Abstract
Protein kinase inhibitors with enhanced selectivity can be designed by optimizing binding interactions with less conserved inactive conformations because such inhibitors will be less likely to compete with ATP for binding and therefore may be less impacted by high intracellular concentrations of ATP. Analysis of the ATP-binding cleft in a number of inactive protein kinases, particularly in the autoinhibited conformation, led to the identification of a previously undisclosed non-polar region in this cleft. This ATP-incompatible hydrophobic region is distinct from the previously characterized hydrophobic allosteric back pocket, as well as the main pocket. Generalized hypothetical models of inactive kinases were constructed and, for the work described here, we selected the fibroblast growth factor receptor (FGFR) tyrosine kinase family as a case study. Initial optimization of a FGFR2 inhibitor identified from a library of commercial compounds was guided using structural information from the model. We describe the inhibitory characteristics of this compound in biophysical, biochemical, and cell-based assays, and have characterized the binding mode using x-ray crystallographic studies. The results demonstrate, as expected, that these inhibitors prevent activation of the autoinhibited conformation, retain full inhibitory potency in the presence of physiological concentrations of ATP, and have favorable inhibitory activity in cancer cells. Given the widespread regulation of kinases by autoinhibitory mechanisms, the approach described herein provides a new paradigm for the discovery of inhibitors by targeting inactive conformations of protein kinases.
Keywords: Drug Design, Kinetics, Protein Phosphorylation, Protein Structure, Receptor Tyrosine Kinase, X-ray Crystallography, FGFR, Kinase Autoinhibition, Non-competitive, Small Molecule Protein Kinase Inhibitor
Introduction
It has long been hypothesized that mapping the spatial rearrangements that take place during the cycling between productive (active) and non-productive (inactive) states of kinases should lead to a better understanding of kinase dynamics, structure, function, and regulation (1–3). Furthermore, targeting kinase inhibitors to the inactive state is attractive because that form is more likely to represent a distinct conformation that may in turn lead to the identification of more selective inhibitors (4). However, despite advances in the field, designing inhibitors that target an inactive conformation of a kinase remains challenging (5) and largely empirical. The work described here provides a generally applicable methodology for the design of inhibitors that preferentially bind to the inactive state of a target kinase through developing a better understanding of the interplay between the conformational transitions that take place upon activation (6).
Previous approaches have focused on the analysis of so-called “type II inhibitors” (7), which induce a distinct DFG-out conformation and occupy an additional hydrophobic pocket created by this rearrangement. The DFG-out conformation is a prerequisite for designing such type II inhibitors. The success of this strategy relies on the fact that despite the conserved nature of the ATP site, there are sufficient adjacent pockets that can be utilized in the design of type II inhibitors. Only a limited number of kinases are predicted to be amenable to this approach (8).
A modeling approach was employed here for targeting inactive kinases that is distinct from the model utilized in the design of type II inhibitors. Efforts were focused on the design of inhibitors that stabilize the autoinhibited state, postulating that mimicking the regulating kinase activity of nature may provide guideposts for inhibitor design (9, 10). In their natural state, kinases employ diverse mechanisms to control activation, and many can adopt a wide spectrum of inactive conformations through the positioning of the kinase loops and/or interaction of the catalytic site with autoinhibitory regulatory elements. However, structural information from crystallographic studies is limited, because autoinhibited kinases are difficult to crystallize, and important regulatory domains are frequently omitted in crystallization experiments.
The recently described co-crystal structure of a small molecule inhibitor of c-Met, ARQ 197, bound to the inactive kinase (11, 44) was employed. In this case, the autoinhibited conformation was stabilized by unusual and extensive hydrophobic interactions. Comparison of this structure with other autoinhibited and inhibitor-bound inactive kinase structures in the public domain led to the identification of networks of hydrophobic regions that are formed by the interaction of three to five non-polar residues. It was hypothesized that in the autoinhibited conformation, these hydrophobic residues interact to form “hydrophobic clusters” that stabilize the inactive state of the kinase and interfere with ATP binding. These conserved hydrophobic clusters were utilized as the basis for the design of novel small molecule kinase inhibitors.
A generalized model of a hypothetical inactive kinase conformation was developed using computational methods that combined structural elements of inactive kinase conformations with the hydrophobic clusters present in the c-Met·ARQ 197 complex. Described herein is the application of this model to identify a series of fibroblast growth factor receptor (FGFR)2 tyrosine kinase family (12) inhibitors. An extensive array of biophysical and biochemical assays were utilized to measure and characterize the interactions of these inhibitors with the inactive kinase, as well as to determine the effect on autophosphorylation and substrate activation of FGFR kinase. Particular attention was paid to carefully controlling the activation state of the kinase as well as elucidating kinetic mechanisms of inhibition. Additionally, the biological activity of these inhibitors in a cell-based assay is described as well as the effect of systematic mutations on inhibition of autophosphorylation.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The gene that encodes the kinase domains of FGFR1 (residues 462–765) or FGFR2 (residues 458–768) was inserted between the BamHI and SalI sites of the pET15b vector and biscistronically cloned at the SalI site with the λ-phosphatase gene that carries the second ribosome binding site. Two mutations were introduced in FGFR1, C488A and C584S, to prevent formation of disulfide-linked oligomers (13). The inactive form of FGFR constructs were co-expressed with λ-phosphatase as His6 fusion proteins that carry a thrombin cleavage site for tag removal. Protein was expressed in BL21(DE3)-RILP Codon Plus competent Escherichia coli cells (Stratagene) with 2× YT medium supplemented with 100 mg/ml of ampicillin. The culture was grown at 25 °C (250 rpm) on a shaker (Innova 43 refrigerated) for 5 h. Growth was monitored by following the A600 of the culture. Once a reading of A600 = 0.5 was obtained the incubator temperature was reduced to 12 °C and growth was continued until A600 = 0.6. The protein expression was induced by adding 250 μm isopropyl 1-thio-β-d-galactopyranoside and the induction was continued at 12 °C for 16 h.
The cell paste was resuspended in 150 ml of lysis buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 0.1% 2-mercaptoethanol) supplemented with 1 mm PMSF, 40 mg of lysozyme, DNase I to 10 μg/ml (stock = 10 mg/ml in 40% glycerol = ×1,000), and lysis was allowed to proceed for 30 min on ice. The lysate was sonicated using a Branson 450D on ice at 60% power. Following sonication, the lysate was mixed with Triton X-100 to a final concentration of 0.5%. Insoluble protein and cell debris were sedimented through a 45-min centrifugation at 40,000 × g at 4 °C. The supernatant was loaded onto a pre-equilibrated nickel-nitrilotriacetic acid-agarose column. The beads were washed with 20 column volumes of buffer containing 25 mm Tris, 0.5 m NaCl, 25 mm imidazole, pH 8.0, 0.1%. Protein was eluted with buffer containing 25 mm Tris, pH 8.0, 100 mm NaCl, and 400 mm imidazole. The concentrated protein was digested with thrombin protease (1:1,000, w/w) at 4 °C for 16 h. The His6 tag was removed by passing the digested sample into a second column of nickel-nitrilotriacetic acid-agarose, the flow-through was collected and concentrated. The protein was further purified on an ion-exchange column using QFF resin followed by size exclusion chromatography on a Superdex 200 column. The peak fraction was concentrated to 10–20 mg/ml. The purity of the FGFR1 and FGFR2 preparations was determined by SDS-PAGE and MS analysis.
