Abstract
Peptidyl-prolyl cis/trans isomerase NIMA-interacting 1 (Pin1) is a unique enzyme that associates with the pSer/Thr-Pro motif and catalyzes cis-trans isomerization. We identified Pin1 in the immunoprecipitates of overexpressed IRS-1 with myc and FLAG tags in mouse livers and confirmed the association between IRS-1 and Pin1 by not only overexpression experiments but also endogenously in the mouse liver. The analysis using deletion- and point-mutated Pin1 and IRS-1 constructs revealed the WW domain located in the N terminus of Pin1 and Ser-434 in the SAIN (Shc and IRS-1 NPXY binding) domain of IRS-1 to be involved in their association. Subsequently, we investigated the role of Pin1 in IRS-1 mediation of insulin signaling. The overexpression of Pin1 in HepG2 cells markedly enhanced insulin-induced IRS-1 phosphorylation and its downstream events: phosphatidylinositol 3-kinase binding with IRS-1 and Akt phosphorylation. In contrast, the treatment of HepG2 cells with Pin1 siRNA or the Pin1 inhibitor Juglone suppressed these events. In good agreement with these in vitro data, Pin1 knock-out mice exhibited impaired insulin signaling with glucose intolerance, whereas adenoviral gene transfer of Pin1 into the ob/ob mouse liver mostly normalized insulin signaling and restored glucose tolerance. In addition, it was also demonstrated that Pin1 plays a critical role in adipose differentiation, making Pin1 knock-out mice resistant to diet-induced obesity. Importantly, Pin1 expression was shown to be up-regulated in accordance with nutrient conditions such as food intake or a high-fat diet. Taken together, these observations indicate that Pin1 binds to IRS-1 and thereby markedly enhances insulin action, essential for adipogenesis.
Keywords: Adipose Tissue, Diabetes, Insulin, Insulin Resistance, Obesity, Pin1
Introduction
Peptidyl-prolyl cis/trans isomerases constitute a special class of enzymes that function not by modifying other peptides covalently but only by producing conformational changes (1–4). Pin1, one of the peptidyl-prolyl cis/trans isomerase isoforms that is distinct from the two major classes of isomerases, the cyclophilins and the FK506-binding proteins (5, 6), was initially cloned as a NIMA kinase interacting protein (7) and specifically recognizes a proline bond preceded by a phosphorylated serine or threonine residue (phospho-Ser/Thr-Pro motif) with a WW domain at its N terminus (8, 9). Then, its C-terminal PPIase domain was shown to catalyze cis-trans isomerization of the target peptidyl-prolyl bonds and thereby modify the actions of target proteins (8, 9). Since its discovery, numerous proteins have been identified as Pin1 substrates (8, 10), implicating it in the regulation of many biological processes. For example, Pin1 inhibits the phosphorylation of Cdc25 and controls the replication checkpoint in the cell cycle (11, 12). Pin1 also stabilizes the tumor suppressor p53 and is abundantly expressed in some malignant tumors, suggesting involvement in malignant transformation (13, 14). On the other hand, Pin1 is also expressed at high levels in most neurons (15, 16). Although much remains to be clarified regarding the role of Pin1 in neurons, it reportedly protects against neurodegeneration via several independent mechanisms (15, 16), as supported by the fact that Pin1 knock-out (KO) mice exhibit abnormal central nervous function (15, 16). In addition, the enzymatic function of Pin1 is reportedly inactivated by oxidative stress modification occurring in the early stages of Alzheimer disease, implicating Pin1 in brain function (16, 17). Therefore, Pin1 has been implicated in the pathogenesis of two major disorders; cancer and Alzheimer disease (9, 10).
Very recently, we reported that Pin1 expression is higher in the fed than in the fasted state and that Pin1 associates with CRTC2, a co-activator of CREB (cAMP-response element-binding protein), and suppresses CRE (cAMP-response element) transcriptional activity (18). Suppressed CRE activity leads to reduced PEPCK expression and gluconeogenesis. In this study we newly identified Pin1 to be a positive regulator of insulin signaling via enhanced insulin-induced insulin receptor substrate-1 (IRS-1)3 phosphorylation. In addition, Pin1 expression was revealed to be markedly increased by high-fat diet feeding. Because a high-fat diet is one of the most common causes of obesity, we also performed experiments to ascertain the effects of Pin1 on adipogenesis in this study. Herein, we present evidence that Pin1 plays a critical role in energy incorporation into the body by enhancing insulin signaling and adipogenesis. Our findings raise the possibility of Pin1 being a promising target molecule for treating diabetes mellitus.
EXPERIMENTAL PROCEDURES
Materials
Affinity-purified antibodies against IRS-1, IRS-2, phosphorylated tyrosine (4G10), and Akt/protein kinase B were prepared as previously described (19). Anti-Pin1 antibody was generated by immunizing rabbits with the peptide QMQKPFEDASFATRTGEMSGPVFTDSGIHIITRTE (amino acids 129–163 of human Pin1). Anti-FLAG tag antibody was purchased from Sigma, and antibodies against the p85 subunit of phosphatidylinositol 3-kinase, phospho-Ser-473 and phospho-Thr-308 of Akt, and anti-actin were from Cell Signaling Technology (Danvers, MA).
Cell Culture
Sf9 cells were grown in TC100 (Invitrogen) medium containing 10% fetal bovine serum (FBS) at 27 °C. HepG2 hepatoma cells and human preadipocyte were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS at 37 °C in 5% (v/v) CO2 in air. Mouse 3T3-L1 fibroblasts were maintained in DMEM containing 10% donor calf serum in an atmosphere of 10% CO2 at 37 °C. Two days after the fibroblasts had reached confluence, differentiation was induced by treating the cells with DMEM containing 4 μg/ml dexamethasone, 200 nm insulin, 0.5 mmol/liter 3-isobutyl-1-methylxanthine, and 10% FBS for 48 h as described previously. Cells were fed DMEM supplemented with 10% FBS every other day and used as mature 3T3-L1 adipocytes at day 8 after the induction of differentiation.
Animals
Pin1 KO mice were generated as previously described (20), and Pin1+/+ and Pin1−/− mice were obtained by breeding Pin1+/− mice. The genotype was determined by PCR and Pin1+/− mice were separated for breeding. Twelve-week-old Pin1+/+ and Pin1−/− mice were fed a standard diet or high-fat diet (HFD) for 4 weeks. The nutrient composition of the standard diet was 54.4% carbohydrate, 23.6% protein, 5.3% fat with a vitamin and mineral mixture, whereas that of the HFD was 7.5% carbohydrate, 24.5% protein, 60% fat with a vitamin and mineral mixture. In other experiments, 12–16-week-old Pin1+/+ and Pin1−/− mice were used.
