for transporters with hexose transport activity, but can grow on minimal medium with maltose as sole carbon source (16). Transformants were grown in liquid medium (maltose) under selective conditions, harvested during logarithmic growth, washed, adjusted to the same cell density and spotted onto agar plates containing minimal medium with different potential carbon sources (all at a final concentration of 2% (w/v)).
Fig. 2 demonstrates that the transformants as well as a control strain with the empty NEV-E vector grew equally well on minimal medium with maltose. As expected, growth of the vector control was not detected or marginal on plates supplemented with glucose, mannose, galactose, or fructose. In contrast, yeast transformants harboring CgHXT1, CgHXT2, CgHXT3, CgHXT5, and CgHXT1s showed good to excellent growth on these different hexoses. On media supplemented with glucose or mannose, CgHXT5-transformed cells grew significantly better than all other transformants. Interestingly, CgHXT1-expressing cells but not CgHXT1s-expressing cells failed to grow on galactose.
FIGURE 2.
Comparative growth analyses of yeast cells expressing one of the five CgHXT cDNAs. Decreasing numbers (left to right) of yeast cells (strain EBY.VW4000) expressing the indicated CgHXT cDNA or harboring the empty expression vector were spotted on agar medium and incubated for 68 h at 29 °C on plates containing the indicated carbon source at a final concentration of 2%.
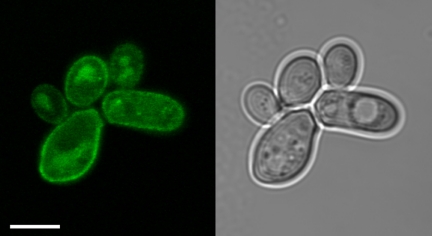
In contrast to all other transformants, CgHXT4-expressing cells were unable to grow on glucose, and also their growth on galactose, mannose, and fructose was only slightly better than that of control cells (Fig. 2). To test, the possibility that CgHXT4 is not targeted to the yeast plasma membrane, we generated a CgHXT4-GFP fusion construct and determined the subcellular localization of the corresponding protein in yeast cells. Fig. 3 shows that CgHXT4-GFP labels the plasma membrane of CgHXT4-GFP-expressing cells demonstrating that the poor complementation of the growth defect cannot be explained by incorrect targeting of the protein.
FIGURE 3.
Subcellular localization of CgHXT4 in baker's yeast. Subcellular localization of CgHXT4-GFP in baker's yeast was determined by confocal microscopy. A single optical section (left) and the corresponding white light image (right) are shown. CgHXT4-GFP fluorescence is observed at the cell surface, putatively the plasma membrane. Scale bar, 5 μm.
We also analyzed the capacity of the different transformants to utilize pentoses, such as xylose, arabinose, and ribose, or hexitols, such as sorbitol and mannitol. On all of these carbon sources the vector control showed some growth and the additional growth-promoting effects of the different CgHXTs were marginal, although minor differences could be observed. In summary, these growth analyses suggested that CgHXT1 to CgHXT5 and CgHXT1s transport preferentially hexoses, although with different preferences for the hexoses offered.
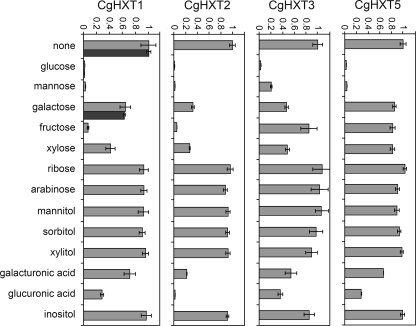
To support these results, we performed quantitative uptake studies with radiolabeled sugars. With none of the radiolabeled substrates tested we could detect measurable transport rates for CgHXT4-expressing cells. With all other transformants, however, we observed significant transport rates for [14C]glucose. Therefore, we studied the uptake of [14C]glucose in the absence or presence of different potential transport competitors (Fig. 4). Clearly and as expected, a 100-fold (CgHXT1, CgHXT2, CgHXT5) or a 50-fold (CgHXT3) excess of unlabeled glucose completely outcompeted the transport of radiolabeled glucose. For CgHXT1, CgHXT2, and CgHXT5 the same almost complete inhibition was observed with a 100-fold excess of mannose, and also for CgHXT3 mannose was the second best competitor, although inhibition was not as efficient as for the other transporters. In addition, fructose was an excellent competitor of glucose transport by CgHXT1 and CgHXT2, and glucuronic acid and to a lesser extent also galacturonic acid were competitors of glucose transport by CgHXT2. This suggested that of the four transporters analyzed, CgHXT2 has the lowest substrate specificity. Accordingly, CgHXT2 also showed the strongest competition by xylose. In contrast, CgHXT5 exhibited the highest substrate specificity, and its strong preference for glucose and mannose (Fig. 4) was reflected by the significantly better growth on these two carbon sources than on all others (Fig. 2). With CgHXT1s, the short form of CgHXT1, we performed competition analyses only with glucose and galactose, which gave the same results as for CgHXT1 (Fig. 4).
