Abstract
Background
Publication of the first diatom genome, that of Thalassiosira pseudonana, established it as a model species for experimental and genomic studies of diatoms. Virtually every ensuing study has treated T. pseudonana as a marine diatom, with genomic and experimental data valued for their insights into the ecology and evolution of diatoms in the world's oceans.
Results
The natural distribution of T. pseudonana spans both marine and fresh waters, and phylogenetic analyses of morphological and molecular datasets show that, 1) T. pseudonana marks an early divergence in a major freshwater radiation by diatoms, and 2) as a species, T. pseudonana is likely ancestrally freshwater. Marine strains therefore represent recent recolonizations of higher salinity habitats. In addition, the combination of a relatively nondescript form and a convoluted taxonomic history has introduced some confusion about the identity of T. pseudonana and, by extension, its phylogeny and ecology. We resolve these issues and use phylogenetic criteria to show that T. pseudonana is more appropriately classified by its original name, Cyclotella nana. Cyclotella contains a mix of marine and freshwater species and so more accurately conveys the complexities of the phylogenetic and natural histories of T. pseudonana.
Conclusions
The multitude of physical barriers that likely must be overcome for diatoms to successfully colonize freshwaters suggests that the physiological traits of T. pseudonana, and the genes underlying those traits, might differ from those of strictly marine diatoms. The freshwater ancestry of T. pseudonana might therefore confound generalizations about the physiological and metabolic properties of marine diatoms. The freshwater component of T. pseudonana's history merits careful consideration in the interpretation of experimental data collected for this important model species.
Keywords: Cyclotella nana, diatom, freshwater, marine, model species, Thalassiosira pseudonana
Background
Diatoms are unicellular photosynthetic algae with secondary, red-algal-derived plastids [1]. With total diversity estimates in the tens to hundreds of thousands of species, diatoms are one of the most diverse lineages of eukaryotes [2] and are critically important to the ecology of both marine and fresh waters. Marine diatoms alone account for roughly one-fifth of global net primary production [3]. Efforts to understand the ecology and evolution of diatoms were catapulted forward when the first diatom genome, from a marine strain of Thalassiosira pseudonana (Hustedt) Hasle et Heimdal, was published in 2004 [4]. The genome revealed unanticipated metabolic pathways and gave a first glimpse into the mosaic nature of diatom nuclear genomes, which contain a mix of genetic material from the stramenopile host cell, diverse bacterial donors, and a succession of green and red algal endosymbionts [4-6].
In addition to its small (32 Mb) genome size, T. pseudonana was chosen for genome sequencing because it is generally considered representative of the large, widespread and predominantly marine genus Thalassiosira [4,7]. Although T. pseudonana has long been used in experimental studies [8-10], its genome sequence established it as the premier model for genome-enabled diatom research [11,12] on topics ranging from nutrient storage and metabolism [13-15] to silica biomineralization [16-19]. Data from these studies are often valued for their insights into the ecology and evolution of marine diatoms and, in turn, their determinant roles in the biogeochemical cycling of nutrients in the world's oceans [12]. Many of these inferences rest on the important assumption that T. pseudonana is a representative model for marine diatoms [20]. We summarize the phylogenetic and natural histories of T. pseudonana and show that it most likely descended from a freshwater ancestor. This portion of its history may have shaped several important physiological traits and the genes underlying those traits, confounding extrapolations to the biology of strictly marine diatoms. Finally, its relatively nondescript form and convoluted taxonomic history have introduced some confusion about the identity of T. pseudonana and, by extension, its phylogenetic and ecological histories. We resolve these issues and use phylogenetic criteria to show that T. pseudonana is more appropriately classified by its original name, Cyclotella nana Hustedt.
Results
The phylogeny and evolutionary ecology of Thalassiosira pseudonana