Crystallization, Data Collection, and Structure Determination
ARQ 069 was dissolved in DMSO to a final concentration of 50 mm and added to FGFR2 or FGFR1 (15 mg/ml) in a 4:1 m ratio. The final DMSO concentration was 2% before crystallization. Crystals of the FGFR2·ARQ 069 complex were grown by sitting-drop vapor diffusion from a solution of 15% polyethylene glycol 4000 and 0.3 m lithium sulfate buffered with 100 mm HEPES at 25 °C. The best crystals were obtained after several rounds of seeding. The crystals were transferred to the cryosolution containing the well solution and 15% glycerol and flash frozen in liquid nitrogen. FGFR1·ARQ 069 complex was crystallized with PEG 10000, 0.3 m (NH4)2SO4, 5% ethylene glycol, and 100 mm MES, pH 6.5, at 4 °C. The crystals were flash frozen in liquid nitrogen after transferring to a cryosolution consisting of well solution and 15% ethylene glycol. The FGFR2·ARQ 069 complex crystals belong to space group P21212, whereas FGFR1·ARQ 069 complex crystals showed a monoclinic space group, C2. Both crystals contained two complexes per asymmetric unit. Data were collected at beamline X29A at the National Synchrotron Light Source (Brookhaven National Laboratory), processed using MOSFLM, and merged using SCALA. The structure of the FGFR·ARQ 069 complex was solved by molecular replacement using MOLREP or Phaser as implemented in the CCP4 software package (14) with the structure of apo-FGFR2 (PDB code 2PSQ) or apo-FGFR1 (PDB code 1FGK) as the search model. The glycine-rich loop and activation loops were deleted in the model to avoid any model biased refinement. The compound and the reorganized glycine-rich loop of FGFR1 were built into 2Fo − Fc and Fo − Fc electron density maps using COOT. The atomic model was refined using Arp/wARP and REFMAC. Data statistics are listed in supplemental Table S1. The structural figures were rendered with PyMol.
Continuous Spectrophotometric Kinase Assay
Autophosphorylation Assay
Kinase activity was monitored using a continuous spectrophotometric assay as described previously (15). In this assay, the consumption of ATP is coupled via the pyruvate kinase/lactate dehydrogenase enzyme pair to the oxidation of NADH, which is monitored through the decrease in absorption at 340 nm. Reactions contained 100 mm Tris, pH 8.0, 10 mm MgCl2, 1 mm phosphoenolpyruvate, 0.28 mm NADH, 89 units/ml of pyruvate kinase, 124 units/ml of lactate dehydrogenase, and 2% DMSO. Reactions were initiated by the addition of ATP to mixtures containing enzyme and various concentrations of ARQ 069. The FGFR2 autophosphorylation reaction was carried out at 0.5 μm enzyme concentration and 1 mm ATP.
Substrate Assay
The substrate phosphorylation reaction was measured with 0.5 μm FGFR2, 50 μm Pyk2 peptide (AGAGSIESDIYAEIPDETC), 1 mm ATP, and 10 mm MgCl2. Reactions were initiated by the addition of ATP to mixtures containing enzyme and various concentrations of ARQ 069. The reaction was monitored by following the decrease in absorbance at 30 °C in a microplate reader (Tecan Safire II).
Substrate Phosphorylation Assays
Compounds were diluted from 30 mm stock solutions in 100% DMSO into a Tris-HCl, pH 7.4, assay buffer containing 0.02 mg/ml of bovine serum albumin (BSA), 10 mm MgCl2, 1 mm EGTA, 0.01% Nonidet P-40, and 10% glycerol, 1 mm DTT, and 0.1 mm Na3VO4. Unphosphorylated recombinant FGFR enzyme, diluted to 600 nm in assay buffer, was preincubated in the presence of various concentrations of ARQ 068 or ARQ 069 for 20 min in a total volume of 40 μl in the wells of a 96-well V-bottom polypropylene microplate (BD Biosciences). 20 μl of a solution containing ATP (final concentration = 500 μm) and biotinylated Pyk2 substrate peptide (Biotin(SC)-AGAGSIESDIYAEIPDETC-NH2 (Midwest Biotech, Fishers, IN) was then added to each well, yielding a final concentration of 2 μm peptide, 400 nm kinase, and a final DMSO concentration of 1%. After a 1-h reaction time, 50 μl of the reaction mixture were added to Nunc MaxisorpTM ELISA plates to which streptavidin (ThermoScientific, Rockford, IL) had been pre-coated (1 μg/ml in Dulbecco's phosphate-buffered saline (PBS)) for at least 24 h at 4 °C, and then blocked with 5% BSA for 2 h at RT. After a 30-min incubation to capture the biotinylated-Pyk2 peptide, the microwells were washed exhaustively with TBST buffer (Tris-buffered saline, 50 mm Tris-HCl, pH 7.4, 0.15 mm NaCl, 0.05% Tween 20). The degree of peptide phosphorylation was immunodetected following an incubation for 1 h with a mouse monoclonal anti-phosphotyrosine antibody (Cell Signaling Technology, Danvers, MA, catalog number 9411) diluted 1:3000 in TBST with 2% BSA, and a subsequent 1-h incubation with alkaline phosphatase-conjugated goat anti-mouse IgG antibody (Promega Corp., Madison, WI) diluted 1:4000 in TBST with 2% BSA. Following the secondary antibody incubation, the plates were washed exhaustively with TBST, and 100 μl of reconstituted Attophos® reagent was added. The fluorescent substrate Attophos (Promega Corp., Madison, WI) was used according to the manufacturer's instructions. The plates were incubated for 2–10 min at RT, and then read in a fluorometer (EnvisionTM, PerkinElmer Life Sciences) at an excitation wavelength of 450 nm and an emission wavelength of 580 nm.
AMP-PNP Competition Assay by Intrinsic Tryptophan Fluorescence Quench
All binding measurements were performed in buffer solution containing 25 mm Tris, pH 7.5, 0.1 m NaCl, and 2% DMSO. A complex of FGFR2 (10 μm) was prepared with various concentrations of ARQ 069 (20, 50, 100, and 200 μm) and incubated on ice for 1 h. AMP-PNP was serially diluted (0–80 μm) with buffer solution (25 mm Tris, pH 7.5, 0.1 m NaCl, and 2% DMSO) into 384-well plates and 10 μl of FGFR2·ARQ 069 complex was added to each well. The volume in each well was made up to 50 μl with buffer, and the complex mixtures were allowed to equilibrate for 1 h at room temperature The final concentration of FGFR2 in each well was 2 μm, and the corresponding final concentrations of ARQ 069 were 5, 10, 20, and 40 μm (5-fold dilutions). To determine the affinity of AMP-PNP to FGFR2 alone, a similar experiment was conducted, in which AMP-PNP was serially diluted at a fixed concentration of FGFR2 (2 μm). To compare the relative affinity of AMP-PNP to FGFR2 and the FGFR2·ARQ 069 complex the intrinsic tryptophan emission was used as a fluorescent probe. The change in intrinsic emission was measured on a Tecan microplate reader at an excitation wavelength of 295 nm and emission wavelength of 345 nm. The magnitude of increase in tryptophan fluorescence (ΔF) was calculated as ΔF = F − F0, where F and F0 are the emission intensities in the presence and absence of AMP-PNP, respectively. Dissociation constants (Kd) were obtained from a fit to the hyperbolic binding equation, ΔF/F0 = Fmax × [AMP-PNP]/([AMP-PNP] + Kd), where Fmax corresponds to the fluorescence intensity at saturation.
Cell-based Assay
Kato III gastric carcinoma cells were obtained from the American Type Tissue Collection (ATCC) and grown in complete RPMI1640 medium containing 10% fetal bovine serum (FBS) (Invitrogen Corp.). Cells were plated in complete medium at a density of 1 × 106 cells per well (9.6 cm2) in six-well tissue culture plates (BD Biosciences). Twenty-four hours following plating, Kato III cells were treated with the indicated concentrations of ARQ 069, to a final concentration of 0.3% DMSO (v/v). Cells were then incubated for 2 h at 37 °C. After 2 h, cells were treated with 150 ng/ml of keratinocyte growth factor, a growth factor that binds to and activates FGFR2b (16) for 15 min. Cells were lysed with E-PAGE buffer (Invitrogen Corp., catalog number EPBUF-01), sonicated three times for 15 s, and then heated for 10 min at 75 °C. Samples (10 μl) were subjected to SDS-PAGE and Western blotting as previously described (17) using antibodies to phosphorylated FGFR (Cell Signaling Technology, Inc., catalog number 3476S), phosphorylated fibroblast receptor substrate (p-FRS, Cell Signaling Technology, catalog number 3864), or β-actin (Sigma, catalog number A5441).
RESULTS
ARQ 523 Was Identified Based on an in Silico Structural Model of Autoinhibited FGFR2
Through an analysis of multiple structures of kinases in the autoinhibited form in comparison to the ARQ 197-bound c-Met crystal structure, a hydrophobic pocket was identified within the kinase cleft distinct from the previously described hydrophobic main pocket and the allosteric back pocket. Importantly, creation of this new hydrophobic environment imposes significant conformational rigidity on the activation segment and glycine-rich loop. As a result, the newly created hydrophobic region, which is not present in active kinase structures, becomes incompatible with ATP binding (supplemental Fig. S1). The three key structural motifs employed in model construction were: 1) the relatively conserved phenylalanine residue of the glycine-rich loop; 2) the phenylalanine residue of the DFG motif; and 3) the aliphatic side chain of the arginine residue from the A-loop.
