Abstract
Membrane PTK7 pseudo-kinase plays an essential role in planar cell polarity and the non-canonical Wnt pathway in vertebrates. Recently, a new N-ethyl-N-nitrosourea-induced mutant named chuzhoi (chz) was isolated in mice. chz embryos have severe birth defects, including a defective neural tube, defective heart and lung development, and a shortened anterior-posterior body axis. The chz mutation was mapped to the Ala-Asn-Pro tripeptide insertion into the junction region between the fifth and the sixth Ig-like domains of PTK7. Unexpectedly, chz reduced membrane localization of the PTK7 protein. We hypothesized and then proved that the chz mutation caused an insertion of an additional membrane type 1 matrix metalloproteinase cleavage site in PTK7 and that the resulting aberrant proteolysis of chz affected the migratory parameters of the cells. It is likely that aberrations in the membrane type 1 matrix metalloproteinase/PTK7 axis are detrimental to cell movements that shape the body plan and that chz represents a novel model system for increasing our understanding of the role of proteolysis in developmental pathologies, including congenital defects.
Keywords: Development, Plasma Membrane, Protein Kinases, Proteolytic Enzymes, Wnt Pathway, MT1-MMP, PTK7, chuzhoi
Introduction
About three of every 100 babies born in the United States have congenital abnormalities. Congenital abnormalities are ranked 34th of the top 50 causes of total death in the United States. Debilitating cardiovascular and nervous system defects jointly represent 40% of birth defects. The death toll of congenital abnormalities (0.58%) roughly equals that of stomach, skin, or oral cancers. Mechanistic research into birth defects is required to more efficiently contribute to prevention and correction of these disorders.
The canonical, β-catenin-dependent and non-canonical, β-catenin-independent Wnt signaling pathways are conserved in eukaryotes and are critical for the diverse events in embryonic development and disease. The non-canonical Wnt/planar cell polarity (PCP)2 signaling controls cell organization within the tissue plane (1, 2). The Wnt/PCP signaling affects the actin cytoskeleton via RhoA GTPase activation through dishevelled associated activator of morphogenesis 1 (DAAM1) and dishevelled activator of morphogenesis 2 (DAAM2). The first PCP signaling events occur at a gastrulation stage of embryogenesis. These events regulate the polarized directed cell movement to accomplish convergent extension for the anterior-posterior body axis elongation, neural tube closure, and craniofacial morphogenesis (3–6). Convergent extension failure results in a shortened anterior-posterior body axis and widened lateral axis (convergent extension phenotype), a defective neural system, and craniofacial abnormalities.
Ubiquitously expressed PTK7 (also called colon carcinoma kinase-4 (CCK-4)) is a Wnt coreceptor and a regulator of PCP in vertebrates (7–13). Membrane PTK7 includes seven extracellular Ig domains, a transmembrane region, and a catalytically inert cytoplasmic tyrosine kinase (PTK) domain (8, 9, 11, 14). PTK7 is evolutionary conserved in humans and mice and also chicken klingon (KLG), Drosophila (Dtrk/Off-track), and hydra (Lemon) (7). It is likely that PTK7 regulates PCP and signaling via homotypic interactions between the extracellular Ig domains and via the cytoplasmic PTK domain, respectively. PTK7 can form multimers (10). PTK7 signaling can be achieved through its interactions with plexins and semaphorins, albeit the precise mechanisms are yet to be determined (15, 16). PTK7 mutant mice that expressed either a 1–114 PTK7 truncation or a cytoplasmic domain deletion died because of severe defects in neural tube closure (10).
Recently, a new N-ethyl-N-nitrosourea-induced mutant named chuzhoi (chz) was isolated and characterized in mice. chz mutant embryos have defective convergent extension, including a broadened midline, a shortened anterior-posterior body axis, abnormal cell polarity in the ear, and defective neural tube and heart and lung development. The cause of chz is the unnatural Ala-Asn-Pro insertion into the junction region between the 5th and the 6th Ig-like domains of the PTK7 ectodomain portion. We determined that the insertion, unexpectedly, significantly reduced membrane localization of the PTK7 protein (17).
Recent studies indicate that invasion-promoting MT1-MMP controls PCP in zebrafish (Danio rerio) development (18). MT1-MMP is a prototypic member of a membrane-anchored MMP subfamily and is distinguished from soluble MMPs by a C-terminal transmembrane domain and a cytoplasmic tail (19, 20). MT1-MMP cleaves extracellular matrix proteins, initiates the activation pathway of soluble MMPs and controls the functionality of cell adhesion and signaling receptors (21–25). Knockout of MT1-MMP has the most significant phenotype among MMP gene knockout mice: MT1-MMP knockout mice are dwarfs and die at adulthood (26). Likewise, a loss of the structurally similar primordial At2-MMP induces dwarfism in Arabidopsis plants (27).
MT1-MMP knockdown affects PCP and directed migration of the mesodermal cells in gastrulation and craniofacial morphogenesis (18). Van Gogh-like 2, a regulator of the non-canonical Wnt pathway, colocalizes with MT1-MMP and redistributes toward the leading edge of polarized human cancer cells (18). It is likely that pericellular proteolysis and PCP converge to promote efficient directed cell migration (28).