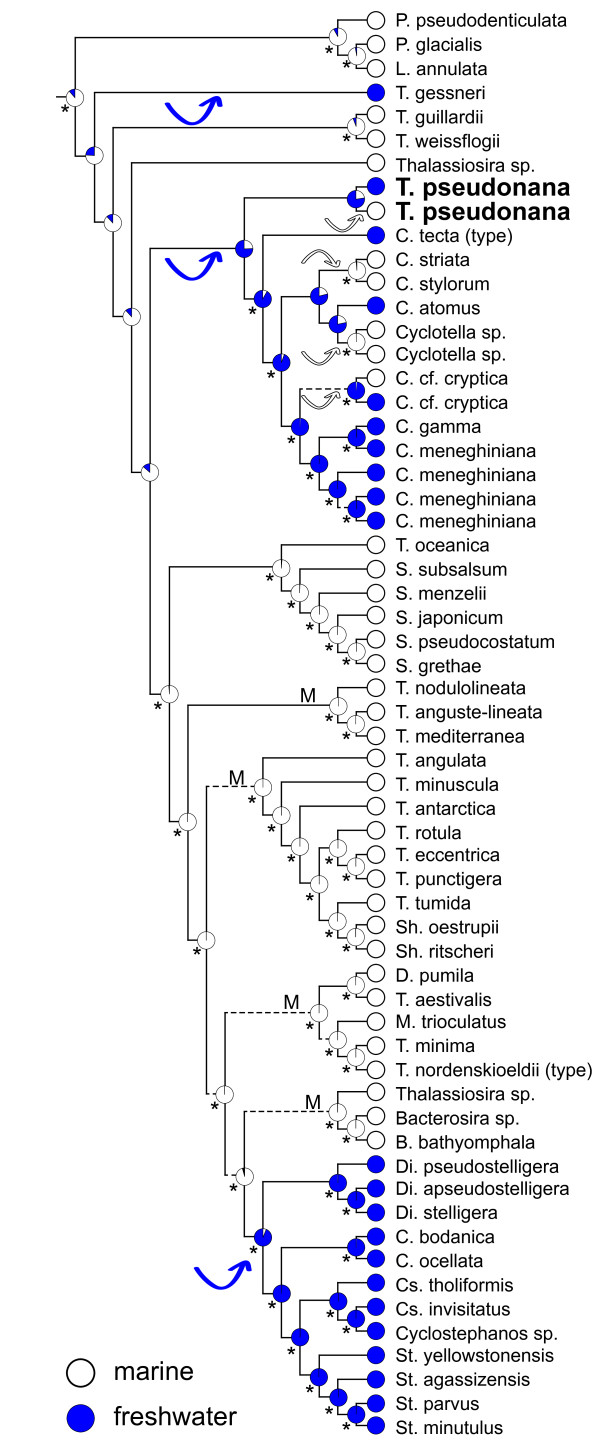
A combined phylogenetic analysis of two chloroplast (psbC and rbcL) and two nuclear (SSU and partial LSU rDNA) genes strongly and unambiguously resolved the phylogenetic position of T. pseudonana as sister to a clade of marine and freshwater species in the genus Cyclotella (Figure 1). This clade includes C. meneghiniana, which has been cited as an exemplar cosmopolitan freshwater diatom [21], and C. tecta, which is the nomenclatural type of the genus Cyclotella (Figure 1). The phylogeny also shows that members of the genus Thalassiosira are distributed across 10 different lineages within the Thalassiosirales (Figure 1). Among these, the lineages that include T. pseudonana and T. nordenskioeldii (the nomenclatural type of the genus Thalassiosira) are only distantly related (Figure 1).
Figure 1.
Phylogeny of the diatom order Thalassiosirales based on a Bayesian analysis of four concatenated genes. Pie graphs at internal nodes show the relative maximum likelihood support for the inferred ancestral habitat type, and asterisks show that the more strongly supported ancestral habitat type is significantly better than the alternative. Arrows represent inferred marine-to-freshwater (blue) and freshwater-to-marine (white) colonization events. Dashed lines identify nodes with <0.95 Bayesian posterior probability support, and lineages containing potentially strong candidates for alternative model marine diatoms are marked with "M." Cyclotella tecta and Thalassiosira nordenskioeldii are the nomenclatural types ("type") for their respective genera. Genus abbreviations are: Bacterosira (B), Cyclotella (C), Cyclostephanos (Cs), Detonula (D), Discostella (Di), Lauderia (L), Minidiscus (M), Porosira (P), Shionodiscus (Sh), Skeletonema (S), Stephanodiscus (St), and Thalassiosira (T). The figure is modified from ref. [25].
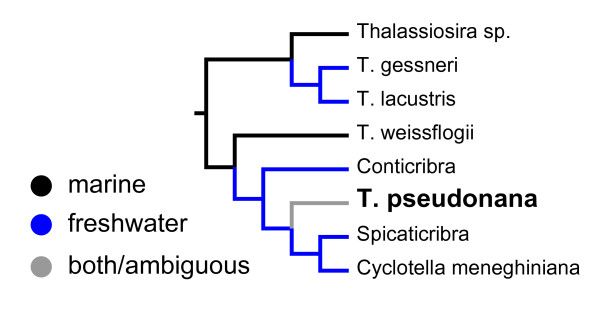
We extended a previous morphologically based phylogenetic analysis of Thalassiosirales [22] to include T. pseudonana, C. meneghiniana, and two recently described genera, Conticribra and Spicaticribra [23,24], both of which have morphological features that suggest a close relationship to the T. pseudonana+C. meneghiniana clade. The full morphological tree is shown in Additional File 1. Like the molecular phylogeny, morphological data also resolve T. pseudonana into a clade with C. meneghiniana (Figure 2). Conticribra tricircularis, which is known only from freshwaters [24], is sister to this clade (Figure 2).
Figure 2.
Phylogenetic analysis of morphological characters places the model marine diatom Thalassiosira pseudonana within a clade of freshwater diatoms. Ancestral states were reconstructed using parsimony. The full tree is shown in Additional File 1.
Molecular phylogenetic analyses included both marine and freshwater strains of T. pseudonana, and these always formed a strongly supported clade [25]. The close relationship of these strains to T. pseudonana strain CCMP1335, which had its genome sequenced [4], is supported by their near-identical chloroplast psbC and rbcL genes and nuclear SSU and partial LSU rDNA genes (Additional file 2). Maximum likelihood reconstructions show that the common ancestor of the clade that includes T. pseudonana, C. meneghiniana, and C. tecta was most likely a freshwater diatom (Figure 1). Although the proportional likelihood strongly supports this conclusion (0.76 vs. 0.24), the difference is not statistically significant (Figure 1). However, morphological data place the monotypic freshwater genus Conticribra as sister to the T. pseudonana+C. meneghiniana clade, which unambiguously optimizes the common ancestor of the T. pseudonana+C. meneghiniana clade as freshwater in this analysis (Figure 2). Finally, the common ancestor of the two T. pseudonana strains was likely freshwater as well (Figure 1).
The identity of Thalassiosira pseudonana
The combination of a relatively nondescript morphology and a series of three name changes over the years [26-29] has led to some uncertainty about the identity of T. pseudonana (Additional file 3). Several species are frequently confused with T. pseudonana, and two of these, T. guillardii and T. oceanica, were initially included under a broader concept of T. pseudonana (Additional file 4; ref. [30]). Resolution of these taxonomic and nomenclatural uncertainties required observation of the original sample (the "type material") from which T. pseudonana—originally named Cyclotella nana—was described (Additional file 5; ref. [29]).
