Summary
We isolated two insertion mutants of Bordetella avium that exhibited a peculiar clumped-growth phenotype and found them to be attenuated in turkey tracheal colonization. The mutants contained transposon insertions in homologues of the wlbA and wlbL genes of Bordetella pertussis. The wlb genetic locus of B. pertussis has been previously described as containing 12 genes involved in lipopolysaccharide (LPS) biosynthesis. Polyacrylamide gel analysis of LPS from B. avium wlbA and wlbL insertion mutants confirmed an alteration in the LPS profile. Subsequent cloning and complementation of the wlbA and wlbL mutants in trans with a recombinant plasmid containing the homologous wlb locus from B. avium eliminated the clumped-growth phenotype and restored the LPS profile to that of wild-type B. avium. Also, a parental level of tracheal colonization was restored to both mutants by the recombinant plasmid. Interestingly, complementation of the wlbA and wlbL mutants with a recombinant plasmid containing the heterologous wlb locus from B. pertussis, B. bronchiseptica, or Bordetella parapertussis eliminated the clumped-growth phenotype and resulted in a change in the LPS profile, although not to that of wild-type B. avium. The mutants also acquired resistance to a newly identified B. avium-specific bacteriophage, Ba1. Complementation of both wlbA and wlbL mutants with the homologous wlb locus of B. avium, but not the heterologous B. pertussis locus, restored sensitivity to Ba1. Complementation of the wlbL mutant, but not the wlbA mutant, with the heterologous wlb locus of Bordetella bronchiseptica or B. parapertussis restored partial sensitivity to Ba1. Comparisons of the LPS profile and phage sensitivity of the mutants upon complementation by wlb loci from the heterologous species and by B. avium suggested that phage sensitivity required the presence of O-antigen. At the mechanistic level, both mutants showed a dramatic decrease in serum resistance and a decrease in binding to turkey tracheal rings in vitro. In the case of serum resistance, complementation of both mutants with the homologous wlb locus of B. avium restored serum resistance to wild-type levels. However, in the case of epithelial cell binding, only complementation of the wlbA mutant completely restored binding to wild-type levels (binding was only partially restored in the wlbL mutant). This is the first characterization of LPS mutants of B. avium at the genetic level and the first report of virulence changes by both in vivo and in vitro measurements
Introduction
Bordetella avium is the causative agent of bordetellosis, a disease of the upper respiratory tract of turkeys (Skeeles and Arp, 1997). As with several other members of the Bordetella genus, B. avium binds preferentially to ciliated tracheal epithelial cells (Arp et al., 1993). Death of the ciliated cells is thought to lead to many of the clinical signs associated with avian bordetellosis, which include excessive oculonasal discharge, sneezing, mouth breathing, conjunctivitis and decreased weight gain. In addition, infected turkeys are more susceptible to secondary infections by other pathogens (Barnes and Hofstad, 1978; Saif et al., 1980). The secondary infection, often by Escherichia coli, is frequently fatal and contributes significantly to losses sustained by the poultry industry from infectious agents (Saif et al., 1980; Skeeles and Arp, 1997).
The pathogenesis of bordetellosis and factors affecting tracheal colonization by B. avium are not fully understood (Gentry-Weeks et al., 1988; Temple et al., 1998). In other more well-studied members of the Bordetellae (Bordetella pertussis and Bordetella bronchiseptica), a number of traits have been associated with virulence (Goodwin and Weiss, 1990; Mooi et al., 1992; Geuijen et al., 1997; Ishikawa and Sato, 1997; Cotter et al., 1998; van den Berg et al., 1999). However, in the case of B. pertussis, the species that causes whooping cough in children, suspected virulence traits have been examined only in experiments utilizing heterologous hosts (Parton et al., 1994; Shahin and Cowell, 1994), leading to difficulties in determining the applicability of the results in guiding prevention strategies. In both B. avium and B. bronchiseptica, traits associated with virulence can be tested directly in experimental infections using the natural host (Monack and Falkow, 1993; Cotter and Miller, 1994; Ackermann et al., 1997; Temple et al., 1998). In instances where suspected virulence factors are shared amongst all Bordetella species, the use of the natural host-pathogen pairings has provided results that, in some cases, contradict those obtained from B. pertussis in heterologous hosts (Weiss et al., 1984; Weiss and Goodwin, 1989; Cotter et al., 1998; Temple et al., 1998).
Apart from providing possible insights into human disease, understanding avian bordetellosis is becoming increasingly important as farming practices world-wide adopt poultry production methods based upon high efficiency feed conversion where the commercial turkey particularly excels (Ensminger, 1980). Unfortunately, commercial turkeys were derived from limited genetic stock (only one breed is recognized) and their susceptibility to infectious agents is, in theory, and as tested experimentally (Temple et al., 1998), very uniform. This lack of natural variability, coupled with our growing reliance on them as a food source, provides incentive for a better understanding of their infectious agents.
We have been interested in discovering factors that affect B. avium virulence. In this report, we describe the isolation of two B. avium mutants defective in the normal biosynthesis of lipopolysaccharide (LPS). Our analysis revealed that these mutants were significantly reduced in colonization and had a number of interesting properties that may allow further detailed analysis of the role of LPS in producing the characteristic signs of bordetellosis.
Results
Discovery, isolation and genetic characterization of LPS mutants
Isolation of an avirulent insertion mutant (strain PAS78) during a screen of signature-tagged transposon mutants (Hensel et al., 1995; P.A.S. et al., unpublished results) first alerted us to the possible importance of a peculiar clumped-growth phenotype (e.g. clumping when grown in broth culture). Subsequent screening of random insertion mutants for this clumped-growth phenotype led to the isolation and characterization of an additional mutant (strain PAS85). Both mutants showed significantly reduced colonization in turkey poults (documented in a later section). In addition to clumping when grown in broth culture, both mutants displayed a ‘rough’ phenotype that consisted of alteration in colony morphology characteristic of some LPS mutants of Salmonella typhimurium (Rick, 1987). These results prompted work to define the genetic defect and to further characterize these mutants.
DNA flanking each insertion was cloned and sequenced. Sequence adjacent to the insertion in strain PAS85 was homologous to the 5′ region of the wlbA gene and the predicted amino acid sequence was 75% identical to WlbA of B. pertussis (EMBL no. X90711). Sequence adjacent to the insertion in strain PAS78 was similar to the middle of the wlbL gene of B. pertussis. The predicted amino acid sequence of this region was 58% identical to WlbL of B. pertussis (EMBL no. X90711). Both the wlbA and wlbL genes are part of the wlb genetic locus consisting of 12 genes (A–L) in B. pertussis that are involved in LPS biosynthesis, specifically the addition of a trisaccharide to the distal portion of the core and O-antigen biosynthesis (Allen and Maskell, 1996; Allen et al., 1998).