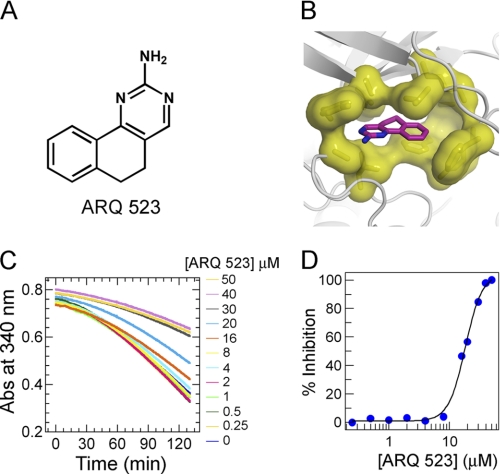
Using this generalized structural template, homology models were built for multiple target kinases and used in silico to identify a variety of inhibitors. In the work described here, the corresponding residues present in the FGFR2 kinase were employed. The resulting FGFR2 homology model was used for virtual screening of compound databases to identify chemical scaffolds suitable for optimization. A promising template, ARQ 523 (5,6-dihydrobenzo[h]quinazolin-2-amine) (Fig. 1A), was identified using docking (ICM) and receptor pharmacophore fitting (Accelrys) (Fig. 1B).
FIGURE 1.
Docking model of ARQ 523 and inhibition of FGFR2 autophosphorylation. A, chemical structure of ARQ 523. B, ARQ 523 docked into the in silico model of FGFR2 kinase. Binding of ARQ 523 requires compatible hydrophobic residues (shown as yellow surfaces). C, kinetics of FGFR2 autophosphorylation measured by a continuous spectrophotometeric enzyme coupled assay. Enzyme activity was followed by measuring the decrease in absorbance at 340 nm. FGFR2 (1 μm) was preincubated with various concentrations of ARQ 523 (0–50 μm) and the reaction was initiated with 1 mm ATP. D, the potency of ARQ 523 (IC50) was determined by fitting the autophosphorylation rate at various inhibitor concentrations.
ARQ 523 Binds to Unphosphorylated FGFR2 and Inhibits Autophosphorylation
To demonstrate that ARQ 523 binds to FGFR2 in solution, intrinsic tryptophan fluorescence quenching was used. Confirmation that FGFR2 was purified in a non-phosphorylated state was provided by mass spectrometric (MS) analysis. Binding measurements provided a Kd of 16 ± 3.9 μm and confirmed the interaction of ARQ 523 with the inactive form of FGFR2. The inhibitory effect of ARQ 523 on FGFR kinase activation was assessed by following the kinetics of the intermolecular autophosphorylation reaction (18–20) in a continuous spectrophotometric assay. The rate of autophosphorylation was reduced in a concentration-dependent manner with increasing levels of inhibitor. An IC50 value of 18 ± 2.9 μm confirmed that ARQ 523 inhibited FGFR2 autophosphorylation (Fig. 1, C and D).
ARQ 069, an Analog of ARQ 523, Binds to FGFR2 in an Enantiospecific Manner
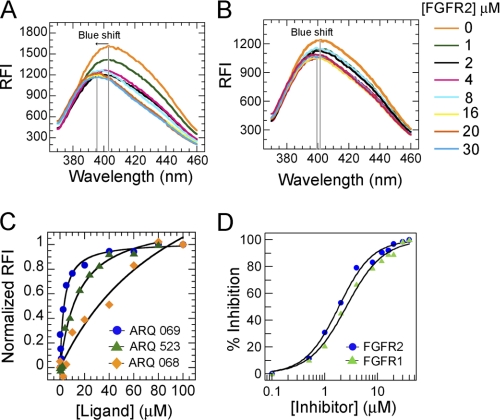
Structural modifications were made based on the in silico model to optimize the potency of ARQ 523 against the inactive form of FGFR kinase. Introduction of a phenyl group at the 6-position of ARQ 523 created a chiral center producing two enantiomers (supplemental Methods Scheme 1): ARQ 068, the R-enantiomer, and ARQ 069, the S-enantiomer. The impact of stereochemistry on inhibitor binding to FGFR2 was measured by indirect affinity mass spectrometry, thermal shift (21, 22), and intrinsic tryptophan fluorescence quench assays (Fig. 2C and supplemental Fig. S2). As predicted from the structural model, ARQ 069 had a significantly higher affinity for the protein relative to either ARQ 068 or ARQ 523. The affinity of the two enantiomers for FGFR2 was also quantitated by a tryptophan fluorescence quench assay. Although ARQ 069 showed an affinity for FGFR2 of 5.2 ± 1.4 μm, ARQ 068 bound poorly (>100 μm).
FIGURE 2.
ARQ 069 binds to a hydrophobic region and enantioselectively inhibits FGFR activity. A, intrinsic fluorescence emission change of ARQ 069 upon binding to inactive FGFR2. The fluorescence emission spectra of ARQ 069 at a concentration of 1 μm were measured at an excitation wavelength of 332 nm in the presence of increasing concentrations of FGFR2 protein. Interaction of FGFR2 with ARQ 069 is accompanied by a shift in the emission maximum followed by a decrease in fluorescence intensity. B, ARQ 068 did not show a significant change in the emission maximum or emission intensity in the presence of increasing FGFR2 concentrations. The concentrations of FGFR2 used to determine the binding of both ligands are shown. C, the affinity of ARQ 523, ARQ 069, and ARQ 068 to FGFR2 was measured by following the intrinsic tryptophan quench and recording the fluorescence emission at 345 nm at various concentrations of ligand. The FGFR2 concentration was held constant at 0.5 μm. D, inhibition of FGFR1 and FGFR2 autophosphorylation by ARQ 069. The autophosphorylation reaction was followed by continuously monitoring the decrease in absorbance at 340 nm. Formation of ADP was coupled with the oxidation of NADH. The kinase domains of FGFR2 or FGFR1 (1 μm) were incubated at increasing concentrations of ARQ 069 (0 to 40 μm), the reaction was initiated with the addition of 1 mm ATP. IC50 values were determined by fitting the rate of autophosphorylation inhibition at various concentrations of ARQ 069.
The nature of the interaction of ARQ 069 and ARQ 068 with FGFR2 was further characterized by measuring the change in intrinsic fluorescence of the two enantiomers. ARQ 069 displayed a blue shift of the emission maximum, from 404 to 393 nm, with a concomitant decrease in the emission intensity. ARQ 068 showed no change in either the emission intensity or the maximum. A blue shift of 11 nm upon addition of FGFR2 to ARQ 069 indicates that the binding involves strong hydrophobic interactions (Fig. 2, A and B). These results collectively are in agreement with the predicted mode of binding and confirm the enantiospecificity of the hydrophobic interaction with FGFR2.
ARQ 069 Targets the Inactive Forms of FGFR1 and FGFR2 Kinases and Inhibits Their Enzymatic Activity
A peptide substrate phosphorylation assay was implemented using an enzyme-linked immunosorbent assay (ELISA) employing a biotinylated Pyk2 peptide to determine the effects of the binding of ARQ 069 on FGFR2 and FGFR1. When preincubated with unphosphorylated FGFR1 or FGFR2 prior to the introduction of ATP and peptidic substrate, ARQ 069 inhibited the enzymatic activity of both FGFR1 (IC50 = 0.84 ± 0.15 μm) and FGFR2 (IC50 = 1.23 ± 0.03 μm). By contrast, when ARQ 069 was preincubated with either phosphorylated FGFR1 or FGFR2, the potency of ARQ 069 in inhibiting Pyk2 phosphorylation was markedly reduced, with IC50 values determined to be greater than 30 and 24.8 μm for FGFR1 and FGFR2, respectively (Table 1). These results demonstrate that ARQ 069 exhibits at least a 20-fold preference for binding to the unphosphorylated, inactive forms of FGFR1 and FGFR2 and that it does so in an enantioselective manner because ARQ 068 does not inhibit either the unphosphorylated or phosphorylated forms of either kinase.