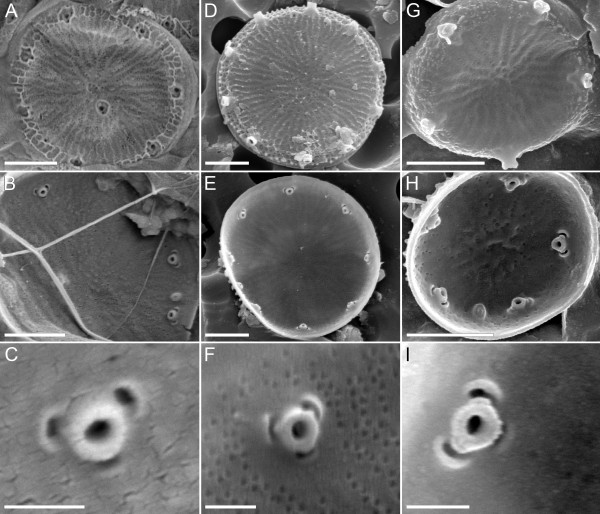
Several small Thalassiosirales were found in the C. nana type material (Additional file 6), but the most common small diatom encountered matches both the original description of C. nana [29] and the traditional concept of T. pseudonana (Figure 3; refs. [28-34]). All of these diatoms, and those used for phylogenetic reconstruction (Figure 1) and genome sequencing [4], are small (less than 10 μm diameter), circular, radially symmetric diatoms with 0-3 central-area strutted processes, numerous marginal-area strutted processes with three satellite pores each, and a labiate process situated between two marginal-area strutted processes (Figure 3 and Additional file 7). Ultrastructural features of the strutted processes are phylogenetically informative [25], and the strutted processes of all these diatoms have prominent, raised cowlings around the satellite pores and a more-or-less prominent siliceous ridge (opercle) on the central strutted process tube, situated directly above the corresponding satellite pore (Figure 3 and Additional file 7).
Figure 3.
Scanning electron micrographs of three ecologically diverse representatives of Thalassiosira pseudonana. (A-C) Marine culture strain CCMP1335 whose genome was sequenced; (D-F) freshwater specimens from the original type collection for the species from the River Wümme, Germany; (G-I) freshwater culture strain ETC1 from Lake Erie, Michigan, used in molecular phylogenetic analyses (Figure 1). The first and second rows show the cell exterior and interior, respectively (scale bar = 1 μm), and the third row shows the interior ultrastructure of the strutted process (scale bar = 200 nm).
Taken together, molecular and morphological evidence lead us to conclude that the model T. pseudonana strain CCMP1335 [4] and the culture strains used in molecular phylogenetic analyses (Figure 1) are all conspecific with specimens originally described as Cyclotella nana (Additional file 4).
Discussion
Robustness of phylogenetic results and ancestral reconstructions
Our conclusions about the ancestry of T. pseudonana rest primarily on the assumption that our analyses have accurately resolved its position within the broader Thalassiosirales phylogeny. As suggested by the numerous taxonomic and nomenclatural uncertainties (Additional files 3 and 4), T. pseudonana has gross morphological similarities to species of Thalassiosira, Cyclotella, and Discostella. In fact, T. pseudonana is commonly confused with several species of Discostella and Thalassiosira (viz., the purely freshwater species D. pseudostelligera and the purely marine species T. guillardii and T. oceanica). We included each of these species in our molecular dataset, and they all resolved well away from T. pseudonana (Figure 1). Cyclotella and Thalassiosira are especially large and morphologically diverse genera, though years of monographic research have led to numerous informally recognized morphological groups within each of them [34-43]. Our molecular dataset includes at least one representative from each of the informally designated groups within both Thalassiosira and Cyclotella. By sampling much of the known range of morphological diversity within these two genera, we have included many of the lineages within the Thalassiosirales phylogeny where one might predict T. pseudonana to fall.
The molecular dataset ultimately resolved T. pseudonana as sister to a clade that includes the C. meneghiniana species complex and the nomenclatural type of the genus Cyclotella, C. tecta. Comparing phylogenies derived from morphological and molecular datasets is a powerful way to detect phylogenetic incongruence, or likewise, to strengthen an existing phylogenetic hypothesis based on just one of the two data types [44]. Despite their different and largely non-overlapping taxon sampling schemes, the molecular and morphological datasets were congruent in their placement of T. pseudonana as sister to Cyclotella (Figures 1 and 2). Moreover, there was also congruence between morphology and molecules for the larger set of taxa common to both datasets, viz: (T. gessneri, (T. weissflogii, (T. pseudonana, C. meneghiniana))).
Our habitat codings were based on the habitat from which each culture originally was isolated. Although an individual culture strain might not capture the full range of genetic and phenotypic variability of its species, coding a culture strain by its natural habitat is conceptually similar to the common practice of representing a species in phylogenetic analyses with gene sequences from a single individual. Thalassiosira pseudonana can tolerate a wide range of salinities [30], so we could have coded each of the strains in our analyses as simply "euryhaline." For our purposes, however, individually coding each strain according to its natural habitat should have been more conservative, in that it allowed for a marine ancestral state reconstruction for both T. pseudonana and the nodes immediately surrounding it.
Although future data might show that some of the T. pseudonana strains considered here represent different biological species, it is not clear that those species would necessarily resolve along marine-freshwater lines. The finding by Guillard and Ryther [30] that three different T. pseudonana clones (3H, 5A and e.p.) maintained high growth rates from 0.5-37‰ salinity supports the hypothesis that T. pseudonana is, in fact, a single species that tolerates a wide range of salinities. Finally, the reconstruction of a freshwater ancestor for T. pseudonana (Figure 1) applies regardless of whether marine and freshwater strains are considered the same, or different, species.