FIGURE 4.
Substrate specificities of the indicated CgHXT transporters. Substrate specificities of CgHXT1, CgHXT2, CgHXT3, and CgHXT5 were determined in yeast cells (strain EBY.VW4000) expressing the respective cDNA. Dark bars added to the CgHXT1 analyses (none, glucose, galactose) show results obtained with the shorter CgHXT1s protein. Relative transport rates of radiolabeled glucose (initial concentration, 0.1 mm for CgHXT1, CgHXT1s, CgHXT3, and CgHXT5; 1 mm for CgHXT2) were determined in the absence (none) or presence of the indicated competitor (competitor concentration was 10 mm; n = 3; ± S.E.).
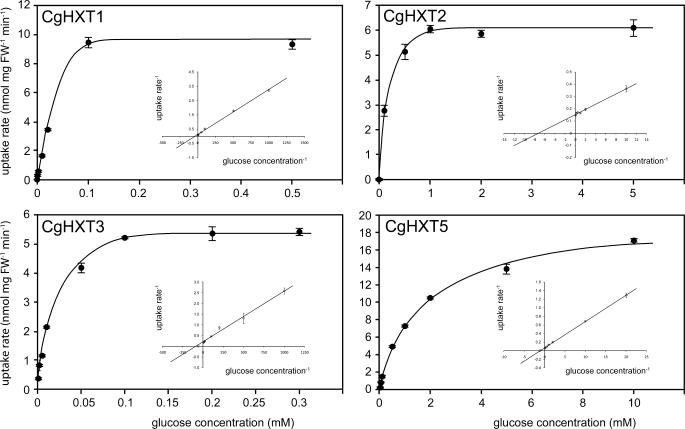
Major differences between the different transporters were observed, when we determined their affinities for glucose (Fig. 5). For CgHXT1 and CgHXT3 we observed very high affinities with Km values of only 20.6 ± 0.3 μm and 13 ± 2 μm, respectively. With a Km value of 136 ± 20 μm, the glucose affinity of CgHXT2 was significantly lower, and with a Km value of 920 ± 120 μm, CgHXT5 can be characterized as a low affinity transporter. This low affinity of CgHXT5 agrees well with the excellent growth of CgHXT5-expressing yeast cells on glucose or mannose containing media, as a correlation of low substrate affinity (LA) and high transport capacity (HC) has been shown for different transport systems (33, 34, 35). In summary, the data of Figs. 2, 4, and 5 suggest that CgHXT5 is a low-affinity/high capacity (LAHC) transporter, which may allow rapid growth at sufficiently high substrate concentrations.
FIGURE 5.
Km values for glucose of the indicated CgHXT proteins. Uptake rates for [14C]glucose yeast cells (strain EBY.VW4000) expressing the respective cDNA were determined at the indicated substrate concentrations at pH 5.0. Insets show double reciprocal (Lineweaver-Burk) plots of the same data (n = 3; ± S.E.).