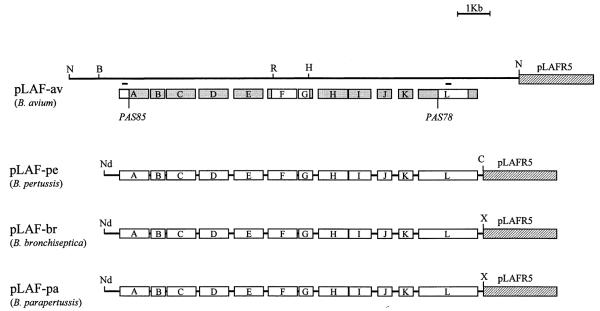
In order to complement our wlbA and wlbL insertion mutants, we cloned a 15 kb region containing the wlbL and wlbA genes of B. avium. The resultant B. avium wlb locus is shown (Fig. 1), where it is compared with similar genetic loci from B. pertussis, B. parapertussis and B. bronchiseptica (Allen et al., 1998). Although we have not sequenced the entire region of the B. avium locus, restriction analysis, Southern blot analysis (data not shown), partial sequence analysis (Fig. 1) and functional analysis (below) indicated the wlb locus of B. avium is similar to the loci of the other Bordetella species. Partial sequence analysis included approximately 600 bp on either end of the cloned segment and the resultant sequence showed no homology to current GenBank sequences. In addition, centre sequence data (wlbF and wlbG) served to confirm the general similarity of the locus with the other species at the protein sequence level.
Fig. 1.
Diagrams of plasmids containing the wlb genetic loci of B. avium (pLAF-av), B. pertussis (pLAF-pe), B. bronchiseptica (pLAF-br) and B. parapertussis (pLAF-pa). Shaded boxes in the B. avium locus indicate proposed genetic structure based upon similarity to other members of the Bordetellae shown. White boxes in the B. avium locus indicate sequenced areas (wlbA, Accession No. AF248033; wlbF, Accession No. AF248035and No. AF248036; wlbG, Accession No. AF248036; wlbL, Accession No. AF248034). Gene organization of the heterologous wlb loci have been redrawn from published work for comparison (Allen et al., 1998). Insertion sites of B. avium mutants are indicated. Lines above wlbA and wlbL genes indicate the location of digoxigenin-labelled PCR probes. Restriction sites are B, BamHI; C, ClaI; R, EcoRI; H, HindIII; Nd, NdeI; N, NotI; X, XbaI.
Trans-complementation of wlbA and wlbL mutant phenotypes with plasmids containing homologous and heterologous wlb genes
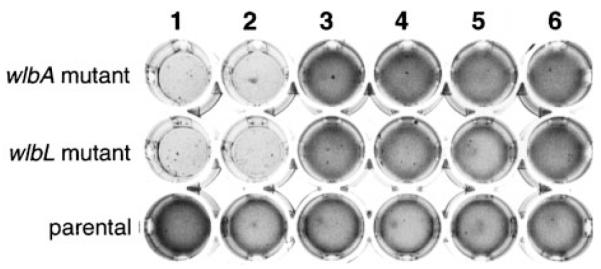
In addition to clumping when grown in broth culture and reduced colonization in turkey poults, wlbA and wlbL insertion mutants displayed an alteration in their LPS profile upon PAGE and a loss of sensitivity to a B. avium bacteriophage, Ba1 (Shelton et al., submitted). The clumped-growth phenotype was readily complemented by plasmids containing the homologous and heterologous wlb loci (Fig. 2). In this analysis, each complementing plasmid contained the wlb locus of B. avium (pLAF-av), B. pertussis (pLAF-pe), B. bronchiseptica (pLAF-br) or B. parapertussis (pLAF-pa) cloned into the broad host range vector, pLAFR5 (refer to Fig. 1 and Table 1 for diagrams and sources of the plasmids respectively). The vector (pLAFR5) was the only plasmid that did not noticeably complement the clumped-growth phenotype. Similar results were seen with the restoration of a smooth colony morphology upon homologous and heterologous wlb loci complementation (data not shown).
Fig. 2.
Clumped-growth phenotype of B. avium wlb mutants. Cultures were grown in a 48 well tissue culture tray at 37°C overnight with rotary shaking. Indicated at left are the genotypes. Lane 1 contains the strains without any recombinant plasmids. Lanes 2–6 contain the plasmids pLAFR5, pLAF-av, pLAF-pe, pLAF-br or pLAF-pa respectively.
Table 1.
Bacteria, bacteriophage and plasmids.
| Bacteria/bacteriophage/ plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Bacteria | ||
| 197N | parental B. avium strain; NalR | Temple et al. (1998) |
| PAS78 | wlbL insertion mutant of B. avium 197N; wlbL::Tn5; KanR, NalR | This study |
| PAS85 | wlbA insertion mutant of B. avium 197N; wlbA::Tn5; KanR, NalR | This study |
| E. coli S17.1(γpir) | SmR | Simon et al. (1983) |
| E. coli DH5α | Life Technologies | |
| Bacteriophage | ||
| Ba1c1 | clear plaque derivative of B. avium phage Ba1 | Shelton et al. (submitted) |
| Plasmids | ||
| pGEM-11z(f) | E. coli cloning vector; ApR | Promega |
| pGEM-av | NotI 15 kb wlb locus of B. avium in pGEM-11zf; ApR | This study |
| pLAFR5 | broad host range vector; TcR | Keen et al. (1988) |
| pLAF-av |
NotI 15 kb wlb locus of B. avium from pGEM-av cloned into pLAFR5; TcR |
This study |
| pLAF-pe | wlb locus of B. pertussis in pLAFR5; TcR | Allen et al. (1998) |
| pLAF-br | wlb locus of B. bronchiseptica in pLAFR5; TcR | Allen et al. (1998) |
| pLAF-pa | wlb locus of B. parapertussis in pLAFR5; TcR | Allen et al. (1998) |
Silver-stained polyacrylamide gel analysis of LPS structure showed complete restoration to the parental LPS profile when either the wlbL or the wlbA mutant was complemented with the homologous wlb locus (pLAF-av), evidenced by restored O-antigen production (compare lane 1 with lane 3, Fig. 3). However, only partial restoration of the profile was seen when either mutant was complemented with the wlb locus from other Bordetella species (lanes 4–6, Fig. 3). Specifically, none of the heterologous regions restored the O-antigen to parental levels, but there was a distinct change in migration of the single, fast-migrating LPS band associated with the mutants. This faster migrating band seen in the mutants [lanes 2 and 2′, Fig. 3; indicated by an asterisk (*)] was raised to a higher migrating (parental) band in the complemented mutants [lanes 4–6 compared with lane 1, Fig. 3; indicated by a double asterisk (**)].
Fig. 3.
