Abstract
A significant amount of evidence shows that microenvironmental signals generated from extracellular matrix (ECM) molecules, soluble factors, and cell–cell adhesion complexes cooperate at the extra- and intracellular level. This synergetic action of microenvironmental cues is crucial for normal mammary gland development and breast malignancy. To explore how the microenvironmental genes coordinate in human breast cancer at the genome level, we have performed gene co-expression network analysis in three independent microarray datasets and identified two microenvironment networks in human breast cancer tissues. Network I represents crosstalk and cooperation of ECM microenvironment and soluble factors during breast malignancy. The correlated expression of cytokines, chemokines, and cell adhesion proteins in Network II implicates the coordinated action of these molecules in modulating the immune response in breast cancer tissues. These results suggest that microenvironmental cues are integrated with gene transcriptional networks to promote breast cancer development.
Introduction
Cell proliferation, apoptosis, invasion and differentiation are regulated by the microenvironmental signals generated from cell–ECM adhesion, cell–cell contacts, and various soluble factors.1,2 In mammary tissue, luminal and basal epithelial cells form bi-layer tubular or acinar structures where basal cells adhere to a basement membrane (BM). The BM is comprised largely of laminins, type IV collagen, entactin/nidogen, and proteoglycans. Outside of the BM, stromal cells, adipocytes, and immune cells can produce a variety of proteins and small molecules to affect epithelial behaviors. During mammary gland branching, alveologenesis, lactation, and involution, the expression and/or activities of collagens, laminin, matrix metalloproteinases (MMPs) and growth factors are tightly regulated both temporally and spatially (reviewed in ref. 3). Integrated signals generated from these microenvironmental cues are crucial for normal mammary gland development and tissue-specific function, and altering the balance of microenvironmental cues is sufficient not only to disrupt normal tissue morphogenesis but also to induce breast malignancy and progression.
Dialogues between a cell and its microenvironment are mainly through three types of transmembrane proteins: growth factor and hormone receptors, ECM receptors, and junction proteins. Growth factors, hormones, and their receptors have been considered major regulators of mammary epithelial cell (MEC) proliferation and differentiation. However, the MECs have distinct responses to soluble factors when adhering to different ECM molecules or when growing at different densities, suggesting that ECM receptors and junction proteins also play central roles in regulating these processes. In fact, a number of studies have shown that these membrane proteins and their downstream signals cooperate to regulate acinar morphogenesis and mammary specific gene expression,4–8 supporting the concept that tissue architecture and function are determined by integrated microenvironmental signals.
In this review, we discuss how ECM molecules, soluble factors, and cadherin complexes cooperate to direct normal and malignant epithelial tissue architecture and function. Using high throughput gene expression profiles, we have identified two transcription networks composed of microenvironment-related genes which integrate microenvironment remodeling at the genome level in human breast cancer.
Integrated microenvironmental signals in normal and malignant mammary gland
Cooperation between ECM and soluble factors
Prolactin, a lactogenic hormone mainly produced in the pituitary gland, is required for the alveologenesis and milk production in mice.9 Canonical prolactin signal transduction is initiated by binding of the hormone to the receptor, which activates transient JAK2-mediated phosphorylation and nuclear translocation of STAT5.10,11 However, in the absence of ECM protein, prolactin induces transient STAT5 activation but fails to activate milk protein expression.7,8,12 Using a three-dimensional (3D) culture model, we and others have shown that laminin-dependent biochemical signals are necessary for the milk protein expression.5,7,8,13 We further demonstrate that laminin cooperates with prolactin to induce a sustained activation of STAT5, and that blocking the sustained activation inhibits chromatin remodeling and milk protein expression.7 Transmission of biochemical and biophysical signals from laminin to a cell is mediated predominantly by integrins, a family of heterodimer proteins composed of α and β subunits.14 Deletion of the β1 subunit in mammary epithelial cells impairs STAT5 nuclear translocation and lactation function,15 suggesting that cooperation between ECM and lactogenic hormones is functionally important in vivo.
Cooperative effects have also been identified from signals generated by integrins and epidermal growth factor receptors (EGFRs) during breast malignancy. Disruption of tissue organization in breast cancer is frequently associated with alteration of EGFR and integrin profiles,6,16,17 implicating crosstalk between the two receptors. In HTM-3522 human breast cancer progression cell lines, both EGFR and β1 integrin levels are increased appreciably in malignant T4-2 cells compared to their non-malignant counterpart S1 cells.6,18 In 3D culture, blocking EGFR or β1 integrin signals with antibodies or chemical inhibitors significantly reduces expression of both proteins and restores polarity in the malignant cells, indicating that β1 integrin and EGFR signals are coupled to modulate tissue organization.6 Another example is from α6β4 integrin and EGFR2. α6β4 integrin is a major component of hemidesmosomes, mediating cell adhesion to laminin-332.19 The cytoplasmic tail of β4 integrin provides a variety of binding sites for cytoskeletal and signaling proteins. EGF treatment induces serine phosphorylation in this region via protein kinase C (PKC), which destabilizes the hemidesmosomes and enhances cell migration.20 In mammary epithelial cells, the β4 integrin-EGFR2 complex enhances activation of the transcription factors STAT3 and c-Jun, which contributes to the disruption of epithelial polarity and hyperproliferation, respectively.4 EGFR2-dependent mammary tumor onset and growth are suppressed by introducing a targeted deletion of the β4 signaling domain.4 A recent study showed that neuregulin-1 binds to integrins αvβ3 and α6β4, and that this interaction is necessary for EGFR phosphorylation, and AKT and Erk1/2 activation in MCF-7 and T47D human breast cancer cells.21 These studies established the importance of integrated signals between ECM and growth factors in determining normal and malignant mammary tissue structure and function.