As a result, it is not entirely surprising that PTK7 and MT1-MMP are functionally linked in eukaryotes (18, 28–30). Cellular MT1-MMP, a key proinvasive proteinase, functions as a principal PTK7 sheddase. MT1-MMP cleaves the PKP621↓LI sequence of the seventh Ig-like domain of membrane PTK7. MT1-MMP proteolysis generates the C-terminal, membrane-tethered (50-kDa) and the N-terminal, soluble (70-kDa) PTK7 1–621 fragments. Cleavage site L622D mutation results in the PTK7 mutant that is resistant to MT1-MMP. Our analysis of the MT1-MMP/PTK7 axis suggests that a balance between the MT1-MMP activity and the membrane/soluble PTK7 species is required for the correct execution of the cell migration program in both human cancer cells and zebrafish (D. rerio). An abnormal balance caused defects in both zebrafish development and directional migration of tumor cells.
On the basis of these data and the protein sequence of PTK7, we then hypothesized that the Ala-Asn-Pro insertion (QVLANPEK↓LK503, the arrow indicates the scissile bond, the insert is underlined) introduced a consensus MMP PXX↓L cleavage site in the PTK7 sequence. Here, we experimentally confirmed our hypothesis and demonstrated that human chz exhibits an additional MT1-MMP cleavage site. It becomes now possible to suggest that the N-terminal, soluble (70-kDa) 1–621 chz fragment readily degrades in vivo. This degradation of soluble chz leads to the aberrant membrane/soluble PTK ratio and to the significantly modified migratory and signaling cell characteristics (17, 29).
Overall, our data, especially if combined with the results of Paudyal el al. (17), in mice suggest that mutational aberrations of the MT1-MMP/PTK7 axis cause the PCP abnormalities during gastrulation and birth defects in eukaryotes. Further analysis is warranted to determine whether PTK7 controls cell polarity in different contexts in the course of normal development and how mutations in PTK7 disrupt PCP in relation to disease, including birth defects in humans.
MATERIALS AND METHODS
Cells and Reagents
Human fibrosarcoma HT1080 cells and human breast carcinoma MCF7 cells were obtained from the ATCC. The cells were grown in DMEM supplemented with 10% FBS. A goat polyclonal antibody AF4499 against the N-terminal 31–199 portion of PTK7 was purchased from R&D Systems. A murine monoclonal 3G4 antibody, MAB1767, against the catalytic domain of MT1-MMP and the GM6001 hydroxamate inhibitor were from Chemicon. A murine monoclonal FLAG M2 antibody, the FLAG M2 antibody, and streptavidin-agarose beads were from Sigma. Rhotekin-Rho binding domain (RBD)-agarose beads and a RhoA monoclonal antibody, ARH01, were from Cytoskeleton. EZ-Link sulfosuccinimidyl 2-(biotinamido)-ethyl-1,3-dithiopropionate and Alexa Fluor 594-phalloidin were acquired from Pierce and Invitrogen, respectively.
Cloning and Mutagenesis
HT1080 cells with the ectopic expression of MT1-MMP (HT-MT1 cells), HT1080 cells in which MT1-MMP was transcriptionally silenced (over 90% silencing) using siRNA (HT-siMT1 cells), and the required scrambled controls were obtained and extensively characterized earlier (31–34). The C-terminally tagged (with a V5 tag) wild type and catalytically inert (E240A) mutant MT1-MMP constructs (29, 32) were used to generate the MCF7-MT1 and MCF7-E240A cells, respectively. HT1080 cells with the ectopic expression of the FLAG-tagged PTK7 constructs, including the full-length wild-type PTK7 (HT-PTK7), the uncleavable PTK7 L622D mutant (HT-L622D), and the N-terminal-soluble, 1–700 ectodomain form of PTK7 (HT-sPTK7), were obtained and characterized earlier (29). The PTK7-FLAG template was used in the PCR reactions with the following primers: forward, 5′-CGTGTCCAAGTGCTGGCTAACCCAGAAAAGCTCAAGTTC-3′; and reverse, 5′-GAACTTGAGCTTTTCTGGGTTAGCCAGCACTTGGACACG-3′ to generate the chz mutant sequence (the mutant sequence is underlined). The resulting chz construct was subcloned into the pcDNA3.1D/V5-His-TOPO directional TOPO expression vector (Invitrogen). HT1080 and MCF7 cells were stably transfected with the chz construct using Lipofectamine 2000 (Invitrogen) to generate HT-chz and MCF7-chz cells, respectively. Stably transfected clones were selected in the presence of G418. The expression of chz was validated by Western blotting with the FLAG and PTK7 antibodies.
Cell Surface Biotinylation
Cell surface proteins were biotinylated by incubating cells for 1 h on ice in PBS containing 0.1 mg/ml EZ-Link sulfosuccinimidyl 2-(biotinamido)-ethyl-1,3-dithiopropionate. Cells were lysed in 20 mm Tris-HCl, 150 mm NaCl, 1% deoxycholate, and 1% octylphenoxypolyethoxyethanol (IGEPAL) (pH 7.4), supplemented with a protease inhibitor mixture set III (Sigma), 1 mm phenylmethylsulfonyl fluoride, and 10 mm EDTA. Biotinylated proteins were precipitated from cell lysates using streptavidin-agarose beads (Sigma). Biotinylated proteins were eluted from the beads using 2× SDS sample loading buffer (125 mm Tris-HCl (pH 6.8), 4% (v/v) SDS, 0.005% bromphenol blue, 20% (v/v) glycerol, 20 mm DTT).