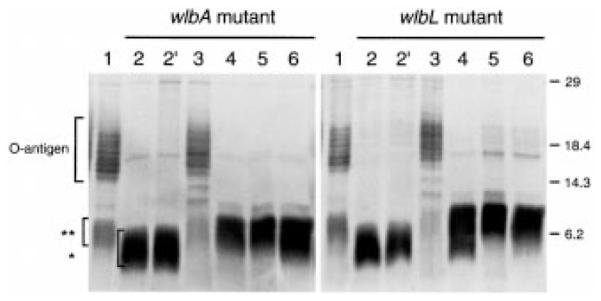
Silver-stained polyacrylamide gel analysis of B. avium LPS mutants and complemented mutants (wlbA, first panel; wlbL, second panel). First panel: lane 1, parental B avium strain 197N; lane 2 and 2′, wlbA mutant strain (PAS85) with no addition or addition of the pLAFR5 vector respectively; lanes 3–6, strain PAS85 complimented with pLAF-av, pLAF-pe, pLAF-br, or pLAF-pa respectively. Second panel: lane 1, parental B. avium strain 197N; lane 2 and 2′, wlbL mutant strain (PAS78) with no addition or addition of the pLAFR5 vector respectively; lanes 3–6, strain PAS78 complimented with pLAF-av, pLAF-pe, pLAF-br, or pLAF-pa, respectively. Molecular weight markers are indicated in kdaltons. Lower (*) and higher (**) migrating lipid-A-associated bands are indicated by brackets. O-antigen bands are indicated by the top bracketed area.
We also found that the loss of sensitivity to a newly identified B. avium-specific bacteriophage, Ba1, in the wlbA and wlbL mutants could be completely restored in both mutants by complementation with the homologous wlb locus (compare rows 1–3, Fig. 4). Phage sensitivity could be partially restored by the heterologous loci only in the wlbL mutant (compare rows 4–6, Fig. 4). Of the heterologous loci complementing the wlbL mutant, the locus from B. pertussis (row 4, Fig. 4) was clearly the least effective.
Fig. 4.
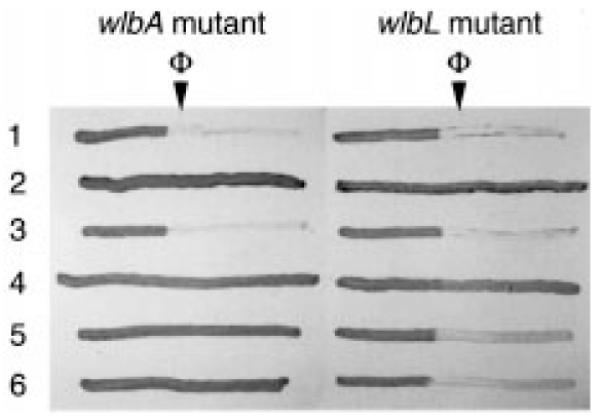
Cross-streak analysis of wlbA (first panel) and wlbL (second panel) mutants and complemented mutants with a clear plaque-forming derivative of bacteriophage Ba1, Ba1c1. Streak 1, parental B. avium strain 197N with the vector plasmid pLAFR5. Streaks 2–6, wlb mutant (PAS85 or PAS78) complemented with pLAFR5, pLAF-av, pLAF-pe, pLAF-br or pLAF-pa respectively. The arrow denotes where phage lysates were vertically streaked prior to making the cross-streaks.
Restored turkey tracheal colonization of wlbA and wlbL mutants complemented in trans by a plasmid containing the B. avium wlb locus
Fifty percent infectious dose (ID50) measurements revealed a pronounced and statistically significant defect in the ability of wlbA and wlbL mutants (strains PAS78 and PAS85) to colonize turkey tracheas when compared with the parental strain (197N; Table 2). This defect in turkey tracheal colonization of the mutants was completely restored by complementation with the homologous wlb genetic locus from B. avium (Table 2). In this experiment, the wlbA and wlbL mutants, harbouring the pLAF-av recombinant plasmid, displayed an ID50 statistically indistinguishable from that of the parental and clearly distinguishable from the mutant harbouring just the vector plasmid (pLAFR5) (Table 2). There was no statistically relevant difference in the average ID50 between the wlbA and wlbL mutants even although the values were noticeably different.
Table 2.
Infectious dose measurements (50%) of LPS mutants in turkey poults.
| Straina | Plasmidb | Relevant propertiesc genotype (phenotype) |
ID50d(× 106) | Significantly different from parentalg strain |
|---|---|---|---|---|
| 197N | – | Parental strain (WlbA+, WlbL+) | 7 ± 8.7 | NA |
| PAS85 | – | wlbA::Tn5 (WlbA−) | 81 ± 73 | Yes |
| pLAFR5 | wlbA::Tn5 (WlbA−) | 110 | Yes | |
| pLAF-av | wlbA::Tn5/wlbA-L (WlbA+) | 9 | No | |
| PAS78 | – | wlbL::Tn5 (WlbL−) | 1900 ± 1100e | Yes |
| pLAFR5 | wlbL::Tn5 (WlbL−) | > 500f | Yes | |
| pLAF-av | wlbL::Tn5/wlbA-L (WlbL+) | 2 | No |
Strain numbers refer to specific insertion mutant isolates.
Plasmid pLAFR5 refers to the vector; plasmid pLAF-av refers to the pLAFR5 vector containing the wlb locus of B. avium.
The wlb genetic mnemonics refer to LPS biosynthesis genes interrupted by an insertion.
Fifty percent infectious dose (ID50) measurements were performed as previously described (in the text). Averages or individual measurements are indicated. When two or more experiments were performed, the standard deviation of the mean is indicated (+/−).
These ID50 values were extrapolated from colonization rates obtained from birds infected at dose of ~107 and ~108 cfu.
The inequality refers to the fact that no birds were infected at any dose given. The numeric value shown is lowest possible ID50 achievable (i.e. it represents the ID50 value generated if all animals were infected at a dose one order of magnitude higher than the highest dose employed).
ID50 measurements of both mutant strains (PAS85 and PAS78) were found to be significantly different (P < 0.05) from the parental strain using the Student’s t-test. Mutants containing either the vector or complementing plasmid were found to be significantly different (P < 0.05) from the parental strain using the Z-test analysis. NA; not applicable.
Resistance to naïve turkey serum and tracheal binding in vitro are significantly altered by wlbA and wlbL lesions
Two in vitro assays that measured (i) serum resistance and (ii) tracheal ring binding were performed to assess possible reasons for the decrease in colonization rate of the LPS mutants seen in vivo. In the case of the serum resistance assay, we took advantage of a preliminary observation that our parental B. avium strain was completely resistant to naïve turkey poult serum, and our prior observation that this strain also attaches to tracheal rings in vitro (Temple et al., 1998). When tested, the two LPS mutants showed a significant decrease in serum resistance and in binding to turkey tracheal rings in vitro (Fig. 5A and B respectively).