In addition to the crosstalk at the biochemical level, growth factor activity can be modulated by ECM molecules through physical interactions. For instance, binding of fibroblast growth factors (FGFs) and vascular endothelial growth factors (VEGFs) to heparin, a component of ECM proteoglycans, mediate sequestration, stabilization and high affinity receptor binding and signaling of the factor.22 Hepatocyte growth factor (HGF) binds to fibronectin (FN) and vitronectin, thus forming complexes with integrins and the HGF receptor in endothelial cells, which leads to increased cell migration.23 In the mammary gland, HGF is produced by stromal cells and acts on epithelial cells to regulate branching morphogenesis.24 Interestingly, deposition of FN also increases appreciably at the branching stage,25 which may enhance HGF activity and stimulate ductal elongation. Furthermore, the interaction between the ECM and soluble factors assists in generating a morphogen gradient. In Drosophila, binding of collagen IV to Dpp creates a gradient distribution of the factor, which is crucial for drosoventral axis development.26 These examples illustrate the capacity of the ECM to act as a reservoir and cofactor of soluble factors in regulating normal tissue development.
Crosstalk of cadherins with growth factors and ECM
Cell–cell contacts in epithelial tissue are predominantly mediated by cadherin, gap, and tight junctions. Cadherin proteins, especially E-cadherin, play important roles in regulating epithelial cell homeostasis. Expression of a dominant negative E-cadherin in mammary epithelial cells results in apoptosis with a concomitant loss of milk production at parturition, indicating that cell–cell adhesion is essential for the function of the mammary gland.27,28 Using a functional micropattern technique, a recent study demonstrates that cell migration requires integrated signals from E-cadherin- and integrin-based adhesion complexes.29 E-cadherin can constrain integrin-based focal adhesion assembly and regulate cell migration along the direction of E-cadherin/ECM interfaces.29 In addition, the interaction of the E-cadherin/catenin complex with EGFR family members represses the cellular response to EGF and inhibits EGFR pro-mitotic activity.30,31 Therefore, cell motility and mitotic activities require the coordinated action of cadherins and other microenvironmental cues.
Cell-cell adhesion mediated by E-cadherin/catenin is a dynamic process that is regulated by ECM molecules and growth factors at various levels, including gene transcription, post-translational modifications, and protein stability. Integrin-linked kinase (ILK), a serine/theronine protein kinase that is activated by the engagement of the ECM, interacts with the cytoplasmic domain of β1 and β3 integrins.32 Overexpression of ILK in a mammary epithelial cell line represses E-cadherin expression and induces the epithelial to mesenchymal transition (EMT), a process associated with mammary tumor invasion and metastasis.33 In the human mammary epithelial cell line PMC42, incubation with EGF led to down-regulation of E-cadherin, which correlated with the enhancement of cell migration and spreading.34 EGF treatment also induces tyrosine phosphorylation on β- and γ-catenin and impairs the adhesive function on the E-cadherin/catenin complexes resulting in scattering of E-cadherin-positive cervical cancer cells.35 This study demonstrated that E-cadherin/catenin stability can be modulated by growth factors through post-translational modifications.
The above examples clearly show that microenvironmental signals cooperate at different levels to direct mammary tissue architecture and epithelial function (Fig. 1). During breast cancer development, these microenvironmental signals often go awry, which, in turn, disrupts normal tissue structure and promotes cancer progression.1 However, the majority of previous studies focused only on one or two microenvironment-related genes or pathways, and it still remains to be determined how the microenvironmental cues integrate at the genome level to promote breast cancer development.
Fig. 1.
Microenvironment signals crosstalk at multiple levels to regulate cellular function. The signals generated from the ECM, soluble factors, and cell–cell contacts cooperate at four different levels: (1) ECM molecules modulate growth factor activity through physical interaction; (2) integrins, receptors of soluble factors, and cadherins form complexes on the cell membrane to cooperatively activate/repress downstream signals; (3) intracellular effectors of integrins, receptors, and cadherins crosstalk with each other; and (4) expression of microenvironment genes can be regulated by various microenvironmental signals.
