Abstract
Background
Peroxiredoxin 6 (PRDX6) is involved in redox regulation of the cell and is thought to be protective against oxidant injury. Little is known about genetic variation within the PRDX6 gene and its association with acute lung injury (ALI). In this study we sequenced the PRDX6 gene to uncover common variants, and tested association with ALI following major trauma.
Methods
To examine the extent of variation in the PRDX6 gene, we performed direct sequencing of the 5' UTR, exons, introns and the 3' UTR in 25 African American cases and controls and 23 European American cases and controls (selected from a cohort study of major trauma), which uncovered 80 SNPs. In silico modeling was performed using Patrocles and Transcriptional Element Search System (TESS). Thirty seven novel and tagging SNPs were tested for association with ALI compared with ICU at-risk controls who did not develop ALI in a cohort study of 259 African American and 254 European American subjects that had been admitted to the ICU with major trauma.
Results
Resequencing of critically ill subjects demonstrated 43 novel SNPs not previously reported. Coding regions demonstrated no detectable variation, indicating conservation of the protein. Block haplotype analyses reveal that recombination rates within the gene seem low in both Caucasians and African Americans. Several novel SNPs appeared to have the potential for functional consequence using in silico modeling. Chi2 analysis of ALI incidence and genotype showed no significant association between the SNPs in this study and ALI. Haplotype analysis did not reveal any association beyond single SNP analyses.
Conclusions
This study revealed novel SNPs within the PRDX6 gene and its 5' and 3' flanking regions via direct sequencing. There was no association found between these SNPs and ALI, possibly due to a low sample size, which was limited to detection of relative risks of 1.93 and above. Future studies may focus on the role of PRDX6 genetic variation in other diseases, where oxidative stress is suspected.
Keywords: Peroxiredoxin, Acute Lung Injury, Oxidant Stress, Genetic Polymorphisms
Background
Acute Respiratory Distress Syndrome (ARDS) and Acute Lung Injury (ALI) affect 100,000-150,000 patients each year in the United States alone [1,2]. ALI is an inflammatory syndrome characterized by acute respiratory failure due to non-cardiogenic pulmonary edema and hypoxemia [3]. Oxidant stress caused by reactive oxygen species (ROS) is thought to be a major contributor to the pathogenesis of ALI. ROS can be generated by inflammatory cells or pulmonary endothelium and cause damage to proteins, DNA, and lipids [4].
The risk of developing ALI/ARDS is not uniformly distributed in the critically ill population, suggesting a genetic influence on outcomes [5]. Peroxiredoxins are a superfamily of non-heme and non-selenium peroxidases that are widely distributed throughout all phyla [6]. The Peroxiredoxin 6 gene (PRDX6) is located on chromosome 1q24 and is approximately 12 Kb in length, containing 5 exons. The Prdx6 protein encoded by PRDX6 is involved in redox regulation of the cell and has been shown in cell and animal models to be protective against oxidative injury [7]. Prdx6 also has phospholipase A2 activity and has an important role in lung surfactant metabolism [7]. The protein product, Prdx6, has been shown to affect the cellular level of H2O2 produced in cells stimulated with platelet-derived growth factor or tumor necrosis factor-α, and modulating signaling induced by those ligands [8], thus indicating that Prdx6 can have an effect on cytokine levels and cell signaling cascades. Recent studies suggest that Prdx6 is only active following heterodimerization with glutathione-S-transferase pi, indicating that there is an important interaction between Prdx6 and GSTpi [6]. Despite these important functions, little is known about genetic variation within the PRDX6 gene [9] and its association with ALI.
In order to determine if variation within PRDX6 is associated with ALI risk in either the African American (AA) or European American (EA) populations, we performed direct sequencing of the 5' UTR, exons, introns, and the 3' untranslated region (UTR) in 48 subjects (25 African Americans and 23 European Americans) and identified 80 variants, many of which have not been previously reported. Eighteen of the eighty variants, along with 19 tagging SNPs selected using HapMap http://hapmap.ncbi.nlm.nih.gov/, were tested for association with ALI using a custom genotyping platform.
Methods
Patient population
Between 1999 and 2006, patients were enrolled in a major trauma cohort study designed to study molecular risks for acute lung injury [10-12]. Participants met the following inclusion criteria: 1) admission to the intensive care unit (ICU) as a result of acute trauma directly from the field or via that hospital's Emergency Department; and 2) have an Injury Severity Score (ISS) ≥ 16 as calculated on the basis of information available during their first 24 hours of hospitalization. The following demographic and clinical variables were collected upon admission to the ICU: age, gender, ISS, blunt mechanism, and acute physiology and chronic health evaluation (APACHE) (Table 1). Exclusion criteria were death or discharge from the ICU within 24 hours of admission, less than 13 years of age, current or past evidence of congestive heart failure (CHF) or recent acute myocardial infarction, severe chronic respiratory disease, morbid obesity, burns on over 30% of the total body surface area, and lung or bone marrow transplant [10].
Table 1.
Clinical data for individuals enrolled in the study by ancestry
| ALI | No ALI | |
|---|---|---|
| African Americans (N = 285) | (N = 71) | (N = 187) |
| Age (± SD) | 36.7 ± 17.5 | 31.9 ± 12.4 |
| Gender (% of males ) | 32 | 84 |
| ISS (± SD) | 25.3 ± 8.9 | 22.8 ± 6.3 |
| Blunt Mechanism (%) (% of Blunt Trauma) |
25 | 59 |
| APACHE (± SD) | 45.4 ± 12.4 | 40.2 ± 13.5 |
| European Americans (N = 269) | (N = 86) | (N = 168) |
| Age (± SD) | 41.8 ± 19.9 | 39.3 ± 18.5 |
| Gender (% of males ) | 27 | 61 |
| ISS (± SD) | 26.9 ± 7.5 | 25.2 ± 7.2 |
| Blunt Mechanism (%) | 4 | 7 |
| APACHE (± SD) | 41.9 ± 14.7 | 35.7 ± 12.8 |
The definition of ALI was in accordance with the American European Consensus Conference (AECC) [3]. ALI and ARDS were defined as: acute onset; bilateral pulmonary infiltrates on chest X-ray consistent with pulmonary edema; absence of evidence of left atrial hypertension; and poor systemic oxygenation, and a ratio of arterial oxygen (PaO2) to the fraction of inspired oxygen (FiO2) less than or equal to 300 for ALI and 200 for ARDS [3]. All chest x-rays were reviewed independently by 2 trained observers. In our population, greater than 85% of subjects meeting criteria for ALI also met criteria for ARDS.
Clinical Data and Biosample Collection
Clinical data were collected by trained study nurses using a standardized research case report form designed for the trauma cohort study. Blood for analysis was obtained from residual blood samples in tubes containing ethyledenediaminetetraacetic acid (EDTA) that had been previously drawn for other clinical purposes. Study personnel collected residual samples each day, centrifuged, and separated the buffy coat layers, which were frozen at -80°C [10]. All clinical and biosample collection protocols were approved by the institutional review board (IRB) at the University of Pennsylvania School of Medicine under a waiver of informed consent.
PRDX6 resequencing
Genomic DNA was extracted from whole blood using Qiagen Qiamp DNA Blood Midi Kits (Qiagen USA) and stored in the provided tris-EDTA buffer. DNA from 25 African American and 23 European American subjects selected from the major trauma cohort, with ALI status equally distributed within each group, were selected for sequencing of PCR fragments, providing a power of 99% to detect minor allele frequencies of at least 5% [13]. PCR primers for 4 Kb upstream of the ATG start site, all exons, introns, and 4 Kb of the 3' UTR were designed using PCRoverlap (Children's Hospital of Philadelphia (CHOP) bioinformatics core) to generate amplicons between 600 and 800 bp that overlapped by at least 100 bp (Additional file 1). Following primer optimization, DNA was amplified and sequenced in the forward and reverse direction using a 3730 automated sequencer at the CHOP Nucleic Acid and Protein Core facility (Philadelphia, PA). Sequencher 4.8 (Gene Codes Ann Arbor, MI) was used to facilitate secondary peak calls and to compare the sequence data to the NCBI reference sequence.