Preparation of Adenoviruses Expressing MEF-tagged IRS Protein, Pin1, and GFP
The myc-TEV (tobacco etch virus)-FLAG (MEF) tag cassette was generated by DNA synthesis and inserted into cloning sites in the mammalian expression vector pcDNA3 (Invitrogen) and termed pcDNA3-MEF, as reported previously (19). To create the N-terminal MEF-tagged IRS-1 construct, human IRS-1 cDNA was inserted into pcDNA3-MEF. Then the coding portion of MEF-tagged IRS-1 was isolated from pcDNA3-MEF-IRS-1. Similarly, the coding regions of MEF-tagged IRS-2 and S434A IRS-1 were prepared. Recombinant adenovirus expressing MEF-tagged IRS-1, MEF-tagged S434A IRS-1, MEF-tagged IRS-2, human Pin1 with a C-terminal HA tag, GFP-tagged Pin1, and GFP were generated, and the adenovirus encoding LacZ served as a control. The recombinant adenoviruses expressing human Pin1 with a C-terminal HA tag and LacZ were used for adenoviral gene transfer into ob/ob mice.
Purification of MEF-tagged IRS-1-containing Complexes from Mouse Livers
Recombinant adenovirus expressing MEF-tagged IRS-1 was purified and concentrated using cesium chloride ultracentrifugation. Adenovirus encoding LacZ served as a control. Male C57B6 mice, 9 weeks of age, obtained from Nippon Bio-Supply Center (Tokyo, Japan), were injected via the tail vein with adenovirus at a dose of 2.5 × 107 plaque-forming units/g of body wt. Four days later, mouse livers were removed and lysed in lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10% (w/v) glycerol, 100 mm NaF, 10 mm EGTA, 1 mm Na3VO4, 1% (w/v) Triton X-100, 5 μm ZnCl2, 2 mm PMSF, 10 μg/ml aprotinin, and 1 μg/ml leupeptin). Lysates were centrifuged at 100,000 × g for 20 min at 4 °C. Supernatants were passed through a 5-μm filter, incubated with 150 μl of Sepharose beads for 60 min at 4 °C, and then passed through a 0.65-μm filter. The filtrated supernatant was mixed with 150 μl of anti-myc-conjugated Sepharose beads for the first immunoprecipitation. After incubation for 90 min at 4 °C, the beads were washed 5 times with 1.5 ml of TNTG buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10% (w/v) glycerol, and 0.1% (w/v) Triton X-100), twice with buffer A (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.1% (w/v) Triton X-100), and finally once with TNT buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 0.1% (w/v) Triton X-100). The washed beads were incubated with 15 units of tobacco etch virus protease (Invitrogen) in 150 μl of TNT buffer to release bound materials from the beads. After incubation for 60 min at room temperature, supernatants were pooled, and the beads were washed twice with 75 μl of buffer A. The resulting supernatants were combined and incubated with 25 μl of FLAG-Sepharose beads for the second immunoprecipitation. After a 60-min incubation at room temperature, the beads were washed 3 times with 500 μl of buffer A, and proteins bound to the FLAG beads were dissociated by incubation with 1 mm synthetic FLAG peptides in buffer A for 120 min at 4 °C. Approximately 3 μg of protein (0.01% of starting materials) were routinely recovered by this procedure. The samples were electrophoresed and subjected to SDS-PAGE and immunoblotting.
Immunoprecipitation and Immunoblotting
The cells were solubilized with Laemmli buffer (0.2 m Tris-HCl, 4% SDS, 10% glycerol, 5% 2-mercaptoethanol, 0.1% bromphenol blue). Equal amounts of protein from whole cell lysates were resolved by SDS-PAGE. Then the proteins were transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA) using an electroblotting apparatus (Mighty Small Transphor; Amersham Biosciences) and subjected to immunoblotting using the Super Signal West Pico Chemiluminescence System (Pierce). The results of several immunoblots were quantitatively analyzed using an LAS-3000 mini (FUJIFILM, Tokyo, Japan).
Supernatants containing equal amounts of protein (2 mg) were incubated with anti-IRS-1 and anti-IRS-2 antibodies (5 μg each) and then with 100 μl of protein A- and G-Sepharose. These immunoprecipitates and cell lysates were boiled in Laemmli sample buffer containing 100 mmol/liter dithiothreitol, electrophoresed, and immunoblotted with anti-IRS-1, anti-IRS-2, anti-p85 (phosphatidylinositol-3-kinase), anti-Pin1, phospho-Akt (Thr-308 and Ser-473), or 4G10 antibody. The bands were quantitatively analyzed using the LAS-3000 mini.
Preparation of Baculovirus-produced Recombinant Proteins
Full-length coding regions of human Pin1, GFP-tagged Pin1, IRS-1, and DsRed-tagged full-length and deletion mutants of IRS-1 were subcloned into pBacPAK9 transfer vector (Clontech, Mountain View, CA), and baculoviruses were prepared according to the manufacturer's instructions. For protein production, Sf9 cells were infected with these baculoviruses and grown for 48 h.
Preparation of Glutathione S-transferase (GST)-Pin1 Fusion Protein
cDNAs encoding full-length human Pin1 and the WW and PPIase domains of Pin1 were subcloned into a pGEX-4T-1 vector (GE Healthcare), which was used to transform Escherichia coli JM105 (Promega, Madison, WI). Transformed cells were grown to an A600 of 0.6 in LB medium supplemented with 0.1 mg/ml ampicillin and stimulated for 3 h with 1.0 mm isopropyl-β-d-thiogalactopyranoside. GST fusion proteins were isolated and purified by affinity chromatography on a glutathione-Sepharose 4B column (GE Healthcare). Glutathione was removed by dialysis against phosphate-buffered saline containing 10 mm dithiothreitol.
RNA Interference
For the knockdown of human Pin1, a validated StealthTM RNAi of human Pin1 (5′-CGGCAACAGCAGCAGUGGUGGCAAA-3′) was used. After RNAi and RNAiMAX in Opti-MEM had been mixed, they were introduced into HepG2. Forty-eight hours later, the cells were stimulated with insulin for the indicated times. Mouse Pin1 RNAi (5′-CACAGTATTTATTGTTCTAA-3′) was purchased from Invitrogen. Similarly, 48 h after introduction into 3T3-L1 cells, differentiation was induced by a mixture including isobutylmethylxanthine, dexamethasone, and insulin.
Intraperitoneal Glucose, Insulin, and Pyruvate Tolerance Tests
Mice were fasted for 14 h followed by blood sampling and intraperitoneal injection of glucose (2 g/kg body wt), insulin (0.75 units/kg body wt), or pyruvate (2 g/kg body wt). Whole venous blood was obtained from the tail vein at the indicated times after the glucose load. Blood glucose was measured with a portable blood glucose monitor.
In Vivo Insulin Stimulation
In brief, mice were anesthetized with pentobarbital sodium. The portal vein was exposed, and 0.4 ml of normal saline (0.9% NaCl) with or without insulin (25 milliunits/g of body wt) were injected. Livers were removed 30 s later, hind limb skeletal muscles 90 s thereafter, and immediately homogenized with a Polytron homogenizer in 6 volumes of solubilization buffer. Both extracts were centrifuged at 15,000 g for 30 min at 4 °C, and the supernatants were used as samples for immunoprecipitation and immunoblotting.