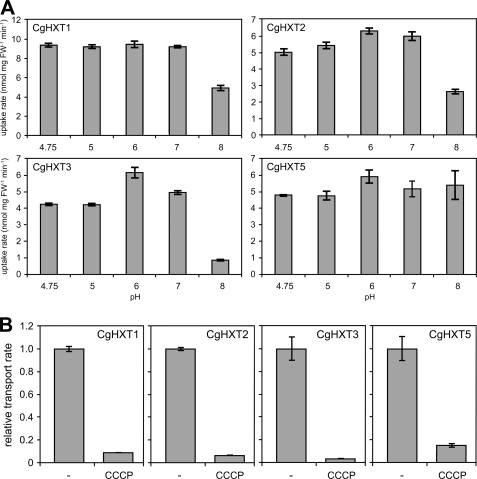
We also studied the pH dependence of CgHXT1, CgHXT2, CgHXT3, and CgHXT5 and the sensitivity of each transporter to cyanide-m-chlorophenylhydrazone (CCCP), an uncoupler of transmembrane proton gradients. Fig. 6A demonstrates that CgHXT1, CgHXT2, and CgHXT3 have their pH optima at neutral and acidic pH-values. At pH 8, the transport capacities of these transporters are clearly reduced. In contrast, CgHXT5 seems to be rather unaffected by pH changes between 4.75 and 8.0. Nevertheless, the transport analyses with or without 50 μm CCCP clearly demonstrate that the activities of all 4 transporters are strongly reduced already at low uncoupler concentrations suggesting that CgHXT1, CgHXT2, CgHXT3, and CgHXT5 might act as energy-dependent H+/hexose symporters. The invariantly high transport rates of CgHXT5 at pH 8, and the strong but compared with the other transporters less pronounced sensitivity of this protein to CCCP may point toward a minor role of the ΔpH for the driving force for this transporter and a more important role of the electrical potential Δψ.
FIGURE 6.
pH dependence and CCCP sensitivities of CgHXT activities. A, transport rates for 14C-labeled glucose were determined at the indicated pH-values in 50 mm sodium phosphate buffer at initial glucose concentrations of 1 mm for CgHXT1, CgHXT2, CgHXT5 or 0.1 mm for CgHXT3 (n = 3; ± S.E.). B, sensitivities to the uncoupler CCCP were studied at pH 5.0 in the absence or presence of 50 μm CCCP (n = 3; ± S.E.).
Expression of the Different CgHXT Genes in C. graminicola
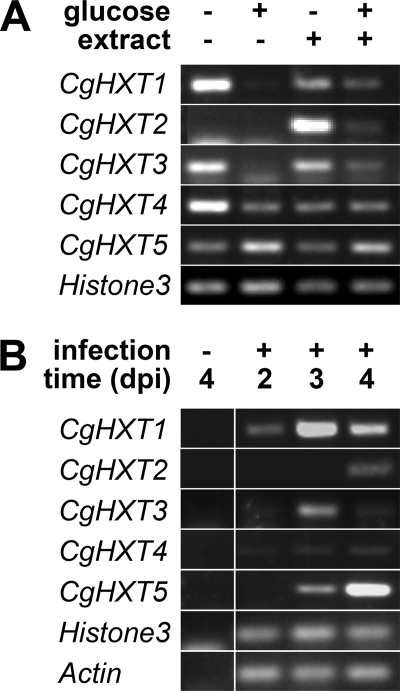
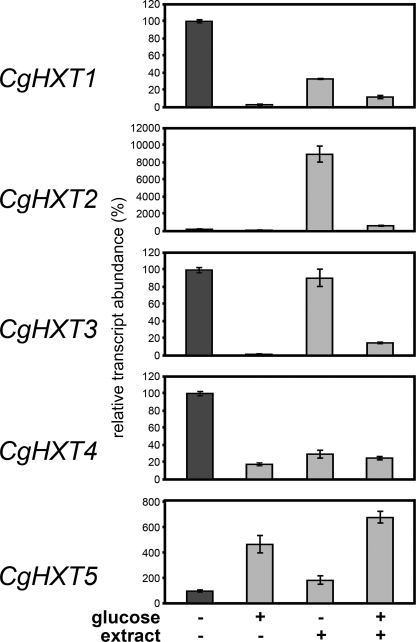
To study the expression of CgHXT1 to CgHXT5 in C. graminicola and to identify factors potentially involved in the regulation of this expression, we determined the respective mRNA abundance in C. graminicola growing on defined substrates in vitro (Figs. 7A and 8) or within infected maize leaves at different days post inoculation (dpi) (Figs. 7B and 9). The semiquantitative and quantitative analyses gave essentially the same results. Figs. 7A and 8 show analyses of CgHXT1 to CgHXT5 mRNA levels in C. graminicola cells incubated in medium that was supplemented with 1.5% glucose, with extracts from uninfected leaves and maize cell walls, or with a combination of both (4). The amounts of cDNA used for the semiquantitative (Fig. 7A) and quantitative RT-PCRs (Fig. 8) were normalized to yield similar amounts of Histone3 mRNA. Clearly, the glucose concentrations used suppressed or reduced the abundance of CgHXT1, CgHXT3, and CgHXT4 transcripts in the absence or presence of maize leaf extract. Interestingly, addition of leaf extract strongly up-regulated the transcript levels of CgHXT2 (about 900-fold as calculated from Fig. 8), the only gene that was neither expressed in the absence nor in the presence of glucose. Leaf extracts had a similar effect as glucose on CgHXT4 and caused a reduction in the mRNA levels of this gene. In contrast to the other transporter genes, CgHXT5 expression was up-regulated by glucose, and addition of maize-leaf extract did not affect this induction (Fig. 7).