SNP genotyping
Novel variants with a minor allele frequency (MAF) of 0.04 or higher and tagging SNPs, from HapMap http://hapmap.ncbi.nlm.nih.gov/, were validated using the SNPlex genotyping system (Applied Biosystems Inc. Foster City, CA). Tagging SNPs were selected using Tagger's pairwise testing methods described by Bakker and colleagues [14]. Genotyping novel variants not only served to test for association, but allowed us to validate those SNPs in a larger population. Tagging SNPs were also selected to provide better coverage of the haplotype structure of PRDX6. SNPlex utilizes an oligonucleotide ligation/PCR assay with universal ZipChute probe detection to perform genotyping of up to 48 SNPs in a single reaction. ZipChute probes were custom designed and detected by capillary electrophoresis using the Applied Biosystems 3130 Analyzer and genotype calls were determined using Gene Mapper 4.0 (Applied Biosystems Foster City, CA).
All genotyping was performed in the University of Pennsylvania's Laboratory for Molecular Epidemiology (LME). Staff was blinded to the disease status and genotyping calls were performed in subsamples by plate. Each plate contained six positive controls to test for concordance. Genotyping calls were performed automatically using the algorithm described by Da La Vega and colleagues [15].
In silico modeling of putative function in SNP sites
We sought to test inferred function in silico using transcription factor binding and mRNA binding tools. TESS is a web-based software tool for locating possible transcription factor binding sites in DNA sequences using weight matrix models. It can also be used for browsing information about relevant transcription factors in the TRANSFAC database [16]. All SNPs discovered within the 5' UTR and the first intron were submitted to TESS as 21 base pair long FASTA sequences with the reference allele of the SNP of interest in the 11th position. A second search was performed using the alternative allele in the 11th position. To eliminate any poor matches due to background noise, transcription factors with log-likelihood scores (La) less than 12, were eliminated. TESS results were compared with experimental transcription factor binding site (TFBS) data registered in the University of California Santa Cruz (UCSC) Genome Browser by the Encyclopedia of DNA Elements (ENCODE) consortium [17]. The ENCODE data were filtered by chromosome and position.
A search query was performed for potential miRNA binding sites in the 3' UTR of PRDX6 using Patrocles http://www.patrocles.org/. Patrocles is an online database containing DNA sequence polymorphisms that are predicted to interrupt miRNA-mediated gene regulation [18]. The search was performed using "PRDX6" as a key word in the target gene id field and miRNA target motifs were defined by Xie et al. [19] and Lewis et al [20].
Statistical Analysis of ALI association
259 African American and 254 European American subjects enrolled in the major trauma cohort were used to test for association of novel variants and tagging SNPs with ALI. Association of each PRDX6 SNP with ALI was determined separately for European Americans and African Americans using an additive model Chi2 test, with a p-value < 0.0014 for African Americans considered significant. Dominant and recessive inheritance models were also tested using Chi2 analysis. Multivariable analyses of potential confounding were performed using logistic regression methods. Power was calculated using the power for genetic association analyses (PGA) [21]. Using PGA, we estimated that a sample size of 250 subjects per race category would provide 80% power to detect relative risks of 2.26 or greater for SNPs with a prevalence of 0.05 or greater and 1.93 or greater for SNPs with a prevalence of 0.10 or greater, assuming a Bonferroni-corrected alpha = 0.0014 for African Americans, and an incidence of ALI = 0.30 (Additional file 2). These statistical analyses were performed using STATA 11 (STATA Data Corp, College Station, TX). Pairwise linkage disequilibrium was evaluated using Haploview http://www.broadinstitute.org/mpg/haploview. Genotypes with a completion rate of 95% or greater were considered for analysis in Haploview. LD was calculated in terms of r2 values and blocks were defined using the default algorithm using the confidence intervals methods of Gabriel and colleagues [22].
Haplotypes were inferred using the standard expectation maximization algorithm in Haploview [23,24] and the following confidence interval (CI) criteria: CI minima for strong LD: 0.7 - 0.98; upper CI maximum for strong LD: 0.98; fraction of strong LD in informative comparisons ≥ 0.95; and exclude markers with minor allele frequency (MAF) < 0.05. Haplotypes were tested for association with ALI first in a global association test, which performed contingency testing using all haplotypes of an LD block compared to no haplotypes, and then as individual haplotypes versus ALI coded in an additive fashion PLINK [25]. Haplotype multiple testing was addressed by applying permutation tests (10,000 permutations).
Results
Identification of novel polymorphisms in PRDX6
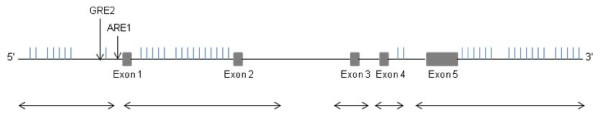
Direct sequencing of the PRDX6 gene in 48 subjects revealed 80 genetic variants, none of which were in coding regions (31 in the 3' UTR, 22 in the 5' UTR and 27 intronic) (Additional file 3). The variants identified via direct sequencing were compared with those registered in the NCBI dbSNP database (Build 130) and 43 were found to be novel SNPs (Table 2). Thirty seven were matched with SNPs catalogued in dbSNP (Build 130) and Genewindow http://genewindow.nci.nih.gov/Welcome based on chromosome position (Table 3). Twenty five of the novel SNPs uncovered had a MAF > 0.04 and were submitted to the NCBI to be catalogued and assigned ss numbers in the submitter records section of dbSNP (Table 2). Novel SNPs were also compared with SNPs registered in the 1000 Genomes database. Thirty six out of thirty seven known SNPs overlapped with SNPs registered in 1000 Genomes, but only sixteen out of forty-three novel SNPs identified via our sequencing effort were also registered in 1000 Genomes (Table 4). Several variants were only observed in one individual. As a quality control measure we present the confidence scores for these genotypes in additional file 4. Confidence scores are reported as the percentage of overlap between heterozygote peaks. Previous studies indicated that two transcription factor binding sites, the ARE1 (-357 to -349) [26] and GRE2 (-750 to -738) (A. Fisher, unpublished observations), may play a role in the regulation of PRDX6. We were unable to sequence the ARE1 region and portions of the intronic regions due to the GC rich content of the flanking sequence (Figure 1). The GRE2 region was successfully sequenced, but showed no variation.
Table 2.