Immunofluorescence
Cells grown on glass coverslips were fixed for 10 min with 4% paraformaldehyde, permeabilized for 5 min with 0.1% Triton X-100, and blocked for 1 h in 1% casein. Cells were stained with a primary antibody (normally at a 1:1000 dilution), followed by a secondary antibody conjugated with Alexa Fluor 488 or Alexa Fluor 594 (Molecular Probes, dilution 1:500). The coverslips were mounted in Vectashield mounting medium with DAPI (Vector Laboratories). Images were acquired using an Olympus BX51 fluorescence microscope equipped with a MagnaFire digital camera (Olympus).
Rho Activation Assay
The Rho activation assay kit (Cytoskeleton) was used to measure cellular GTP-RhoA. GTP-RhoA was precipitated from the cell lysates using rhotekin-RBD beads. The precipitated samples were analyzed by Western blotting with a RhoA monoclonal antibody.
Invasion Assay
The invasion assay was performed in triplicate in wells of a 24-well Transwell plate with an 8-μm pore size membrane (32). The membranes of the Transwell inserts were coated with type I collagen (2.5 μg/well, BD Biosciences). Cells (1 × 105/well) were placed into DMEM (0.1 ml) in the upper chamber. The 10% FBS-containing DMEM (used as a chemoattractant, 0.6 ml) was placed in the lower chamber. Serum-free DMEM (0.6 ml) was used as a control. Cells were allowed to invade for 4 h. The cells were stained for 10 min with 0.2% crystal violet/20% methanol (0.3 ml). The cells on the upper membrane surface were removed with a cotton swab. The dye from the cells that migrated onto the lower surface of the membrane was extracted with 1% SDS (0.25 ml). The resulting A570 nm was measured using a plate reader.
Gelatin Substrate Zymography of MMP-2 and Western Blotting of RhoA, PTK7, and MT1-MMP
Basic protocols for these techniques have been described previously (29). The purified MMP-2 proenzyme (100 ng/ml) was added to MCF7 cells 12–14 h prior to the gelatin zymography analysis (35). For the Western blotting analysis of sPTK7, cells were incubated for 16 h in serum-free DMEM (20 ml/15-cm dish). The medium was then collected and concentrated 50-fold using Amicon Ultra-15 centrifugal filter units with a 30-kDa cutoff (Millipore). The concentrated samples (20 μl each) were used in Western blotting with a PTK7 antibody.
Modeling of the PTK7 and chz Structures
The three-dimensional structure of PTK7 was modeled by threading the PTK7 peptide sequence on the known structures of the homologues using MODWEB software (36, 37). The six N-terminal Ig domains of PTK7 (residues 28–588) were built using titin (PDB code 3B43) as a template. Because of their high (38%) sequence identity, the seventh Ig domain of PTK7 (residues 594–684) was built using the 22–105 fragment of Kiaa1556 (PDB code 2DM7). The 703–778 transmembrane region and the 789–1072 kinase domain of PTK7 were built using Hla (PDB code 1SYS) and a 258–525 fragment of Src (PDB code 2BDF; 38% sequence identity), respectively. The modeled fragment structures were merged to generate the structural model of PTK7 and chz. The structure of the catalytic domain of MT1-MMP was derived directly from PDB codes 1BQQ and 1BUV. The structures were then visualized using PyMOL software (DeLano Scientific).
RESULTS
PTK7 and chz Modeling
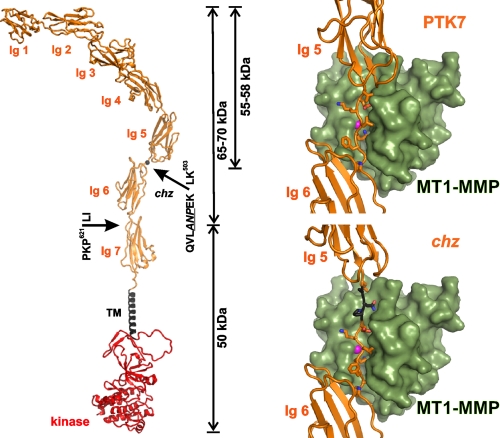
To visualize the structure of chz, we modeled the three-dimensional structures of PTK and chz (Fig. 1). Our structural modeling suggests that the conventional MT1-MMP PKP621↓LI cleavage site is localized in the seventh Ig-like domain of PTK7. The Ala-Asn-Pro insertion that potentially results in the creation of the additional Pro-Glu-Lys↓Leu-Lys503 MMP cleavage site in the chz mutant sequence is localized in the junction region between the fifth and the sixth Ig-like domains.
FIGURE 1.
A structure model of the full-length membrane PTK7. Left panel, Ig domains 1–7, the transmembrane (TM), and kinase domains are shown in orange, black, and red, respectively. Cleavage of the PKP621↓LI sequence in the seventh Ig-like domain and of the QVLANPEK↓LK503 chz in the junction region between the fifth and the sixth Ig-like domains, respectively, is shown by the arrows. The mutant residues are underlined. Right panel, a structural model of the QVLEK↓LK503 original PTK7 and the QVLANPEK↓LK503 chz mutant sequences in the catalytic cleft of the MT1-MMP catalytic domain. Green, MT1-MMP; orange, PTK7/chz; black, Ala-Asn-Pro insert in chz; magenta ball, active site zinc.