Microenvironment gene networks in human breast cancer
To understand how microenvironmental signals integrate at the genome level in the normal and malignant mammary gland, we first need to obtain global information of the microenvironmental cues. Unlike intracellular signaling proteins, which can be modulated by phosphorylation and acetylation, the activities of microenvironment molecules are largely determined at the transcriptional and translational levels. With the advent of DNA microarray technology, expression profiles have been generated from more than a thousand breast cancer tissues, and the majority of these profiles are available in public databases, such as the Gene Expression Omnibus (GEO) datasets, where we can retrieve information regarding the microenvironment gene transcriptome. Next, we need to identify sets of genes that act together to integrate microenvironment remodeling. Since gene pairs with high correlation are usually hypothesized to be biologically relevant and to interact directly in signaling pathways, one method of finding candidate gene sets is to identify networks of genes that have significantly correlated mRNA levels.36 Gene co-expression network analysis is a systems biology method using correlation statistics as pairwise similarity measurements between gene expression profiles followed by either direct correlation thresholding or a combination of significance level tests with correlation thresholding. In a gene co-expression network, graph nodes represent genes and edges represent gene associations. This approach illustrates the strong relationship which connects transcripts' regulatory patterns to the functional organization of the cell. Further, through Gene Ontology and KEGG pathway analyses of network genes, enriched categories provide hints to biological processes and the molecular function of the unknown genes based on the functionally known gene. Using correlation and linkage analysis, a recent study has identified gene networks associated with skin tumor susceptibility.37 Taking advantage of published microarray data and the recent progress in the capabilities of analyzing transcriptional networks, we decided to generate microenvironment gene networks to investigate how microenvironmental signals are integrated in human breast cancer.
Generation of the microenvironment gene networks in human breast cancer
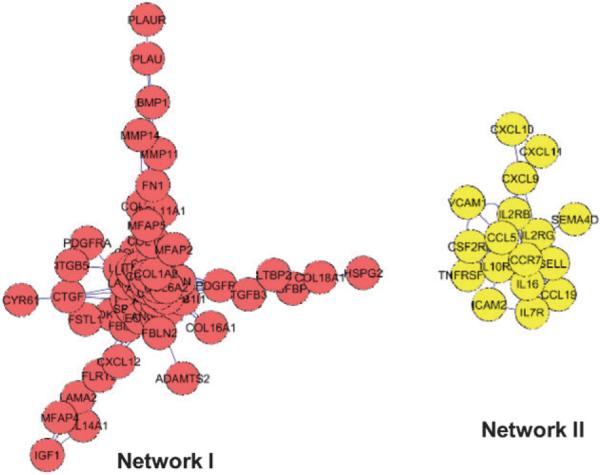
To identify the microenvironmental genes whose mRNA levels are significantly correlated in human breast cancer tissues, we first generated a gene list containing about seven hundreds microenvironment-related genes, encoding ECM proteins and receptors, ECM remodeling enzymes, soluble factors and receptors as well as junction proteins (Table S1‡). Microenvironment genes whose mRNA levels are significantly correlated in human breast cancer microarray datasets were selected to construct microenvironment gene networks (Fig. S1‡). We performed gene co-expression network analysis in three independent breast cancer microarray datasets, and the most conserved gene networks are shown in Fig. 2, which was generated using Cytoscape. In the networks, significantly correlated transcripts are drawn as nodes connected by an edge. We identified two microenvironment gene networks from human breast cancer tissues. One network contains 64 genes (Network I), the majority of which are ECM microenvironment-related genes and soluble factor genes. The other network is composed of 17 genes (Network II), mainly encoding cytokines/chemokines and their receptors (Table 1).
Fig. 2.
Two microenvironment gene networks were identified using correlation and linkage analysis of three microarray datasets generated from human breast cancer tissues. Significantly correlated transcripts are drawn as nodes connected by an edge in the networks. Network I contains 64 genes of which the majority are ECM- and soluble factor-related genes. Network II is composed of 17 genes, and 13 of them encode cytokines, chemokines and their receptors.
Table 1.
Genes with correlated expression in Network I and II
| Category | Number | Gene symbol |
|---|---|---|
| Network I | ||
| ECM protein | 36 | THBS2, COL6A1, SPON1, BGN, COL5A1, LAMA4, HSPG2, COL1A2, COL1A1, COL6A3, MFAP4, CYR61, COL8A2, COL3A1, EFEMP2, FBLN2, COL10A1, LUM, COL6A2, COL18A1, LAMB1, SPARC, ECM2, FBN1, COL5A2, COMP, COL16A1, NID2, FN1, LAMA2, COL11A1, DCN, COL14A1 |
| ECM remodeling-related | 12 | LOXL1, MMP11, MMP14, ADAM12, SERPIN1, SERPINF1, PLAU, LOX, MMP2, PCOLCE, ADAMTS2, PLAUR |
| ECM receptor | 2 | ITBG5, ITGBL1 |
| Soluble factors and receptors | 14 | PDGFRL, CXCL12, IGF1, CTGF, FSTL1, TGFB3, PDGFRA, IGFBP7, LTBP2, PDGFRB, ANGPTL2, BMP1,DKK3, MFAP2 |
| Network II | ||
| Soluble factors and receptors | 14 | CXCL11, TNFRSF1B, SEMA4D, IL7R, IL10RA, CCR7, IL2RG, CXCL9, CXCL10, CCL5, IL16, CSF2RB, CCL19 |
| Cell–cell adhesion | 3 | VCAM1, SELL, ICAM2 |
Correlated gene expression usually reflects co-regulation and/or associated biological function. For instance, mRNA levels of genes A, B, and C are significantly correlated and form a network (Fig. 3); the correlation may be due to co-regulation of genes A, B, and C, or A and B may be regulated by gene C (Fig. 3A). In another scenario, proteins translated from gene A and B may form complexes or have a physical interaction, and correlated expression can ensure that the proteins are produced at the required ratio (Fig. 3B). The third possibility is that gene A and B are components of the same signaling pathway, and correlated expression may facilitate the coordinated action of this pathway (Fig. 3C).