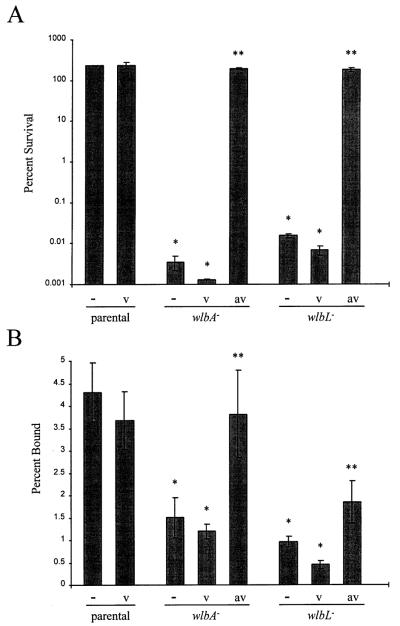
Fig. 5.
Measurements of serum resistance (A) and tracheal ring binding (B) by parent, mutants and complemented strains. Strains are indicated below the solid lines as follows: parental, B. avium 197N; wlbA−, wlbA mutant PAS85; wlbL−, wlbL mutant PAS78. Abbreviations along the x-axis refer to the following: −, indicates strain containing no plasmid; v, indicates strain containing vector plasmid pLAFR5; av, indicates strain containing recombinant plasmid pLAF-av. Single asterisk indicates significant difference (P < 0.05) from parent B. avium 197N. Double asterisk indicates significant difference (P < 0.05) from mutant containing pLAFR5 as determined by Student’s t-test. Error bars indicate standard error. Refer to text for a description of the assays.
In the case of serum resistance, parental B. avium was resistant to naïve turkey serum as demonstrated by greater than 100% survival in 50% serum (Fig. 5A). The wlbA and wlbL mutants showed a decrease in resistance to serum as demonstrated by 0.004% and 0.015% survival rates respectively. Complementation of both mutants with the homologous wlb locus of B. avium, but not vector, restored serum resistance to parental levels.
In the case of tracheal ring binding, the two LPS mutants showed a significant decrease in the percentage of bacteria bound compared with the parental strain (Fig. 5B). Upon complementation with the homologous wlb locus of B. avium, attachment was significantly increased in both mutants when compared with the mutant containing the vector alone. However, only the wlbA mutant was restored to parental levels.
Discussion
Two transposon insertion mutants of B. avium were identified that (i) exhibited clumping when grown in broth, (ii) had altered LPS polyacrylamide gel profiles, (iii) were significantly reduced in turkey poult colonization, (iv) were resistant to Ba1 bacteriophage, (v) showed a decrease in serum resistance, and (vi) showed a decrease in turkey tracheal epithelial cell attachment. The insertions interrupted genes associated with LPS biosynthesis. One mutant had an insertion in a gene similar to the wlbA gene of B. pertussis and the other had an insertion in a gene similar to the wlbL gene at the same genetic locus. Cloning of the entire B. avium wlb genetic locus (encoding genes wlbA–wlbL) and partial sequencing indicated similarity of the B. avium wbl locus to the other Bordetella species where the wbl sequence has been obtained (B. pertussis, B. bronchiseptica and B. parapertussis: Keen et al., 1988; Allen and Maskell, 1996; Allen et al., 1998).
Trans-complementation of B. avium wlbA and wlbL mutants with both homologous and heterologous wlb loci revealed that homologous genes from B. avium as well as heterologous genes from B. pertussis, B. bronchiseptica and Bordetella parapertussis could readily complement the clumped-growth phenotype in both mutants. Complementation of the clumped-growth phenotype appeared to correlate with the shift of a faster migrating LPS band in the mutants to a slower migrating band comparable to the lower band in the parent. Considering that the wlb genetic locus is responsible for the addition of a trisaccharide to the lipid-A-core region in the other Bordetella species (Peppler, 1984; Caroff et al., 1990; Preston et al., 1996; Maskell and Allen, 1997), we tentatively conclude that the wlbA and wlbL gene products of heterologous plasmids affect oligosaccharide additions to the mutant lipid-A-core region and that these additions are sufficient to complement the clumped-growth phenotype.
In contrast to the clumped-growth phenotype, restoration of B. avium-specific bacteriophage (Ba1) sensitivity was more dependent upon having the homologous genes, especially with respect to complementing the wlbA mutant. That is, in the wlbA mutant, phage sensitivity was, at best, poorly restored regardless of the heterologous locus employed. In the wlbL mutants, partial phage sensitivity was evident upon heterologous complementation by B. bronchiseptica and B. parapertussis wlb loci, but not as evident upon complementation by the B. pertussis genetic locus. Interestingly, the degree to which phage sensitivity was restored by the heterologous loci correlated well with the degree to which O-antigen was restored (compare appropriate regions of the gels in Fig. 2 with the cross-streaks in Fig. 3). These data suggest that phage Ba1 may bind to a portion of the O-antigen. Further examination of phage resistant phenotypes (e.g. directly selecting for phage resistant mutants) will probably provide a more sophisticated understanding of the relationship between LPS and phage sensitivity. Also, more evidence obtained from competitive binding assays are needed to show, unequivocally, that phage interact directly with O-antigen.
The precise structural defect in LPS biosynthesis in the wlbA and wlbL mutants was not determined. However, from previous studies of B. pertussis LPS structure, the wlbL gene encodes a protein similar to enzymes typically involved in the synthesis of 6-deoxy and dideoxy sugars (Allen and Maskell, 1996). In B. pertussis, this corresponds to synthesis of N-acetyl-N-methylfucosamine (FucNAcMe), which is one of the trisaccharides attached to the lipid-A-core LPS. The wlbA gene encodes a putative dehydrogenase. Allen and Maskell (1996) propose that this gene product most probably is involved in the biosynthesis of 2,3-dideoxy-2,3-di-N-acetylmannosa-minuronic acid (2,3-diNAcManA), which is also one of the trisaccharides attached to the lipid-A-core LPS of B. pertussis. One might reason that the specificity of the enzymatic reaction of the wlbL gene product [over that of the dehydrogenase (the wlbA gene product)] may account for the differences in the degree of heterologous complementation found in wlbA and wlbL mutants. Another reason, other than specificity, may involve differing degrees of polarity in the wlbA and wlbL insertions. That is, the transposon insertion in wlbA may have polar effects on downstream wlb genes and be more difficult to complement due to the possibility of numerous heterologous gene products necessary for effective complementation. In contrast, the wlbL insertion, at the end of the locus, may not affect other relevant genes.