Novel SNPs discovered via direct sequencing
| Novel SNPs | Region | Chr. Position | SNP | 5' Flanking Sequence | 3' Flanking Sequence | MAF in EA | MAF in AA | ss# (dbSNP) |
|---|---|---|---|---|---|---|---|---|
| PRDX6_171709541 | 5' UTR | 171709541 | G/C | CTTCAAGGTTC | ACCCTTATAGC | 0.04 | 0.02 | ss217326279 |
| PRDX6_171709910 | 5' UTR | 171709910 | G/T | ATGATCATTTTT | GAAATATACAG | 0.00 | 0.10 | ss217326288 |
| PRDX6_171710327 | 5' UTR | 171710327 | C/T | ACCCTAGCCCC | TGTGCTGGCA | 0.04 | 0.00 | ss217326273 |
| PRDX6_171710490 | 5' UTR | 171710490 | C/T | TGCACTGCGGA | GCAGGGACCT | 0.00 | 0.06 | ss217326283 |
| PRDX6_171710775 | 5' UTR | 171710775 | G/C | CTTATGGCTGG | GTGAGACATG | 0.00 | 0.02 | |
| PRDX6_171710821 | 5' UTR | 171710821 | C/T | ACTGCACTGAG | TTGTGTAAAGT | 0.00 | 0.10 | ss217326292 |
| PRDX6_171711029 | 5' UTR | 171711029 | C/T | ACTCAGAGACC | GGGTCCTCCG | 0.00 | 0.02 | |
| PRDX6_171712345 | 5' UTR | 171712345 | A/T | ATGGTTCATAA | AGAAAGGGGA | 0.87 | 0.64 | ss217326296 |
| PRDX6_171713694 | intron | 171713694 | G/T | TCACTTCCCCG | AGTGCCCAGG | 0.00 | 0.04 | ss217326300 |
| PRDX6_171713738 | intron | 171713738 | G/T | CCTCCGTTCTG | TGCTCCCTGG | 0.00 | 0.02 | |
| PRDX6_171713872 | intron | 171713872 | C/T | GCACAAAATGT | TAAAACCACTA | 0.00 | 0.12 | ss217326323 |
| PRDX6_171713919 | intron | 171713919 | G/C | AAAGACTTTTTG | AGCCGCCTCC | 0.02 | 0.00 | |
| PRDX6_171714107 | intron | 171714107 | C/T | CCAGGACACGT | TCCCCAACTTT | 0.00 | 0.04 | ss217326304 |
| PRDX6_171714984 | intron | 171714984 | C/T | GATCAAAAGTG | TTATCAGGGAG | 0.04 | 0.04 | ss217326307 |
| PRDX6_171715019 | intron | 171715019 | A/G | AGGAACACGGT | TATCTGCATTT | 0.00 | 0.10 | ss217326318 |
| PRDX6_171715596 | intron | 171715596 | A/G | GGGAGGGAAG | TGAACTGGCTT | 0.00 | 0.02 | |
| PRDX6_171716007 | intron | 171716007 | A/G | AAACCTTGGGA | GTGGCAGCCG | 0.00 | 0.04 | ss217326311 |
| PRDX6_171716032 | intron | 171716032 | G/C | TAAGTAGGAAG | TGCCCTTGTCT | 0.00 | 0.02 | |
| PRDX6_171716554 | intron | 171716554 | A/C | AGAAGCCAAGT | AACTTTAATTTT | 0.00 | 0.02 | |
| PRDX6_171716572 | intron | 171716572 | A/T | TCAACTTTAATT | TAAATAGAAGA | 0.00 | 0.02 | |
| PRDX6_171716582 | intron | 171716582 | A/G | TTACATATAAAT | ATAGAAACCTA | 0.00 | 0.04 | ss217326315 |
| PRDX6_171716584 | intron | 171716584 | A/T | ACATATAAATAG | AGAAACCTATT | 0.00 | 0.02 | |
| PRDX6_171716603 | intron | 171716603 | A/G | AACCTATTTATT | ATTACATAATTT | 0.00 | 0.02 | |
| PRDX6_171723151 | Intron | 171723151 | C/T | AAAGCTAGCAT | TGGAGAAGAA | 0.00 | 0.02 | |
| PRDX6_171723403 | Intron | 171723403 | C/T | CTTGATTAGTCT | AGCACCTGTAG | 0.00 | 0.02 | |
| PRDX6_171723889 | 3'UTR | 171723889 | G/T | AAAACTCAAAT | GGATCTCTGCA | 0.00 | 0.04 | |
| PRDX6_171723918 | 3'UTR | 171723918 | A/G | GCTTGTGACCA | GTCATATTTGT | 0.00 | 0.02 | |
| PRDX6_171724000 | 3'UTR | 171724000 | G/C | TAACTGTCCTAT | TCCTCTCCTGT | 0.00 | 0.04 | ss217326227 |
| PRDX6_171724128 | 3'UTR | 171724128 | G/T | TTTTTTTAATAT | TGATCACAGAA | 0.00 | 0.04 | ss217326232 |
| PRDX6_171724182 | 3'UTR | 171724182 | A/G | CATATTCTTTTA | TCTTGATCACA | 0.00 | 0.04 | ss217326236 |
| PRDX6_171724286 | 3'UTR | 171724286 | A/T | TTGCTATAAAAA | TTTGTGATAAG | 0.00 | 0.02 | |
| PRDX6_171724949 | 3'UTR | 171724949 | C/T | ACTCTACTAATA | CAGGTTTAGAA | 0.26 | 0.00 | ss217326270 |
| PRDX6_171725122 | 3'UTR | 171725122 | G/T | GGACCTGCTTC | TTGTAGTTTGC | 0.00 | 0.04 | ss217326239 |
| PRDX6_171725183 | 3'UTR | 171725183 | C/T | GGGATCATCGC | GTCTCATAAGG | 0.00 | 0.04 | ss217326242 |
| PRDX6_171725257 | 3'UTR | 171725257 | A/T | CCTCCCAAAGG | CATCCAAATAC | 0.00 | 0.04 | ss217326247 |
| PRDX6_171725485 | 3'UTR | 171725485 | C/T | CCTGCCTCAGC | GAGCAGCTGG | 0.00 | 0.06 | ss217326250 |
| PRDX6_171725681 | 3'UTR | 171725681 | G/T | ATATTTTTATTG | TAGAATAATGT | 0.00 | 0.02 | |
| PRDX6_171725826 | 3'UTR | 171725826 | A/G | TCTGGGGAATG | TTTGAAAGAGA | 0.04 | 0.00 | ss217326254 |
| PRDX6_171727146 | 3'UTR | 171727146 | G/C | CTGTGATTCCT | TTGTGGTCTTG | 0.02 | 0.00 | |
| PRDX6_171727831 | 3'UTR | 171727831 | C/T | ATGCATGGGAT | ATTATCCTCTA | 0.04 | 0.02 | ss217326261 |
| PRDX6_171728416 | 3'UTR | 171728416 | G/T | CCTCATTAGGG | CTCTTAGCCCT | 0.04 | 0.02 | ss217326266 |
| PRDX6_171728455 | 3'UTR | 171728455 | G/C | AATCGGGAGGC | TGTTAACAGGT | 0.00 | 0.04 | ss217326257 |
| PRDX6_171729049 | 3'UTR | 171729049 | C/T | GTTCTTAAACTA | AATAGCATGAG | 0.00 | 0.02 | |
Table 3.
Known SNPs discovered via direct sequencing
| Knowns SNPs | Region | Chr. Position | SNP | MAF in EA (discovery) | MAF in AA (discovery) | MAF in EA (dbSNP) | MAF in AA (dbSNP) |
|---|---|---|---|---|---|---|---|
| rs13376447 | 5' UTR | 171709896 | A/G | 0.00 | 0.06 | 0.01 | 0.09 |
| rs10753081 | 5' UTR | 171710154 | C/T | 0.39 | 0.30 | 0.73 | 0.21 |
| rs34282688 | 5' UTR | 171710819 | C/T | 0.07 | 0.00 | NA | NA |
| rs9425722 | 5' UTR | 171711268 | C/T | 1.02 | 0.70 | 0.04 | 0.24 |
| rs35152701 | 5' UTR | 171711269 | A/G | 0.02 | 0.00 | 0.00 | 0.00 |
| rs12739142 | 5' UTR | 171711278 | A/G | 0.04 | 0.00 | 0.02 | 0.04 |
| rs4354572 | 5' UTR | 171711459 | C/T | 1.02 | 0.66 | 0.00 | 0.04 |
| rs34619706 | 5' UTR | 171711670 | A/G | 0.09 | 0.02 | 0.07 | 0.04 |
| rs4382766 | 5' UTR | 171711699 | C/T | 0.54 | 0.28 | 0.30 | 0.50 |
| rs13376392 | 5' UTR | 171711701 | C/T | 0.00 | 0.06 | 0.00 | 0.04 |
| rs11576174 | 5' UTR | 171712301 | G/T | 0.04 | 0.00 | 0.15 | 0.00 |
| rs34977864 | 5' UTR | 171712466 | G/T | 0.00 | 0.06 | 0.00 | 0.10 |
| rs35441546 | 5' UTR | 171712652 | C/T | 0.00 | 0.06 | 0.00 | 0.03 |
| rs35133735 | 5' UTR | 171712738 | C/T | 0.00 | 0.06 | 0.00 | 0.00 |
| rs6671141 | intron | 171713557 | G/T | 0.26 | 0.26 | 0.24 | 0.30 |
| rs35749242 | intron | 171714171 | A/G | 0.00 | 0.12 | 0.00 | 0.03 |
| rs35918328 | intron | 171714199 | A/G | 0.00 | 0.02 | 0.00 | 0.10 |
| rs35899698 | intron | 171714279 | C/T | 0.00 | 0.10 | 0.00 | 0.00 |
| rs33942654 | intron | 171714884 | A/G | 0.26 | 0.18 | 0.23 | 0.33 |
| rs35679908 | intron | 171715013 | A/G | 0.00 | 0.02 | 0.00 | 0.00 |
| rs9425723 | intron | 171715118 | A/G | 0.33 | 0.62 | 0.23 | 0.63 |
| rs9425724 | intron | 171715123 | A/G | 0.33 | 0.54 | 0.23 | 0.53 |
| rs7540065 | intron | 171715659 | A/G | 0.33 | 0.56 | 0.23 | 0.53 |
| rs35244306 | intron | 171716247 | C/T | 0.00 | 0.14 | 0.00 | 0.07 |
| rs4611 | 3'UTR | 171723640 | C/T | 0.33 | 0.50 | 0.77 | 0.47 |
| rs3833536 | 3'UTR | 171724172 | C/- | 0.00 | 0.00 | 0.05 | 0.04 |
| rs7314 | 3'UTR | 171724222 | A/G | 0.20 | 0.50 | 0.23 | 0.52 |
| rs36005931 | 3'UTR | 171724224 | A/G | 0.00 | 0.04 | 0.00 | 0.10 |
| rs2000 | 3'UTR | 171724457 | A/G | 0.04 | 0.00 | 0.03 | 0.00 |
| rs34129563 | 3'UTR | 171724720 | G/C | 0.11 | 0.00 | 0.02 | 0.03 |
| rs9425727 | 3'UTR | 171725216 | G/C | 0.02 | 0.14 | 0.00 | 0.04 |
| rs35358649 | 3'UTR | 171725429 | C/T | 0.02 | 0.00 | 0.00 | 0.00 |
| rs35547740 | 3'UTR | 171725569 | (-/T) | 0.00 | 0.04 | 0.00 | 0.07 |
| rs6702835 | 3'UTR | 171725723 | A/G | 0.33 | 0.48 | 0.23 | 0.54 |
| rs60587131 | 3'UTR | 171726777 | G/C | 0.00 | 0.02 | 0.05 | NA |
| rs57032935 | 3'UTR | 171726836 | G/C | 0.15 | 0.22 | 0.18 | 0.31 |
| rs6664925 | 3'UTR | 171729033 | A/G | 0.00 | 0.18 | 0.00 | 0.16 |
Table 4.