To summarize, phylogenetic analyses place T. pseudonana as sister to a predominately freshwater clade of diatoms in the genus Cyclotella. The congruence between the molecular and morphological datasets provides especially strong support for this result [44]. Ancestral state reconstructions showed that the common ancestor of this clade was either unequivocally freshwater (morphological data, parsimony analysis) or likely freshwater (molecular data, likelihood analysis). Like any hypothesis, the results are potentially subject to change as new data become available or different character codings are used, but with respect to T. pseudonana, phylogenetic results and ancestral state reconstructions appear to be at least moderately robust to taxon sampling, data type, and optimality criterion.
Is Thalassiosira pseudonana a good model for marine diatoms?
The tremendous importance of marine diatoms in global carbon fixation and marine food webs understandably compelled Armbrust et al. [4] to choose a marine diatom as the first for whole-genome sequencing [7]. Thalassiosira pseudonana belongs to the large and predominantly marine genus Thalassiosira and so was reasonably considered representative of that genus and of marine diatoms more broadly [7]. Subsequent phylogenetic analyses showed, however, that members of the genus Thalassiosira are spread across some 10 distinct evolutionary lineages (Figure 1). The extent of independent, unshared evolutionary history distinguishing these 10 lineages from one another is such that no single model species can possibly capture all of the diverse ecological, metabolic, and genomic properties of "Thalassiosira"—a prediction increasingly borne out by experimental and genomic data [45-47]. Of course, no one would expect uniformity across such a broad range of species diversity, but their shared classification in the genus Thalassiosira falsely suggests that the taxa in these 10 lineages constitute a more biologically coherent group than the phylogeny reveals them to be (Figure 1). The persistence of this, or any, misinformative classification represents a failure by systematists—ourselves included—to provide a phylogenetically based, and therefore biologically informative, alternative. Such an alternative would facilitate, for example, ecologically based selection of model species.
Salinity preference provides a glaring example of a trait that varies considerably across the 10 lineages of Thalassiosira, and Thalassiosirales as a whole (Figure 1). With a distribution that spans both marine and freshwater habitats, T. pseudonana embodies this variation. The evolutionary history of Thalassiosirales includes just a few major freshwater diversifications [25], and T. pseudonana marks an early divergence in one of them (Figure 1). In fact, phylogenetic analyses and formal mapping of habitat preference onto phylogenetic trees show that, 1) T. pseudonana is part of a large clade that is likely ancestrally freshwater (Figures 1 and 2), and 2) as a species, T. pseudonana is likely ancestrally freshwater as well (Figure 1). If so, these data suggest that marine strains of T. pseudonana represent recent recolonizations of higher salinity habitats by a derived freshwater ancestor (Figures 1 and 2).
Parker et al. [20] keenly raised the question as to how well our current selection of model microalgae represent their corresponding dominant forms in the marine environment. The data presented here suggest that the substantial freshwater component of T. pseudonana's phylogenetic history might confound its use of as a model for marine diatoms. For example, there is great interest in understanding how diatoms sequester, store, and use iron because it limits their growth over vast offshore regions of the world's oceans [14,15,46,47]. By contrast, iron generally does not limit diatom growth in freshwater [48] and coastal ecosystems [49]. Experimental data show that coastal marine strains of T. pseudonana and T. weissflogii require more iron to maintain a maximal growth rate than a marine strain of the pennate diatom, Phaeodactylum tricornutum, and the oceanic diatom, T. oceanica, respectively [14,47]. As currently understood, these differences reflect either vastly different iron uptake [14] and storage [15] architectures between entirely different classes of diatoms (pennates vs. non-pennates), or slight modifications of shared iron uptake [46] or photosynthetic [47] architectures between closely related species. Some of these differences might, at least in part, reflect that T. pseudonana descended from a freshwater ancestor that experienced different (perhaps relaxed) constraints on its iron uptake and utilization machinery. Extending studies like that of Kustka et al. [14] to include strictly freshwater pennate and non-pennate diatoms along with strictly marine pennate and non-pennates might help tease apart the relative importance of shared history versus shared ecology in the evolution of iron metabolism in diatoms. In this context, "strictly" refers to diatoms that occur exclusively in either marine or freshwaters and whose immediate ancestor shared a similar distribution (e.g., Figure 1, "M").
Among the several hundred species of Thalassiosirales, which might provide a more representative model for marine diatoms? The phylogeny provides a powerful, predictive guide for selecting candidates. In this case, one might first consider the five strictly marine clades (Figure 1, "M") and then winnow down the candidate pool based on practical (e.g., genome size and amenability to cell culture) and ecological (e.g., oceanic vs. offshore) criteria.
What is Thalassiosira pseudonana?
The diatom originally described as Cyclotella nana from the River Wümme has also gone by the names T. pseudonana [28], C. pseudonana [27], and most recently, Discostella nana [26] (Additional file 3). Like grammar, common usage tends to prevail, and this diatom became universally known as T. pseudonana. Importantly, none of the name changes was based on direct observations of the type material, and none either had [27,28] or took advantage of [26] the insights of a strongly supported phylogenetic hypothesis for the Thalassiosirales [25]. A combination of evidence (Figure 3, Additional files 3 and 4) shows that the diatom originally described by Hustedt [29] as C. nana corresponds to the traditional concept of T. pseudonana [29-34]. Furthermore, phylogenetic analyses show that T. pseudonana is part of a strongly supported clade that includes C. tecta, which is the nomenclatural type of the genus Cyclotella (Figure 1). By contrast—short of creating a genus that includes nearly the whole of Thalassiosirales—a genus that would include both T. pseudonana and T. nordenskioeldii (the type species of Thalassiosira) cannot be reconciled with the phylogeny (Figure 1). We are therefore compelled to resurrect the original name of T. pseudonana, Cyclotella nana [29], and deprecate T. pseudonana [28], C. pseudonana [27], and D. nana [26] as synonyms.