Glucose Uptake in Isolated Skeletal Muscle
Mice were anesthetized, and soleus muscles were dissected out and rapidly cut into 2–3-mg strips. Muscle strips were incubated in a shaking water bath at 35 °C for 60 min in a 20-ml flask containing 2.0 ml Krebs-Henseleit bicarbonate buffer supplemented with 8 mm glucose, 32 mm mannitol, and 0.1% bovine serum albumin (BSA) (radioimmunoassay grade). Flasks were gassed continuously with 95% O2, 5%CO2 throughout the experiment. The muscles were then incubated for 20 min in oxygenated KRB buffer in the presence or absence of human insulin (2 milliunits/ml) and then incubated for 20 min at 29 °C in 1.5 ml of KRB buffer containing 8 mm 2-deoxy-d- [1,2-3H(N)]glucose (2.25 μCi/ml), 32 mm [14C]mannitol (0.3 μCi/ml), 2 mm sodium pyruvate, and 0.1% BSA. After the incubation, muscles were rapidly blotted, weighed, and solubilized with 1 ml of Soluene 350 (PerkinElmer Life Sciences). The samples were counted in a liquid scintillation counter. 2-Deoxy-[3H]glucose uptake rates were corrected for extracellular trapping using [14C]mannitol counts.
Quantitative Real-time Reverse Transcription-PCR
Total RNA was extracted from 3T3-L1 adipocytes or mouse liver and epididymal adipose tissue using Sepasol reagent (Nakalai Tesche, Kyoto, Japan). First-strand cDNAs were synthesized using PrimeScript reverse transcriptase with oligo(dT). Real-time PCR was performed using SYBR Green PCR master mix (Invitrogen) on an ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA). The primers were designed as follows: SREBP1 forward: GGAGCCATGGATTGCACATT; SREBP1 reverse, GGCCCGGGAAGTCACTGT; SREBP2 forward, TAACCCCTTGACTTCCTTGCT; SREBP2 reverse, TGCTCTTAGCCTCATCCTCAA; ACC forward, TCTCTGGCTTACAGGATGGTTTG; ACC reverse, GAGTCTATTTTCTTTCTGTCTCGACCTT; FAS forward, GCTGCGGAAACTTCAGGAAAT; FAS reverse, AGAGACGTGTCACTCCTGGACTT; SCD forward, GTCAAAGAGAAGGGCGGAAAAC; SCD reverse, AAGGTGTGGTGGTAGTTGTGGAAG.
Statistical Analysis
Results are expressed as the means ± S.E., and significance was assessed using one way analysis of variance unless otherwise indicated.
RESULTS
Increased Pin1 Expression in Mouse Liver, Muscle, and Fat Tissue with High-fat Diet Feeding
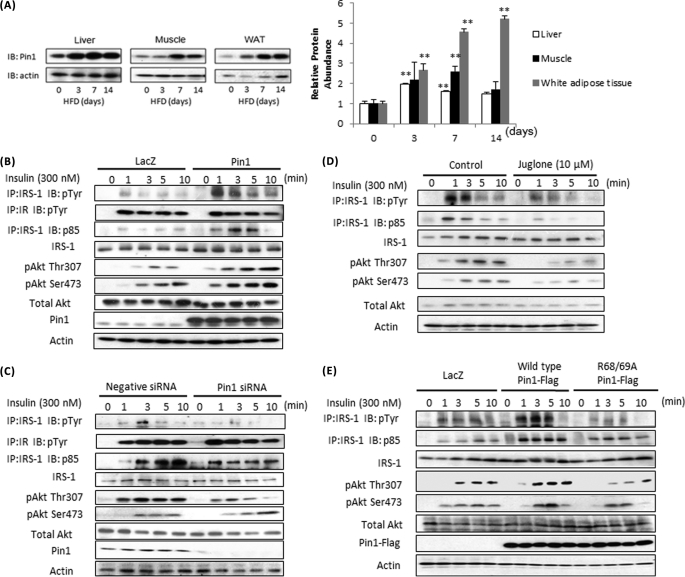
We previously reported that Pin1 protein is increased in the fed state (19). In this study we speculated that Pin1 expression might be regulated by altered nutrient conditions and examined Pin1 expression alteration in mice fed a HFD. As shown in Fig. 1A, HFD feeding markedly increased Pin1 protein in liver, muscle, and epididymal fat tissue. Pin1 amounts in other tissues such as the brain were not significantly altered (supplemental Fig. 1).
FIGURE 1.
Pin1 enhances insulin signaling. A, mice were fed a control diet or HFD for the indicated periods. Liver, muscle, and adipose tissue lysates were prepared, then immunoblotted (IB) with anti-Pin1 or anti-actin antibody. B and C, Pin1 enhances insulin-induced IRS-1 phosphorylation and its downstream signaling. B, LacZ or Pin1 was overexpressed in HepG2 cells. IP, immunoprecipitates. C, HepG2 cells were treated with control siRNA or Pin1 siRNA. In both experiments, at the indicated times after initiating insulin stimulation, lysates were prepared from HepG2 cells. Then phosphorylation levels and protein amounts of IRS-1, p85 associated with IRS-1 and Akt Thr-307 and Ser-473 phosphorylations, and the Pin1 were determined. D, the Pin1 inhibitor Juglone attenuated the insulin-induced IRS-1 phosphorylation in HepG2 cells. HepG2 cells were treated with or without 10 μm Juglone for 30 min. At the indicated times after initiating insulin stimulation, lysates were prepared from HepG2 cells. Then phosphorylation levels and protein amounts of IRS-1, p85 associated with IRS-1, Akt Thr-307 and Ser-473 phosphorylations were determined. E, wild-type, but not the inactive mutant of Pin1, enhanced insulin-induced IRS-1 phosphorylation and its downstream signaling. LacZ, wild-type Pin1, or R68A/R69A-mutated Pin1 was overexpressed in HepG2 cells. At the indicated times after initiating insulin stimulation, lysates were prepared from HepG2 cells. Then, phosphorylation levels and protein amounts of IRS-1, p85 associated with IRS-1, and Akt Thr-307 and Ser-473 phosphorylations were determined. A representative immunoblot from four independent experiments is shown.
Enhancing Effect of Pin1 on Insulin Signaling
Because Pin1 expression was affected by nutrient conditions, we examined the possible involvement of Pin1 in glucose and lipid metabolism regulation. First, to elucidate the effect of Pin1 on insulin signaling, HA-tagged Pin1 was overexpressed in HepG2 cells (Fig. 1B and supplemental Fig. 2A). The overexpressed Pin1 amount was ∼5-fold that of endogenous Pin1. In this state, insulin-induced IRS-1 phosphorylation and p85 association with IRS-1 and Akt phosphorylation were all markedly enhanced, whereas IRS-1 and Akt protein amounts were unchanged. Pin1 gene suppression using siRNA attenuated these events (Fig. 1C and supplemental Fig. 2B). Similar data were obtained using the Pin1-specific inhibitor Juglone, which has no effects on the activities of other prolyl isomerases, such as FKBP (FK506-binding protein) and cyclophilin A (21) (Fig. 1D and supplemental Fig. 2C). In addition, overexpression of the inactive mutant of Pin1 (R68A/R69A), in which Arg-68 and Arg-69 are replaced by Ala, failed to enhance insulin signaling (Fig. 1E and supplemental Fig. 2D). Thus, Pin1 expression level and activity regulate the efficiency of insulin-induced IRS-1 phosphorylation.