Comparison of PRDX6 discovery SNPs with 1000 Genomes database
| PRDX6 SNPs | 1000 Genomes Comparison | PRDX6 SNPs | 1000 Genomes Comparison |
|---|---|---|---|
| PRDX6_171709910 | Matched | PRDX6_171709541 | Not Matched |
| PRDX6_171713872 | Matched | PRDX6_171710327 | Not Matched |
| PRDX6_171714107 | Matched | PRDX6_171710490 | Not Matched |
| PRDX6_171723151 | Matched | PRDX6_171710775 | Not Matched |
| PRDX6_171723918 | Matched | PRDX6_171710821 | Not Matched |
| PRDX6_171724000 | Matched | PRDX6_171711029 | Not Matched |
| PRDX6_171724128 | Matched | PRDX6_171712345 | Not Matched |
| PRDX6_171724182 | Matched | PRDX6_171713694 | Not Matched |
| PRDX6_171725122 | Matched | PRDX6_171713738 | Not Matched |
| PRDX6_171725183 | Matched | PRDX6_171713919 | Not Matched |
| PRDX6_171725257 | Matched | PRDX6_171714984 | Not Matched |
| PRDX6_171725826 | Matched | PRDX6_171715019 | Not Matched |
| PRDX6_171727146 | Matched | PRDX6_171715596 | Not Matched |
| PRDX6_171727831 | Matched | PRDX6_171716007 | Not Matched |
| PRDX6_171728416 | Matched | PRDX6_171716032 | Not Matched |
| PRDX6_171728455 | Matched | PRDX6_171716554 | Not Matched |
| rs10753081 | Matched | PRDX6_171716572 | Not Matched |
| rs11576174 | Matched | PRDX6_171716582 | Not Matched |
| rs12739142 | Matched | PRDX6_171716584 | Not Matched |
| rs13376392 | Matched | PRDX6_171716603 | Not Matched |
| rs13376447 | Matched | PRDX6_171723403 | Not Matched |
| rs2000 | Matched | PRDX6_171723889 | Not Matched |
| rs33942654 | Matched | PRDX6_171724286 | Not Matched |
| rs34129563 | Matched | PRDX6_171724949 | Not Matched |
| rs34282688 | Matched | PRDX6_171725485 | Not Matched |
| rs34619706 | Matched | PRDX6_171725681 | Not Matched |
| rs34977864 | Matched | PRDX6_171729049 | Not Matched |
| rs35133735 | Matched | rs3833536 | Not Matched |
| rs35152701 | Matched | ||
| rs35244306 | Matched | ||
| rs35358649 | Matched | ||
| rs35441546 | Matched | ||
| rs35547740 | Matched | ||
| rs35679908 | Matched | ||
| rs35749242 | Matched | ||
| rs35899698 | Matched | ||
| rs35918328 | Matched | ||
| rs36005931 | Matched | ||
| rs4354572 | Matched | ||
| rs4382766 | Matched | ||
| rs4611 | Matched | ||
| rs57032935 | Matched | ||
| rs60587131 | Matched | ||
| rs6664925 | Matched | ||
| rs6671141 | Matched | ||
| rs6702835 | Matched | ||
| rs7314 | Matched | ||
| rs7540065 | Matched | ||
| rs9425722 | Matched | ||
| rs9425723 | Matched | ||
| rs9425724 | Matched | ||
| rs9425727 | Matched | ||
Figure 1.
PRDX6 gene diagram with novel SNP positions. A schematic of the PRDX6 gene is presented with vertical lines and arrows above the gene indicating novel SNPs and regions of interest, respectively. The horizontal arrows below the schematic are representative of the regions of PRDX6 that were successfully sequenced.
In Silico function of novel SNPs in PRDX6
The TESS results showed several potential transcription factor binding motifs in both the reference and alternative sequence. The reference and alternative sequences were submitted as independent queries and transcription factors were returned for 19 positions in the reference sequence and 21 positions in the alternative sequence (Table 5). Twenty seven out of twenty nine sequences submitted were shown to create, abolish, or change a transcription factor binding site. Fourteen of these SNPs were novel. Comparison of the transcription factors returned from the TESS query with the data from ENCODE showed that only 3 of these putative transcription factor binding sites have been tested by the ENCODE consortium, SP1, GATA-1, and c-Myc. ENCODE data for SP1, GATA-1, and c-Myc revealed that there is no evidence of binding affinity with the sequence results from the PRDX6 gene when filtered for PRDX6 and A549 cells.
Table 5.
Potential transcription factor binding sites within the PRDX6 gene
| SNP | Location | Base Change | Reference Sequence | Alternative Sequence |
|---|---|---|---|---|
| PRDX6_171709541 | 5' UTR | G/C | n/a | Sp1, CACCC binding factor, PuF |
| rs13376447 | 5' UTR | A/G | GATA-1 | GATA-1 |
| PRDX6_171709910 | 5' UTR | G/T | n/a | IL-6 RE-BP |
| rs10753081 | 5' UTR | C/T | GATA-3 | GATA-1 |
| PRDX6_171710327 | 5' UTR | C/T | C/EBPbeta | C/EBPbeta |
| PRDX6_171710775 | 5' UTR | G/C | c-Myb | n/a |
| rs9425722 | 5' UTR | C/T | n/a | c-Myc |
| rs35152701 | 5' UTR | A/G | n/a | TMF, TBP, TFIID |
| rs12739142 | 5' UTR | A/G | IPF1, Isl-1, IPF1 | IPF1 |
| rs4354572 | 5' UTR | C/T | C/EBPbeta | AFP1, ATBF1-B |
| rs11576174 | 5' UTR | G/T | H4TF-1 | ETF, TMF, TFIID, TBP |
| PRDX6_171712345 | 5' UTR | A/T | n/a | SRY, TCF-4E |
| PRDX6_171712345 | 5' UTR | A/T | n/a | SRY |
| rs35441546 | 5' UTR | C/T | Sp1, ETF | CACCC-binding factor, Sp1 |
| rs35133735 | 5' UTR | C/T | n/a | CACCC-binding factor, Sp1 |
| rs6671141 | intron | G/T | GR alpha, PR, PR A, GR beta | n/a |
| PRDX6_171713738 | intron | G/T | n/a | TEF-1 |
| PRDX6_171713872 | intron | C/T | n/a | H4TF-1 |
| rs35918328 | intron | A/G | IPF1 | n/a |
| rs33942654 | intron | A/G | c-Myb | n/a |
| PRDX6_171714984 | intron | C/T | TBP | GATA-1 |
| PRDX6_171715019 | intron | A/G | GATA-3 | GATA-1 |
| rs9425723 | intron | A/G | TBP, TFIID | AP-1, c-Jun |
| rs9425724 | intron | A/G | USF1 | AP-1, AP-4, CCK-1a, c-Myc, CREB, Max1, USF1 |
| PRDX6_171715596 | intron | A/G | c-Myb, c-Ets-2 | n/a |
| PRDX6_171716007 | intron | A/G | n/a | Sp1, C/EBPbeta, CACCC-binding factor |
| PRDX6_171716572 | intron | A/T | TMF, TFIID, ETF, TBP | n/a |
| PRDX6_171716582 | intron | A/G | GATA-1, GATA-3 | n/a |
| PRDX6_171716584 | intron | A/T | GATA-1, GATA-3 | SRY |
*Transcription factor binding site modeling was performed using TESS.