Conclusions
Publication of the T. pseudonana genome established this species as a model for experimental and genomic studies of diatoms [4], virtually all of which treat T. pseudonana as a marine diatom. Given the central roles of marine diatoms in global primary production, marine food webs, and the biogeochemical cycling of important nutrients, there is much to gain in understanding how marine diatoms interact with their environment. This understanding will come in large part from the development and study of model organisms [20] whose traits invariably have been shaped by their phylogeny. Phylogenetic analyses of both morphological and molecular data show that T. pseudonana likely descended from a freshwater ancestor. In some cases, this unusual history might confound extrapolations to strictly marine diatoms. Interestingly, the other diatom with a fully sequenced genome, P. tricornutum, also occurs in freshwater, brackish and marine habitats [50] and so might not be an ideal ecological model for marine diatoms either [20]. Additional genomes from ecologically diverse diatoms will bear these issues out and show, for example, whether the peculiar evolutionary histories of T. pseudonana and P. tricornutum account for some portion of the unusually large genomic divergence between the two species [6].
With few exceptions, diatoms are generally classified into genera that are uniformly marine or freshwater in distribution [51]. Although originally described from a predominantly freshwater river and classified in what is traditionally considered a freshwater genus (Cyclotella), an unfortunate series of nomenclatural changes ultimately landed T. pseudonana in a predominantly marine genus. Although the full extent to which T. pseudonana's name overshadowed the nuances of its ecology is a matter of speculation, T. pseudonana highlights the continued importance of taxonomy and systematics in the post-genomic era and echoes a view espoused by Linnaeus some 260 years ago in his landmark botanical text, Philosophia Botanica: "If you do not know the names of things, the knowledge of them is lost too." Beyond its name, a better appreciation of the complex phylogenetic and natural histories of T. pseudonana has the potential to shape and improve our understanding of diatom ecology and evolution more generally.
Methods
We examined isolectotype material for C. nana (= T. pseudonana) to resolve several taxonomic and nomenclatural uncertainties (Additional files 3, 4 and 5). The sample is sediment collected by Friedrich Hustedt [29] from the River Wümme near Bremen, Germany, and corresponds to lectotype slide BRM 380/36 from the diatom collection at the Alfred Wegener Institute (Additional file 5).
All diatom samples were soaked in 30% hydrogen peroxide overnight to dissolve organic matter, then rinsed with distilled water several times and concentrated onto filter pads. Dried filter pads were affixed to scanning electron microscope (SEM) stubs with double-sided adhesive carbon discs, and coated with 15 nm of Iridium using a Cressington 208 Bench Top Sputter Coater. SEM observations were made with a Zeiss Supra 40 VP.
Alverson et al. [25] used two chloroplast (psbC and rbcL) and two nuclear genes (SSU and partial LSU rDNA) to reconstruct the phylogeny of Thalassiosirales. The 82-taxon matrix was nearly complete, with one of the four genes missing for just three taxa [25]. The final tree (Figure 1) was based on a Bayesian analysis of all four concatenated genes (5,102 characters total). The protein-coding psbC and rbcL genes were treated as a single marker, with first and second codon positions (GTR + G + I) modeled separately from third codon positions (GTR + G + I) [52]. Separate GTR + G + I models were applied to the LSU rDNA gene and SSU rDNA loops, and the Doublet + G + I model was applied to the SSU rDNA stems [25]. Additional information about the data matrix, partitions, and exact model specifications can be found in the publicly available data file (identifier doi:10.5061/dryad.8661 at http://datadryad.org/). We performed four independent Bayesian runs, each of which used four Markov chains and was run for a total 40 million generations. The slide and cump analyses in the AWTY diagnostic software tool [53] were used to assess the stationarity of each run, and the AWTY compare analysis was used to determine whether the stationary phases of the four independent runs had sampled from the same posterior distribution. The latter analysis showed that that one of the four runs sampled a slightly different posterior distribution than the other three runs, so the final consensus tree (Figure 1) was based on the combined posterior distributions of the three similar runs.
Habitat (marine or freshwater) was scored based the natural provenance of the culture. The collection site for each culture is listed in Alverson et al. [25]. We scored strains collected from brackish and other low-salinity habitats as marine, thus biasing towards marine rather than freshwater ancestral reconstructions (all other things been equal). Maximum likelihood reconstructions used the Asymmetrical Markov k-state 2-parameter model as implemented in the Mesquite software package.
Several new freshwater species with gross morphological similarity to T. pseudonana and relatives have been described [23,24] since the publication of Alverson et al. [25]. We added these taxa to the morphological data matrix of Julius and Tanimura [22], and used the branch and bound search algorithm in PAUP* to find the most parsimonious trees for this matrix. Character descriptions are available in Julius and Tanimura [22]. All data matrices are available from Dryad (http://datadryad.org/) using identifiers doi:10.5061/dryad.8661 and doi:10.5061/dryad.8663.
Authors' contributions
AJA conceived the study, performed phylogenetic analyses, captured and analyzed SEM images, and drafted the manuscript. BB gathered, translated, and interpreted historical data on Cyclotella nana and helped in the interpretation of all results. MLJ gathered the morphological data used for phylogenetic analyses and helped in the interpretation of all results. ECT participated in study design and coordination, helped capture and analyze SEM images, and helped in the interpretation of all results. All authors read and approved the final manuscript.