The original QVLEKLK503 PTK7 sequence does not appropriately fit into the catalytic cleft of the MT1-MMP catalytic domain. In contrast, the QVLANPEK↓LK503 chz mutant sequence matches well the structural parameters of the active site cleft of MT1-MMP. The P1′ Leu-502 of the putative Lys-Leu scissile bond appears proximal to the active site zinc (Fig. 1).
Cell Surface chz Is Efficiently Degraded
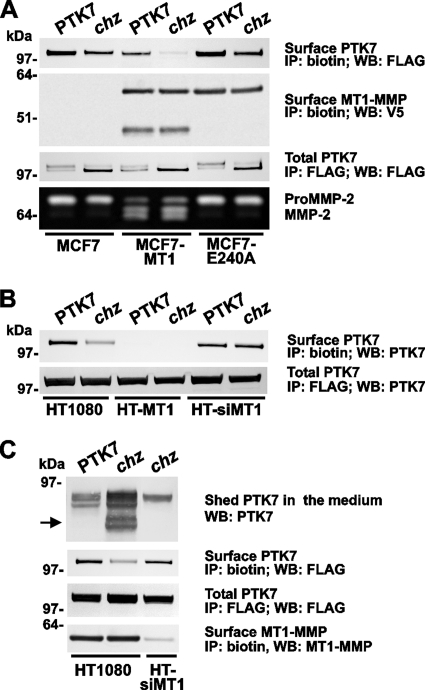
To determine the expression levels and potential link of PTK7 with MT1-MMP, the FLAG-tagged PTK7 and chz constructs were designed. Breast carcinoma MCF-7 cells were stably transfected with the wild-type PTK7 and chz constructs. In addition, PTK7 and chz were coexpressed in MCF-7 cells with the wild-type MT1-MMP and the catalytically inactive E240A MT1-MMP mutant (MCF7-MT1 and MCF7-E240A cells, respectively). To facilitate the identification of MT1-MMP, the proteinase constructs were tagged with a V5 tag. We specifically selected MCF7 cells for our studies because the parental cell line is deficient in MT1-MMP (38). As expected, gelatin zymography of the medium aliquots confirmed the ability of MCF7-MT1 cells to transform the proenzyme of MMP-2 into the activated MMP-2 forms, whereas parental MCF7 cells and MCF7-E240 cells were inactive in this assay (Fig. 2A).
FIGURE 2.
Chz is efficiently cleaved by MT1-MMP. A, cell surface, biotin-labeled, and total cell lysate immunoprecipitated samples (surface and total, respectively) of MCF7 cells expressing the FLAG-tagged PTK7/chz constructs and of MCF7-MT1 and MCF7-E240 cells that coexpressed the FLAG-tagged PTK7/chz constructs with the V5-tagged MT1-MMP and the catalytically inert E240 MT1-MMP mutant constructs, respectively, were analyzed by Western blotting. Bottom panel, the ability of cellular MT1-MMP to activate the added MMP-2 proenzyme (100 ng/ml) was analyzed with gelatin zymography. B, cell surface, biotin-labeled, and total cell lysate samples (surface and total, respectively) of HT1080, HT-MT1, and HT-siMT1 cells expressing the FLAG-tagged PTK7/chz constructs were analyzed by Western blotting. C, conditioned medium aliquots and cell surface, biotin-labeled, and total cell lysate immunoprecipitated samples (surface and total, respectively) of HT1080 cells expressing the FLAG-tagged PTK7/chz constructs and of HT-siMT1 cells expressing the FLAG-tagged chz construct were analyzed by Western blotting. Equal amounts of total protein (50 μg/lane) were analyzed in A, B, and C. The arrow indicates the abnormal cleavage product in chz. IP, immunoprecipitation with the FLAG antibody and streptavidin beads. WB, Western blotting with the FLAG, V5, PTK7, and MT1-MMP antibodies. For the beads and antibodies used, see panel labels.
Cells were surface-biotinylated and then lysed. Both biotin-labeled, cell surface-associated chz and MT1-MMP were precipitated using streptavidin beads. Cells expressing PTK7 were used as controls in our experiments. The precipitated chz and MT1-MMP samples were analyzed by Western blotting with the FLAG and V5 antibodies, respectively. MT1-MMP was represented by the enzyme form on the surface of MCF7-E240A and MCF7-MT1 cells. The latter also exhibited the 45-kDa self-proteolysis degradation form that was absent in MCF7-E240 cells. The levels of cell surface chz inversely correlated with the cell surface MT1-MMP activity. Thus, minor levels of chz as compared with PTK7 were detected on the surface of MCF7-MT1 cells (Fig. 2A).
To corroborate these data, we determined the levels of total cellular PTK7 and chz in MCF7, MCF7-MT1, and MCF7-E240A cells (Fig. 2A). For these purposes, cells were lysed, and chz was pulled down from the cell extracts using the FLAG antibody beads. The precipitates were analyzed by Western blotting with the FLAG antibody. The chz construct was efficiently synthesized by MCF7, MCF7-MT1, and MCF7-E240A. The presence of the two isoforms of PTK7/chz we observed in the cell extracts is not precisely clear as of now. The levels of cellular chz exceeded those of PTK7 in all three lines. The cell surface levels of chz, however, were low, especially in MCF7-MT1 cells.