A Patrocles miRNA database search for PRDX6 revealed eight SNPs in the 3' UTR of PRDX6 as potential miRNA binding sites (Table 6). Of the eight SNPs returned from the search query, three matched SNPs from this study (rs4611, rs36005931, and rs2000). rs4611 and rs36005931 are located within octamers that have been conserved among several species, but do not correspond to a known miRNA. The G allele of rs2000 is part of an octamer capable of binding miR-942. A literature search for miR-942 returned only sequence data, with no known function to date.
Table 6.
PRDX6 SNPs thought to be miRNA target sites
| SNP ID | Chromosome Position | Base Change | Ancestral | Derived | miRNA |
|---|---|---|---|---|---|
| rs4611 | 171723640 | T/C | TTGGTGCT | ||
| CTGGTGCT | |||||
| rs15268 | 171723695 | C/A | AGCAATTA | hsa-miR-302f | |
| ATTACATA | hsa-miR-380 | ||||
| rs3211528 | 171724201 | G/A | CTGGGGGA | hsa-miR-361-3p | |
| rs36005931 | 171724224 | A/G | GTGCCTTC | ||
| TGTGCCTT | |||||
| rs35820016 | 171724277 | T/A | TTTTGCTA | hsa-miR-548p | |
| TTGCAATA | |||||
| rs1804053 | 171724351 | G/A | ATGTAGCA | hsa-miR-221 | |
| hsa-miR-222 | |||||
| rs11544001 | 171724433 | A/G | GTGCATGA | ||
| TGCATGAA | |||||
| TGTGCATG | |||||
| rs2000 | 171724457 | G/A | AGAGAAGA | hsa-miR-942 | |
*miRNA target site modeling was performed using Patrocles.
Association of PRDX6 with ALI
The trauma cohort described in Table 1 was genotyped for 37 PRDX6 SNPs using SNPlex. All SNPs were tested for Hardy-Weinberg equilibrium (Additional file 5). Chi2 analysis of incidence of ALI compared to genotype using an additive model showed no significant association between any of the SNPs in this study and ALI (Table 7). Dominant and recessive models failed to demonstrate an association between our SNPs of interest and ALI (Additional file 6). The genotype concordance rate based on assay positive controls was 100% and the frequency of missing genotypes is presented in Table 7. Logistic regression analysis after adjustment for age and ISS showed no association between ALI and our SNPs (Table 8).
Table 7.
Association of 37 PRDX6 genotypes and risk of ALI using and additive model in a population of African and European Americans with major trauma
| European Americans | African Americans | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Source | MAF (ALI) | MAF (non-ALI) | Missing Genotype Frequency | p value | MAF (ALI) | MAF (non-ALI) | Missing Genotype Frequency | p value |
| hCV1948447 | Tagging | 0.22 | 0.20 | 0.016 | 0.538 | 0.06 | 0.05 | 0.019 | 0.507 |
| hCV25599136 | Tagging | 0.00 | 0.00 | 0.161 | NA | 0.03 | 0.03 | 0.230 | 0.842 |
| hCV25599144 | Tagging | 0.00 | 0.01 | 0.399 | 0.999 | 0.00 | 0.00 | 0.296 | NA |
| hCV9040425 | Tagging | 0.27 | 0.25 | 0.021 | 0.739 | 0.38 | 0.35 | 0.019 | 0.725 |
| hCV9040434 | Tagging | 0.25 | 0.24 | 0.407 | 0.838 | 0.31 | 0.29 | 0.307 | 0.537 |
| Position23855054 | Sequencing | 0.00 | 0.00 | 0.012 | NA | 0.09 | 0.08 | 0.019 | 0.535 |
| Position23855203 | Sequencing | 0.00 | 0.00 | 0.679 | NA | 0.10 | 0.17 | 0.662 | 0.268 |
| Position23859396 | Sequencing | 0.00 | 0.00 | 0.012 | NA | 0.04 | 0.02 | 0.019 | 0.302 |
| Position23863249 | Sequencing | 0.00 | 0.00 | 0.012 | NA | 0.03 | 0.03 | 0.023 | 0.577 |
| PRDX6_171709910 | Sequencing | 0.03 | 0.01 | 0.436 | 0.192 | 0.04 | 0.06 | 0.494 | 0.505 |
| PRDX6_171711459 | Sequencing | 0.00 | 0.00 | 0.021 | NA | 0.11 | 0.18 | 0.043 | 0.088 |
| PRDX6_171713872 | Sequencing | 0.00 | 0.01 | 0.037 | 0.999 | 0.08 | 0.05 | 0.070 | 0.331 |
| PRDX6_171715019 | Sequencing | 0.00 | 0.00 | 0.029 | 0.999 | 0.05 | 0.05 | 0.058 | 0.879 |
| PRDX6_171724949 | Sequencing | 0.22 | 0.19 | 0.029 | 0.455 | 0.07 | 0.05 | 0.043 | 0.481 |
| rs10753081 | Sequencing | 0.34 | 0.29 | 0.008 | 0.246 | 0.37 | 0.34 | 0.000 | 0.676 |
| rs2000 | Tagging | 0.05 | 0.05 | 0.045 | 0.966 | 0.02 | 0.02 | 0.031 | 0.793 |
| rs33942654 | Sequencing | 0.23 | 0.20 | 0.021 | 0.321 | 0.34 | 0.29 | 0.054 | 0.138 |
| rs34129563 | Sequencing | 0.07 | 0.03 | 0.021 | 0.080 | 0.01 | 0.01 | 0.039 | 0.559 |
| rs34619706 | Sequencing | 0.08 | 0.08 | 0.037 | 0.900 | 0.03 | 0.01 | 0.047 | 0.287 |
| rs35244306 | Sequencing | 0.00 | 0.01 | 0.021 | 0.999 | 0.11 | 0.08 | 0.039 | 0.517 |
| rs35749242 | Sequencing | 0.00 | 0.01 | 0.021 | 0.999 | 0.08 | 0.06 | 0.039 | 0.516 |
| rs4354572 | Sequencing | 0.00 | 0.00 | 0.354 | NA | 0.11 | 0.17 | 0.358 | 0.251 |
| rs4382766 | Sequencing | 0.33 | 0.29 | 0.025 | 0.312 | 0.35 | 0.34 | 0.047 | 0.830 |
| rs4916362 | Tagging | 0.34 | 0.29 | 0.021 | 0.308 | 0.35 | 0.32 | 0.019 | 0.670 |
| rs57032935 | Sequencing | 0.23 | 0.19 | 0.037 | 0.296 | 0.34 | 0.29 | 0.082 | 0.131 |
| rs6671141 | Sequencing | 0.23 | 0.19 | 0.210 | 0.421 | 0.38 | 0.33 | 0.163 | 0.170 |
| rs6699179 | Tagging | 0.00 | 0.00 | 0.214 | NA | 0.01 | 0.00 | 0.171 | 0.999 |
| rs6702828 | Tagging | 0.00 | 0.00 | 0.012 | NA | 0.01 | 0.00 | 0.027 | 0.778 |
| rs6702835 | Sequencing | 0.27 | 0.25 | 0.008 | 0.606 | 0.42 | 0.37 | 0.000 | 0.493 |
| rs7314 | Sequencing | 0.27 | 0.25 | 0.045 | 0.666 | 0.34 | 0.30 | 0.078 | 0.571 |
| rs7367963 | Tagging | 0.34 | 0.29 | 0.029 | 0.284 | 0.35 | 0.31 | 0.019 | 0.574 |
| rs7521536 | Tagging | 0.24 | 0.20 | 0.025 | 0.360 | 0.32 | 0.29 | 0.043 | 0.320 |
| rs7529377 | Tagging | 0.24 | 0.20 | 0.016 | 0.331 | 0.28 | 0.26 | 0.027 | 0.321 |
| rs7540065 | Sequencing | 0.27 | 0.25 | 0.021 | 0.726 | 0.37 | 0.35 | 0.039 | 0.997 |
| rs912767 | Tagging | 0.24 | 0.20 | 0.016 | 0.331 | 0.29 | 0.26 | 0.027 | 0.240 |
| rs9425722 | Sequencing | 0.00 | 0.00 | 0.342 | NA | 0.08 | 0.17 | 0.335 | 0.048 |
| rs9425725 | Tagging | 0.00 | 0.00 | 0.012 | NA | 0.13 | 0.20 | 0.027 | 0.091 |
Table 8.