Supplementary Material
Strict consensus of eight most parsimonious trees from a phylogenetic analysis of 32 morphological characters for select Thalassiosirales.
Pairwise differences among Thalassiosira pseudonana culture strains at four loci.
The nomenclatural history of Thalassiosira pseudonana.
The identity of Thalassiosira pseudonana.
Hustedt's observations on the type locality of Cyclotella nana (= T. pseudonana).
Scanning electron micrographs of thalassiosiroid diatoms in the type material (Additional file 4) of Cyclotella nana (= Thalassiosira pseudonana): Stephanodiscus hantzschii Grunow (A, B), Stephanodiscus minutulus (Kützing) Cleve & Möller (C, D), Discostella pseudostelligera (Hustedt) Houk & Klee (E-H), Cyclotella striata (Kützing) Grunow in Cleve & Grunow (I), Cyclotella atomus Hustedt (J), Cyclotella meneghiniana Kützing (K,L), Cyclostephanos invisitatus (Hohn & Hellerman) Theriot, Stoermer & Håkansson (M), Shionodiscus sp. (N), Thalassiosira sp. 1 (O), Thalassiosira sp. 2 (P). Scale bar = 2 μM.
Scanning electron micrographs showing exterior (A, B) and interior (C, D) views of Thalassiosira pseudonana (marine strain NEPC709) from Alverson et al. [25]. Scale bar = 2 μM.
Contributor Information
Andrew J Alverson, Email: alversoa@indiana.edu.
Bánk Beszteri, Email: Bank.Beszteri@AWI.DE.
Matthew L Julius, Email: mljulius@stcloudstate.edu.
Edward C Theriot, Email: etheriot@austin.utexas.edu.
Acknowledgements
We thank Alex Wolfe, Klaus Valentin, Davide Pisani, and two anonymous reviewers for critical comments. AJA was supported by an NIH Ruth L. Kirschstein NRSA Postdoctoral Fellowship (1F32GM080079-01A1). MLJ was partially supported by NSF grant DEB-0628554. ECT was supported by the Jane and Roland Blumberg Centennial Professorship in Molecular Evolution and NSF EF-0629410.
References
- Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009;19(2):R81–R88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- Mann DG, Droop SJM. Biodiversity, biogeography and conservation of diatoms. Hydrobiologia. 1996;336(1-3):19–32. doi: 10.1007/BF00010816. [DOI] [Google Scholar]
- Nelson DM, Treguer P, Brzezinski MA, Leynaert A, Queguiner B. Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem Cy. 1995;9(3):359–372. doi: 10.1029/95GB01070. [DOI] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M. et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science. 2004;306(5693):79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science. 2009;324(5935):1724–1726. doi: 10.1126/science.1172983. [DOI] [PubMed] [Google Scholar]
- Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP. et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456(7219):239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Armbrust EV, Rynearson TA, Jenkins BD. In: Genomics and Evolution of Microbial Eukaryotes. Katz LA, Bhattacharya D, editor. Oxford: Oxford Univeristy Press; 2006. Genomic insights into diatom evolution and metabolism; pp. 201–213. [Google Scholar]
- Guillard RRL, Kilham P, Jackson TA. Kinetics of silicon-limited growth in the marine diatom Thalassiosira pseudonana Hasle and Heimdal (Cyclotella nana Hustedt) J Phycol. 1973;9:233–237. [Google Scholar]
- Nelson DM, Goering JJ, Kilham SS, Guillard RRL. Kinetics of silicic-acid uptake and rates of silica dissolution in marine diatom Thalassiosira pseudonana. J Phycol. 1976;12(2):246–252. [Google Scholar]
- Paasche E. Silicon and the ecology of marine plankton diatoms. I Thalassiosira pseudonana (Cyclotella nana) grown in a chemostat with silicate as limiting nutrient. Mar Biol (Berl) 1973;19:117–126. doi: 10.1007/BF00353582. [DOI] [Google Scholar]
- Poulsen N, Chesley PM, Kroger N. Molecular genetic manipulation of the diatom Thalassiosira pseudonana (Bacillariophyceae) J Phycol. 2006;42(5):1059–1065. doi: 10.1111/j.1529-8817.2006.00269.x. [DOI] [Google Scholar]
- Bowler C, Vardi A, Allen AE. Oceanographic and biogeochemical insights from diatom genomes. Annu Rev Mar Sci. 2010;2:333–365. doi: 10.1146/annurev-marine-120308-081051. [DOI] [PubMed] [Google Scholar]
- Parker MS, Armbrust EV. Synergistic effects of light, temperature, and nitrogen source on transcription of genes for carbon and nitrogen metabolism in the centric diatom Thalassiosira pseudonana (Bacillariophyceae) J Phycol. 2005;41(6):1142–1153. doi: 10.1111/j.1529-8817.2005.00139.x. [DOI] [Google Scholar]
- Kustka AB, Allen AE, Morel FMM. Sequence analysis and transcriptional regulation of iron acquisition genes in two marine diatoms. J Phycol. 2007;43(4):715–729. doi: 10.1111/j.1529-8817.2007.00359.x. [DOI] [Google Scholar]
- Marchetti A, Parker MS, Moccia LP, Lin EO, Arrieta AL, Ribalet F, Murphy MEP, Maldonado MT, Armbrust EV. Ferritin is used for iron storage in bloom-forming marine pennate diatoms. Nature. 2009;457(7228):467–470. doi: 10.1038/nature07539. [DOI] [PubMed] [Google Scholar]