Fig. 3.
Diagram showing how correlated gene expression is associated with functional integration. The expression of gene A, B, and C is significantly correlated, and a network is established based on the correlation. (a) Correlated gene expression may be caused by coordinated transcriptional regulation. Transcription of gene A and B is regulated by C, or gene A, B, and C are all controlled by factor D. (b) Proteins with physical interactions may have correlated expression: (i) protein A and B form a heterodimer; (ii) protein A is a ligand of receptor B; (iii) protein A is a substrate of enzyme B. (c) Proteins A, B and C are components of a common signaling pathway. Correlated gene expression indicates coordinated action of the signaling pathway.
In fact, we observed all three scenarios in the microenvironment gene networks. In Network I, PDGFRB mRNA levels are correlated with the mRNA levels of multiple ECM protein genes (Fig. 4A). It has been shown that the PDGF pathway is involved in the pathogenesis of fibrotic diseases,38 and downregulation of PDGFB and PDGFRB attenuates type I collagen expression.39 Thus, the connection of PDGFR and ECM protein genes in the network suggests that the PDGF pathway may control ECM protein production during breast cancer development. We also found that expression of lysyl oxidase (LOX) and LOXL1 were directly linked with that of collagen and other ECM protein genes (Fig. 4B). LOXs are involved in covalent intra- and intermolecular crosslinking of collagen.40 A recent study showed that LOX-mediated collagen crosslinking modulates tissue stiffness and fibrosis to enhance growth factor signaling and breast malignancy.41 The correlated expression between LOX and collagen genes most likely represents the functional linkage of the enzymes and substrates. In the same network, mRNA levels of COL1A1 and COL1A2 were significantly correlated (Fig. 4B). The products of these two genes assemble and form type I collagen fibers. This type of correlation is most likely to ensure that the subunits of a protein complex are produced at a certain ratio.
Fig. 4.
(a) A gene network shows that mRNA levels of PDGFR and ECM-related genes are significantly correlated in human breast cancer. (b) A network shows that transcripts of LOX genes are significantly correlated with collagen genes. (c) A network composed of TGFβ- and ECM-related genes may represent the reciprocal regulation between the TGF-β pathway and the ECM microenvironment.
Function analysis of the gene networks
Invasion and metastasis of breast cancer are associated with extensive remodeling of the ECM microenvironment.1 Among the 64 genes in Network I, the majority encodes ECM microenvironment-related proteins such as collagens, laminins, integrins, MMPs, and other types of ECM remodeling enzymes, indicating that ECM remodeling is integrated at the transcriptional level in breast malignancy. To determine if any specific signaling pathways are associated with ECM remodeling, we performed the KEGG pathway analysis. We found that the TGF-β pathway was significantly enriched in Network I. In the canonical TGF-β signal pathway, binding of TGF-β to its receptor activates Smad transcription factors, which further elicits a broad range of responses such as differentiation, apoptosis, and EMT, and microenvironment remodeling (reviewed in ref. 42). In Network I, the activities of the TGF-β pathway are correlated with the expression of ECM proteins, MMPs, and integrins (Fig. 4), suggesting that this pathway directs ECM remodeling during breast cancer development and progression.
Signal transduction between TGF-β and the ECM microenvironment is bidirectional. TGF-β is produced and secreted in a latent form and remains in the ECM forming a complex with latency associated peptides (LAPs) and latent TGF-β binding protein (LTBP) until being further processed and released in its active form.43,44 LAP contains an Arg-Gly-Asp (RGD) motif which can recruit the latent TGF-β complexes to αv integrin, and binding induces conformational changes that facilitate the release of active TGF-β.45 Thrombospondin (THBS), a matricellular glycoprotein, can activate TGF-β by binding to the latent complex to prevent its association with the mature factor.46 MMP9 and MMP2 can cleave the latent form and release the activated TGF-β.47 These studies indicate that TGF-β activation is modulated by the ECM microenvironment. Consistent with these results, we also found that multiple MMPs, THBSs and integrin genes were linked with the TGF-β pathway in the Network (Fig. 4c). Therefore, this gene network represents the reciprocal regulation between the TGF-β pathway and the ECM microenvironment.