These results suggest the existence of the efficient degradation of cell surface chz rather than the low synthesis of chz by the cells and that this degradation of cell surface chz is MT1-MMP-dependent. Because breast carcinoma MCF7 cells do not efficiently invade the matrix layers in vitro, it is exceedingly difficult to observe the effects of PTK7, chz, and MT1-MMP on the invasion of this cell type.
To support our data further and to determine the potential effect of PTK7 and chz on cell invasion, we employed highly migratory and invasive fibrosarcoma HT1080 cells, which synthesize MT1-MMP naturally. First, we determined the status of cellular chz in HT1080 cells. As controls, we used HT-siMT1 cells with the transcriptional silencing of MT1-MMP and also HT-MT1 cells with the forced expression of MT1-MMP (Fig. 2B). To measure cell surface chz, the cells were surface-biotinylated and lysed. Biotin-labeled material was captured using streptavidin beads. To determine the total cell chz, untreated cells were lysed, and the FLAG-tagged constructs were captured on FLAG antibody beads. Both biotin-labeled and total cell samples were then analyzed by Western blotting with the antibody against the PTK7 ectodomain. There was no difference in the levels of total PTK7 and chz in HT1080, HT-MT1, and HT-siMT1 cells. Cell surface chz was diminished in HT1080 cells as compared with PTK7. Both PTK7 and chz were predominantly shed by MT1-MMP activity in HT-MT1 cells. As a result, there was no detectable PTK7 and chz in the HT-MT1 cell surface samples. Transcriptional knockout of MT1-MMP restored the cell surface PTK7 and chz in HT-siMT1 cells.
In agreement, as detected by Western blotting with the FLAG antibody, there was no difference in the total levels of cellular PTK7 and chz in HT1080 and HT-siMT1 cells. Consistent with other assays, minor cell surface levels of chz were observed in HT1080 cells as compared with PTK7. The surface levels of chz significantly increased in HT-siMT1 cells. Because MT1-MMP is a principal sheddase of membrane PTK7 (29), we also assessed the levels of the shed chz ectodomain in the concentrated medium samples using Western blotting with the PTK7 antibody. MT1-MMP proteolysis released the soluble, 65- to 70-kDa PTK7 1–621 ectodomain into the medium (Fig. 2C). As compared with PTK7, extensive shedding of chz was observed in HT1080 cells. This shedding was greatly diminished in HT-siMT1 cells. The presence of the additional, low molecular weight fragments (55–58 kDa) of chz directly supported the presence of the additional MT1-MMP cleavage site in the ectodomain sequence. The molecular mass of these fragments (55–58 kDa) correlated with the expected size of the N-terminal, 1–500 proteolytic fragment of chz.
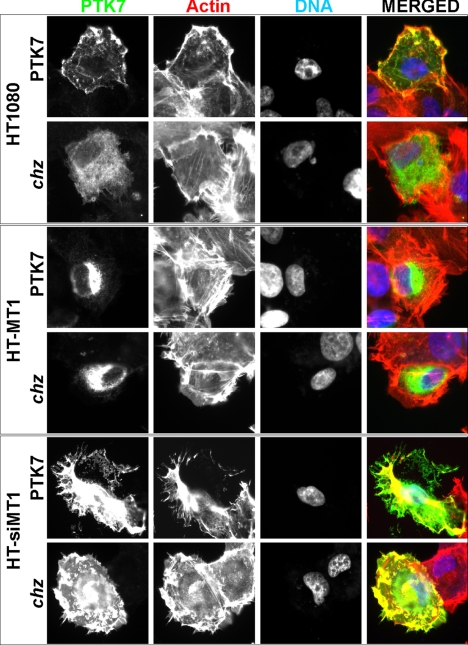
To visualize the relations between chz and MT1-MMP, we immunostained the permeabilized HT1080, HT-MT1, and HT-siMT1 cells using the PTK7 antibody (Fig. 3). There was an abundant immunoreactivity of both PTK7 and especially chz in the plasma membrane and intracellular compartments of HT-siMT1 cells. In turn, in HT1080 cells, only PTK7 but not chz was detected on the plasma membrane. Both PTK7 and chz were not detected on the plasma membrane in HT-MT1 cells, thus supporting the clearance of PTK7 and especially chz by the plasma membrane MT1-MMP activity. Immunostaining of actin revealed the rearrangements in the actin cytoskeleton in the cells we tested.
FIGURE 3.
Immunoreactivity of PTK7 and chz in HT1080 cells. Staining of HT1080, HT-MT1, and HT-siMT1 cells expressing the PTK7/chz constructs using the PTK7 antibody (green) and phalloidin (actin, red). Blue, nuclei (DAPI).
Chz Affects Cell Invasion
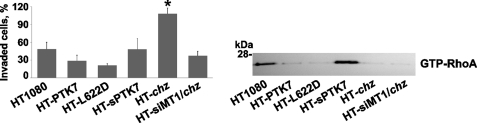
According our data and that of others, PTK7 is directly involved in the regulation of cell migration (29, 39–43). Cell surface levels of PTK7 appear to correlate inversely with tumor aggressiveness and metastatic potential (29). Because of these earlier data and because actin cytoskeleton dynamics are also associated with cell migration (44, 45), we next analyzed the invasive capacity of HT1080, HT-PTK7, HT-L622D, HT-sPTK7, HT-chz, and HT-siMT1/chz cells.