Multivariate analysis adjusted for age and injury severity score
| African Americans | European Americans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | ORtrend | Ptrend | ORdom | Pdom | ORrec | Prec | ORtrend | Ptrend | ORdom | Pdom | ORrec | Prec |
| hCV1948447 | 1.33 | 0.507 | 1.19 | 0.705 | 3.87E+09 | 0.999 | 1.16 | 0.538 | 1.06 | 0.840 | 2.41 | 0.204 |
| hCV25599136 | 1.15 | 0.842 | 1.15 | 0.842 | NA | NA | NA | NA | NA | NA | NA | NA |
| hCV25599144 | NA | NA | NA | NA | NA | NA | 4.19E-10 | 0.999 | 4.19E-10 | 0.999 | NA | NA |
| hCV9040425 | 1.08 | 0.725 | 1.08 | 0.781 | 1.13 | 0.760 | 1.08 | 0.739 | 1.04 | 0.885 | 1.32 | 0.593 |
| hCV9040434 | 1.17 | 0.537 | 1.30 | 0.442 | 1.04 | 0.947 | 1.06 | 0.838 | 0.94 | 0.871 | 2.05 | 0.365 |
| Position23855054 | 1.25 | 0.535 | 1.39 | 0.398 | 2.07E-09 | 0.999 | NA | NA | NA | NA | NA | NA |
| Position23855203 | 0.56 | 0.268 | 0.60 | 0.374 | 1.52E-09 | 0.999 | NA | NA | NA | NA | NA | NA |
| Position23859396 | 1.73 | 0.302 | 2.17 | 0.190 | 1.70E-09 | 0.999 | NA | NA | NA | NA | NA | NA |
| Position23863249 | 1.36 | 0.577 | 1.61 | 0.431 | 1.70E-09 | 0.999 | NA | NA | NA | NA | NA | NA |
| PRDX6_171709910 | 0.66 | 0.505 | 0.72 | 0.636 | 1.74E-09 | 0.999 | 5.13 | 0.192 | 5.13 | 0.192 | NA | NA |
| PRDX6_171711459 | 0.58 | 0.088 | 0.61 | 0.146 | 2.02E-09 | 0.998 | NA | NA | NA | NA | NA | NA |
| PRDX6_171713872 | 1.49 | 0.331 | 1.69 | 0.229 | 1.06E-09 | 0.999 | 1.28E-09 | 0.999 | 1.28E-09 | 0.999 | NA | NA |
| PRDX6_171715019 | 0.93 | 0.879 | 1.01 | 0.989 | 1.07E-09 | 0.999 | 1.77E-09 | 0.999 | 1.77E-09 | 0.999 | NA | NA |
| PRDX6_171724949 | 1.35 | 0.481 | 1.21 | 0.681 | 4.18E+09 | 0.999 | 1.20 | 0.455 | 1.10 | 0.740 | 2.50 | 0.186 |
| rs10753081 | 1.09 | 0.676 | 1.19 | 0.543 | 0.98 | 0.970 | 1.28 | 0.246 | 1.34 | 0.288 | 1.44 | 0.439 |
| rs2000 | 1.21 | 0.793 | 1.21 | 0.793 | NA | NA | 0.98 | 0.966 | 1.25 | 0.661 | 1.30E-09 | 0.999 |
| rs33942654 | 1.39 | 0.138 | 1.78 | 0.057 | 1.05 | 0.923 | 1.27 | 0.321 | 1.14 | 0.640 | 2.96 | 0.103 |
| rs34129563 | 1.73 | 0.559 | 1.73 | 0.559 | NA | NA | 2.16 | 0.080 | 2.09 | 0.115 | 3.42E+09 | 0.999 |
| rs34619706 | 2.13 | 0.287 | 2.13 | 0.287 | NA | NA | 0.96 | 0.900 | 0.89 | 0.773 | 2.32 | 0.554 |
| rs35244306 | 1.25 | 0.517 | 1.41 | 0.349 | 1.34E-09 | 0.999 | 1.29E-09 | 0.999 | 1.29E-09 | 0.999 | NA | NA |
| rs35749242 | 1.30 | 0.516 | 1.43 | 0.399 | 1.09E-09 | 0.999 | 1.29E-09 | 0.999 | 1.29E-09 | 0.999 | NA | NA |
| rs4354572 | 0.65 | 0.251 | 0.70 | 0.386 | 1.85E-09 | 0.999 | NA | NA | NA | NA | NA | NA |
| rs4382766 | 0.96 | 0.830 | 0.94 | 0.838 | 0.94 | 0.892 | 1.24 | 0.312 | 1.26 | 0.409 | 1.50 | 0.394 |
| rs4916362 | 1.09 | 0.670 | 1.19 | 0.551 | 0.99 | 0.987 | 1.24 | 0.308 | 1.28 | 0.381 | 1.44 | 0.436 |
| rs57032935 | 1.40 | 0.131 | 1.81 | 0.053 | 1.07 | 0.888 | 1.29 | 0.296 | 1.16 | 0.600 | 2.98 | 0.101 |
| rs6671141 | 1.37 | 0.170 | 1.43 | 0.256 | 1.64 | 0.261 | 1.24 | 0.421 | 1.21 | 0.549 | 1.86 | 0.398 |
| rs6699179 | 1.14E+09 | 0.999 | 1.14E+09 | 0.999 | NA | NA | NA | NA | NA | NA | NA | NA |
| rs6702828 | 1.59 | 0.778 | 1.59 | 0.778 | NA | NA | NA | NA | NA | NA | NA | NA |
| rs6702835 | 1.15 | 0.493 | 1.78 | 0.063 | 0.61 | 0.242 | 1.12 | 0.606 | 1.12 | 0.686 | 1.28 | 0.632 |
| rs7314 | 1.13 | 0.571 | 1.26 | 0.437 | 1.02 | 0.963 | 1.10 | 0.666 | 1.07 | 0.820 | 1.38 | 0.542 |
| rs7367963 | 1.12 | 0.574 | 1.13 | 0.676 | 1.26 | 0.589 | 1.26 | 0.284 | 1.30 | 0.347 | 1.45 | 0.432 |
| rs7521536 | 1.24 | 0.320 | 1.47 | 0.191 | 1.01 | 0.977 | 1.24 | 0.360 | 1.11 | 0.705 | 2.95 | 0.104 |
| rs7529377 | 1.25 | 0.321 | 1.31 | 0.360 | 1.40 | 0.506 | 1.26 | 0.331 | 1.14 | 0.651 | 2.93 | 0.106 |
| rs7540065 | 1.00 | 0.997 | 1.03 | 0.922 | 0.95 | 0.896 | 1.08 | 0.726 | 1.04 | 0.883 | 1.35 | 0.570 |
| rs912767 | 1.30 | 0.240 | 1.40 | 0.250 | 1.40 | 0.505 | 1.26 | 0.331 | 1.14 | 0.651 | 2.93 | 0.106 |
| rs9425722 | 0.44 | 0.048 | 0.44 | 0.065 | 1.97E-09 | 0.999 | NA | NA | NA | NA | NA | NA |
| rs9425725 | 0.60 | 0.091 | 0.63 | 0.154 | 2.03E-09 | 0.998 | NA | NA | NA | NA | NA | NA |
Haplotype Analysis
Haplotype blocks were created for both African and European Americans using 27 and 28 SNP markers, respectively. Haplotype blocks were created for a region spanning 100.6 kb of chromosome 1. For African Americans, there were 14 SNP markers with genotyping completion rate of less than 95% and were thus excluded from the haplotype analysis. For European Americans, there were 9 SNPs with a genotype completion rate of less than 95% and were excluded from the haplotype analysis.