Cytokines and chemokines mediate immune and inflammatory responses, which are functionally important for breast cancer development and progression. In Network II, genes encoding cytokines, chemokines, and their receptors are significantly enriched (11 out of 17), and mRNA levels of chemokines are significantly correlated with the expression of cytokines and their receptors. For instance, CCL5 expression correlated with the expression of IL2RA, IL2RG and IL10RA in the network. It has been reported that CCL5 and IL2 cooperate to induce the proliferation and activation of certain natural-killer (NK) cells,48 but it remains to be validated whether this cooperation occurs in breast cancer to modulate NK cell activities. In addition, CCL5 has been shown to be produced from bone-marrow-derived human mesenchymal stem cells and to stimulate breast cancer cell motility, invasion and metastasis in a paracrine manner.49 Therefore, CCL5 may have distinct functions in modulating activities of NK cells and breast cancer cells during the malignancy. In Network II, CXCL9 and CCR7 have correlated expression with IL2R, IL16, and IL7R in the network. A significant amount of evidence suggests that cytokines can induce the production of chemokines. For instance, IL2 stimulates the expression of a number of chemokines and their receptors, including CCL3, CCL4, CCR2, CCR4, CCR5, and CCR8.50–53 Thus, the correlated expression of cytokine and chemokine genes in Network II suggests the crosstalk of these two types of soluble factors. Further investigation of the crosstalk in breast cancer is crucial to our understanding of immune response in cancer development and progression.
The identification of TGFβ-ECM and cytokine-chemokines networks indicates that the co-expression network analysis is robust and efficient in characterizing coordinated microenvironmental signals. In addition to these two well-characterized networks, we also discovered a number of novel connections. For instance, in Network I, ANGPTL2 has correlated expression with various ECM protein genes (Fig. S2‡), suggesting a functional connection between ANGPTL2 and ECM remodeling. In network II, we found that expression of cell adhesion proteins (SELL (selectin L), VCAM1 (vascular cell adhesion molecule-1), and ICAM2 (intercellular adhesion molecule 2)) is associated with cytokine and chemokine genes. SELL encodes a cell surface adhesion molecule that belongs to the selectin family. This protein is required for binding and subsequent rolling of leucocytes on endothelial cells, which facilitates their migration into secondary lymphoid organs and inflammation sites.54 VCAM1 is a cell surface adhesion protein produced by endothelial cells after cytokine treatment, which mediates the adhesion of various immune cells to vascular endothelium and functions in leukocyte-endothelial cell signal transduction.55 ICAM2 is an intercellular adhesion molecule, and the ICAM2/LFA-1 interaction may stimulate T cell aggregation and NK cell activity.56 Treatment with anti-ICAM-2 mAb in vivo eradicates transplanted colon carcinomas, and this process is dependent on the induction of tumor-specific cytotoxic T cell-mediated immune responses.57 Although the function of VCAM1, ICAM2, and SELL in breast cancer is not well-understood, our findings demonstrate that the mRNA levels of these three genes were associated with expression of chemokines and cytokines. Based on correlated expression and functional connections among those genes, we propose that cell adhesion molecules and cytokines/chemokines may cooperate to control the immune and inflammatory responses during breast cancer development.
Conclusions
With the advent of genetic animal and three-dimensional culture models, it has become clear that breast cancer development requires integrated microenvironmental signals. Given the progress in expression profile analysis and access to an abundant amount of published microarray data, we attempted to delineate how microenvironmental signals are integrated at the genome level in human breast cancer. In our exploratory study, we found that mRNA levels of microenvironment genes are significantly correlated and form transcription networks in human breast cancer. These networks represent the coordinated regulation and cooperative effects of microenvironmental cues in breast malignancy.
The microenvironment of breast cancer is largely determined by the crosstalk of malignant epithelial cells with cancer-associated stromal and immune cells. It would be interesting to analyze what types of cells in cancer tissue express the components of microenvironment gene networks using in situ hybridization and immunohistochemistry analyses, and to determine whether the networks represent the crosstalk between the epithelial, stromal, and immune compartment utilizing 3D co-culture assays and tissue-specific transgenic mouse models.
Supplementary Material
Insight, innovation, integration.
It has become clear that integrated microenvironment cues are crucial for normal and malignant tissue development. However, how the microenvironmental signals coordinate largely remains to be determined. Given the progress in expression profile analysis and access to an abundant amount of published microarray data, we attempted to delineate how microenvironmental signals are integrated at the genome level in human breast cancer. Using gene co-expression network analysis, we have identified two microenvironment networks in human breast cancer tissues. These networks represent the coordinated regulation and cooperative effects of ECM molecules, soluble factors, and cell-adhesion complexes in breast malignancy.
Acknowledgements
R.X. is deeply grateful to Dr Mina J. Bissell (Lawrence Berkeley National Laboratory) for her training and support. We thank Dr Nathan L. Vanderford for scientific editing. This work was supported by grants from NCI (P30 CA147886), Office of Biological & Environmental Research, of the U.S. Department of Energy (DE-AC02-05CH11231), Laboratory Directed Research & Development Program (LDRD), National Institutes of Health, National Cancer Institute grants (R01 CA116481 to JHM).