Association of Pin1 with IRS-1
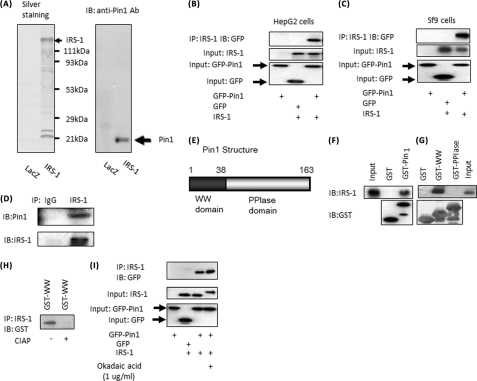
Because Pin1 markedly enhanced insulin-induced IRS-1 phosphorylation, we considered a direct association of Pin1 with IRS-1. First, we used an MEF-tagged purification system, i.e. IRS-1 fused with N-terminal triple tags consisting of an myc tag, tobacco etch virus protease cleavage sequence, and FLAG tag overexpressed in mouse liver. Sequential affinity purification was performed using three steps; myc tag antibody immunoprecipitation, elution by tobacco etch virus protease digestion, and finally, FLAG tag antibody immunoprecipitation. Purified IRS-1 containing complexes were electrophoresed and then silver-stained, which demonstrated bait proteins of IRS-1 and many others including 14-3-3 proteins. Immunoblotting using Pin1 antibody indicated the presence of Pin1 in IRS-1 complexes (Fig. 2A). To confirm the association between IRS-1 and Pin1, IRS-1 and either GFP-Pin1 or GFP was co-overexpressed in HepG2 or Sf-9 cells. As shown in Fig. 2, B and C, GFP-Pin1, but not GFP alone, was detected in IRS-1 immunoprecipitates. Furthermore, Pin1 was detected in immunoprecipitates with anti-IRS-1 antibody, but not control IgG, from mouse liver (Fig. 2D). Thus, the association between IRS-1 and Pin1 is physiological.
FIGURE 2.
A, Pin1 association with IRS-1 is shown. IRS-1 with the N-terminal MEF tag was overexpressed in mouse liver using adenovirus gene transfer, and IRS-1-containing complexes were purified. The samples were electrophoresed, then silver-stained (left panel) and immunoblotted (IB) with anti-Pin1 antibody (Ab, right panel). B, IRS-1 or control LacZ was overexpressed with GFP or GFP-Pin1 in HepG2 cells. The cell lysates were immunoprecipitated (IP) with anti-IRS-1 antibody followed by immunoblotting with anti-GFP antibody. C, IRS-1 was overexpressed with GFP or GFP-Pin1 in Sf9 cells. The cell lysates were immunoprecipitated with anti-IRS-1 antibody followed by immunoblotting with anti-GFP antibody. D, the mouse liver cell lysates were immunoprecipitated with anti-IRS-1, and the immunoprecipitates were immunoblotted with anti-Pin1 and anti-IRS-1. E, Pin1 structure. GST-Pin1, GST-Pin1 WW domain, and GST-Pin1 PPIase domain were prepared. With incubation, these GST-proteins were conjugated to beads and cell lysates from IRS-1 overexpressing Sf-9 cells. F, GST-Pin1, but not GST alone, bound to IRS-1 in vitro. G, GST-WW, but not the GST-PPIase domain, bound IRS-1. H, lysates of Sf-9 cells overexprressing IRS-1 were treated with or without 40 units/ml alkaline phosphatase. Then the cell lysates were incubated with GST-WW domain and immunoprecipitated with anti-IRS-1 antibody. The immunoprecipitates were electrophoresed and immunoblotted with anti-GST antibody. I, HepG2 cells overexpressing IRS-1 or control LacZ were overexpressed with GFP or GFP-Pin1, then incubated with or without 1 μg/ml okadaic acid for 1 h. The cell lysates were immunoprecipitated with anti-IRS-1 antibody followed by immunoblotting with anti-GFP antibody.
To identify the Pin1 domain responsible for the association with IRS-1, GST-Pin1, the GST-Pin1 WW domain, and the GST-Pin1 PPIase domain were prepared (Fig. 2E). These GST proteins were conjugated to beads and incubated with cell lysates from IRS-1-overexpressing Sf-9 cells. GST-Pin1, but not GST alone, bound to IRS-1 in vitro (Fig. 2F). This pulldown system experiment revealed the GST-WW domain, but not that of GST-PPIase, to bind IRS-1 (Fig. 2G). In addition, treating IRS-1 with alkaline phosphatase completely abolished the association with Pin1 (Fig. 2H), whereas okadaic acid treatment significantly increased this association (Fig. 2I), suggesting involvement of serine and/or threonine phosphorylation in IRS-1.
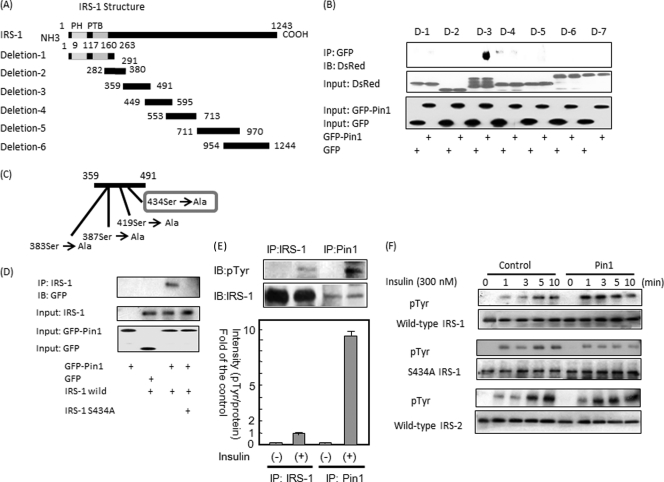
Subsequently, six DsRed-tagged IRS-1 deletion (N termini) mutants (Fig. 3A) and GFP-tagged Pin1 were co-overexpressed in Sf-9 cells. As shown in Fig. 3B, the IRS-1 deletion mutant 3 containing amino acids 359–491 was immunoprecipitated with GFP-tagged Pin1. There are four serine-proline motifs in this portion (Fig. 3C). Each of these serine residues was replaced with alanine, and the mutant not associating with Pin1 was examined. As shown in Fig. 3D, IRS-1 with serine 434 replaced by alanine did not bind Pin1, indicating that the association between IRS-1 and Pin1 is mediated via the phosphoserine 434-containing motif in IRS-1 and the WW domain in Pin1. Ser-434 is in the SAIN (Shc and IRS-1 NPXY binding) domain, which has been suggested to be involved in the association with the activated insulin receptor (22, 23). IRS-1 contains many serine/threonine residues that are heavily phosphorylated even under basal conditions, and Ser-434 is probably one such residue.
FIGURE 3.