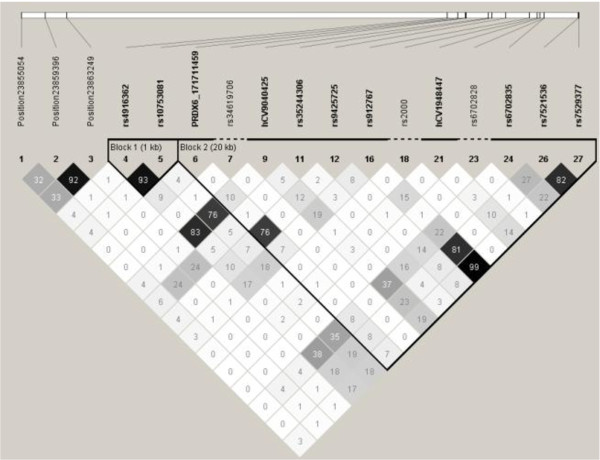
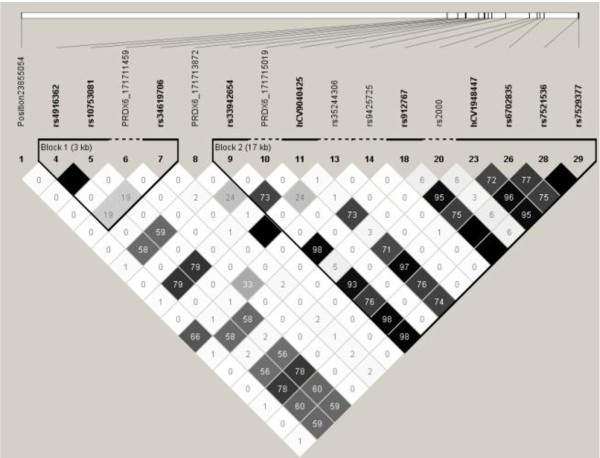
In African Americans, there were 2 blocks (block1 = rs4916362 and rs10753081; block2 = PRDX6_171711459, rs34619706, hCV9040425, rs35244306, rs9425725, rs912767, rs2000, hCV1948447, rs6702828, rs6702835, rs7521536, rs7529377) (Figure 2). In European Americans, there were 2 blocks (block1 = rs4916362, rs10753081, PRDX6_171711459, rs34619706; block2 = rs33942654, PRDX6_171715019, hCV9040425, rs35244306, rs9425725, rs912767, rs2000, hCV1948447, rs6702835, rs7521536, rs7529377) (Figure 3). The haplotype structure of PRDX6 appears to be in low LD, in both subgroups. Haplotype analyses of association with ALI did not reveal any significant associations above and single SNP analyses (Table 9).
Figure 2.
Haplotype Structure of 17 PRDX6 SNPs in African Americans with r2 Values (N = 259).
Figure 3.
Haplotype Structure of 17 PRDX6 SNPs in European Americans with r2 Values (N = 254).
Table 9.
Haplotype Analysis among African and European Americans
| African Americans | European Americans | ||||||
|---|---|---|---|---|---|---|---|
| Block | Haplotype | Population Frequencies | p value | Block | Haplotype | Population Frequencies | p value |
| Block 1 | GT | 0.654 | 0.719 | Block 1 | ACA | 0.690 | 0.520 |
| AC | 0.330 | 0.833 | GTA | 0.230 | 0.479 | ||
| GC | 0.016 | 0.563 | GTG | 0.080 | 0.999 | ||
| Block 2 | TGTTACGTG | 0.282 | 0.449 | Block 2 | GGACGTG | 0.734 | 0.883 |
| TATTGCACA | 0.212 | 0.386 | AAGTACA | 0.202 | 0.634 | ||
| CATCACATG | 0.153 | 0.083 | GAACATG | 0.050 | 0.297 | ||
| TACTACGTG | 0.099 | 0.485 | |||||
| TGTTACATG | 0.072 | 0.455 | |||||
| TATTACATG | 0.055 | 0.935 | |||||
| TATTGTACA | 0.051 | 0.894 | |||||
| TATTACACG | 0.036 | 0.969 | |||||
| TATCACATG | 0.028 | 0.953 | |||||
Discussion
Prdx6 is a member of the thiol-specific antioxidant protein family and in overexpressing cell and mouse models has been shown to be protective against oxidant stress which null models show sensitivity to oxidants [7,9,27]. Thus, PRDX6 is a suitable candidate gene for ALI risk. The extent of genetic variation within PRDX6 remains largely unknown, therefore we performed direct sequencing of the PRDX6 gene, and identified novel variants for future study. We also tested the newly discovered SNPs and tagging SNPs for association with ALI using our trauma cohort, and did not demonstrate an association with trauma-related ALI.
We identified 43 novel variants among African American and European American subjects with either ALI or control status. None of the 43 SNPs identified were in coding regions which may indicate that the Prdx6 protein is highly conserved across phyla. Approximately 19 kb on chromosome 1 was sequenced in order to achieve adequate coverage of the PRDX6 gene and flanking 5' and 3' UTRs. Special attention was given to the GRE2 and ARE1 regions -749 to -737 and -357 to -349, respectively. The ARE1 within the PRDX6 promoter was shown to play a role in regulation of transcription and to be inducible under conditions of oxidative stress [26] and the GRE2 may be capable of binding transcription factors under oxidative stress conditions [28]. Due to the GC rich content of the region surrounding the ARE1, we were unable to optimize PCR reaction conditions in a way to prime through the secondary structure. The GRE2 region was sequenced, but no variation was noted. The GC rich region within the PRDX6 promoter might warrant further investigation since methylation of DNA cytosine residues are often found in the sequence context CpG. Several new sequencing approaches are emerging that target methylation sites using restriction enzyme treatment followed by sequence by synthesis [29].
In addition to comparing our results with NCBI's dbSNP, we compared our novel and known SNPs with the resequencing data registered in 1000 Genomes. The 1000 Genomes project aims to find most genetic variants with frequencies of at least 1%. Thus far three sequencing projects contribute to the database, low coverage sequencing of 179 individuals from 4 populations, high coverage sequencing of 2 mother-father-child trios, and exon targeting sequencing of 697 individuals from 7 populations [30]. Although 1000 Genomes aims to identify over 95% of variation in any individual, 27 of our novel SNPs and 1 previously recorded SNP are not present in the database, signifying a need for resequencing of extreme phenotypes, such as ALI cases.
Novel and previously recorded SNPs in the 5' UTR and first intron of PRDX6 were submitted to TESS to determine their likelihood of being in transcription factor binding sites. We found 19 motifs in the reference sequences that are capable of binding known transcription factors and 21 in the alternative sequence. A comparison between the results of the reference sequence search and the alternative revealed that in most cases, the SNP of interest changes the motif enough to cause a different transcription factor to bind that site or can cause a binding site to disappear and vice versa. After comparison with the ENCODE data, we found that our sequences have not yet been shown to bind the three overlapping transcription factors tested in ENCODE experimentally.
Known SNPs validated in the sequencing effort were compared using a Patrocles search query for miRNA target sites within PRDX6 to determine if any of our SNPs were in putative target sites for miRNAs. Three of the eight SNPs returned from the search corresponded with our known SNPs. Only one of the three SNPs was found to have a corresponding known miRNA (miR-942). Some miRNAs are known to control the expression of genes at the posttranscriptional level [31]. However, very limited data are available on miR-942.
We performed an association study for ALI using newly uncovered SNPs and SNPs selected from Hapmap and NCBI's dbSNP and observed no significant association between any of the SNPs in this study and ALI. This lack of association may be due to several causes. First, the detectable effect size is modest because of sample size limitations. We genotyped 513 subjects to test for an association between our selected SNPs and ALI, but this sample size was inadequate to detect relative risks below 1.93 and 1.69 for alleles with MAFs of 0.05 and 0.10, respectively. Second, our analyses were limited to patients with severe trauma. Thus, our study did not evaluate a possible association with other causes of ALI such as sepsis. Finally, it is possible that PRDX6 genetic variation may not modify the risk of ALI.
The genotype data were used to construct haplotype blocks to better assess the PRDX6 gene structure. Haplotype analysis plays an important role in association studies between genotype and phenotype, since SNPs found to be in strong LD can capture most of the genetic variation across fairly large regions [24]. The haplotype blocks constructed from our genotype data did not show strong linkage disequilibrium using confidence intervals, therefore tagging SNP strategies in future studies should be approached with caution.
Our resequencing data did not show any variation in the coding region of PRDX6. Had nonsynonymous SNPs been discovered, it would have prompted us to investigate whether any of these SNPs had any effect on protein structure, which could cause a loss of function in Prdx6. Since we cannot make a connection between coding region SNPs and conformational changes in the protein, we examined regulatory effects. We found several promoter SNPs that change the sequence of potential TFBSs based on conservation data. We were unable to confirm that these sequences were in fact TFBSs due to the lack of available data. However if any of our promoter SNPs showed a significant association with ALI or another phenotype perhaps using a larger sample size, future studies using promoter constructs could offer more information on upregulation of PRDX6. We also found several SNPs in the 3'UTR. It is possible that one or more of these SNPs is responsible for changing an miRNA binding site, thus repressing protein translation.