Consistent with our earlier results (29), the ectopic expression of PTK and especially the uncleavable L622D PTK7 mutant suppressed cell migration (Fig. 4). On the contrary, the expression of chz enhanced cell migration ∼2-fold relative to the original, highly migratory HT1080 cells. In HT-siMT1 cells, the stimulatory effect of chz was not observed, again suggesting the presence of a link between MT1-MMP activity and the chz functionality.
FIGURE 4.
chz stimulates cell invasion but does not activate GTP-RhoA. Left panel, cell invasion through a type 1 collagen matrix by HT1080 cells transfected with the indicated MT1-MMP and PTK7/chz constructs. FBS (10%) was used as a chemoattractant. The error bars represent mean ± S.D. *, p < 0.05. The experiments were repeated multiple times with comparable results. Right panel, GTP-RhoA pull-down assay in HT1080 cells transfected with the indicated MT1-MMP and PTK7/chz constructs. The samples (1 mg of total protein each) were precipitated using rhotekin beads. The precipitates were analyzed by Western blotting with the RhoA antibody.
To shed more light on the effects of chz, we determined the activation status of cellular RhoA GTPase (Fig. 4). In agreement with our previous observations (29), only the HT-sPTK7 and original HT1080 cells exhibited significant levels of activated GTP-RhoA. In other cells we tested, including HT-chz and HT-siMT1/chz, the GTP-RhoA levels were similarly low. On the basis of these results, we suggest that GTP-Rho is not critical for the yet to be determined PTK7 pathway that drives cell locomotion.
DISCUSSION
Ubiquitously expressed PTK7 pseudo-kinase is an essential component of the non-canonical Wnt/PCP signaling pathway, arguably one of the most important cellular pathways in eukaryotes (46, 47). At its most basic level, polarity may be considered as the generation of asymmetry within a single cell, whether the result of the asymmetry is a directed movement that leads to cellular migration or to the relocalization of specific multicellular structures. The nature of its specific binding partners and the multifunctional role of PTK7 in cell migration are yet to be elucidated. It is likely that in different cell systems, PTK7 is involved in the interactions with Plexin1 (41), receptor for activated protein kinase C (RACK1) (40), vascular endothelial growth factor receptor 1 (FLT-1) (39), disheveled (dsh) (43), Van Gogh-like 2 (VANGL2) (17, 30) and β-catenin (48).
Polarized cell locomotion also correlates well with pericellular proteolysis and extracellular matrix remodeling, both of which involve the action of MT1-MMP (49–51). Our results suggest that the Wnt/PCP pathway and pericellular proteolysis converge at PTK7 to promote efficient directed cell migration both in normal development and cancer (29).
PTK7 is a major cleavage target of MT1-MMP in the plasma membrane. MT1-MMP directly cleaves the PKP621↓LI sequence in an exposed region of PTK7, generating the N-terminal, soluble PTK7 ectodomain. Because PTK7 readily oligomerizes through its ectodomain (10, 52), membrane PTK7 forms a complex either with itself or with the solubilized ectodomain or with some yet to be identified adaptor protein(s) (2, 13, 39–41, 43, 48). It is likely that these different complex types elicit the dissimilar downstream signaling events and that these signaling events differentially regulate multiple cell functions, including PCP and directional motility, in processes as diverse as cancer cell invasion and embryonic cell migration during gastrulation in eukaryotes (13, 18, 28, 30). In agreement, the soluble PTK7 ectodomain inhibited angiogenesis in vitro and in vivo in a dominant-negative fashion by competing with full-length PTK7 (12). As a result, the ratio between the soluble and the membrane PTK7 appears to be an important factor in the Wnt/PCP regulation. The importance of PTK7 is also illustrated by the fact that PTK7 mutant mice that express a 1–114 PTK7 truncation die perinatally because of severe defects in neural tube closure. Overexpression of the mutant PTK7 lacking its cytoplasmic domain resulted in similar abnormalities (10).
From these perspectives, identification of the chz PTK7 mutant represents a step forward to a better understanding of PTK7 functionality. The chz mutation results in the Ala-Asn-Pro insertion into the PTK7 sequence and causes a significant reduction in membrane localization of the PTK7 protein (17). Our experimental results firmly determined that the three-amino-acid insert incorporated an additional MT1-MMP cleavage site into the PTK7 ectodomain. In fact, because of the chz mutation, the consensus PXX↓L MMP cleavage site is reconstituted in the PTK7 protein. Regardless of the primary importance of MT1-MMP in the cleavage of chz, however, it cannot be ruled out that certain additional soluble and membrane-type MMPs also contribute to the aberrant cleavage of chz in other cells/tissue systems.
It is now clear that because of the aberrant MT1-MMP proteolysis, the soluble N-terminal 1–621 70-kDa fragment is unstable in chz. In a model migration system, the forced expression of chz significantly stimulated cancer cell invasion. Accordingly, it is now tempting to hypothesize that chz affected the precisely balanced, polarized, directed cell movement in the course of gastrulation in mice and that these aberrations in shaping the body plan resulted in the multiple phenotypic defects in the chz mutant animals (17).