A and B, the constructs of IRS-1 deletion mutants are shown. Baculoviruses expressing these six mutants with the N-terminal DsRed tag were prepared. These six deletion mutants were overexpressed with GFP or GFP-Pin1 in Sf-9 cells. The cell lysates were immunoprecipitated (IP) with anti-GFP antibody followed by immunoblotting (IB) with anti-DsRed antibody. The upper immunoblot shows that mutant 3 binds to GFP-Pin1, but not to GFP alone. PH, pleckstrin homology; PTB, phosphotyrosine binding. C and D, shown are the orientations of four candidate Ser/Pro motifs in mutant 3 involved in the association with Pin1. Wild-type IRS-1 or IRS-1 S434A was overexpressed with GFP-Pin1 or GFP in Sf-9 cells. The cell lysates were immunoprecipitated with anti-IRS-1 antibody followed by immunoblotting with anti-GFP. The upper panel shows that, unlike the wild type, IRS-1 S434A does not associate with Pin1. E, HepG2 cells were stimulated with insulin for 3 min, then cell lysates were immunoprecipitated with anti-IRS-1 or anti-Pin1 antibody. Both samples were electrophoresed and immunoblotted with anti-Tyr(P) (pTyr) or anti-IRS-1 antibody (upper panel). The IRS-1 phosphorylation level and protein amount were calculated as the means ± S.E. of three samples (*, p < 0.05). F, MEF-tagged IRS-1, MEF-tagged S434A IRS-1, and MEF-tagged IRS-2 were overexpressed with control LacZ or Pin1 in HepG2 cells. Cell lysates were then immunoprecipitated with anti-FLAG-tag antibody and immunoblotted with anti-Tyr(P) and anti-FLAG-tag antibodies.
Furthermore, to test whether or not enhanced IRS-1 phosphorylation is really mediated by direct association between IRS-1 and Pin1, insulin-induced phosphorylation levels of IRS-1 were compared between immunoprecipitation with IRS-1 antibody versus anti-Pin1 antibody using HepG2 cells. There was much less IRS-1 in the anti-Pin1 antibody immunoprecipitate than in the anti-IRS-1 immunoprecipitate, whereas insulin-induced IRS-1 phosphorylation was far greater with anti-Pin1 antibody immunoprecipitated IRS-1 (Fig. 3E). The tyrosine phosphorylation level/IRS-1 protein ratio was extremely high in the anti-Pin1 antibody immunoprecipitate as compared with the whole IRS-1 ratio. In addition, the effects of Pin1 on insulin-induced phosphorylations of S434A-mutated IRS-1 unable to associate with Pin1 as well as on IRS-2 were examined (Fig. 3F). FLAG-tagged wild-type, S434A IRS-1, or FLAG-tagged IRS-2 were co-overexpressed with Pin1 or LacZ in the HepG2 cells. The insulin-induced tyrosine phosphorylation of S434A IRS-1 was unaffected by Pin1 overexpression, whereas that of wild-type IRS-1 was markedly enhanced (Fig. 3F). It is noteworthy that IRS-2 phosphorylation is not significantly altered by Pin1. The phosphorylation of endogenous IRS-2 was also unaffected by Pin1 overexpression, although IRS-2 binds to Pin1 when overexpressed in HepG2 cells (supplemental Fig. 3). Our observations suggest that association of Pin1 with IRS-1 markedly enhances insulin-induced IRS-1 phosphorylation and its downstream signaling.
Insulin Resistance and Glucose Intolerance in Pin1 KO Mice
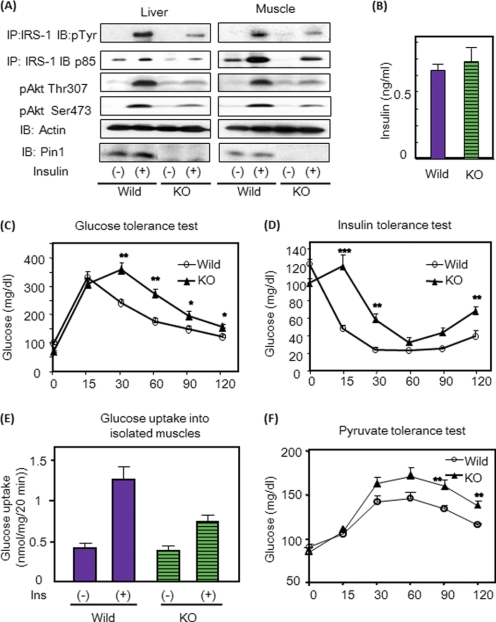
To elucidate the in vivo role of Pin1 in metabolism, the insulin signaling and sensitivity of Pin1 KO mice (20) were investigated. Insulin signaling was investigated first. Injecting insulin via the portal vein revealed insulin-induced IRS-1 tyrosine phosphorylation, the association of p85 with IRS-1 and Akt phosphorylation weaker in Pin1 KO liver and muscle than in those of wild-type mice (Fig. 4A and supplemental Fig. 4). Glucose and insulin tolerance tests demonstrated the insulin resistance of Pin1 KO mice (Fig. 4, C and D), whereas fasting serum insulin concentrations did not differ (Fig. 4B). The insulin-induced glucose uptake into isolated soleus muscle was also decreased in Pin1 KO mice as compared with controls, indicating muscle insulin resistance (Fig. 4E). The pyruvate tolerance test showed higher blood glucose concentrations in Pin1 KO mice, suggesting hepatic insulin resistance in these mice (Fig. 4F). Thus, in Pin1 KO mice, IRS-1 phosphorylation and downstream events are attenuated, and insulin resistance is present in both liver and muscle.
FIGURE 4.
Pin1 KO mice exhibit insulin resistance. A, insulin was injected into control and Pin1 KO mice, and IRS-1 and Akt phosphorylations were determined. IB, immunoblot. B, fasting insulin concentrations in control and Pin1 KO mice are shown. C, glucose tolerance test results are shown. 2 mg/kg glucose was injected into control and Pin1 KO mice, and blood glucose concentrations were measured as indicated. D, insulin tolerance test results are shown. 0.75 units/kg insulin was injected intraperitoneally into control, and Pin1 KO mice and blood glucose concentrations were measured as indicated. E, shown is glucose uptake into isolated muscle. F, pyruvate tolerance test results are shown. 2 mg/kg pyruvate was injected into control, and Pin1 KO mice and blood glucose concentrations were measured, as indicated. The results are presented as the means (±S.E.) of six independent experiments (*, p < 0.05; **, p < 0.01).
Hepatic Pin1 Overexpression Improves Insulin Resistance in ob/ob Mice
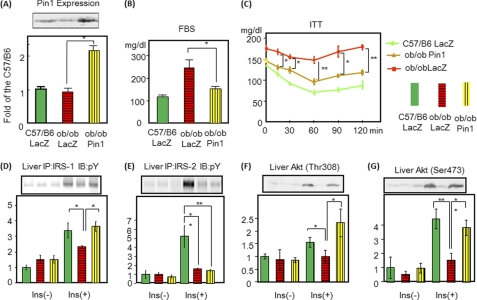
Interestingly, Pin1 expression was not altered in ob/ob mice as compared with lean controls. The reason is unclear, but we speculate that lack of a Pin1 expression response would exacerbate lipid and glucose metabolism abnormalities in ob/ob mice. We examined whether Pin1 overexpression might improve insulin resistance in ob/ob mice. Adenovirus for expressing Pin1 was injected intravenously. Adenovirus-mediated gene expression is reportedly limited to the liver (24) if adenovirus is injected into the bloodstream. Indeed, 72 h after injection, the amount of Pin1 was increased in the liver but not in muscle or adipose tissue (Fig. 5A and supplemental Fig. 5). In ob/ob mice, insulin-induced tyrosine phosphorylations of IRS-1 and IRS-2 and phosphatidylinositol 3-kinase/Akt activations were markedly impaired as reported previously (25) (Fig. 5, D–G). Pin1 overexpression normalized the decreased insulin-induced IRS-1, but not IRS-2, phosphorylation (Fig. 5, D and E). Hepatic Akt phosphorylations were also restored by Pin1 overexpression (Fig. 5, F and G). As reflected by improved insulin-signaling, glucose tolerance improved, i.e. fasting glucose concentrations (Fig. 5B) and expression levels of PEPCK and glucose-6-phosphatase were normalized (supplemental Fig. 6). The insulin tolerance test response was also improved (Fig. 5C). Thus, Pin1 overexpression improves insulin sensitivity in ob/ob mice.