LPS has been shown to be important for virulence in a variety of bacterial pathogens (Sigel et al., 1980; Waldor et al., 1994; Sandlin et al., 1995; Bilge et al., 1996; Brown and Curtiss, 1996; Licht et al., 1996; Chiang and Mekalanos, 1999). In the Bordetellae, there have been no examinations of the virulence of LPS mutants in vivo; although, in B. pertussis, in vitro assays have indicated the importance of LPS in epithelial cell cytotoxicity (Flak and Goldman, 1999) and phagocytosis (Weingart and Weiss, 2000). Our results clearly show a significant defect in tracheal colonization of our wlbA and wlbL mutants, and restoration of parental levels of colonization upon genotypic complementation. There was no statistically significant difference between the wlbA and wlbL mutants in their ability to effect tracheal colonization. This correlated with our observation that the LPS profiles of the two mutants were indistinguishable after PAGE. Nevertheless, further colonization studies and more detailed structural analyses will probably reveal differences.
In order to relate the effects of wlbA and wlbL mutations more closely to traits associated with virulence, we examined two parameters: resistance to naïve serum and tracheal attachment. These traits have been correlated with virulence in vivo (Fernandez and Weiss, 1994; Temple et al., 1998). With regard to serum sensitivity, the wlbL and wlbA mutants were both dramatically more sensitive to naïve turkey serum. This sensitivity probably decreases the ability of the mutants to initiate colonization (tracheal secretions contain serum; Suresh et al., 1994) and later may enhance the clearance of the microorganism (the resolution of experimental avian bordetellosis is completely correlated with the onset of humoral immunity; Suresh et al., 1994; Suresh and Arp, 1995). Attachment of the mutants to tracheal rings was also significantly affected. Because B. avium, like all Bordetella species, is dramatically ciliatropic (Arp and Fagerland, 1987), we infer that these mutants are impaired in binding to ciliated tracheal epithelial cells and thus would be at a disadvantage in colonizing normal turkey tracheas. Both of these effects may contribute to the decrease in colonization of turkey poults that we observed in vivo.
In addition to establishing the importance for the wlb locus in virulence, the present work suggests several avenues for further investigation. For example, the use of phage resistance as a selection for avirulent mutants may provide better understanding of the role phage receptors in virulence. Clearly, phage have been instrumental in uncovering and relating features of the bacterial cell surface to virulence (Raleigh and Signer, 1982; Nnalue et al., 1990). In addition, the present understanding of Bordetellae LPS structure may be exploited by using heterologous loci (Di Fabio et al., 1992; Preston et al., 1996; Maskell and Allen, 1997) in more refined complementation tests. The most important aspect of future LPS analysis will be to determine how alterations in its structure affect changes in colonization, serum resistance and tracheal cell binding.
Taken together, the above results indicate a role for LPS in B. avium virulence and phage sensitivity, thus providing a framework and tools for further genetic and biochemical studies of B. avium LPS as an important virulence factor in bordetellosis.
Experimental procedures
Bacterial strains and growth conditions
All bacterial strains, bacteriophage and plasmids employed in this study are listed in Table 1. Brain–heart infusion (BHI) medium (Difco) and Stainer–Scholte medium were employed under B. avium growth conditions that have been described previously (Temple et al., 1998). Antibiotics were added at the concentrations of 40 μg ml−1, kanamycin; 20 μg ml−1, tetracycline; 100 or 800 μg ml−1, ampicillin; or 25 μg ml−1, nalidixic acid when appropriate. All E. coli strains were grown in Luria (L) broth or agar (Miller, 1972) at 37°C overnight.
To screen for clumping in broth culture, B. avium was grown in 1.0 ml of BHI broth in a 48-well cell culture cluster (Costar) with rotary shaking overnight at 37°C.
Tn5 insertion mutagenesis
Random-insertion mutants were generated by a modification of the method of Temple et al., 1998. Conjugation was performed between E. coli S17.1 (γpir) containing pUTKm2 (de Lorenzo et al., 1990; Hensel et al., 1995) and B. avium 197N. Next, 100 μl of a 10-fold concentrated overnight culture of B. avium was spread onto selective medium (BHI containing kanamycin and nalidixic acid) to create a recipient lawn. Overnight cultures of S17.1γpir containing pUTKm2 plasmids were serially diluted and 20 μl was spotted onto the recipient lawn. Plates were incubated for 48 h at 37°C and resultant independent exconjugants were isolated. Single insertions and the loss of the plasmid were monitored by Southern analysis and sensitivity to 800 μg ml−1 ampicillin. Resultant insertion mutants were grown separately in 1.0 ml of BHI broth containing kanamycin in 48-well cell culture clusters.
Infectious dose determinations
The 50% infectious dose (ID50) measurement of each mutant was performed as previously described (Temple et al., 1998) and approved under North Carolina State University Animal Care Protocol 97-129-A. Briefly, three sets of 10 turkey poults (1 week old) were inoculated with approximately 106, 107 or 108 colony-forming units (cfu) of a mutant B. avium strain. Inocula was determined by vigorous resuspension of the bacteria prior to plating dilutions. At the same time, three sets of 10 turkey poults were inoculated with approximately 105, 106 or 107 cfu of the parental B. avium strain (197N). As a negative control, 10 turkey poults were inoculated with phosphate buffered saline. Prior to inoculation, four poults were removed and tested for prior exposure to B. avium. Fourteen days post-inoculation, one tracheal swab was collected from each turkey, expressed onto lactose MacConkey agar, and streaked for isolation. The cultures were incubated at 37°C for 48 h and the number of birds colonized (presence of any B. avium) was recorded. Isolates were confirmed by testing for appropriate antibiotic resistance and phenotype. ID50s were calculated using a Reed and Muench analysis (Reed and Muench, 1938).
In the case of complementation, ID50 determinations were carried out essentially as above. Briefly, three sets of 10 turkey poults were inoculated with approximately 106, 107 or 108 cfu of B. avium wlbA mutant PAS85 or wlbL mutant PAS78 complemented with the plasmid vector (pLAFR5), and another three sets at the same approximate doses with PAS85 or PAS78 complemented with the recombinant plasmid containing the wlb locus of B. avium (pLAF-av). In these experiments, one set of 10 turkey poults was inoculated with approximately 107 cfu of parental B. avium 197N containing the plasmid vector (pLAFR5) as a positive infection control. Colonizing bacteria were scored after plating directly onto lactose MacConkey agar (no antibiotic selection) and BHI agar containing tetracycline and nalidixic acid to differentiate plasmid-containing B. avium from those that had lost the plasmid.