TABLE 1.
Biochemical inhibition of FGFR1 and FGFR2 by ARQ 069 and ARQ 068
ARQ 069 preferentially inhibits unphosphorylated forms of FGFR1 and FGFR2 over phosphorylated forms. ARQ 068 poorly inhibits both forms of FGFR isoforms.
| Compound | Unphosphorylated |
Phosphorylated |
||
|---|---|---|---|---|
| FGFR1 | FGFR2 | FGFR1 | FGFR2 | |
| ARQ 069 | 0.84 ± 0.15 μm | 1.23 ± 0.03 μm | >30 μm | 24.8 ± 3.8 μm |
| ARQ 068 | >30 μm | >30 μm | >30 μm | >30 μm |
ARQ 069 Inhibits FGFR2 and FGFR1 Autophosphorylation through a Mechanism That Is Not ATP Dependent
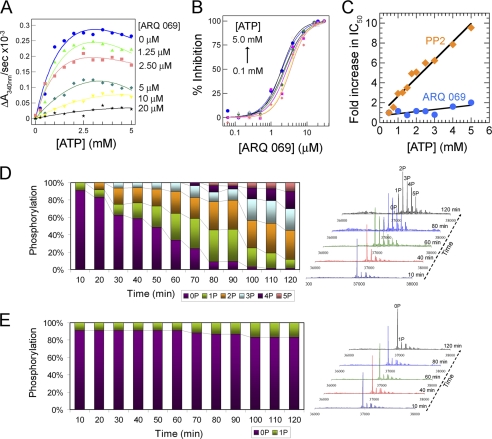
FGFR1 and FGFR2 were independently incubated with increasing concentrations of ARQ 069, and the inhibition of autophosphorylation measured in the presence of 1 mm ATP. Both FGFR1 and FGFR2 autophosphorylation were inhibited with IC50 values of 2.8 ± 1.3 and 1.9 ± 0.9 μm, respectively (Fig. 2D). Next, the effect of ATP concentration on the apparent potency of ARQ 069 inhibition of FGFR2 activation was evaluated. The rate of FGFR2 autophosphorylation in the absence of inhibitor increased with ATP concentration, and plateaued at 1.4 mm ATP (Fig. 3A). At ARQ 069 concentrations of 20 μm, over 90% of autophosphorylation was abolished, and further increases in ATP concentration, even up to 5 mm, had no effect (Fig. 3B). When the autophosphorylation inhibition potency (IC50) of ARQ 069 was plotted as a function of ATP concentration, the inhibition was shown to be ATP-independent (Fig. 3C). The kinetic behavior of ARQ 069 was compared with the previously characterized, non-ATP competitive Src inhibitor, PP2 (23), which also inhibits FGFR2. Interestingly, PP2 was ATP competitive against FGFR2 as reflected by a linear reduction of autophosphorylation inhibition with increasing ATP concentration.
FIGURE 3.
Inhibition of FGFR2 autophosphorylation by ARQ 069 is not dependent on ATP concentration. A, FGFR2 (1 μm) was preincubated at the indicated concentrations of ARQ 069. Autophosphorylation reaction rates were evaluated across a wide range of ATP concentrations. The reaction was followed by a continuous spectrometric assay measuring ADP formation coupled to the NADH oxidation reaction, which monitors a decrease in absorbance at 340 nm. Activation of FGFR2 increased hyperbolically with increases in ATP concentrations and the rate of autoactivation was dramatically reduced upon preincubation with ARQ 069. Above a 20 μm concentration of ARQ 069, the inhibition of FGFR2 activation does not change significantly up to the highest ATP concentration of 5 mm. B, dose-response curves were derived from the same experiments. ARQ 069 retained its ability to inhibit FGFR2 autophosphorylation at all concentrations of ATP as indicated by similar IC50 values. C, in a similar experiment, the non-ATP competitive inhibitor of Src, PP2, showed ATP competitive inhibition kinetics with FGFR2 kinase as reflected by a linear dependence of IC50 values as a function of ATP. ARQ 069 behaves kinetically as non-ATP competitive when tested in a similar ATP concentration range as used for PP2. D, FGFR2 (2 μm) autophosphorylation was also monitored by ESI-MS as a function of time at 5 mm ATP concentration. The right panel shows the deconvolted MS spectra at representative time points. E, in a similar autophosphorylation experiment at 5 mm ATP concentration, FGFR2 preincubated with 20 μm ARQ 069 showed strong phosphorylation inhibition as reflected by the weak autophosphorylation levels. Together these results support an ATP-independent mechanism of inhibition of FGFR2. Details of the MS experiments for D and E can be found under “supplemental Methods”.
Time-dependent mass spectrometric analysis of the phosphorylation status of FGFR2 during the autophosphorylation reaction confirmed that inhibition by ARQ 069 was sustained at high ATP concentrations. In the absence of the inhibitor, phosphorylation of up to five tyrosine residues was detected (Fig. 3D and supplemental Fig. S5). However, preincubation of FGFR2 with 20 μm ARQ 069 resulted in the complete inhibition of FGFR2 phosphorylation even at ATP concentrations of 5 mm (Fig. 3E). These results show that inhibition of FGFR2 autophosphorylation by ARQ 069 is not ATP-dependent even at physiological concentrations of ATP.
ARQ 069 Interacts with FGFR2 to Preclude Binding of ATP
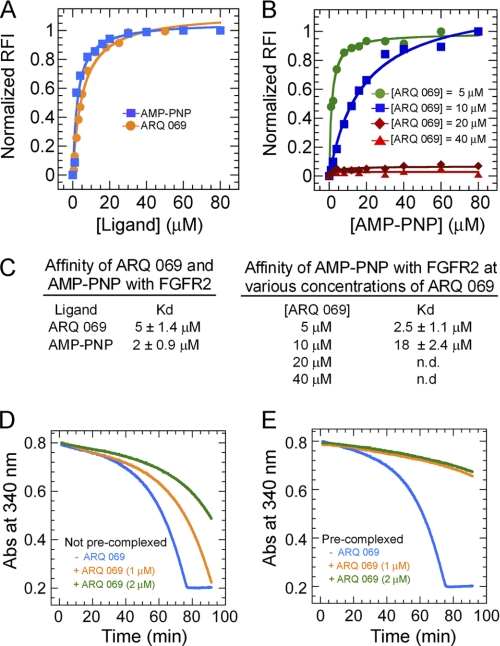
The mechanism of inhibition of FGFR2 autophosphorylation was investigated by means of competitive binding experiments with AMP-PNP, a non-hydrolyzable analog of ATP. Binding was monitored by intrinsic tryptophan fluorescence quench. The Kd measured for AMP-PNP and ARQ 069 binding to FGFR2 was 2 and 5 μm, respectively (Fig. 4, A and C). The competition between these two ligands was examined by incubating increasing concentrations of AMP-PNP with a pre-formed complex of FGFR2 and various concentrations of ARQ 069 (Fig. 4B). As shown in Fig. 4C, the binding affinity of AMP-PNP for FGFR2 decreased with increasing concentrations of preincubated ARQ 069. Furthermore, at concentrations greater than 20 μm ARQ 069, AMP-PNP binding to FGFR2 was not detected. These results demonstrate that at low inhibitor concentrations, sufficient sites are available for binding of AMP-PNP, but at higher inhibitor concentrations, the binding site is no longer available for ATP analog binding. Indirect affinity mass spectrometry displacement experiments confirmed this concentration-dependent exclusion of AMP-PNP binding by ARQ 069. Finally, when a pre-formed complex of FGFR2 and ARQ 069 was treated with a 10-fold excess of AMP-PNP, the amount of FGFR2-bound ARQ 069 remained unchanged, even after an incubation period of 1 h (supplemental Fig. S3). These findings collectively demonstrate that ARQ 069 recognizes a conformation of FGFR2 that is not compatible with AMP-PNP binding (see the following section).
FIGURE 4.