- Sumper M, Brunner E. Silica biomineralisation in diatoms: The model organism Thalassiosira pseudonana. ChemBioChem. 2008;9(8):1187–1194. doi: 10.1002/cbic.200700764. [DOI] [PubMed] [Google Scholar]
- Frigeri LG, Radabaugh TR, Haynes PA, Hildebrand M. Identification of proteins from a cell wall fraction of the diatom Thalassiosira pseudonana: Insights into silica structure formation. Mol Cell Proteomics. 2005;5:182–193. doi: 10.1074/mcp.M500174-MCP200. [DOI] [PubMed] [Google Scholar]
- Mock T, Samanta MP, Iverson V, Berthiaume C, Robison M, Holtermann K, Durkin C, BonDurant SS, Richmond K, Rodesch M. et al. Whole-genome expression profiling of the marine diatom Thalassiosira pseudonana identifies genes involved in silicon bioprocesses. P Natl Acad Sci USA. 2008;105(5):1579–1584. doi: 10.1073/pnas.0707946105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamatrakoln K, Hildebrand M. Analysis of Thalassiosira pseudonana silicon transporters indicates distinct regulatory levels and transport activity through the cell cycle. Eukaryot Cell. 2007;6(2):271–279. doi: 10.1128/EC.00235-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MS, Mock T, Armbrust EV. Genomic insights into marine microalgae. Annu Rev Genet. 2008;42:619–645. doi: 10.1146/annurev.genet.42.110807.091417. [DOI] [PubMed] [Google Scholar]
- Finlay BJ, Monaghan EB, Maberly SC. Hypothesis: The rate and scale of dispersal of freshwater diatom species is a function of their global abundance. Protist. 2002;153(3):261–273. doi: 10.1078/1434-4610-00103. [DOI] [PubMed] [Google Scholar]
- Julius ML, Tanimura Y. Cladistic analysis of plicated Thalassiosira (Bacillariophyceae) Phycologia. 2001;40(2):111–122. doi: 10.2216/i0031-8884-40-2-111.1. [DOI] [Google Scholar]
- Johansen J, Kociolek P, Lowe R. Spicaticribra kingstonii, gen. nov. et sp. nov. (Thalassiosiraceae, Bacillariophyta) from Great Smoky Mountains National Park, USA. Diatom Res. 2008;23(2):367–375. [Google Scholar]
- Stachura-Suchoples K, Williams DM. Description of Conticribra tricircularis, a new genus and species of Thalassiosirales, with a discussion on its relationship to other continuous cribra species of Thalassiosira Cleve (Bacillariophyta) and its freshwater origin. Eur J Phycol. 2009;44(4):477–486. doi: 10.1080/09670260903225431. [DOI] [Google Scholar]
- Alverson AJ, Jansen RK, Theriot EC. Bridging the Rubicon: Phylogenetic analysis reveals repeated colonizations of marine and fresh waters by thalassiosiroid diatoms. Mol Phylogenet Evol. 2007;45(1):193–210. doi: 10.1016/j.ympev.2007.03.024. [DOI] [PubMed] [Google Scholar]
- Chang TP, Chang-Scheider H. Zentrische kieselalgen in Kemptener seen [Centric diatoms in lakes of Kempten] Ber Deut Bot Ges. 2008;78:5–15. [Google Scholar]
- Chang TP, Steinberg C. Identifizierung von nanoplanktischen Kieselalgen (Centrales, Bacillariophyceae) in der Rott und im Rott-Stausee (Bayern, Bundesrepublik Deutschland) [Identification of nanoplanktonic diatoms (Centrales, Bacillariophyceae) in River Rott and Rott Reservoir (Bavaria, F.R.G.)] Arch Protistenkd. 1989;137(2):111–129. [Google Scholar]
- Hasle GR, Heimdal BR. Some species of the centric diatom genus Thalassiosira studied in the light and electron microscopes. Beihefte zur Nova Hedwigia. 1970;31:543–581. [Google Scholar]
- Hustedt F. Die Diatomeenflora des Flußsystems der Weser im Gebiet der Hansestadt Bremen [Diatom flora of the tributaries of the Weser near the city of Bremen] Abh Naturw Ver Bremen. 1957;34:181–440. [Google Scholar]
- Guillard RRL, Ryther JH. Studies of marine planktonic diatoms. I Cyclotella nana Hustedt and Detonula confervacea (Cleve) Gran. Can J Microbiol. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- Belcher JH, Swale EMF. Species of Thalassiosira (Diatoms, Bacillariophyceae) in the plankton of English rivers. Br Phycol J. 1977;12(3):291–297. doi: 10.1080/00071617700650311. [DOI] [Google Scholar]
- Hasle GR. The marine, planktonic diatoms Thalassiosira oceanica sp. nov. and T. partheneia. J Phycol. 1983;19(2):220–229. doi: 10.1111/j.0022-3646.1983.00220.x. [DOI] [Google Scholar]
- Hustedt F Die Diatomeenflora der Unterweser von der Lesummündung bis Bremerhaven mit Berücksichtigung des Unterlaufs der Hunte und Geeste [Diatom flora of the lower Weser from the Lesum estuary to Bremerhaven, with consideration of the lower Hunte and Geeste] Veröff Inst Meeresforsch 1959613–176.21575020 [Google Scholar]
- Lowe RL. Comparative ultrastructure of the valves of some Cyclotella species (Bacillariophyceae) J Phycol. 1975;11(4):415–424. [Google Scholar]
- Fryxell GA, Hasle GR. Thalassiosira eccentrica (Ehrenb.) Cleve, T. symmetrica sp. nov., and some related centric diatoms. J Phycol. 1972;8:297–317. [Google Scholar]