FIGURE 7.
Semiquantitative RT-PCR analyses of CgHXT1 to CgHXT5 expression levels. A, expression levels of the Histone3 gene under the same conditions is shown as control. RT-PCRs were performed on total RNA isolated from axenically grown C. graminicola grown for 3 days at 23 °C in the presence of the indicated additions (glucose, 1.5%; extract, maize cell walls plus soluble extract from maize leaves as described under “Experimental Procedures.” One of three biological repeats is shown. Transcript levels of the histone H3 gene under the same conditions is shown as control. B, transcript abundances of CgHXT1 to CgHXT5 were studied in C. graminicola-infected maize leaves at 2, 3, or 4 dpi. mRNA levels of the C. graminicola actin and histone H3 genes under the same conditions are shown as controls. RT-PCRs were performed on total RNA isolated from uninfected or infected maize leaves (maize plus fungal RNA) at the indicated times (one of three biological repeats is shown).
FIGURE 8.
Quantitative RT-PCR analyses of CgHXT1 to CgHXT5 transcript levels in axenically grown C. graminicola. mRNA levels of CgHXT1 to CgHXT5 were studied in C. graminicola grown for 3 days at 23 °C in the presence of the indicated additions (glucose, 1.5%; extract, maize cell walls plus soluble extract from maize leaves as described under “Experimental Procedures.”). mRNA levels in C. graminicola cells incubated without glucose and extract (dark bars) were set to 100%. Histone3 transcript levels served as controls. The RNA were obtained from 3 biological repeats (± S.E.) and were different from those shown in Fig. 7A.
FIGURE 9.
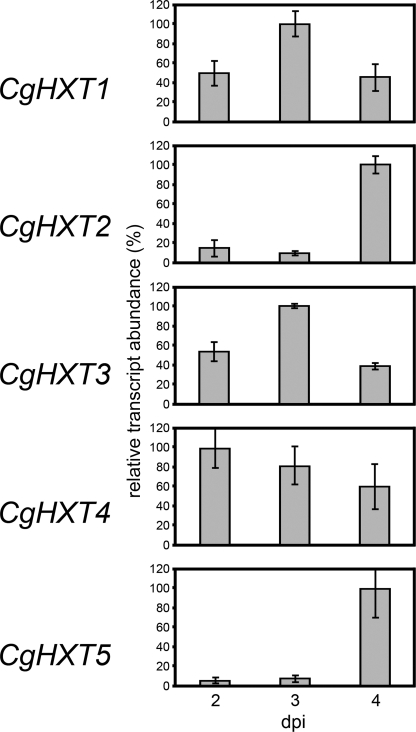
Quantitative RT-PCR analyses of CgHXT1 to CgHXT5 transcript levels in C. graminicola-infected maize leaves. mRNA levels of CgHXT1 to CgHXT5 were studied at 2, 3, or 4 dpi. Actin and Histone3 transcript levels served as controls. RT-PCRs were performed on total RNA isolated from healthy or infected maize leaves (maize plus fungal RNA) at the indicated times. The RNA were obtained from three biological repeats (± S.E.).
Figs. 7B and 9 show semiquantitative and quantitative RT-PCR analyses of CgHXT1 to CgHXT5 mRNA levels in C. graminicola-infected maize leaves. Whereas the transcript levels of CgHXT4 were not significantly altered during the infection, CgHXT1 and CgHXT3 mRNA abundances were clearly (about 2-fold) up-regulated at 3 dpi and fell back to the initial levels at 4 dpi. In contrast, mRNA levels of CgHXT2 and even more clearly of CgHXT5 were drastically up-regulated, i.e. 10–20-fold increased, exclusively at 4 dpi, when the fungus had already initiated its destructive, necrotrophic development. The amounts of cDNA used for the semiquantitative (Fig. 7B) and quantitative RT-PCRs (Fig. 9) were normalized to yield similar amounts of actin and histone3 mRNA. Note that the fungus-specific mRNAs were not detected in uninfected maize leaves.