Interaction of ARQ 069 with FGFR2 by an ATP-independent mechanism that exhibits slow off-rate kinetics. A, both ARQ 069 and AMP-PNP show similar binding affinities. B, competitive ligand binding of ARQ 069 and AMP-PNP with FGFR2. FGFR2 (0.5 μm) was preincubated with increasing concentrations of ARQ 069 and titrated with the non-hydrolyzable ATP analog, AMP-PNP. As the concentration of ARQ 069 reaches saturating concentrations (above 20 μm), AMP-PNP failed to displace FGFR2-bound ARQ 069. C, affinity values derived from these experiments are shown. AMP-PNP can effectively displace ARQ 069 (5 μm) bound to FGFR2 at 5 μm. However, at a concentration of 10 μm of ARQ 069 the affinity of AMP-PNP to FGFR2 decreased by an order of magnitude. At saturating concentrations above 20 μm ARQ 069, AMP-PNP was unable to displace the inhibitor from FGFR2. D, the rate of autophosphorylation was measured for FGFR2 (1 μm) at two concentrations of ARQ 069 (1 and 2 μm) in the presence of 4 mm ATP concentration. E, FGFR2 (100 μm) was pre-complexed with ARQ 069 at two concentrations (100 and 200 μm) and diluted by 100-fold into the assay reaction mixture. The final concentrations of enzyme and inhibitor were identical to the control experiment (i.e. minimal complexing time) shown in panel D. The reaction was followed in the presence of 4 mm ATP and the rate of activation was measured from the slope of the curve at identical intervals. The inhibition of pre-complexed ARQ 069/FGFR2 was 7-fold higher than un-complexed FGFR2 and was not reduced over time by a high ATP concentration.
Inhibition of FGFR2 by ARQ 069 follows a Slow Dissociation Mechanism
The rate of dissociation (off-rate) of ARQ 069 from FGFR2 was determined by pre-addition of ARQ 069 to FGFR2 and equilibration for 30 min at room temperature. The resulting enzyme-inhibitor complex was diluted (100-fold) into the autophosphorylation assay. FGFR2 activation at any time point is directly related to the rate of dissociation of the enzyme-inhibitor complex. In the control reaction, no pre-mixing of the inhibitor and FGFR2 was performed.
As expected, when ARQ 069 was equilibrated for 30 min with FGFR2 prior to the addition of ATP, a 7-fold higher inhibition of phosphorylation was observed (Fig. 4, D and E). Thus, slow dissociation kinetics likely contributes to the observed mixed-type mechanism of inhibition.
Kinase Selectivity of ARQ 069
As ARQ 069 represented the first step in optimization of the potency of this in silico selected scaffold, it was important to determine the selectivity of this promising but early lead molecule. ARQ 069 was tested in a conventional panel of 96 human kinases at a concentration of 10 μm using well established microfluidics methodology. Of the nine kinases that were inhibited by 30% or greater, four were representatives of the FGFR kinase family: FGFR1, FGFR2, FGFR2 (N549H), and FGFR3. The other five kinases were Aurora C, c-Raf-1, Aurora A, VEGFR2 (KDR), and CK1δ. The remaining kinases (n = 87) were all inhibited by less than 30%. These results were encouraging and supported further optimization of the series.
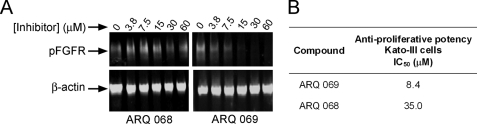
ARQ 069 Inhibits the Phosphorylation of FGFR and Exerts an Anti-proliferative Effect in an FGFR2-dependent Human Gastric Cancer Cell Line in an Enantioselective Manner
The proliferation of Kato III cells in vitro has been shown to be driven in large measure by overexpression of the most abundantly expressed FGFR family member, FGFR2 (24). Following exposure to ARQ 069, Kato III cancer cells were stimulated by the FGFR2-specific keratinocyte growth factor ligand (25), and the degree of phosphorylation of FGFR measured using a pan-anti-phospho-FGFR antibody. During 2 h of exposure to ARQ 069 (Fig. 5A) the degree of phosphorylation of FGFR (predominantly FGFR2) was reduced in a concentration-dependent manner, without decreasing β-actin, the latter being a surrogate marker of cellular viability and integrity.
FIGURE 5.
Inhibition of cellular FGFR phosphorylation by ARQ 069 in Kato III gastric carcinoma cells. A, Kato III human gastric carcinoma cells were plated and treated for 2 h with the indicated concentrations of ARQ 069 and ARQ 068. Ten minutes after stimulation by keratinocyte growth factor, cells were harvested, lysed, and subjected to SDS-PAGE and Western blotting using an antibody to phosphorylated FGFR. An antibody to β-actin served as a loading control. Because this cell line is known to harbor predominantly FGFR2, with a very low abundance of FGFR1, FGFR3, and FGFR4 (data not shown), the signal generated by the anti-phosphorylated FGFR is considered to be largely phosphorylated FGFR2 (24). ARQ 069 inhibited FGFR phosphorylation with an IC50 value of 9.7 μm, whereas ARQ 068 completely failed to inhibit FGFR phosphorylation at concentrations up to 60 μm. B, anti-proliferative activity of FGFR inhibitors in Kato III cells. Kato III gastric carcinoma cells were seeded in 96-well plates overnight (2,000 cells/well) in culture medium with 10% FBS. The next day, cells were treated with different concentrations of the indicated compounds for 72 h at 37 °C. Cells were then incubated and stained for 4 h with methane thiosulfonate (MTS) reagent (final concentration of 0.5 mg/ml) (Promega) per well, lysed, and color development was quantitated by spectrophotometry at λ = 450 nm. The concentration of kinase inhibitor required to inhibit 50% of cell growth (GI50) was calculated using ExcelFitTM. Although ARQ 069 showed anti-proliferative activity in this FGFR2-dependent cell line, ARQ 068 was a much weaker inhibitor.
To test the enantioselective nature of the inhibition, the activity of ARQ 068 was also determined. As expected, ARQ 069 inhibited FGFR phosphorylation in Kato III cells with an IC50 of 9.7 μm, whereas ARQ 068 failed to inhibit FGFR phosphorylation at concentrations up to 60 μm. When Kato III cells were treated with the enantiomers for 72 h in a standard MTS cytotoxicity assay, the same rank order in anti-proliferative potencies was observed (see Fig. 5B).
Structural Basis for ARQ 069 Recognition of the Inactive Conformation of FGFR2
Previous structural studies have shown that FGFR2 can adopt an autoinhibited conformation with a triad of residues forming a hydrogen bond network near the hinge region. Phosphorylation of A-loop tyrosines induces a break in this hydrogen bond network, or “molecular brake” leading to kinase activation (supplemental Fig. S4B). The unphosphorylated form of the FGFR2 kinase domain was co-crystallized with ARQ 069, and the structure solved to a resolution of 2.1 Å (Fig. 6A and supplemental Table S1). Comparison of the FGFR2·ARQ 069 complex structure with previously solved apo-FGFR2 (26) showed that FGFR2 adopts nearly identical configurations in the presence or absence of ARQ 069 with an overall root mean square deviation of 0.283 Å (supplemental Fig. S4A). However, further analysis of the FGFR2·ARQ 069 complex structure in comparison with the apo-FGFR2 structure indicated that both structures form strong crystallographic dimers, which are mediated by key structural elements such as the glycine-rich loop, activation loop, and the αC-helix (supplemental Fig. S4C). This was an unexpected finding, because the FGFR2 kinase domain was purified in the monomeric state and did not dimerize in solution. Furthermore, as ARQ 069 was optimized, additional co-crystal structures of analogs with much improved potency and stronger binding to inactive FGFR2 were determined. However, no clear structural basis for the increased potency was apparent. Together these observations led to the conclusion that it would be difficult to find experimental conditions for co-crystallization of these inhibitors with FGFR2 that would demonstrate an artifact-free structure. Indeed, despite extensive crystallization experiments, FGFR2 could not be crystallized in an alternative crystal packing, and therefore the structure of the FGFR1·ARQ 069 complex was determined as a surrogate in hope that it would yield co-crystal structures more representative of those found in solution.
FIGURE 6.