Abbreviations
- ECM
extracellular matrix
- MMP
matrix metalloproteinase
- BM
basement membrane
- EGFR
epidermal growth factor receptor
- FGF
fibroblast growth factor
- HGF
hepatocyte growth factor
- PDGF
platelet-derived growth factor
- PKC
protein kinase C
- LOX
lysyl oxidase
- EMT
epithelial-mesenchymal transition
- LAPs
latency associated peptides
- LTBP
latent TGF-β binding protein
Footnotes
Published as part of an Integrative Biology themed issue in honour of Mina J. Bissell: Guest Editor Mary Helen Barcellos-Hoff.
Electronic supplementary information (ESI) available: Table S1, Fig. S1 and S2. See DOI: 10.1039/c0ib00087f
References
- 1.Bissell MJ, Radisky D. Putting tumours in context. Nat. Rev. Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LaBarge MA, Nelson CM, Villadsen R, Fridriksdottir A, Ruth JR, Fridriksdottir A, Ruth JR, Stampfer MR, Petersen OW, Bissell MJ. Human mammary progenitor cell fate decisions are products of interactions with combinatorial microenvironments. Integr. Biol. 2009;1:70–79. doi: 10.1039/b816472j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu R, Boudreau A, Bissell MJ. Tissue architecture and function: dynamic reciprocity via extra- and intra-cellular matrices. Cancer Metastasis Rev. 2009;28:167–176. doi: 10.1007/s10555-008-9178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo W, Pylayeva Y, Pepe A, Yoshioka T, Muller WJ, et al. Beta 4 integrin amplifies ErbB2 signaling to promote mammary tumorigenesis. Cell. 2006;126:489–502. doi: 10.1016/j.cell.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 5.Akhtar N, Streuli CH. Rac1 links integrin-mediated adhesion to the control of lactational differentiation in mammary epithelia. J. Cell Biol. 2006;173:781–793. doi: 10.1083/jcb.200601059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang F, Weaver VM, Petersen OW, Larabell CA, Dedhar S, et al. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu R, Nelson CM, Muschler JL, Veiseh M, Vonderhaar BK, et al. Sustained activation of STAT5 is essential for chromatin remodeling and maintenance of mammary-specific function. J. Cell Biol. 2009;184:57–66. doi: 10.1083/jcb.200807021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Streuli CH, Edwards GM, Delcommenne M, Whitelaw CB, Burdon TG, et al. Stat5 as a target for regulation by extracellular matrix. J. Biol. Chem. 1995;270:21639–21644. doi: 10.1074/jbc.270.37.21639. [DOI] [PubMed] [Google Scholar]
- 9.Goffin V, Binart N, Touraine P, Kelly PA. Prolactin: the new biology of an old hormone. Annu. Rev. Physiol. 2002;64:47–67. doi: 10.1146/annurev.physiol.64.081501.131049. [DOI] [PubMed] [Google Scholar]
- 10.Gouilleux F, Wakao H, Mundt M, Groner B. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J. 1994;13:4361–4369. doi: 10.1002/j.1460-2075.1994.tb06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson CJ, Burdon TG. Prolactin signal transduction mechanisms in the mammary gland: the role of the Jak/Stat pathway. Rev. Reprod. 1996;1:1–5. doi: 10.1530/ror.0.0010001. [DOI] [PubMed] [Google Scholar]
- 12.Xu R, Spencer VA, Bissell MJ. Extracellular matrix-regulated gene expression requires cooperation of SWI/SNF and transcription factors. J. Biol. Chem. 2007;282:14992–14999. doi: 10.1074/jbc.M610316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roskelley CD, Desprez PY, Bissell MJ. Extra-cellular matrix-dependent tissue-specific gene expression in mammary epithelial cells requires both physical and biochemical signal transduction. Proc. Natl. Acad. Sci. U. S. A. 1994;91:12378–12382. doi: 10.1073/pnas.91.26.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 15.Naylor MJ, Li N, Cheung J, Lowe ET, Lambert E, et al. Ablation of beta1 integrin in mammary epithelium reveals a key role for integrin in glandular morphogenesis and differentiation. J. Cell Biol. 2005;171:717–728. doi: 10.1083/jcb.200503144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gui GP, Wells CA, Browne PD, Yeomans P, Jordan S, et al. Integrin expression in primary breast cancer and its relation to axillary nodal status. Surgery. 1995;117:102–108. doi: 10.1016/s0039-6060(05)80236-3. [DOI] [PubMed] [Google Scholar]