Cloning and sequence analysis of insertion mutants
B. avium DNA adjacent to the transposon was cloned by taking advantage of the gene encoding neomycin phosphotransferase, conferring kanamycin resistance, within the transposon. Chromosomal DNA from the wlbL mutant (strain PAS78) was digested with BglII and ligated into pLitmus28 (New England Biolabs) digested with BglII. The ligation mixture was introduced into competent E. coli DH5α by transformation and transformants were selected on L agar containing kanamycin and 100 μg ml−1 ampicillin. One of the resulting transformants harboured a cloned DNA segment containing a portion of the transposon and downstream flanking DNA. The clone was sequenced at the UNC-CH Automated DNA Sequencing Facility on a Model 377 DNA Sequencer (Perkin Elmer, Applied Biosystems Division), using the ABI Prism™ Dye Terminator Cycle Sequencing Ready Reaction Kit with AmpliTaq™ DNA polymerase, FS (Perkin Elmer, Applied Biosystems Division), using primer P7 (5′-dGCA CTT GTG TAT AAG AGT CAG; Hensel et al., 1995). Chromosomal DNA from the wlbA mutant (strain PAS85) was digested with NotI and ligated into pBluescript (Stratagene) digested with NotI. The ligation mixture was introduced into competent E. coli DH5α by transformation and transformants were selected on L agar containing kanamycin and 100 μg ml−1 ampicillin. One of the resultant transformants contained most of the transposon and the upstream flanking region. The clone was sequenced as above using primer P6 (5′-dCCT AGG CGC CCA GAT CTG AT; Hensel et al., 1995).
LPS isolation and SDS–PAGE
LPS extraction from all strains were performed as previously described (Hitchcock and Brown, 1983; Apicella et al., 1994). Briefly, overnight broth cultures of B. avium were washed in PBS and diluted to 0.4 OD units ml−1 (approximately 4 × 108 cells ml−1). Next, 1 ml of cells was pelleted, resuspended in 100 μl lysis buffer (1 M Tris, pH 6.8, 6% SDS, 20% glycerol) and incubated at 95°C for 5–10 min. After cooling, 0.25 unit μl−1 DNase I (Sigma or Boehringer Mannheim) and 1 μg μl−1 RNase A (Sigma) were added and samples were incubated at room temperature for 10 min. Proteinase K (Sigma) was added (0.50 μg μl−1), samples were incubated at 55°C for 2 h and the resultant lysate was stored at 4°C. In some cases, the lysate was further treated by the addition of 900 μl cold acetone, incubated on ice for 1 h followed by centrifugation. The supernatant was discarded and the pellet containing LPS was resuspended in 50–100 μl of sample loading buffer. LPS was separated by 18% SDS–PAGE (Laemmli, 1970; Ausubel et al., 1995) and stained by the silver staining technique of Tsai and Frasch (1982).
Cloning of the B. avium wlb locus
Oligonucleotides that anneal adjacent to the PAS78 and PAS85 insertion mutations were used to generate 200 bp digoxygenin-labelled PCR products within the coding regions of wlbL and wlbA respectively. DNA of the parental strain, 197N, was digested with NotI and resultant fragments were separated by agarose gel electrophoresis. After transfer to a nylon membrane, a 15 kb fragment showed homology to both the wlbL and wlbA dig-PCR probes (data not shown). NotI digested DNA fragments, from strain 197N, ranging in size from 12 to 19 kb were gel purified, ligated into pGEM11Z(f) digested with NotI, and introduced into competent E. coli DH5α by transformation. One hundred transformants were patched onto L-agar containing ampicillin and grown overnight at 37°C. Colony lifts were performed as previously described (Ausubel et al., 1995). Hybridization to the wlbL probe revealed one positive clone. Further analysis, by the PCR and Southern blot hybridization, revealed the presence of both wlbA and wlbL genes. This clone contained a 15 kb insert and was designated pGEM-av.
The B. avium 15 kb DNA fragment from pGEM-av was subsequently subcloned into the broad host range vector pLAFR5 as follows. The plasmid (pGEM-av) was digested with NotI and the 5′ overhanging ends were filled-in using Klenow polymerase to create blunt ends. BclI linkers were ligated onto the blunt ends followed by complete digestion with BclI. The resultant 15 kb fragment was gel purified after agarose gel electrophoresis and ligated into pLAFR5 digested with BamHI. The ligation mixture was introduced into competent E. coli DH5α by transformation. This clone contained a 15 kb insert and is designated as pLAF-av.
Complementation analysis
The plasmid pLAF-av contains the wlb locus of B. avium 197N and was constructed as described above. The plasmids pLAF-pe, pLAF-br, pLAF-pa and pLAFR5 contain the entire 12 kb wlb locus of B. pertussis (pLAF-pe), B. bronchiseptica (pLAF-br) or B. parapertussis (pLAF-pa) in the broad host range vector pLAFR5. These plasmids have been previously described (Allen et al., 1998; Allen and Maskell, 1996; Keen et al., 1988) and were kindly provided by A. Preston, Centre for Veterinary Science, University of Cambridge, UK. The pLAF-av, pLAF-pe, pLAF-br, pLAF-pa and pLAFR5 broad host range plasmids were maintained in E. coli XL1-blue or DH5α and were introduced into E. coli S17.1(γpir) by transformation prior to conjugation with B. avium. All E. coli strains containing plasmids were grown in L broth or agar containing tetracycline at 37°C. All B. avium strains containing the complementing plasmids were grown in BHI broth or on BHI agar containing tetracycline at 37°C.
Ba1 sensitivity
Sensitivity to B. avium bacteriophage Ba1 (Shelton et al. submitted) was determined by cross-streaking (Silhavy et al., 1984). A clear plaque mutant Ba1 bacteriophage (Ba1c1; Shelton et al. submitted) was spread down the middle of a BHI-agar plate containing tetracycline and nalidixic acid. B. avium colonies were then picked from an agar plate with a sterile tooth pick and streaked perpendicularly across the bacteriophage streak. Plates were incubated overnight at 37°C.
Serum resistance assay
Bacteria were grown in BHI broth with the appropriate antibiotic at 37°C overnight with shaking. The overnight culture was diluted 1:100 and grown to mid-logarithmic phase. Cell density was determined by measuring the optical density at 600 nm. Cells were washed two times in PBS and diluted to approximately 107 cells ml−1 in PBS prior to use. Each reaction contained 16 μl of diluted cells, 64 μl of PBS and 80 μl of naïve turkey poult serum. A 10 μl aliquot was removed immediately to determine the initial viable bacterial cell count. The mixture was then incubated at 37°C for 1 h. Prior to plating, the cells were collected by centrifugation and resuspended in 150 μl of PBS. Bacterial cell counts were determined by plating 10-fold serial dilutions onto BHI agar plates. Each strain was tested in triplicate bactericidal assays. At the same time, each strain was also incubated with heat inactivated serum as a control. The degree of serum resistance was quantified by determining the percent of the initial population that survived the 1 h serum treatment. In all cases, heat inactivation negated any bactericidal effect (data not shown).