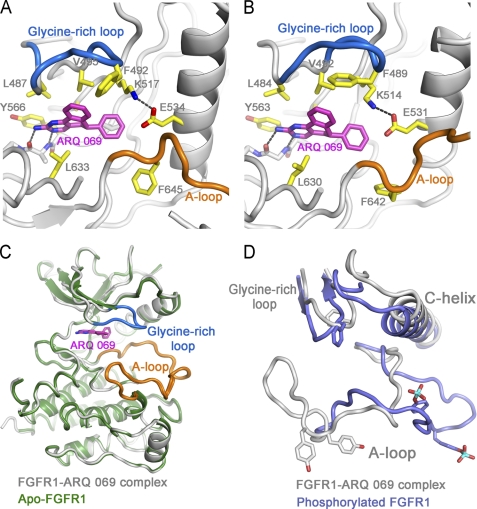
Crystal structure of FGFR2 and FGFR1 bound to ARQ 069. A, the binding mode of ARQ 069 with FGFR2. B, the binding mode of ARQ 069 with FGFR1. The residues that are in close proximity to ARQ 069 are colored in yellow. The hinge and salt bridge interactions are depicted as dotted lines. The overall binding mode of ARQ 069 is very similar in both FGFR1 and FGFR2, however, they show distinct glycine-rich loop conformations highlighted in marine blue. C, superposition of ARQ 069-bound FGFR1 with apo-FGFR1 structure (PDB code 1FGK). Both structures are identical except that the glycine-rich loop is disordered in the apo-FGFR1 structure. D, superposition of ARQ 069-bound FGFR1 with the phosphorylated form of FGFR1 (PDB ID 3GQI). Schematic representations of key structural elements are shown for both structures.
ARQ 069 Binds to the Autoinhibited Conformation of FGFR1
The overall FGFR1 structure is nearly identical to the previously published autoinhibited form with a root mean square deviation of only 0.260 Å (13) (Fig. 6B and supplemental Table S1). The aminopyrimidine group makes two hydrogen bond interactions with the hinge region. The 5,6-dihydrobenzo[h]quinazolin-2-amine core of ARQ 069 is sandwiched in a hydrophobic cleft between residues Val492, Leu484, Ala512, Tyr563 (from above), and Leu630 (below). The plane of the phenyl ring of ARQ 069 is perpendicular to the 5,6-dihydrobenzo[h]quinazolin-2-amine core and occupies the main pocket. This phenyl ring makes hydrophobic contacts with the gatekeeper residue (Val561) and the methionine residue of the α-helix (Met535), and this hydrophobic pocket is capped with the aliphatic side chain of the catalytic lysine residue (Lys514) (Fig. 6A). The most striking feature of the FGFR1·ARQ 069 structure is the conformation of the A-loop, because this segment is virtually identical to that in the autoinhibited FGFR1 structure and appears to interfere with substrate binding but not with ATP binding (13) (Fig. 6C). Moreover, the αC-helix and glycine-rich loop adopt conformations that are very distinct from the phosphorylated form of FGFR1 (Fig. 6D).
Interestingly, one major distinguishing feature between apo-FGFR1 and the FGFR1·ARQ 069 complex is the significant difference found within the glycine-rich loop. When the inhibitor binds, the phenylalanine (Phe489) of the glycine-rich loop makes a downward movement and establishes van der Waals interactions with the fused phenyl ring of ARQ 069. This glycine-rich loop structure is not observed in apo-FGFR1 (Fig. 6C). Computational modeling of the binding of ARQ 069 into the active FGFR1 structure suggests that the position of the phenyl ring in the ATP-binding cleft imposes steric conflict between the glycine-rich loop and ARQ 069 (Fig. 7A). Presumably, the interaction between ARQ 069 and the phenyl ring of the glycine-rich loop serves as an anchor to stabilize the conformation of the glycine-rich loop and contribute to the preference of ARQ 069 for the inactive form of FGFR1.
FIGURE 7.
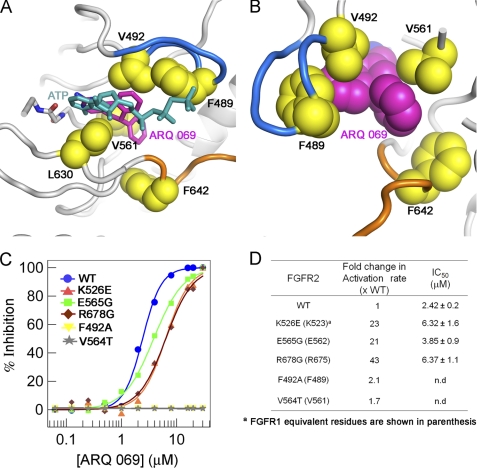
The non-conserved hydrophobic residues are key determinants of ARQ 069 binding to FGFR kinase. A, the ARQ 069 interaction with FGFR1 requires a rearrangement of hydrophobic residues in the ATP-binding cleft that re-orients the hydrophobic residues (yellow) creating a non-polar environment that is not compatible with ATP binding. The downward conformation of the glycine-rich loop in FGFR1 is mediated by hydrophobic interaction between Phe489, Val492, and ARQ 069. The FGFR1·ARQ 069 complex structure was superimposed with AMP-ACP-bound phosphorylated FGFR1 (PDB ID 3GQI) and ATP is shown in sticks. B, non-polar interaction of ARQ 069 with two non-conserved hydrophobic residues (Phe489 and Val561). C, mutational analysis of ARQ 069 interaction with FGFR2. The effect of activating mutations (K526E, E565G, and R678G) and non-conserved hydrophobic residue (F492A and V564T) mutations on FGFR2 autophosphorylation inhibition by ARQ 069 was measured. The potency of autoactivation was measured using a continuous spectrophotometeric assay to derive IC50 values. D, comparison of the activation rate (as measured by substrate phosphorylation) with the potency of FGFR2 inhibition by ARQ 069 for the various mutant proteins. Activating mutant proteins show a significant increase in the activation rate and autophosphorylation remains inhibited by ARQ 069. However, mutation of ARQ 069 interacting residues F492A and V564T in FGFR2 prevents inhibition of autophosphorylation by ARQ 069.
Hydrophobic Residues Play a Key Role in the Interaction of ARQ 069 with FGFR2
To further probe the hydrophobic interaction of ARQ 069 with non-polar residues in FGFR2 two mutations were made. The co-crystal structure of the FGFR1·ARQ 069 complex indicated that the two non-conserved hydrophobic residues (Val564 and Phe492 residues in FGFR2) in FGFR1 participate in non-polar interactions with ARQ 069 (Fig. 7B). To confirm this, Val564 was mutated to threonine, and Phe492 was mutated to alanine. Three additional mutations were made to assess the effects of activating mutations of FGFR2. Three activating mutants were selected, K526E (the hinge region), E565G (the αC-helix), and R678G (interacts with the A-loop), as each mutation maps to a distinct location in the kinase domain. Each was selected to better understand whether constitutive activation of FGFR2 prevents ARQ 069 from recognizing the autoinhibited form of the kinase.
To confirm that each mutant remained catalytically competent the activation rate of the five mutant proteins was tested (unphosphorylated forms) in the substrate phosphorylation assay using Pyk2 as substrate. As shown in Fig. 7D, and as expected, the activation rate was significantly elevated for the enzymes containing the activating mutations. The degree of activation of V564T and F492A was not significantly altered compared with wild type. The ability of ARQ 069 to inhibit the autophosphorylation of each of the mutants was assessed. The inhibitory activity of ARQ 069 was measured using the autophosphorylation assay. Interestingly, the activating mutants showed less than a 3-fold decrease in potency as reflected by the increased IC50 values and as expected the V564T and F492A mutants showed an increased resistance to inhibition by ARQ 069 (Fig. 7, C and D). These results clearly demonstrate that the hydrophobic residues in the autoinhibited form of FGFR2 are critical for interaction with ARQ 069 and confirm the initial hypothesis that the hydrophobic clusters in kinases can be utilized to design inhibitors for inactive kinases.
DISCUSSION
For more than 25 years it has been recognized that kinases exist in active and/or inactive states in cells. A mechanism by which this transformation takes place both in the absence of small molecule inhibitors (3) and in their presence (27) is emerging. NMR spectroscopy (28) and deuterium exchange studies (29) have shown dramatic conformational transitions in kinases upon ATP binding indicating that these differences may be exploitable for the structure-based design of inhibitors.