- Fryxell GA, Hasle GR. The genus Thalassiosira: some species with a modified ring of central strutted processes. Beiheft zur Nova Hedwigia. 1977;54:67–98. [Google Scholar]
- Fryxell GA, Hasle GR. The genus Thalassiosira: species with internal extensions of the strutted processes. Phycologia. 1979;18(4):378–393. doi: 10.2216/i0031-8884-18-4-378.1. [DOI] [Google Scholar]
- Hasle GR. Some Thalassiosira species with one central process. Norwegian Journal of Botany. 1978;25:77–110. [Google Scholar]
- Hasle GR, Fryxell GA. The genus Thalassiosira: some species with a linear areola array. Nova Hedwigia Beih. 1977;54:14–60. [Google Scholar]
- Hasle GR, Lange CB. Freshwater and brackish water Thalassiosira (Bacillariophyceae) taxa with tangentially undulated valves. Phycologia. 1989;28(1):120–135. doi: 10.2216/i0031-8884-28-1-120.1. [DOI] [Google Scholar]
- Håkansson H. A compilation and evaluation of species in the general Stephanodiscus, Cyclostephanos and Cyclotella with a new genus in the family Stephanodiscaceae. Diatom Res. 2002;17(1):1–139. [Google Scholar]
- Loginova LP. In: Proceedings of the 10th International Symposium on Recent and Fossil Diatoms. Simola H, editor. Koenigstein: Otto Koeltz Science Publishers; 1990. Classification of the diatom genus Cyclotella; pp. 37–46. [Google Scholar]
- Serieyssol KK. In: Proceedings of the Sixth Symposium on Recent and Fossil Diatoms. Ross R, editor. Koenigstein: Otto Koeltz Science Publishers; 1980. Cyclotella species of late Miocene age from St. Bauzile, France; pp. 27–42. [Google Scholar]
- Pisani D, Benton MJ, Wilkinson M. Congruence of morphological and molecular phylogenies. Acta Biotheor. 2007;55(3):269–281. doi: 10.1007/s10441-007-9015-8. [DOI] [PubMed] [Google Scholar]
- von Dassow P, Petersen TW, Chepurnov VA, Armbrust EV. Inter- and intraspecific relationships between nuclear DNA content and cell size in selected members of the centric diatom genus Thalassiosira (Bacillariophyceae) J Phycol. 2008;44(2):335–349. doi: 10.1111/j.1529-8817.2008.00476.x. [DOI] [PubMed] [Google Scholar]
- Maldonado MT, Allen AE, Chong JS, Lin K, Leus D, Karpenko N, Harris SL. Copper-dependent iron transport in coastal and oceanic diatoms. Limnol Oceanogr. 2006;51(4):1729–1743. doi: 10.4319/lo.2006.51.4.1729. [DOI] [Google Scholar]
- Strzepek RF, Harrison PJ. Photosynthetic architecture differs in coastal and oceanic diatoms. Nature. 2004;431(7009):689–692. doi: 10.1038/nature02954. [DOI] [PubMed] [Google Scholar]
- Wetzel RG. Limnology: Lake and River Ecosystems. Third. San Diego: Academic Press; 2001. Iron, sulfur, and silica cycles; pp. 289–331. [Google Scholar]
- Sunda WG, Huntsman SA. Iron uptake and growth limitation in oceanic and coastal phytoplankton. Mar Chem. 1995;50(1-4):189–206. doi: 10.1016/0304-4203(95)00035-P. [DOI] [Google Scholar]
- De Martino A, Meichenin A, Shi J, Pan KH, Bowler C. Genetic and phenotypic characterization of Phaeodactylum tricornutum (Bacillariophyceae) accessions. J Phycol. 2007;43(5):992–1009. doi: 10.1111/j.1529-8817.2007.00384.x. [DOI] [Google Scholar]
- Round FE, Sims PA. In: Proceedings of the Sixth Symposium on Recent and Fossil Diatoms. Ross R, editor. Hirschberg: Otto Koeltz Science Publishers; 1980. The distribution of diatom genera in marine and freshwater environments and some evolutionary considerations; pp. 301–320. [Google Scholar]
- Shapiro B, Rambaut A, Drummond AJ. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol. 2006;23(1):7–9. doi: 10.1093/molbev/msj021. [DOI] [PubMed] [Google Scholar]
- Wilgenbusch JC, Warren DL, Swofford DL. AWTY: A system for graphical exploration of MCMC convergence in Bayesian phylogenetic inference. 2004. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strict consensus of eight most parsimonious trees from a phylogenetic analysis of 32 morphological characters for select Thalassiosirales.
Pairwise differences among Thalassiosira pseudonana culture strains at four loci.
The nomenclatural history of Thalassiosira pseudonana.
The identity of Thalassiosira pseudonana.
Hustedt's observations on the type locality of Cyclotella nana (= T. pseudonana).
Scanning electron micrographs of thalassiosiroid diatoms in the type material (Additional file 4) of Cyclotella nana (= Thalassiosira pseudonana): Stephanodiscus hantzschii Grunow (A, B), Stephanodiscus minutulus (Kützing) Cleve & Möller (C, D), Discostella pseudostelligera (Hustedt) Houk & Klee (E-H), Cyclotella striata (Kützing) Grunow in Cleve & Grunow (I), Cyclotella atomus Hustedt (J), Cyclotella meneghiniana Kützing (K,L), Cyclostephanos invisitatus (Hohn & Hellerman) Theriot, Stoermer & Håkansson (M), Shionodiscus sp. (N), Thalassiosira sp. 1 (O), Thalassiosira sp. 2 (P). Scale bar = 2 μM.
Scanning electron micrographs showing exterior (A, B) and interior (C, D) views of Thalassiosira pseudonana (marine strain NEPC709) from Alverson et al. [25]. Scale bar = 2 μM.