Intriguingly, the forced expression of the soluble PTK7 1–700 ectodomain but not of chz significantly up-regulated cellular GTP-RhoA, suggesting that the invasion-promoting function of chz does not require RhoA activity.
Taken together, our novel data in a combination with our results and those of others (17, 18, 28, 29) imply that the MT1-MMP/PTK7 axis plays an important role in multiple cell locomotion situations, including morphogenetic cell movements, that shape the body plan and that the aberrations of either of MT1-MMP or PTK7 or both in gastrulation may result in multiple developmental diseases, including congenital defects. Accordingly, we believe that further studies into the MT1-MMP and PTK7 roles in PCP and their potential link to diseases such as birth defects are warranted to design a means for birth defect prognostics and correction.
This work was supported, in whole or in part, by National Institutes of Health Grants CA83017 and CA77470 (to A. Y. S.).
- PCP
- planar cell polarity
- PTK7
- protein tyrosine kinase 7
- chz
- chuzhoi
- MT1-MMP
- membrane type-1 metalloproteinase
- MMP
- metalloproteinase
- sPTK7
- soluble PTK7.
REFERENCES
- 1. Simons M., Mlodzik M. (2008) Annu. Rev. Genet. 42, 517–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Katoh M. (2005) Oncol. Rep. 14, 1583–1588 [PubMed] [Google Scholar]
- 3. Roszko I., Sawada A., Solnica-Krezel L. (2009) Semin. Cell Dev. Biol. 20, 986–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dale T. C. (1998) Biochem. J. 329, 209–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yin C., Ciruna B., Solnica-Krezel L. (2009) Curr. Top. Dev. Biol. 89, 163–192 [DOI] [PubMed] [Google Scholar]
- 6. Goodrich L. V. (2008) Neuron 60, 9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grassot J., Gouy M., Perrieèe G., Mouchiroud G. (2006) Mol. Biol. Evol. 23, 1232–1241 [DOI] [PubMed] [Google Scholar]
- 8. Jung J. W., Ji A. R., Lee J., Kim U. J., Lee S. T. (2002) Biochim. Biophys. Acta 1579, 153–163 [DOI] [PubMed] [Google Scholar]
- 9. Jung J. W., Shin W. S., Song J., Lee S. T. (2004) Gene 328, 75–84 [DOI] [PubMed] [Google Scholar]
- 10. Lu X., Borchers A. G., Jolicoeur C., Rayburn H., Baker J. C., Tessier-Lavigne M. (2004) Nature 430, 93–98 [DOI] [PubMed] [Google Scholar]
- 11. Park S. K., Lee H. S., Lee S. T. (1996) J. Biochem. 119, 235–239 [DOI] [PubMed] [Google Scholar]
- 12. Shin W. S., Maeng Y. S., Jung J. W., Min J. K., Kwon Y. G., Lee S. T. (2008) Biochem. Biophys. Res. Commun. 371, 793–798 [DOI] [PubMed] [Google Scholar]
- 13. Yen W. W., Williams M., Periasamy A., Conaway M., Burdsal C., Keller R., Lu X., Sutherland A. (2009) Development 136, 2039–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mossie K., Jallal B., Alves F., Sures I., Plowman G. D., Ullrich A. (1995) Oncogene 11, 2179–2184 [PubMed] [Google Scholar]
- 15. Whitford K. L., Ghosh A. (2001) Neuron 32, 1–3 [DOI] [PubMed] [Google Scholar]
- 16. Toyofuku T., Zhang H., Kumanogoh A., Takegahara N., Suto F., Kamei J., Aoki K., Yabuki M., Hori M., Fujisawa H., Kikutani H. (2004) Genes Dev. 18, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paudyal A., Damrau C., Patterson V. L., Ermakov A., Formstone C., Lalanne Z., Wells S., Lu X., Norris D. P., Dean C. H., Henderson D. J., Murdoch J. N. (2010) BMC Dev. Biol. 10, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Coyle R. C., Latimer A., Jessen J. R. (2008) Exp. Cell Res. 314, 2150–2162 [DOI] [PubMed] [Google Scholar]
- 19. Egeblad M., Werb Z. (2002) Nat. Rev. Cancer 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 20. Itoh Y. (2006) IUBMB Life 58, 589–596 [DOI] [PubMed] [Google Scholar]
- 21. Morrison C. J., Butler G. S., Rodríguez D., Overall C. M. (2009) Curr. Opin. Cell Biol. 21, 645–653 [DOI] [PubMed] [Google Scholar]
- 22. Rodríguez D., Morrison C. J., Overall C. M. (2010) Biochim. Biophys. Acta 1803, 39–54 [DOI] [PubMed] [Google Scholar]