FIGURE 5.
Hepatic overexpression of Pin1 normalizes the hyperglycemia with insulin resistance in ob/ob mice. LacZ or Pin1 adenovirus was injected via the tail vein. A, after 72 h, liver cell lysates were prepared, and the Pin1 expression level was determined by immunoblotting with anti-Pin1 antibody. The upper panel is a representative blot, and the lower graph shows the quantitative analysis (n = 6). B, shown are fasting blood glucose concentrations in C57/B6, ob/ob overexpressing LacZ, and ob/ob overexpressing Pin1 mice. C, insulin tolerance tests (ITT) for the three groups are shown. D and E, tyrosine phosphorylations of hepatic IRS-1 and IRS-2 are shown. Mice were injected with insulin via the portal vein, and phosphorylations of IRS-1 and IRS-2 were examined as reported previously. IP, immunoprecipitation; pY, Tyr(P). F and G, Akt phosphorylations at Thr-308 and Ser-473 were also examined in the livers of all three groups. n = 6 for each group and representative blots are shown in the upper panels. The results are presented as graphs with means (±S.E.). *, p < 0.05; **, p < 0.05.
Critical Role of Pin1 in Adipose Differentiation
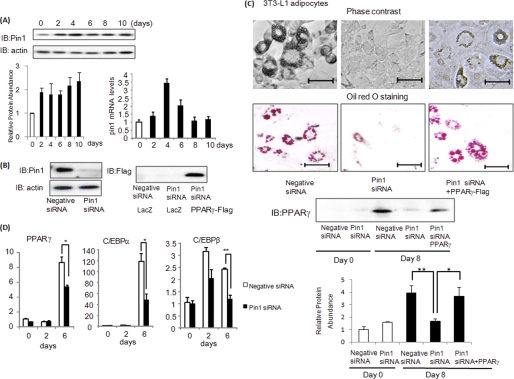
Given the known role of insulin in adipose differentiation (26) and the increased Pin1 expression in WAT with HFD feeding, we speculated that Pin1 plays a critical role in diet-induced adipogenesis. First, Pin1 expression was shown to be up-regulated in 3T3-L1 cells during adipose differentiation (Fig. 6A). Then, the effect of Pin1 gene silencing using siRNA on adipose differentiation of 3T3-L1 adipocytes or human preadipocytes was examined (Fig. 6, B and C, and supplemental Fig. 7). Adipose differentiation of these cells was markedly impaired with suppressed inductions of PPARγ, c/EBPα, and C/EBPβ, known as master regulator genes of adipose differentiation (27–29) (Fig. 6D). In addition, expression of PPARγ protein, a marker of mature adipocytes, was reduced by Pin1 knockdown (Fig. 6C). Compensatory overexpression of PPARγ overcomes the suppressive effect of Pin1 siRNA on adipose differentiation (Fig. 6C).
FIGURE 6.
Pin1 plays a critical role in adipose differentiation. A, 3T3-L1 preadipocytes were induced to differentiate into adipocytes, and Pin1 protein and mRNA levels were determined at the indicated periods after initiation of differentiation. IB, immunoblots. B, 3T3-L1 preadipocytes were treated with Pin1 siRNA. The amounts of Pin1 and actin protein were determined with immunoblotting. C, 3T3-L1 preadipocytes were treated with control or Pin1 siRNA,or in combination with Pin1 siRNA and FLAG-tagged PPARγ adenovirus gene transfer. These cells were then induced to differentiate into adipocytes. Microscopic observations and oil red O staining results at 8 days are shown. Expression of PPARγ protein was detected as a marker of adipocyte maturity. Scale bars = 50 μm. D, before and 2 or 6 days after the induction of differentiation, PPARγ, c/EBPα, and c/EBPβ mRNA levels of 3T3-L1 cells were determined by real-time PCR. *, p < 0.05; **, p < 0.05.
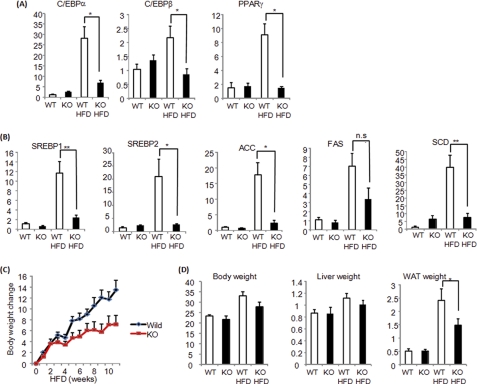
Next, the adipose tissue of Pin1 KO mice was investigated. In normal mouse epididymal adipose tissue, PPARγ, c/EBPα, and CEBP/β as well as SREBP1, SREBP2, acetyl-CoA carboxylase, fatty acid synthase, and stearoyl-CoA-desaturase were markedly up-regulated by HFD feeding, which contributed to the enlargement of adipose tissues. In contrast, in Pin1 KO mouse adipose tissues, these increased gene expressions were significantly suppressed (Fig. 7, A and B), thereby markedly suppressing adipose tissue enlargement and weight gain on the HFD (Fig. 7, C and D). Thus, we concluded that Pin1 is a key regulator of diet-induced adipogenesis as Pin1 KO mice were clearly resistant to HFD-induced obesity.
FIGURE 7.
Pin1 KO mice are resistant to HFD-induced obesity. A, PPARγ, c/EBPα, and c/EBPβ mRNA levels in the adipose tissues of wild-type or Pin1 KO mice, fed a control diet or HFD for 4 weeks, were determined. B, SREBP1, SREBP2, ACC, FAS, and SCD mRNA levels in adipose tissues from wild-type or Pin1 KO mice, fed a control or HFD for 4 weeks, were determined by real-time PCR. C, weight gain after HFD feeding was compared between wild-type and Pin1 KO mice. D, the weights of the liver and adipose tissue were compared between wild-type and Pin1 KO mice, which had been fed either a control diet or HFD for 4 weeks. ACC, acetyl-CoA carboxylase; FAS, fatty acid synthase; SCD, stearoyl-CoA-desaturase.
DISCUSSION
Pin1 amounts increased according to nutrient state, as shown by marked elevation on a high-fat as compared with a normal diet. The Pin1 level was also higher in the fed than in the fasted state (19). Whereas previous studies have shown that Pin1 expression generally correlates with cell proliferative potential in normal tissues (7, 12) and is further up-regulated in many human cancers (30, 31), our findings are the first suggesting a relationship between Pin1 protein and nutrient conditions.