GenBankTM accession numbers: CgHXT1 cDNA, FN433101; CgHXT1s cDNA, FN433106; CgHXT2 cDNA, FN433102; CgHXT3 cDNA, FN433103; CgHXT4 cDNA, FN433104; CgHXT5 cDNA, FN433105; CgHXT1 gene, FN433645; CgHXT2 gene, FR746061; CgHXT3 gene, FR746062; CgHXT4 gene, FR746063; CgHXT5 gene, FR746064.
DISCUSSION
The ascomycete C. graminicola is a hemibiotrophic fungus and, therefore, switches from an initial biotrophic growth phase during initial host colonization to a necrotrophic phase that leads to the destruction of the host (1, 36). This developmental switch is paralleled by a transition from apoplastic growth to the feeding on cytoplasmic contents after breaching the host's plasma membranes and, thus, is also attended by a change in the available carbon sources and their concentrations. Finally, during the biotrophic growth phase it is important for the fungus to avoid the production of signaling molecules and/or to secrete effectors that contribute to suppress defense responses by the host plant (4, 37). This precautionary measure is probably unnecessary after the transition to necrotrophic growth. The change in lifestyle, i.e. the biotrophy-necrotrophy transition, will affect the availability of sugars and result in up- or down-regulation of fungal genes for plasma membrane-localized sugar transporters at different growth phases.
CgHXT1 to CgHXT5 Represent Different Types of Transporters That Are Also Found in Other Pathogens
Our BLAST searches in publicly available genomic sequences from C. graminicola identified five genes encoding proteins with homology to fungal hexose transporters. RT-PCR analyses on fungal RNA lead to the identification of the corresponding cDNAs revealing the genomic organization of the respective genes (Fig. 1A) and confirming predicted protein sequences (Table 2). Interestingly, the identified proteins shared only 37–50% identical amino acids. These identity values are significantly lower than those known from baker's yeast HXT proteins (Hxt1p to Hxt13p), which share between 75 and 87% identical amino acids. Even the yeast Gal2p galactose transporter shares 65–70% identical amino acids with the different yeast HXT proteins. Similarly low identity values (30–35%) were found only in comparisons of yeast HXT proteins and yeast Snf2p or Rgt2p glucose sensors.
All this suggested that CgHXT1 to CgHXT5 might represent transporters from five independent subgroups. In fact, phylogenetic analyses confirmed this hypothesis demonstrating that the C. graminicola transporters (i) are on five clearly separated branches, (ii) that these branches do not include baker's yeast HXT transporters or glucose sensors, and (iii) that these branches include closely related but mostly uncharacterized homologs from several other pathogens (Fig. 1B).
This demonstrates that the identified CgHXT transporters are encoded by genes that were formed early in fungal evolution and maintained during the development of genera and species with highly divergent developmental approaches (necrotrophic, hemibiotrophic, saprophytic). Obviously, this differs from the situation in baker's yeast, where all hexose transporter genes seem to be products of recent gene duplications.
CgHXT1 to CgHXT5 Have Different Functional Properties
For one of the genes, CgHXT1, two alternatively spliced forms were identified that had the 4th intron of the CgHXT1 gene either removed yielding the shorter CgHXT1s mRNA or not removed yielding the CgHXT1 mRNA. Compared with the amount of CgHXT1 mRNA, the relative proportion of CgHXT1s mRNA is extremely low, and therefore this shorter mRNA was not quantified in qRT-PCR analyses (not shown). However, as the reading frames downstream from the alternative splice site was the same in both mRNAs, and as the residues missing in the CgHXT1s protein were from the cytoplasmic C terminus, we expected that both splice variants might encode functionally active proteins.
This was confirmed in expression analyses of CgHXT1 and CgHXT1s cDNAs in baker's yeast (Fig. 2). Both cDNAs complemented the growth defect of a hexose transport-deficient yeast strain (EBY.VW4000) on glucose, mannose and fructose, and CgHXT1s (but not CgHXT1) even complemented the growth defect on galactose (Fig. 2). Interestingly, quantitative transport studies with glucose-grown CgHXT1 and CgHXT1s cells (Fig. 4) yielded identical competition data for galactose. This shows that the different cytoplasmic C termini of CgHXT1 and CgHXT1s do not affect the substrate specificity of the proteins, which is expected for a sequence difference near the C terminus. The different C termini might, however, affect the stability of the two proteins on different carbon sources (glucose or galactose) and, thus, explain the clearly different growth properties on galactose as sole carbon source.