- 23. Strongin A. Y. (2010) Biochim. Biophys. Acta 1803, 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hotary K., Li X. Y., Allen E., Stevens S. L., Weiss S. J. (2006) Genes Dev. 20, 2673–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sabeh F., Ota I., Holmbeck K., Birkedal-Hansen H., Soloway P., Balbin M., Lopez-Otin C., Shapiro S., Inada M., Krane S., Allen E., Chung D., Weiss S. J. (2004) J. Cell Biol. 167, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Holmbeck K., Bianco P., Caterina J., Yamada S., Kromer M., Kuznetsov S. A., Mankani M., Robey P. G., Poole A. R., Pidoux I., Ward J. M., Birkedal-Hansen H. (1999) Cell 99, 81–92 [DOI] [PubMed] [Google Scholar]
- 27. Golldack D., Popova O. V., Dietz K. J. (2002) J. Biol. Chem. 277, 5541–5547 [DOI] [PubMed] [Google Scholar]
- 28. Jessen J. R. (2009) Zebrafish 6, 21–28 [DOI] [PubMed] [Google Scholar]
- 29. Golubkov V. S., Chekanov A. V., Cieplak P., Aleshin A. E., Chernov A. V., Zhu W., Radichev I. A., Zhang D., Dong P. D., Strongin A. Y. (2010) J. Biol. Chem. 285, 35740–35749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cantrell V. A., Jessen J. R. (2010) Cancer Lett. 287, 54–61 [DOI] [PubMed] [Google Scholar]
- 31. Golubkov V. S., Boyd S., Savinov A. Y., Chekanov A. V., Osterman A. L., Remacle A., Rozanov D. V., Doxsey S. J., Strongin A. Y. (2005) J. Biol. Chem. 280, 25079–25086 [DOI] [PubMed] [Google Scholar]
- 32. Golubkov V. S., Chekanov A. V., Savinov A. Y., Rozanov D. V., Golubkova N. V., Strongin A. Y. (2006) Cancer Res. 66, 10460–10465 [DOI] [PubMed] [Google Scholar]
- 33. Rozanov D. V., Savinov A. Y., Williams R., Liu K., Golubkov V. S., Krajewski S., Strongin A. Y. (2008) Cancer Res. 68, 4086–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sounni N. E., Rozanov D. V., Remacle A. G., Golubkov V. S., Noel A., Strongin A. Y. (2010) Int. J. Cancer. 126, 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Golubkov V. S., Cieplak P., Chekanov A. V., Ratnikov B. I., Aleshin A. E., Golubkova N. V., Postnova T. I., Radichev I. A., Rozanov D. V., Zhu W., Motamedchaboki K., Strongin A. Y. (2010) J. Biol. Chem. 285, 27726–27736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Eswar N., Eramian D., Webb B., Shen M. Y., Sali A. (2008) Methods Mol. Biol. 426, 145–159 [DOI] [PubMed] [Google Scholar]
- 37. Pieper U., Eswar N., Webb B. M., Eramian D., Kelly L., Barkan D. T., Carter H., Mankoo P., Karchin R., Marti-Renom M. A., Davis F. P., Sali A. (2009) Nucleic Acids Res. 37, D347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deryugina E. I., Bourdon M. A., Jungwirth K., Smith J. W., Strongin A. Y. (2000) Int. J. Cancer 86, 15–23 [DOI] [PubMed] [Google Scholar]
- 39. Lee H. K., Chauhan S. K., Kay E., Dana R. (2011) Blood (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wehner P., Shnitsar I., Urlaub H., Borchers A. (2011) Development 138, 1321–1327 [DOI] [PubMed] [Google Scholar]
- 41. Wagner G., Peradziryi H., Wehner P., Borchers A. (2010) Biochem. Biophys. Res. Commun. 402, 402–407 [DOI] [PubMed] [Google Scholar]
- 42. Prebet T., Lhoumeau A. C., Arnoulet C., Aulas A., Marchetto S., Audebert S., Puppo F., Chabannon C., Sainty D., Santoni M. J., Sebbagh M., Summerour V., Huon Y., Shin W. S., Lee S. T., Esterni B., Vey N., Borg J. P. (2010) Blood 116, 2315–2323 [DOI] [PubMed] [Google Scholar]
- 43. Shnitsar I., Borchers A. (2008) Development 135, 4015–4024 [DOI] [PubMed] [Google Scholar]
- 44. Parsons J. T., Horwitz A. R., Schwartz M. A. (2010) Nat. Rev. Mol. Cell Biol. 11, 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. (2003) Science 302, 1704–1709 [DOI] [PubMed] [Google Scholar]
- 46. Neth P., Ries C., Karow M., Egea V., Ilmer M., Jochum M. (2007) Stem Cell Rev. 3, 18–29 [DOI] [PubMed] [Google Scholar]
- 47. Polakis P. (2000) Genes Dev. 14, 1837–1851 [PubMed] [Google Scholar]
- 48. Puppo F., Thomé V., Lhoumeau A. C., Cibois M., Gangar A., Lembo F., Belotti E., Marchetto S., Lécine P., Prébet T., Sebbagh M., Shin W. S., Lee S. T., Kodjabachian L., Borg J. P. (2011) EMBO Rep. 12, 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wolf K., Friedl P. (2009) Clin. Exp. Metastasis 26, 289–298 [DOI] [PubMed] [Google Scholar]
- 50. Itoh Y., Seiki M. (2006) J. Cell. Physiol. 206, 1–8 [DOI] [PubMed] [Google Scholar]
- 51. Seiki M., Yana I. (2003) Cancer Sci. 94, 569–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pulido D., Campuzano S., Koda T., Modolell J., Barbacid M. (1992) EMBO J. 11, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]