We also demonstrated that Pin1 is a positive regulator of insulin signaling via enhanced insulin-induced IRS-1 phosphorylation, based on a data series obtained using in vitro and in vivo overexpression of Pin1, gene silencing, a specific inhibitor, and Pin1 KO mice. The principal insulin receptor substrates, IRS-protein family members such as IRS-1 and IRS-2, adaptor proteins from the activated insulin receptor, are tyrosine-phosphorylated and thereby activate phosphatidylinositol 3-kinase/Akt. Pin1 binds to the Ser-434-containing motif of IRS-1, which is in the SAIN domain, via its WW domain. Because the SAIN domain reportedly plays an important role in the association with the insulin receptor (21, 22), it is speculated that Pin1 modifies the conformation of the SAIN domain and thereby enhances IRS-1 tyrosine phosphorylation by the insulin receptor. Similar enhanced phosphorylation of signal transduction protein via association with Pin1 has been reported for STAT3 (32).
Regarding the downstream signaling from the insulin receptor, the phosphatidylinositol 3-kinase/Akt pathway activation is essential for almost all insulin-induced glucose and lipid metabolism activities, e.g. glucose uptake, glycogen synthesis, suppression of glucose output, and triglyceride synthesis. Thus, in the muscles of Pin1 KO mice showing impaired Akt activation, glucose incorporation into muscle with insulin stimulation was decreased.
Insulin-induced phosphatidylinositol 3-kinase/Akt activations also play a role in adipogenesis (25, 33). It was shown that Pin1-deficient preadipocytes clearly fail to differentiate into adipocytes. This suppression was accompanied by insufficient inductions of PPARγ, c/EBPα, and c/EBPβ. On the other hand, overexpression of PPARγ reversed the Pin1 siRNA-induced suppression of adipose differentiation. Although the molecular mechanisms underlying insulin-induced induction of adipogenic genes has yet to be fully elucidated, previous reports have shown the essential action of SREBPs for adipogenesis to probably be via regulation of its downstream PPARγ (34, 35). Indeed, in adipose tissue from Pin1 KO mice, SREBPs and downstream gene expressions were decreased. These observations indicate that, as a consequence of impaired adipose differentiation, Pin1 KO mice are resistant to HFD-induced obesity.
Based on our data series, we can speculate as to the physiological significance of Pin1 with respect to metabolic regulation. When nutrients enter the body, excess glucose, lipids, and amino acids are stored mainly in the liver, muscle, and adipose tissue. Although insulin is the critical hormone for this process, we consider Pin1 to be involved in the mechanisms enhancing insulin sensitivity in peripheral tissues. Increased Pin1 expression functions to increase insulin sensitivity, thereby maintaining homeostasis in response to food or excess nutrient intake. Intriguingly, Pin1 enhances the signal from IRS-1 but not that from IRS-2. Although IRS-2 bound to Pin1 when both were overexpressed in HepG2 cells (supplemental Fig. 3), no association of endogenous IRS-2 and Pin1 was detected (data not shown). Thus, it is likely that IRS-2 associates with Pin1 far less efficiently than does IRS-1, which might account for the different effects of Pin1 on IRS-1 and IRS-2. Although several studies have revealed different roles for IRS-1 and IRS-2 (36, 37), we speculate that selective enhancement of IRS-1 signaling may contribute to greater hepatic lipid synthesis, as IRS-1 phosphorylation lasts much longer than that of IRS-2. Indeed, constitutive Akt overexpression induces marked lipid accumulation in the liver with blood glucose lowering (24). Thus, increased Pin1 expression is considered to be a physiologically beneficial response to excess nutrient intake.
However, excessive energy intake eventually causes fatty liver and obesity, which lead to the development of metabolic syndrome. In this condition, several kinases including mammalian target of rapamycin (mTOR), inhibitory κB kinase β, c-Jun N-terminal kinase, extracellular signal regulated kinase, and S6 kinase, induce the phosphorylations of serine 307, 302 and 612 of IRS-1, leading to reduced insulin-induced IRS-1 tyrosine phosphorylations and thereby contributing to insulin resistance (38–43). Thus, the efficiency of IRS-1 tyrosine phosphorylations appears to be up-regulated by increased Pin1 association and down-regulated by the aforementioned multiple serine phosphorylations. We speculate that both phenomena take place under conditions of energy excess, but when prolonged, the latter down-regulating mechanism becomes dominant.
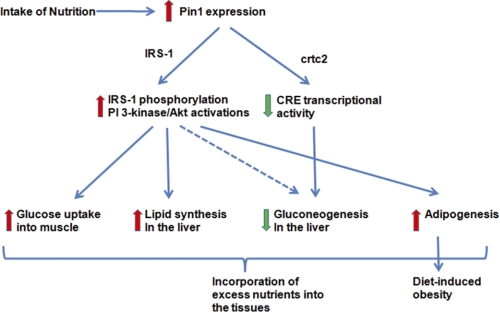
In conclusion, we have demonstrated the important role of Pin1 as a positive modulator of insulin signaling as well as an inducer of obesity. Thus, Pin1 might be regarded as somewhat of a double-edged sword in that it increases insulin sensitivity but also promotes obesity (Fig. 8). However, suppression of Pin1 activity specifically in adipocytes might be a novel preventive treatment for obesity. Otherwise, if hepatic lipid accumulation is not severe, agents increasing Pin1 expression or enzymatic activity, i.e. targeting Pin1, may hold promise for treating Type 2 diabetes mellitus by improving hepatic insulin sensitivity.
FIGURE 8.
Schema show the role of Pin1 in glucose and lipid metabolism in response to nutrient intakes.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–7.
- IRS-1
- insulin receptor substrate-1
- HFD
- high-fat diet
- SAIN
- Shc and IRS-1 NPXY binding
- PPAR
- peroxisome proliferator-activated receptor
- MEF
- myc-TEV (tobacco etch virus)-Flag.