Quantitative transport analyses with CgHXT2 to CgHXT5 expressing yeast cells showed that CgHXT2, CgHXT3 and CgHXT5 perfectly complemented the growth defect of EBY.VW4000 cells on glucose or other monosaccharides. In contrast, CgHXT4, the longest of the analyzed proteins, complemented this defect only poorly. Determination of the Km values for glucose of the four functional transporters characterized CgHXT1 and CgHXT3 as high affinity transporters, CgHXT2 as a transporter with medium affinity and CgHXT5 as low affinity transporter (Fig. 5).
When we tested the sensitivities of the different transporters to CCCP, an uncoupler of transmembrane proton gradients, they all were highly CCCP-sensitive (Fig. 6B) suggesting that all 4 transporters act as energy-dependent H+-symporters. The CgHXT1, CgHXT2, and CgHXT5 proteins transport their substrates over a wide pH range (pH 4.7 to pH 8.0) and show only some (CgHXT1 and CgHXT2) or no (CgHXT5) reduction in their transport activity at alkaline pH (Fig. 6A). A clear preference for slightly acidic pH values was observed only for CgHXT3, a transporter that is active primarily during the biotrophic growth phase, when the hyphae grow in the more acidic plant apoplast (Fig. 6A).
CgHXT1 to CgHXT5 show specific expression patterns in different developmental phases of C. graminicola. Comparative expression analyses of CgHXT1 to CgHXT5 in C. graminicola hyphae at different time points after maize-leaf infection revealed interesting differences fitting surprisingly well to the functional characteristics of the encoded proteins.
In infected maize leaves, the genes for the high-affinity transporters CgHXT1 and CgHXT3 were expressed under all growth conditions, but clearly up-regulated during biotrophic development (Figs. 7B and 9). Moreover, in in vitro cultures, the abundance of CgHXT1 and CgHXT3 mRNA was clearly reduced in the presence of extracellular glucose (Fig. 7A), which may explain the down-regulation of CgHXT1 and CgHXT3 expression during necrotrophic development in infected maize leaves (4 dpi in Figs. 7A and 8). In contrast, CgHXT5 mRNA levels were increased in glucose-supplemented in vitro cultures (Figs. 7A and 8), which might explain the massive up-regulation of CgHXT5 mRNA levels during necrotrophic development in infected maize leaves. Moreover, of the C. graminicola transporters characterized, CgHXT5 is the transporter with the highest glucose uptake capacity as suggested by its comparatively low Km and by the high uptake rates obtained with CgHXT5-expressing baker's yeast cells (Fig. 5).
In summary, these results indicate that glucose might represent a regulator of CgHXT1, CgHXT3, and CgHXT5 gene expression in hyphae growing both under sterile in vitro conditions and in infected maize leaves. Moreover, they suggest that LAHC transporters are preferred during necrotrophic development whereas HALC transporters seem to be used during biotrophic development. This latter observation is supported by the massive up-regulation of CgHXT2 mRNA levels during necrotrophic development (Figs. 7B and 9). The CgHXT2 gene encodes a transporter with medium substrate affinity (Fig. 5), and the mRNA levels seem to be up-regulated by a factor(s) different from glucose, which rather seems to have a negative effect on CgHXT2 mRNA abundance.
It is tempting to speculate that xylose or uronic acids might be involved in this up-regulation of CgHXT2 mRNA levels. These molecules are released from plant cell walls during enzymatic degradation and affect CgHXT2-driven glucose transport substantially (Fig. 4). They could account both for the high CgHXT2 mRNA abundance in the presence of cell wall extract (Fig. 7A and 8) and for the induction of CgHXT2 and CgHXT5 expression during the necrotrophic phase (Figs. 7B and 9).
CgHXT4 Might Be a Sugar Sensor
CgHXT4 mRNA levels were low (Fig. 7B) and not significantly altered during the different developmental stages in infected maize leaves (Fig. 9). Increased CgHXT4 mRNA levels were observed only in carbon-starved hyphae, but both glucose and maize extract lead to a massive reduction of CgHXT4 mRNA levels (Fig. 7A). These mostly low mRNA levels, the lack of measurable transport activities in CgHXT4-expressing yeast cells (although the protein is targeted to the plasma membrane; Fig. 3), and the very poor complementation of the growth defect of the EBY.VW4000 yeast mutant might point toward a role of CgHXT4 as glucose sensor rather than as a glucose transporter.