- 17.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 18.Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrinblocking antibodies. J. Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margadant C, Frijns E, Wilhelmsen K, Sonnenberg A. Regulation of hemidesmosome disassembly by growth factor receptors. Curr. Opin. Cell Biol. 2008;20:589–596. doi: 10.1016/j.ceb.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Wilhelmsen K, Litjens SH, Kuikman I, Margadant C, van Rheenen J, et al. Serine phosphorylation of the integrin beta4 subunit is necessary for epidermal growth factor receptor induced hemidesmosome disruption. Mol. Biol. Cell. 2007;18:3512–3522. doi: 10.1091/mbc.E07-04-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ieguchi K, Fujita M, Ma Z, Davari P, Taniguchi Y, et al. Direct binding of the EGF-like domain of neuregulin-1 to integrins ({alpha}v{beta}3 and {alpha}6{beta}4) is involved in neuregulin-1/ErbB signaling. J. Biol. Chem. 2010;285:31388–31398. doi: 10.1074/jbc.M110.113878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vlodavsky I, Miao HQ, Medalion B, Danagher P, Ron D. Involvement of heparan sulfate and related molecules in sequestration and growth promoting activity of fibroblast growth factor. Cancer Metastasis Rev. 1996;15:177–186. doi: 10.1007/BF00437470. [DOI] [PubMed] [Google Scholar]
- 23.Rahman S, Patel Y, Murray J, Patel KV, Sumathipala R, et al. Novel hepatocyte growth factor (HGF) binding domains on fibronectin and vitronectin coordinate a distinct and amplified Met-integrin induced signalling pathway in endothelial cells. BMC Cell Biol. 2005;6:8. doi: 10.1186/1471-2121-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brinkmann V, Foroutan H, Sachs M, Weidner KM, Birchmeier W. Hepatocyte growth factor/scatter factor induces a variety of tissue-specific morphogenic programs in epithelial cells. J. Cell Biol. 1995;131:1573–1586. doi: 10.1083/jcb.131.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodward TL, Mienaltowski AS, Modi RR, Bennett JM, Haslam SZ. Fibronectin and the alpha(5)beta(1) integrin are under developmental and ovarian steroid regulation in the normal mouse mammary gland. Endocrinology. 2001;142:3214–3222. doi: 10.1210/endo.142.7.8273. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Harris RE, Bayston LJ, Ashe HL. Type IV collagens regulate BMP signalling in Drosophila. Nature. 2008;455:72–77. doi: 10.1038/nature07214. [DOI] [PubMed] [Google Scholar]
- 27.Delmas V, Pla P, Feracci H, Thiery JP, Kemler R, et al. Expression of the cytoplasmic domain of E-cadherin induces precocious mammary epithelial alveolar formation and affects cell polarity and cell-matrix integrity. Dev. Biol. 1999;216:491–506. doi: 10.1006/dbio.1999.9517. [DOI] [PubMed] [Google Scholar]
- 28.Boussadia O, Kutsch S, Hierholzer A, Delmas V, Kemler R. E-cadherin is a survival factor for the lactating mouse mammary gland. Mech. Dev. 2002;115:53–62. doi: 10.1016/s0925-4773(02)00090-4. [DOI] [PubMed] [Google Scholar]
- 29.Borghi N, Lowndes M, Maruthamuthu V, Gardel ML, Nelson WJ. Regulation of cell motile behavior by crosstalk between cadherin- and integrin-mediated adhesions. Proc. Natl. Acad. Sci. U. S. A. 2010;107:13324–13329. doi: 10.1073/pnas.1002662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perrais M, Chen X, Perez-Moreno M, Gumbiner BM. E-cadherin homophilic ligation inhibits cell growth and epidermal growth factor receptor signaling independently of other cell interactions. Mol. Biol. Cell. 2007;18:2013–2025. doi: 10.1091/mbc.E06-04-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian X, Karpova T, Sheppard AM, McNally J, Lowy DR. E-cadherin-mediated adhesion inhibits ligand-dependent activation of diverse receptor tyrosine kinases. EMBO J. 2004;23:1739–1748. doi: 10.1038/sj.emboj.7600136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Legate KR, Montanez E, Kudlacek O, Fassler R. ILK, PINCH and parvin: the tIPP of integrin signalling. Nat. Rev. Mol. Cell Biol. 2006;7:20–31. doi: 10.1038/nrm1789. [DOI] [PubMed] [Google Scholar]
- 33.McPhee TR, McDonald PC, Oloumi A, Dedhar S. Integrin-linked kinase regulates E-cadherin expression through PARP-1. Dev. Dyn. 2008;237:2737–2747. doi: 10.1002/dvdy.21685. [DOI] [PubMed] [Google Scholar]
- 34.Ackland ML, Newgreen DF, Fridman M, Waltham MC, Arvanitis A, et al. Epidermal growth factor-induced epitheliomesenchymal transition in human breast carcinoma cells. Lab. Invest. 2003;83:435–448. doi: 10.1097/01.lab.0000059927.97515.fd. [DOI] [PubMed] [Google Scholar]
- 35.Moon HS, Choi EA, Park HY, Choi JY, Chung HW, et al. Expression and tyrosine phosphorylation of E-cadherin, beta- and gamma-catenin, and epidermal growth factor receptor in cervical cancer cells. Gynecol. Oncol. 2001;81:355–359. doi: 10.1006/gyno.2001.6163. [DOI] [PubMed] [Google Scholar]