Tracheal adherence assay
Adherence assays were performed essentially as previously described (Temple et al., 1998) Briefly, B. avium to be tested for attachment were adjusted to a constant OD600 and serial dilutions were plated to determine the initial bacterial cell count. Approximately 1 × 107 cfu were added to tissue culture wells containing three tracheal rings obtained from 26-day-old turkey embryos, and incubated at 42°C for 3 h with constant rocking. After incubation, the rings were washed and each ring was placed into a separate tube containing 1.0 ml of PBS with 1% Triton X-100, incubated at 4°C for 1–2 h, then vortexed for 1 min to distribute the bacteria. Bacterial cell counts were determined by plating 10-fold serial dilutions onto BHI agar plates. The numbers of cfu/tracheal ring were used to calculate the percent bacteria bound.
Statistical analysis
The significance of mean differences were determined using Student’s t-test with the aid of the Microsoft Excel statistical analysis software (version 4). In some cases, significant differences were determined by the Z-test, which assumes equal variances (Microsoft Excel version 4). In the cases described herein, the Z-test was employed when we asked the probability of a single mutant ID50 value falling within two standard deviations of the mean ID50 calculated using the parental strain. Standard deviation of the mean was calculated using Microsoft Excel STDEV function. Standard error was calculated as the standard deviation divided by the square root of the number of experiments.
Acknowledgements
We thank C. Altier for a critical reading of this manuscript. This work was supported by grants from the NIH (R15 AI/OD3773-01A1 and AI-23695), the USDA (950 934 and 99-35204-7743), D. University, and the State of North Carolina. Also, the generous co-operation and support given by British United Turkeys of America were invaluable and much appreciated.
References
- Ackermann MR, Register KB, Gentry-Weeks C, Gwaltney SM, Magyar T. A porcine model for the evaluation of virulence of Bordetella bronchiseptica. J Comp Pathol. 1997;116:55–61. doi: 10.1016/s0021-9975(97)80043-6. [DOI] [PubMed] [Google Scholar]
- Allen A, Maskell D. The identification, cloning and mutagenesis of a genetic locus required for lipopolysaccharide biosynthesis in Bordetella pertussis. Mol Microbiol. 1996;19:37–52. doi: 10.1046/j.1365-2958.1996.354877.x. [DOI] [PubMed] [Google Scholar]
- Allen AG, Thomas RM, Cadisch JT, Maskell DJ. Molecular and functional analysis of the lipopolysaccharide biosynthesis locus wlb from Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Mol Microbiol. 1998;29:27–38. doi: 10.1046/j.1365-2958.1998.00878.x. [DOI] [PubMed] [Google Scholar]
- Apicella MA, Griffiss JM, Schneider H. Isolation and characterization of lipopolysaccharides, lipooligosaccharides and lipid A. Methods Enzymol. 1994;235:242–252. doi: 10.1016/0076-6879(94)35145-7. [DOI] [PubMed] [Google Scholar]
- Arp LH, Fagerland JA. Ultrastructural pathology of Bordetella avium infection in turkeys. Vet Pathol. 1987;24:411–418. doi: 10.1177/030098588702400508. [DOI] [PubMed] [Google Scholar]
- Arp LH, Huffman EL, Hellwig DH. Glycoconjugates as components of receptors for Bordetella avium on the tracheal mucosa of turkeys. Am J Vet Res. 1993;54:2027–2030. [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DE, Seidman JG, Smith JA, et al. Current Protocols in Molecular Biology. John Wiley & Sons; New York: 1995. [Google Scholar]
- Barnes HJ, Hofstad MS. Factors involved in respiratory disease of turkeys in Iowa. J Am Vet Med Assoc. 1978;173:889–897. [Google Scholar]
- van den Berg BM, Beekhuizen H, Willems RJ, Mooi FR, van Furth R. Role of Bordetella pertussis virulence factors in adherence to epithelial cell lines derived from the human respiratory tract. Infect Immun. 1999;67:1056–1062. doi: 10.1128/iai.67.3.1056-1062.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilge SS, Vary JC, Jr, Dowell SF, Tarr PI. Role of the Escherichia coli 0157:H7 O side chain in adherence and analysis of an rfb locus. Infect Immun. 1996;64:4795–4801. doi: 10.1128/iai.64.11.4795-4801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PK, Curtiss R., III Unique chromosomal regions associated with virulence of an avian pathogenic Escherichia coli strain. Proc Natl Acad Sci USA. 1996;93:11149–11154. doi: 10.1073/pnas.93.20.11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroff M, Chaby R, Karibian D, Perry J, Deprun C, Szabo L. Variations in the carbohydrate regions of Bordetella pertussis lipopolysaccharides: Electrophoretic, serological, and structural features. J Bacteriol. 1990;172:1121–1128. doi: 10.1128/jb.172.2.1121-1128.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang SL, Mekalanos JJ. rfb mutations in Vibrio cholerae do not affect surface production of toxin-coregulated pili but still inhibit intestinal colonization. Infect Immun. 1999;67:976–980. doi: 10.1128/iai.67.2.976-980.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PA, Miller JF. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect Immun. 1994;62:3381–3390. doi: 10.1128/iai.62.8.3381-3390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PA, Yuk MH, Mattoo S, Akerley BJ, Boschwitz J, Relman DA, et al. Filamentous hemagglutinin of Bordetella bronchiseptica is required for efficient establishment of tracheal colonization. Infect Immun. 1998;66:5921–5929. doi: 10.1128/iai.66.12.5921-5929.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fabio JL, Caroff M, Karibian D, Richards JC, Perry MB. Characterization of the common antigenic lipopolysaccharide O-chains produced by Bordetella bronchiseptica and Bordetella parapertussis. FEMS Microbiol Lett. 1992;76:275–282. doi: 10.1016/0378-1097(92)90348-r. [DOI] [PubMed] [Google Scholar]
- Ensminger ME. Poultry Science. The Interstate Press; Danville, IL: 1980. Poultry feeding standards, ration formulation, and feeding programs; pp. 149–188. [Google Scholar]
- Fernandez RC, Weiss AA. Cloning and sequencing of a Bordetella pertussis serum resistance locus. Infect Immun. 1994;62:4727–4738. doi: 10.1128/iai.62.11.4727-4738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flak TA, Goldman WE. Signaling and cellular specificity of airway nitric oxide production in pertussis. Cell Microbiol. 1999;1:51–60. doi: 10.1046/j.1462-5822.1999.00004.x. [DOI] [PubMed] [Google Scholar]