Our structural analysis of a number of kinases in the inactive and more specifically in the autoinhibited state show a cleft lined with non-polar residues organized into clusters that make the site hydrophobic and incompatible with nucleotide binding. Comparison of the ARQ 197-c-Met co-crystal structure with those of apo-kinases in the autoinhibited state led to the hypothesis that non-polar residues observed in the autoinhibited conformation could be exploited in the design of inhibitors. The aggregation of these hydrophobic clusters is likely facilitated by several regulatory mechanisms, such as a DFG-in/out flip (30), positioning of hydrophobic residues into the ATP-binding pocket (31), and interaction with autoinhibitory elements like the juxtamembrane domain (32). To our knowledge, there has been no previously described systematic approach to exploit the presence of hydrophobic clusters in the ATP-binding cleft as the basis for the design of inhibitors.
The ATP-binding pocket described in previously reported structures of unphosphorylated apo-FGFR2, phosphorylated FGFR2 bound to an ATP analog, the gain-of-function, loss-of-function, and mutant FGFR2 structures are quite similar (26, 33). On the basis of this structural information it is difficult to describe how FGFR activation takes place and to find support for conformational dynamics playing a key role in FGFR autoinhibition and activation. Indeed, the observation of a crystallization artifact with unphosphorylated FGFR2 impeded the validation of the homology model of FGFR2. Nonetheless, the FGFR2 model-driven optimization of ARQ 523 to ARQ 069 and the selectivity of ARQ 069 toward the inactive form of FGFR2 do support the use of the homology model of the autoinhibited FGFR2. Although not all the predicted structural features of the homology model were present in the crystal structure of the FGFR1·ARQ 069 complex, FGFR1 bound to ARQ 069 does adopt the overall autoinhibited conformation previously reported (13).
The results of the crystal structure, together with mutational analysis, validated the initial hypothesis that the non-conserved hydrophobic residues aggregate to form hydrophobic clusters in the inactive state and participate in non-polar interactions with inhibitors. The two non-conserved hydrophobic residues Val561 and Phe489 (FGFR1 numbering) are critical for interaction with ARQ 069. Interestingly, upon binding to ARQ 069, the phenylalanine of the glycine-rich loop of FGFR1 adopts a downward conformation, and Val492 participates in a non-polar interaction with the fused phenyl ring of ARQ 069 (corresponding to Val495) in FGFR2. This interaction is not observed in the apo-FGFR1 or apo-FGFR2 structures. Modeling of ARQ 069 binding to the ATP analog-bound FGFR1 structure shows a number of steric conflicts between the flexible glycine-rich loop and ARQ 069. This structural difference could contribute to the selectivity of ARQ 069 for the inactive forms of FGFR1 and FGFR2. A number of crystal structures have been reported for FGFR1 bound to small molecule inhibitors (34, 35). Surprisingly, all of these structures show FGFR1 in a conformation very similar to that resulting from ARQ 069 binding, except for the position of the glycine-rich loop. A similar downward glycine-rich loop conformation has, however, been observed in several other kinase-inhibitor complexes and has been suggested to play a role in stabilizing the inactive state of kinases (2, 4, 36, 37).
The structural incompatibility of ATP binding to the conformation of ARQ 069-bound FGFR1 or FGFR2 does not explain the non-competitive kinetic profile of ARQ 069 at high ATP concentrations. ARQ 069 was thus profiled extensively to characterize its mechanism of inhibition.
The conformational dynamics of kinases, between inactive and active states, is known to be influenced by the concentration of ATP. Indeed below the Kd of ARQ 069 and with an excess of AMP-PNP, FGFR2 is more likely to adopt an active conformation. However, above saturating concentrations, ARQ 069 essentially locks FGFR2 in an inactive conformation that prevents AMP-PNP binding.
A number of earlier studies on the steady-state kinetic measurement of kinase autoactivation have indicated that the Km of ATP for the inactive form is significantly higher than for the activated or phosphorylated form, and that upon autophosphorylation, the Km for both ATP and peptide substrate decrease and kcat increases (38, 39). This is also the case for FGFR2, where the Km for the inactive form is at least 10-fold higher than for the active form (19, 26). This suggests that inactive conformations of FGFR2 do not provide ready access for ATP binding, consistent with the observation that ARQ 069 exhibits non-ATP competitive kinetics at physiological ATP concentrations. Although the binding affinities of ARQ 069 and AMP-PNP to FGFR2 are similar, ARQ 069 is a relatively more potent inhibitor. AMP-PNP being the significantly weaker inhibitor (data not shown) strongly suggests that in solution, the ARQ 069-binding conformation is significantly different from that of the active kinase, as it would otherwise be expected that AMP-PNP and ARQ 069 should have similar inhibitory action.
Other inhibitors that bind in the ATP-binding cleft and display complex mechanisms of inhibition have been described (3, 23, 40, 41). Determination of steady-state kinetic parameters in kinase assays for the inactive form of a protein kinase frequently involves two separate reactions, the autophosphorylation reaction (activation step) followed by a substrate phosphorylation reaction. The type I inhibitors that bind both inactive and active forms are competitive against ATP in these assays. Inhibitors that target the inactive conformation may not show purely competitive kinetics against ATP. For example, certain p38 inhibitors, which bind to the hinge and occupy the allosteric back pocket, exhibit partially ATP-competitive kinetics (42, 43). The slow dissociation of these inhibitors could contribute to the observed non-competitive kinetics. When targeting the inactive conformation, an important assumption made in these assays is that the inhibitory activity is expected to propagate from the autophosphorylation reaction to the substrate phosphorylation reaction. In such complex assays, a number of kinetic parameters affect the determination of the mechanism of inhibition, such as kcat/Km of ATP and substrate and the affinity of the inhibitor for and off-rate of the inhibitor from the inactive form of the kinase. For example, sunitinib, which has previously been shown to be an ATP-competitive inhibitor with the isolated kinase domains of VEGFR2 and PDGFR, was reported recently to target the autoinhibited conformation of c-Kit with full potency, even at physiological ATP concentrations (36). Consistent with these reports, the observed mixed-type binding mode of ARQ 069 with the inactive form of FGFR2 could be attributed, at least in part, to slow dissociation of the enzyme-inhibitor complex and is consistent with potent inhibition at physiological ATP concentrations.
The first step in the optimization of ARQ 523 produced ARQ 069, which was characterized as an FGFR inhibitor that targets the autoinhibited, inactive conformation of FGFR2 leading to good inhibition of substrate phosphorylation. Importantly, ARQ 069 inhibited FGFR2 autophosphorylation in cancer cells within the same range of potency as its biochemical potency in the presence of millimolar concentrations of ATP. In addition, because ARQ 069 demonstrates cellular activity in only a few hours, this suggests that the abundance of the inactive form in these cells is sufficient for significant abrogation of FGFR2 signaling to be achieved during this time frame.
In summary, a new understanding of the role of hydrophobic residues within the ATP-binding cleft of inactive kinases has been utilized to discover novel kinase inhibitors. Unlike type II inhibitors, the inhibitors described here do not require interaction with a gatekeeper residue or interaction with catalytic lysine/glutamic acid residues to access the allosteric back pocket to stabilize the inactive conformation. This knowledge was applied to identify a novel molecular scaffold that binds to FGFR1 and FGFR2 kinases and inhibits FGFR kinase activity in cells. This approach can be generalized to the design of inhibitors for other kinase targets in the autoinhibited conformation. The implementation of this new approach may provide a new generation of kinase inhibitors that exhibit a higher degree of selectivity across the kinome and access an as yet unexploited chemical space for their design. Through this approach it is expected that novel inhibitors can be discovered for a wide range of therapeutically relevant kinase targets.
Supplementary Material
Acknowledgments
We thank Dr. Susan Taylor for critical reading of this manuscript and the ArQule chemistry and analytical chemistry staff for the preparation and purification of ARQ 068 and ARQ 069. In addition, we are grateful to ArQule's research team members for their critical comments on the manuscript.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Scheme 1, “Methods,” Table S1, and Figs. S1–S5.
The atomic coordinates and structure factors (codes 3RI1 and 3RHX) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- FGFR
- fibroblast growth factor receptor
- AMP-PNP
- 5′-adenylyl-β,γ-imidodiphosphate
- DMSO
- dimethyl sulfoxide
- ARQ 523
- 5,6-dihydrobenzo[h]quinazolin-2-amine.