- 36.Quigley D, Balmain A. Systems genetics analysis of cancer susceptibility: from mouse models to humans. Nat. Rev. Genet. 2009;10:651–657. doi: 10.1038/nrg2617. [DOI] [PubMed] [Google Scholar]
- 37.Quigley DA, To MD, Perez-Losada J, Pelorosso FG, Mao JH, et al. Genetic architecture of mouse skin inflammation and tumour susceptibility. Nature. 2009;458:505–508. doi: 10.1038/nature07683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonner JC. Regulation of PDGF and its receptors in fibrotic diseases. Cytokine Growth Factor Rev. 2004;15:255–273. doi: 10.1016/j.cytogfr.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 39.Borkham-Kamphorst E, Stoll D, Gressner AM, Weiskirchen R. Antisense strategy against PDGF B-chain proves effective in preventing experimental liver fibrogenesis. Biochem. Biophys. Res. Commun. 2004;321:413–423. doi: 10.1016/j.bbrc.2004.06.153. [DOI] [PubMed] [Google Scholar]
- 40.Yamauchi M, Shiiba M. Lysine hydroxylation and cross-linking of collagen. Methods Mol. Biol. 2008;446:95–108. doi: 10.1007/978-1-60327-084-7_7. [DOI] [PubMed] [Google Scholar]
- 41.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Padua D, Massague J. Roles of TGFbeta in metastasis. Cell Res. 2009;19:89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 43.Barcellos-Hoff MH. Latency and activation in the control of TGF-beta. J. Mammary Gland Biol. Neoplasia. 1996;1:353–363. [PubMed] [Google Scholar]
- 44.Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. J. Cell Sci. 2003;116:217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- 45.Munger JS, Harpel JG, Giancotti FG, Rifkin DB. Interactions between growth factors and integrins: latent forms of transforming growth factor-beta are ligands for the integrin alphavbeta1. Mol. Biol. Cell. 1998;9:2627–2638. doi: 10.1091/mbc.9.9.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crawford SE, Stellmach V, Murphy-Ullrich JE, Ribeiro SM, Lawler J, et al. Thrombospondin-1 is a major activator of TGF-beta1 in vivo. Cell. 1998;93:1159–1170. doi: 10.1016/s0092-8674(00)81460-9. [DOI] [PubMed] [Google Scholar]
- 47.Yu Q, Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev. 2000;14:163–176. [PMC free article] [PubMed] [Google Scholar]
- 48.Maghazachi AA, Al-Aoukaty A, Schall TJ. CC chemokines induce the generation of killer cells from CD56+ cells. Eur. J. Immunol. 1996;26:315–319. doi: 10.1002/eji.1830260207. [DOI] [PubMed] [Google Scholar]
- 49.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 50.Robertson MJ. Role of chemokines in the biology of natural killer cells. J. Leukocyte Biol. 2002;71:173–183. [PubMed] [Google Scholar]
- 51.Zhang T, Guo CJ, Douglas SD, Metzger DS, O'Brien CP, et al. Alcohol suppresses IL-2-induced CC chemokine production by natural killer cells. Alcohol.: Clin. Exp. Res. 2005;29:1559–1567. doi: 10.1097/01.alc.0000179364.32003.9f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inngjerdingen M, Damaj B, Maghazachi AA. Expression and regulation of chemokine receptors in human natural killer cells. Blood. 2001;97:367–375. doi: 10.1182/blood.v97.2.367. [DOI] [PubMed] [Google Scholar]
- 53.Nieto M, Navarro F, Perez-Villar JJ, del Pozo MA, Gonzalez-Amaro R, et al. Roles of chemokines and receptor polarization in NK-target cell interactions. J. Immunol. 1998;161:3330–3339. [PubMed] [Google Scholar]
- 54.Carlos TM, Harlan JM. Leukocyte-endothelial adhesion molecules. Blood. 1994;84:2068–2101. [PubMed] [Google Scholar]
- 55.Barreiro O, Yanez-Mo M, Serrador JM, Montoya MC, Vicente-Manzanares M, et al. Dynamic interaction of VCAM-1 and ICAM-1 with moesin and ezrin in a novel endothelial docking structure for adherent leukocytes. J. Cell Biol. 2002;157:1233–1245. doi: 10.1083/jcb.200112126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li R, Nortamo P, Kantor C, Kovanen P, Timonen T, et al. A leukocyte integrin binding peptide from intercellular adhesion molecule-2 stimulates T cell adhesion and natural killer cell activity. J. Biol. Chem. 1993;268:21474–21477. [PubMed] [Google Scholar]
- 57.Melero I, Gabari I, Corbi AL, Relloso M, Mazzolini G, et al. An anti-ICAM-2 (CD102) monoclonal antibody induces immune-mediated regressions of transplanted ICAM-2-negative colon carcinomas. Cancer Res. 2002;62:3167–3174. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.