REFERENCES
- 1. Fischer G., Wittmann-Liebold B., Lang K., Kiefhaber T., Schmid F. X. (1989) Nature 337, 476–478 [DOI] [PubMed] [Google Scholar]
- 2. Liu J., Farmer J. D., Jr., Lane W. S., Friedman J., Weissman I., Schreiber S. L. (1991) Cell 66, 807–815 [DOI] [PubMed] [Google Scholar]
- 3. Choi J., Chen J., Schreiber S. L., Clardy J. (1996) Science 273, 239–242 [DOI] [PubMed] [Google Scholar]
- 4. Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002) Mol. Cell 10, 457–468 [DOI] [PubMed] [Google Scholar]
- 5. Schreiber S. L. (1991) Science 251, 283–287 [DOI] [PubMed] [Google Scholar]
- 6. Kunz J., Hall M. N. (1993) Trends Biochem. Sci. 18, 334–338 [DOI] [PubMed] [Google Scholar]
- 7. Lu K. P., Hanes S. D., Hunter T. (1996) Nature 380, 544–547 [DOI] [PubMed] [Google Scholar]
- 8. Wulf G., Finn G., Suizu F., Lu K. P. (2005) Nat. Cell Biol. 7, 435–441 [DOI] [PubMed] [Google Scholar]
- 9. Lu K. P., Zhou X. Z. (2007) Nat. Rev. Mol. Cell Biol. 8, 904–916 [DOI] [PubMed] [Google Scholar]
- 10. Takahashi K., Uchida C., Shin R. W., Shimazaki K., Uchida T. (2008) Cell. Mol. Life Sci. 65, 359–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou X. Z., Kops O., Werner A., Lu P. J., Shen M., Stoller G., Küllertz G., Stark M., Fischer G., Lu K. P. (2000) Mol. Cell 6, 873–883 [DOI] [PubMed] [Google Scholar]
- 12. Winkler K. E., Swenson K. I., Kornbluth S., Means A. R. (2000) Science 287, 1644–1647 [DOI] [PubMed] [Google Scholar]
- 13. Zacchi P., Gostissa M., Uchida T., Salvagno C., Avolio F., Volinia S., Ronai Z., Blandino G., Schneider C., Del Sal G. (2002) Nature 419, 853–857 [DOI] [PubMed] [Google Scholar]
- 14. Zheng H., You H., Zhou X. Z., Murray S. A., Uchida T., Wulf G., Gu L., Tang X., Lu K. P., Xiao Z. X. (2002) Nature 419, 849–853 [DOI] [PubMed] [Google Scholar]
- 15. Lu P. J., Wulf G., Zhou X. Z., Davies P., Lu K. P. (1999) Nature 399, 784–788 [DOI] [PubMed] [Google Scholar]
- 16. Pastorino L., Sun A., Lu P. J., Zhou X. Z., Balastik M., Finn G., Wulf G., Lim J., Li S. H., Li X., Xia W., Nicholson L. K., Lu K. P. (2006) Nature 440, 528–534 [DOI] [PubMed] [Google Scholar]
- 17. Liou Y. C., Sun A., Ryo A., Zhou X. Z., Yu Z. X., Huang H. K., Uchida T., Bronson R., Bing G., Li X., Hunter T., Lu K. P. (2003) Nature 424, 556–561 [DOI] [PubMed] [Google Scholar]
- 18. Nakatsu Y., Sakoda H., Kushiyama A., Ono H., Fujishiro M., Horike N., Yoneda M., Ohno H., Tsuchiya Y., Kamata H., Tahara H., Isobe T., Nishimura F., Katagiri H., Oka Y., Fukushima T., Takahashi S., Kurihara H., Uchida T., Asano T. (2010) J. Biol. Chem. 285, 33018–33027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ichimura T., Yamamura H., Sasamoto K., Tominaga Y., Taoka M., Kakiuchi K., Shinkawa T., Takahashi N., Shimada S., Isobe T. (2005) J. Biol. Chem. 280, 13187–13194 [DOI] [PubMed] [Google Scholar]
- 20. Fujimori F., Takahashi K., Uchida C., Uchida T. (1999) Biochem. Biophys. Res. Commun. 265, 658–663 [DOI] [PubMed] [Google Scholar]
- 21. Hennig L., Christner C., Kipping M., Schelbert B., Rücknagel K. P., Grabley S., Küllertz G., Fischer G. (1998) Biochemistry 37, 5953–5960 [DOI] [PubMed] [Google Scholar]
- 22. Gustafson T. A., He W., Craparo A., Schaub CD., O'Neill T. J. (1995) Mol. Cell. Biol. 15, 2500–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yenush L., Makati K. J., Smith-Hall J., Ishibashi O., Myers M. G., Jr., White M. F. (1996) J. Biol. Chem. 271, 24300–24306 [DOI] [PubMed] [Google Scholar]
- 24. Ono H., Shimano H., Katagiri H., Yahagi N., Sakoda H., Onishi Y., Anai M., Ogihara T., Fujishiro M., Viana A. Y., Fukushima Y., Abe M., Shojima N., Kikuchi M., Yamada N., Oka Y., Asano T. (2003) Diabetes 52, 2905–2913 [DOI] [PubMed] [Google Scholar]
- 25. Kerouz N. J., Hörsch D., Pons S., Kahn C. R. (1997) J. Clin. Invest. 100, 3164–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakae J., Kitamura T., Kitamura Y., Biggs W. H., 3rd, Arden K. C., Accili D. (2003) Dev. Cell 4, 119–129 [DOI] [PubMed] [Google Scholar]
- 27. Umek R. M., Friedman A. D., McKnight S. L. (1991) Science 251, 288–292 [DOI] [PubMed] [Google Scholar]
- 28. Tanaka T., Yoshida N., Kishimoto T., Akira S. (1997) EMBO J. 16, 7432–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Spiegelman B. M. (1998) Diabetes 47, 507–514 [DOI] [PubMed] [Google Scholar]
- 30. Ryo A., Nakamura M., Wulf G., Liou Y. C., Lu K. P. (2001) Nat. Cell Biol. 3, 793–801 [DOI] [PubMed] [Google Scholar]
- 31. Wulf G. M., Ryo A., Wulf G. G., Lee S. W., Niu T., Petkova V., Lu K. P. (2001) EMBO J. 20, 3459–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lufei C., Koh T. H., Uchida T., Cao X. (2007) Oncogene 26, 7656–7664 [DOI] [PubMed] [Google Scholar]
- 33. Rosen E. D., MacDougald O. A. (2006) Nat. Rev. Mol. Cell Biol. 7, 885–896 [DOI] [PubMed] [Google Scholar]
- 34. Tontonoz P., Kim J. B., Graves R. A., Spiegelman B. M. (1993) Mol. Cell. Biol. 13, 4753–4759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiang L., Wang Q., Yu Y., Zhao F., Huang P., Zeng R., Qi R. Z., Li W., Liu Y. (2009) PLoS One 4, e6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Withers D. J., Gutierrez J. S., Towery H., Burks D. J., Ren J. M., Previs S., Zhang Y., Bernal D., Pons S., Shulman G. I., Bonner-Weir S., White M. F. (1998) Nature 391, 900–904 [DOI] [PubMed] [Google Scholar]
- 37. Taniguchi C. M., Emanuelli B., Kahn C. R. (2006) Nat. Rev. Mol. Cell Biol. 7, 85–96 [DOI] [PubMed] [Google Scholar]
- 38. Hotamisligil G. S., Peraldi P., Budavari A., Ellis R., White M. F., Spiegelman B. M. (1996) Science 271, 665–668 [DOI] [PubMed] [Google Scholar]
- 39. Hirosumi J., Tuncman G., Chang L., Görgün C. Z., Uysal K. T., Maeda K., Karin M., Hotamisligil G. S. (2002) Nature 420, 333–336 [DOI] [PubMed] [Google Scholar]
- 40. Pirola L., Johnston A. M., Van Obberghen E. (2004) Diabetologia 47, 170–184 [DOI] [PubMed] [Google Scholar]
- 41. Werner E. D., Lee J., Hansen L., Yuan M., Shoelson S. E. (2004) J. Biol. Chem. 279, 35298–35305 [DOI] [PubMed] [Google Scholar]
- 42. Ozcan U., Yilmaz E., Ozcan L., Furuhashi M., Vaillancourt E., Smith R. O., Görgün C. Z., Hotamisligil G. S. (2006) Science 313, 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morino K., Neschen S., Bilz S., Sono S., Tsirigotis D., Reznick R. M., Moore I., Nagai Y., Samuel V., Sebastian D., White M., Philbrick W., Shulman G. I. (2008) Diabetes 57, 2644–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.