REFERENCES
- 1. Huse M., Kuriyan J. (2002) Cell 109, 275–282 [DOI] [PubMed] [Google Scholar]
- 2. Nolen B., Taylor S., Ghosh G. (2004) Mol. Cell. 15, 661–675 [DOI] [PubMed] [Google Scholar]
- 3. Cheetham G. M. (2004) Curr. Opin. Struct. Biol. 14, 700–705 [DOI] [PubMed] [Google Scholar]
- 4. Johnson L. N. (2009) Q. Rev. Biophys. 42, 1–40 [DOI] [PubMed] [Google Scholar]
- 5. Cowan-Jacob S. W., Möbitz H., Fabbro D. (2009) Curr. Opin. Cell Biol. 21, 280–287 [DOI] [PubMed] [Google Scholar]
- 6. Engh R. A., Bossemeyer D. (2002) Pharmacol. Ther. 93, 99–111 [DOI] [PubMed] [Google Scholar]
- 7. Liu Y., Gray N. S. (2006) Nat. Chem. Biol. 2, 358–364 [DOI] [PubMed] [Google Scholar]
- 8. Kufareva I., Abagyan R. (2008) J. Med. Chem. 51, 7921–7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hubbard S. R. (2002) Front. Biosci. 7, d330–340 [DOI] [PubMed] [Google Scholar]
- 10. Peterson J. R., Golemis E. A. (2004) J. Cell. Biochem. 93, 68–73 [DOI] [PubMed] [Google Scholar]
- 11. Munshi N., Jeay S., Li Y., Chen C. R., France D. S., Ashwell M. A., Hill J., Moussa M. M., Leggett D. S., Li C. J. (2010) Mol. Cancer Ther. 9, 1544–1553 [DOI] [PubMed] [Google Scholar]
- 12. Turner N., Grose R. (2010) Nat. Rev. Cancer. 10, 116–129 [DOI] [PubMed] [Google Scholar]
- 13. Mohammadi M., Schlessinger J., Hubbard S. R. (1996) Cell 86, 577–587 [DOI] [PubMed] [Google Scholar]
- 14. CCP4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 15. Barker S. C., Kassel D. B., Weigl D., Huang X., Luther M. A., Knight W. B. (1995) Biochemistry 34, 14843–14851 [DOI] [PubMed] [Google Scholar]
- 16. Eswarakumar V. P., Lax I., Schlessinger J. (2005) Cytokine Growth Factor Rev. 16, 139–149 [DOI] [PubMed] [Google Scholar]
- 17. Szwaya J., Bruseo C., Nakuci E., McSweeney D., Xiang X., Senator D., France D., Chen C. R. (2007) J. Biomol. Screen. 12, 159–166 [DOI] [PubMed] [Google Scholar]
- 18. Furdui C. M., Lew E. D., Schlessinger J., Anderson K. S. (2006) Mol. Cell. 21, 711–717 [DOI] [PubMed] [Google Scholar]
- 19. Lew E. D., Furdui C. M., Anderson K. S., Schlessinger J. (2009) Sci. Signal. 2, ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bae J. H., Boggon T. J., Tomé F., Mandiyan V., Lax I., Schlessinger J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2866–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lo M. C., Aulabaugh A., Jin G., Cowling R., Bard J., Malamas M., Ellestad G. (2004) Anal. Biochem. 332, 153–159 [DOI] [PubMed] [Google Scholar]
- 22. Pantoliano M. W., Petrella E. C., Kwasnoski J. D., Lobanov V. S., Myslik J., Graf E., Carver T., Asel E., Springer B. A., Lane P., Salemme F. R. (2001) J. Biomol. Screen. 6, 429–440 [DOI] [PubMed] [Google Scholar]
- 23. Karni R., Mizrachi S., Reiss-Sklan E., Gazit A., Livnah O., Levitzki A. (2003) FEBS Lett. 537, 47–52 [DOI] [PubMed] [Google Scholar]
- 24. Kunii K., Davis L., Gorenstein J., Hatch H., Yashiro M., Di Bacco A., Elbi C., Lutterbach B. (2008) Cancer Res. 68, 2340–2348 [DOI] [PubMed] [Google Scholar]
- 25. Eswarakumar V. P., Horowitz M. C., Locklin R., Morriss-Kay G. M., Lonai P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12555–12560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen H., Ma J., Li W., Eliseenkova A. V., Xu C., Neubert T. A., Miller W. T., Mohammadi M. (2007) Mol. Cell. 27, 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao B., Smallwood A., Yang J., Koretke K., Nurse K., Calamari A., Kirkpatrick R. B., Lai Z. (2008) Protein Sci. 17, 1791–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Masterson L. R., Mascioni A., Traaseth N. J., Taylor S. S., Veglia G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 506–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee T., Hoofnagle A. N., Resing K. A., Ahn N. G. (2005) J. Mol. Biol. 353, 600–612 [DOI] [PubMed] [Google Scholar]
- 30. Kornev A. P., Haste N. M., Taylor S. S., Eyck L. F. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17783–17788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Richards M. W., O'Regan L., Mas-Droux C., Blot J. M., Cheung J., Hoelder S., Fry A. M., Bayliss R. (2009) Mol. Cell. 36, 560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hubbard S. R. (2004) Nat. Rev. Mol. Cell Biol. 5, 464–471 [DOI] [PubMed] [Google Scholar]
- 33. Lew E. D., Bae J. H., Rohmann E., Wollnik B., Schlessinger J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19802–19807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mohammadi M., McMahon G., Sun L., Tang C., Hirth P., Yeh B. K., Hubbard S. R., Schlessinger J. (1997) Science 276, 955–960 [DOI] [PubMed] [Google Scholar]
- 35. Ravindranathan K. P., Mandiyan V., Ekkati A. R., Bae J. H., Schlessinger J., Jorgensen W. L. (2010) J. Med. Chem. 53, 1662–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gajiwala K. S., Wu J. C., Christensen J., Deshmukh G. D., Diehl W., DiNitto J. P., English J. M., Greig M. J., He Y. A., Jacques S. L., Lunney E. A., McTigue M., Molina D., Quenzer T., Wells P. A., Yu X., Zhang Y., Zou A., Emmett M. R., Marshall A. G., Zhang H. M., Demetri G. D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1542–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Taylor S. S., Yang J., Wu J., Haste N. M., Radzio-Andzelm E., Anand G. (2004) Biochim. Biophys. Acta 1697, 259–269 [DOI] [PubMed] [Google Scholar]
- 38. Knight Z. A., Shokat K. M. (2005) Chem. Biol. 12, 621–637 [DOI] [PubMed] [Google Scholar]
- 39. Wodicka L. M., Ciceri P., Davis M. I., Hunt J. P., Floyd M., Salerno S., Hua X. H., Ford J. M., Armstrong R. C., Zarrinkar P. P., Treiber D. K. (2010) Chem. Biol. 17, 1241–1249 [DOI] [PubMed] [Google Scholar]
- 40. Dubreuil P., Letard S., Ciufolini M., Gros L., Humbert M., Castéran N., Borge L., Hajem B., Lermet A., Sippl W., Voisset E., Arock M., Auclair C., Leventhal P. S., Mansfield C. D., Moussy A., Hermine O. (2009) PLoS One 4, e7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wong T. W., Lee F. Y., Yu C., Luo F. R., Oppenheimer S., Zhang H., Smykla R. A., Mastalerz H., Fink B. E., Hunt J. T., Gavai A. V., Vite G. D. (2006) Clin. Cancer Res. 12, 6186–6193 [DOI] [PubMed] [Google Scholar]
- 42. Montalban A. G., Boman E., Chang C. D., Ceide S. C., Dahl R., Dalesandro D., Delaet N. G., Erb E., Gibbs A., Kahl J., Kessler L., Lundström J., Miller S., Nakanishi H., Roberts E., Saiah E., Sullivan R., Wang Z., Larson C. J. (2008) Bioorg. Med. Chem. Lett. 18, 5456–5459 [DOI] [PubMed] [Google Scholar]
- 43. Frantz B., Klatt T., Pang M., Parsons J., Rolando A., Williams H., Tocci M. J., O'Keefe S. J., O'Neill E. A. (1998) Biochemistry 37, 13846–13853 [DOI] [PubMed] [Google Scholar]
- 44. Eathiraj S., Palma R., Volckova E., Hirschi M., France D. S., Ashwell M. A., Chan T. C. K. (2011) J. Biol. Chem. 286, 20666–20676 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.