In fact, the CgHXT4 gene encodes the longest of the five proteins (Table 2), and with a predicted cytoplasmic C terminus of 100 amino acids, it has the longest C terminus of the 5 proteins characterized (predicted C termini for CgHXT1, CgHXT2, CgHXT3, and CgHXT5 are 61, 83, 57, and 48 amino acids, respectively). Elongated C termini were also found in the baker's yeast glucose sensors Snf3p and Rgt2p (31, 38) and in the RCO3 glucose transporter from N. crassa, which was also discussed as glucose sensor (39). The RCO3 C terminus contains a glutamine-rich region (39), and a glutamine/asparagine-rich region is also found in the C terminus of CgHXT4. Such glutamine/asparagine-rich regions were repeatedly discussed as mediators of specific protein-protein interactions (40, 41).
In the phylogenetic tree shown in Fig. 1B, RCO3 (accession no. EAA36477 in Fig. 1B) and CgHXT4 are found on the same branch. One of the other proteins on this branch (XP_389518 from G. zeae) also has an elongated C terminus (81 amino acids) and this C terminus also contains an asparagine-rich domain. Finally, The CgHXT4 branch is in close vicinity to the Snf3p/Rgt2p branch, which (together with the other arguments) supports the hypothesis that CgHXT4 might be a sensor of the nutrient status in C. graminicola. Of course, detailed analyses of the CgHXT4 protein and of its role in C. graminicola development and virulence will be necessary.
In summary, our data provide a first characterization of transporters and/or sensors involved in nutrient uptake during the different developmental stages of a hemibiotrophic fungus. Although our screen identified only five transporter genes, we understand that additional transporter genes, e.g. other HXT proteins or α-glucoside transporters (AGTs), might be involved, and further analyses will be necessary to identify and characterize the respective proteins. Moreover, for a complete understanding of the physiological importance of these transporters, C. graminicola mutants in the respective genes need to be generated, and the effects of these mutations on fungal virulence needs to be analyzed.
This work was supported by Deutsche Forschungsgemeinschaft Grants FOR666-DE 403–16 (to H. B. D.) and FOR666-SA 382-21 (to N. S.).
- CCCP
- carbonylcyanide m-chlorophenylhydrazone
- HXT
- hexose transporter
- ORF
- open reading frame
- RGT
- restores glucose transport
- SNF
- sucrose non-fermenting.
REFERENCES
- 1. Münch S., Lingner U., Floss D. S., Ludwig N., Sauer N., Deising H. B. (2008) J. Plant Physiol. 165, 41–51 [DOI] [PubMed] [Google Scholar]
- 2. Horbach R., Navarro-Quesada A. R., Knogge W., Deising H. B. (2011) J. Plant Physiol. 168, 51–62 [DOI] [PubMed] [Google Scholar]
- 3. Mendgen K., Hahn M. (2002) Trends Plant Sci. 7, 352–356 [DOI] [PubMed] [Google Scholar]
- 4. Krijger J. J., Horbach R., Behr M., Schweizer P., Deising H. B., Wirsel S. G. (2008) Mol. Plant-Microbe Interact. 21, 1325–1336 [DOI] [PubMed] [Google Scholar]
- 5. Voegele R. T., Struck C., Hahn M., Mendgen K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8133–8138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wahl R., Wippel K., Goos S., Kämper J., Sauer. N. (2010) PLoS Biol. 8, e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schüssler A., Martin H., Cohen D., Fitz M., Wipf D. (2006) Nature 444, 933–936 [DOI] [PubMed] [Google Scholar]
- 8. Fang W., St. Leger R. J. (2010) Plant Physiol. 154, 1549–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marger M. D., Saier M. H., Jr. (1993) Trends Biochem. Sci. 18, 13–20 [DOI] [PubMed] [Google Scholar]
- 10. Sauer N., Ludwig A., Knoblauch A., Rothe P., Gahrtz M., Klebl F. (2004) Plant J. 40, 120–130 [DOI] [PubMed] [Google Scholar]