- Gentry-Weeks CR, Cookson BT, Goldman WE, Rimler RB, Porter SB, Curtiss R., III Dermonecrotic toxin and tracheal cytotoxin, putative virulence factors of Bordetella avium. Infect Immun. 1988;56:1698–1707. doi: 10.1128/iai.56.7.1698-1707.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuijen CA, Willems RJ, Bongaerts M, Top J, Gielen H, Mooi FR. Role of the Bordetella pertussis minor fimbrial subunit, FimD, in colonization of the mouse respiratory tract. Infect Immun. 1997;65:4222–4228. doi: 10.1128/iai.65.10.4222-4228.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin MS, Weiss AA. Adenylate cyclase toxin is critical for colonization and pertussis toxin is critical for lethal infection by Bordetella pertussis in infant mice. Infect Immun. 1990;58:3445–3447. doi: 10.1128/iai.58.10.3445-3447.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- Hitchcock PJ, Brown TM. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Sato W. Role of Bordetella bronchiseptica sialic acid-binding hemagglutinin as a putative colonization factor. J Vet Med Sci. 1997;59:43–44. doi: 10.1292/jvms.59.43. [DOI] [PubMed] [Google Scholar]
- Keen NT, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Licht TR, Krogfelt KA, Cohen PS, Poulsen LK, Urbance J, Molin S. Role of lipopolysaccharide in colonization of the mouse intestine by Salmonella typhimurium studied by in situ hybridization. Infect Immun. 1996;64:3811–3817. doi: 10.1128/iai.64.9.3811-3817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V, Herrero M, Jakubzik U, Timmis KN. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskell D, Allen A. Molecular biology of lipopolysaccharide biosynthesis in Salmonella and Bordetella. Biochem Soc Trans. 1997;25:850–856. doi: 10.1042/bst0250850. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1972. [Google Scholar]
- Monack DM, Falkow S. Cloning of Bordetella bronchiseptica urease genes and analysis of colonization with a urease-negative mutant strain in a guinea-pig model. Mol Microbiol. 1993;10:545–553. doi: 10.1111/j.1365-2958.1993.tb00926.x. [DOI] [PubMed] [Google Scholar]
- Mooi FR, Jansen WH, Brunings H, Gielen H, van der Heide HG, Walvoort HC, et al. Construction and analysis of Bordetella pertussis mutants defective in the production of fimbriae. Microb Pathog. 1992;12:127–135. doi: 10.1016/0882-4010(92)90115-5. [DOI] [PubMed] [Google Scholar]
- Nnalue NA, Newton S, Stocker BA. Lysogenization of Salmonella choleraesuis by phage 14 increases average length of O-antigen chains, serum resistance and intraperitoneal mouse virulence. Microb Pathog. 1990;8:393–402. doi: 10.1016/0882-4010(90)90026-m. [DOI] [PubMed] [Google Scholar]
- Parton R, Hall E, Wardlaw AC. Responses to Bordetella pertussis mutant strains and to vaccination in the coughing rat model of pertussis. J Med Microbiol. 1994;40:307–312. doi: 10.1099/00222615-40-5-307. [DOI] [PubMed] [Google Scholar]
- Peppler MS. Two physically and serologically distinct lipopolysaccharide profiles in strains of Bordetella pertussis and their phenotype variants. Infect Immun. 1984;43:224–232. doi: 10.1128/iai.43.1.224-232.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston A, Mandrell RE, Gibson BW, Apicella MA. The lipooligosaccharides of pathogenic gram negative bacteria. Crit Rev Microbiol. 1996;22:139–180. doi: 10.3109/10408419609106458. [DOI] [PubMed] [Google Scholar]
- Raleigh EA, Signer ER. Positive selection of nodulation-deficient Rizobium phaseoli. J Bacteriol. 1982;151:83–88. doi: 10.1128/jb.151.1.83-88.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty percent end points. Am J Hyg. 1938;27:293–299. [Google Scholar]
- Rick PD. Lipopolysaccharide biosynthesis. In: Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia Coli and Salmonella Typhimurium Cellular and Molecular Biology. American Society for Microbiology; Washington, DC: 1987. pp. 648–662. [Google Scholar]
- Saif YM, Moorhead PD, Dearth RN, Jackwood DJ. Observations on Alcaligenes faecalis infection in turkeys. Avian Dis. 1980;24:665–684. [PubMed] [Google Scholar]
- Sandlin RC, Lampel KA, Keasler SP, Goldberg MB, Stolzer AL, Maurelli AT. Avirulence of rough mutants of Shigella flexneri: requirement of O antigen for correct unipolar localization of IcsA in the bacterial outer membrane. Infect Immun. 1995;63:229–237. doi: 10.1128/iai.63.1.229-237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahin RD, Cowell JL. Mouse respiratory infection models for pertussis. Methods Enzymol. 1994;235:47–58. doi: 10.1016/0076-6879(94)35130-9. [DOI] [PubMed] [Google Scholar]
- Sigel SP, Lanier S, Baselski VS, Parker CD. In vivo evaluation of pathogenicity of clinical and environmental isolates of Vibrio cholerae. Infect Immun. 1980;28:681–687. doi: 10.1128/iai.28.3.681-687.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy TT, Berman ML, Enquist LW. Experiments with Gene Fusions. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1984. [Google Scholar]
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. BIO/Technology. 1983;1:784–791. [Google Scholar]
- Skeeles JK, Arp LH. Bordetellosis (turkey coryza) In: Calnek BW, Barnes HJ, Beard CW, McDougal LR, Saif YM, editors. Diseases of Poultry. Iowa State University Press; Ames, IA: 1997. pp. 275–288. [Google Scholar]
- Suresh P, Arp LH. Effect of passively administered immunoglobulin G on the colonization and clearance of Bordetella avium in turkeys. Vet Immunol Immunopathol. 1995;49:229–239. doi: 10.1016/0165-2427(95)05467-7. [DOI] [PubMed] [Google Scholar]
- Suresh PL, Arp LH, Huffman EL. Mucosal and systemic humoral immune response to Bordetella avium in experimentally infected turkeys. Avian Dis. 1994;38:225–230. [PubMed] [Google Scholar]
- Temple LM, Weiss AA, Walker KE, Barnes HJ, Christensen VL, Miyamoto DM, et al. Bordetella avium virulence measured in vivo and in vitro. Infect Immun. 1998;66:5244–5251. doi: 10.1128/iai.66.11.5244-5251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C-M, Frasch CE. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- Waldor MK, Colwell R, Mekalanos JJ. The Vibrio cholerae 0139 serogroup antigen includes an O-antigen capsule and lipopolysaccharide virulence determinants. Proc Natl Acad Sci USA. 1994;91:11388–11392. doi: 10.1073/pnas.91.24.11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingart CL, Weiss AA. Bordetella pertussis virulence factors affect phagocytosis by human neutrophils. Infect Immun. 2000;68:1725–1739. doi: 10.1128/iai.68.3.1735-1739.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss AA, Goodwin MSM. Lethal infection by Bordetella pertussis mutants in the infant mouse model. Infect Immun. 1989;57:3757–3764. doi: 10.1128/iai.57.12.3757-3764.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A, Hewlett EL, Meyers GA, Falkow S. Pertussis toxin and extracytoplasmic adeylate cyclase as virulence factors of Bordetella pertussis. J Infect Dis. 1984;150:219–222. doi: 10.1093/infdis/150.2.219. [DOI] [PubMed] [Google Scholar]