- 11. Glazebrook J. (2005) Annu. Rev. Phytopathol. 43, 205–227 [DOI] [PubMed] [Google Scholar]
- 12. Kliebenstein D. J., Rowe H. C. (2008) Plant Sci. 174, 551–556 [Google Scholar]
- 13. Behr M., Humbeck K., Hause G., Deising H. B., Wirsel S. G. (2010) Mol. Plant Microbe Interact. 23, 879–892 [DOI] [PubMed] [Google Scholar]
- 14. Hanahan D. (1983) J. Mol. Biol. 166, 557–580 [DOI] [PubMed] [Google Scholar]
- 15. Münch S., Ludwig N., Floss D. S., Sugui J. A., Koszucka A. M., Voll L. M., Sonnewald U., Deising H. B. (2011) Mol. Plant Pathol. 12, 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wieczorke R., Krampe S., Weierstall T., Freidel K., Hollenberg C. P., Boles E. (1999) FEBS Lett. 464, 123–128 [DOI] [PubMed] [Google Scholar]
- 17. Sauer N., Stolz J. (1994) Plant J. 6, 67–77 [DOI] [PubMed] [Google Scholar]
- 18. Gietz D., St Jean A., Woods R. A., Schiestl R. H. (1992) Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meyer S., Melzer M., Truernit E., Hümmer C., Besenbeck R., Stadler R., Sauer N. (2000) Plant J. 24, 869–882 [DOI] [PubMed] [Google Scholar]
- 20. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 21. Guindon S., Gascuel O. (2003) Systematic Biology 52, 696–704 [DOI] [PubMed] [Google Scholar]
- 22. Page R. D. M. (1996) Comput. Appl. Biosci. 12, 357–358 [DOI] [PubMed] [Google Scholar]
- 23. Lee S. H., Han Y. K., Yun S. H., Lee Y. W. (2009) Eukaryot. Cell 8, 1155–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zheng Y., Zheng W., Lin F., Zhang Y., Yi Y., Wang B., Lu G., Wang Z., Wu W. (2011) Mol. Plant Microbe. Interact. 24, 13–17 [DOI] [PubMed] [Google Scholar]
- 25. Qin J. C., Gao J. M., Zhang Y. M., Yang S. X., Bai M. S., Ma Y. T., Laatsch H. (2009) Steroids 74, 786–790 [DOI] [PubMed] [Google Scholar]
- 26. Li H., Sang J., Li R., Liu Y., Zhang J. (2010) Eur. J. Dermatol. 20, 378–380 [DOI] [PubMed] [Google Scholar]
- 27. Korripally P., Tiwari A., Haritha A., Kiranmayi P., Bhanoori M. (2010) Fungal Genet. Biol. 47, 237–245 [DOI] [PubMed] [Google Scholar]
- 28. Tournas V. (1994) Crit. Rev. Microbiol. 20, 243–263 [DOI] [PubMed] [Google Scholar]
- 29. Brun S., Malagnac F., Bidard F., Lalucque H., Silar P. (2009) Mol. Microbiol. 74, 480–496 [DOI] [PubMed] [Google Scholar]
- 30. Maier A., Völker B., Boles E., Fuhrmann G. F. (2002) FEMS Yeast Res. 2, 539–550 [DOI] [PubMed] [Google Scholar]
- 31. Ozcan S., Dover J., Rosenwald A. G., Wölfl S., Johnston M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 12428–12432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnston M., Kim J. H. (2005) Biochem. Soc. Trans. 33, 247–252 [DOI] [PubMed] [Google Scholar]
- 33. Delrot S., Bonnemain J. L. (1981) Plant Physiol. 67, 560–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Russel J. B. (1990) Nature 56, 3304–3307 [Google Scholar]
- 35. Leonardi M. G., Casartelli M., Parenti P., Giordana B. (1998) Am. J. Physiol. Regul. Integr. Comp. Physiol. 274, R1372–R1375 [DOI] [PubMed] [Google Scholar]
- 36. Horbach R., Graf A., Weihmann F., Antelo L., Mathea S., Liermann J. C., Opatz T., Thines E., Aguirre J., Deising H. B. (2009) Plant Cell 21, 3379–3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. El Gueddari N. E., Rauchhaus U., Moerschbacher B. M., Deising H. B. (2002) New Phytol. 156, 103–112 [Google Scholar]
- 38. Ozcan S., Johnston M. (1999) Microbiol. Mol. Biol. Rev. 63, 554–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Madi L., McBride S. A., Bailey L. A., Ebbole D. J. (1997) Genetics 146, 499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Michelitsch M. D., Weissman J. S. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11910–11915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Okazawa H. (2007) Protein Rev. 6, 451–463 [Google Scholar]