Our study has several limitations. One potential limitation of this study is the number of genotype call failures. Ten and nine markers for African Americans and European Americans respectively were eliminated from our analysis since they were under the 95% completion rate cut-off. This high rate of genotype failure was due to difficulties with consistent assay performance rather than DNA quality. If these genotypes had been obtained, it is a possible that an association may have been observed. Also, we did not adjust our results for ancestry informative markers (AIMs). Instead our population was stratified based on skin color, which may not be an adequate proxy for population admixture effects. Another possible limitation is a candidate gene approach that focused on a single gene: PRDX6. ALI risk may be considered a complex phenotype, and thus likely is not fully explained by a variation in a single gene [10]. Finally, we only tested for association in patients with ALI from severe trauma. Thus, it is possible that PRDX6 may play a role in the initiation or severity of ALI after other insults, including sepsis, or in determining recovery from ALI.
PRDX6 has been shown to play a role not only in ALI, but other diseases as well. A recent studied demonstrated that PRDX6 promotes lung cancer metastasis and invasion via phospholipase A2 activity in mice [32]. Another publication reported that PRDX6 transfected breast cancer cells metastasized more readily to the lungs when compared with control cells [26]. It is possible that our novel SNPs may function in lung cancer as well as ALI. The interaction between GSTpi and PRDX6 is another interesting subject for future studies. GSTpi expression is elevated in tumors from a variety of cancers, including lung cancer, compared to normal tissue [33]. Testing gene-gene interactions between PRDX6 and GSTpi would be an interesting future direction both in ALI and other diseases such as cancer.
Conclusion
In conclusion, this study revealed novel SNPs within the important anti-oxidant PRDX6 gene and its 5' and 3' flanking regions via direct sequencing. Several of these variants have putative function and may be useful for future gene association studies. Although there was no association discovered between our novel and tagging SNPs with trauma-related ALI, future studies may focus on the role of PRDX6 variation in other at risk groups, as well as other diseases.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MR carried out the sequencing and genotyping analysis and drafted the manuscript. RA participated in the design of the study, supervision of laboratory assays, and interpretation of data. NM participated in the design of the study, interpretation of data, and manuscript editing. ML performed statistical analyses. RF performed the statistical analyses. PNL participated in the design of the study, collection of data, and manuscript editing. RG performed statistical analyses. SB performed the statistical analyses. ARL performed statistical analyses. SIF participated in the design of the study and interpretation of data. ABF participated in the design of the study, interpretation of data, and manuscript editing. SMA participated in the design of the study, and interpretation of data. JDC participated in the design of the study, data collection, interpretation of the data, manuscript drafting and manuscript editing. All authors read and approved the final manuscript
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
PCR primers and cycling conditions.
Detectable relative risk vs. disease allele frequency.
Sequencing variants separated by race and case-control status.
Confidence score for sequencing genotypes with only one variant.
Hardy-Weinberg equilibrium values.
Dominant and recessive models in African Americans and European Americans.
Contributor Information
Melanie Rushefski, Email: meldoran@mail.med.upenn.edu.
Richard Aplenc, Email: raplenc@mail.med.upen.edu.
Nuala Meyer, Email: Nuala.Meyer@uphs.upenn.edu.
Mingyao Li, Email: mingyao@mail.med.upenn.edu.
Rui Feng, Email: ruifeng@mail.med.upenn.edu.
Paul N Lanken, Email: Paul.Lanken@uphs.upenn.edu.
Robert Gallop, Email: rgallop@wcupa.edu.
Scarlett Bellamy, Email: bellamys@mail.med.upenn.edu.
A Russell Localio, Email: rlocalio@mail.med.upenn.edu.
Sheldon I Feinstein, Email: sif@mail.med.upenn.edu.
Aron B Fisher, Email: abf@exchange.upenn.edu.
Steven M Albelda, Email: albelda@mail.med.upenn.edu.
Jason D Christie, Email: jchristi@mail.med.upenn.edu.
Acknowledgements
This work was supported by HL60290, HL079063, The Doris Duke Charitable Foundation
References
- Neff M, Rubenfeld G. Clinical Epidemiology of Acute Lung Injury. Seminars in Respiratory and Critical Care Medicine. 2001;22:237–246. doi: 10.1055/s-2001-15781. [DOI] [PubMed] [Google Scholar]
- Rubenfeld GD. Epidemiology of acute lung injury. Critical Care Medicine. 2003;31:S276–84. doi: 10.1097/01.CCM.0000057904.62683.2B. [DOI] [PubMed] [Google Scholar]
- Bernard GR, Artigas A, Brigham KL, Carlet J, Falke K, Hudson L, Lamy M, Legall JR, Morris A, Spragg R. The American European consensus conference on ARDS: definitions, mechanisms, relevant outcomes and clinical trials coordination. Am J Resp Crit Care Med. 1994;149:818–824. doi: 10.1164/ajrccm.149.3.7509706. [DOI] [PubMed] [Google Scholar]
- Gunther A, Walmrath D, Grimminger F, Seeger W. Pathophysiology of Acute Lung Injury. Seminars in Respiratory and Critical Care Medicine. 2001;22:247–258. doi: 10.1055/s-2001-15782. [DOI] [PubMed] [Google Scholar]
- Moss M, Mannino DM. Race and gender differences in acute respiratory distress syndrome deaths in the United States: an analysis of multiple-cause mortality data (1979-1996) Critical Care Medicine. 2003;30:1679–1685. doi: 10.1097/00003246-200208000-00001. [DOI] [PubMed] [Google Scholar]
- Manevich Y, Feinstein SI, Fisher AB. Activation of the antioxidant enzyme 1-CYS peroxiredoxin requires glutathionylation mediated by heterodimerization with πGST. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3780–3785. doi: 10.1073/pnas.0400181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schremmer B, Manevich Y, Feinstein SI, Fisher AB. Peroxiredoxins in the lung with emphasis on peroxiredoxin VI. Sub-cellular Biochemistry. 2007;44:317–344. doi: 10.1007/978-1-4020-6051-9_15. [DOI] [PubMed] [Google Scholar]
- Rhee GS, Chae HZ, Kim K. Peroxiredoxins: A historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radical Biology & Medicine. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- Manevich Y, Sweitzer T, Pak JH, Feinstein SI, Muzykantov V, Fisher AB. 1-Cys peroxiredoxin overexpression protects cells against phospholipid peroxidation-mediated membrane damage. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11599–604. doi: 10.1073/pnas.182384499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JD, Ma SF, Aplenc R, Li M, Lanken PN, Shah CV, Fuchs B, Albelda SM, Flores C, Garcia JGN. Variation in the myosin light chain kinase gene is associated with development of acute lung injury after major trauma. Critical Care Medicine. 2008;36:2794–2800. doi: 10.1097/CCM.0b013e318186b843. [DOI] [PubMed] [Google Scholar]
- Shah CV, Localio AR, Lanken PN, Kahn JM, Bellamy S, Gallop R, Finkel B, Gracias VH, Fuchs BD, Christie JD. The impact of development of acute lung injury on hospital mortality in critically ill trauma patients. Critical Care Medicine. 2008;36:2309–2315. doi: 10.1097/CCM.0b013e318180dc74. [DOI] [PubMed] [Google Scholar]
- Meyer NJ, Li M, Feng R, Bradfield J, Gallop R, Bellamy S, Fuchs BD, Lanken PN, Albelda SM, Rushefski M, Aplenc R, Abramova H, Atochina Vasserman EN, Beers MF, Calfee CS, Cohen MJ, Pittet JF, Christiani DC, O'Keefe GE, Ware LB, May AK, Wurfel MM, Hakonarson H, Christie JD. ANGPT2 Genetic Variant is Associated with Trauma-Associated Acute Lung Injury and Altered Plasma Angiopoietin-2 Isoform Ratio. American Journal of Respiratory and Critical Care Medicine. 2011. [DOI] [PMC free article] [PubMed]
- Kruglyak L, Nickerson DA. Variation is the spice of life. Nature Genetics. 2001;27:234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- De Bakker PIW, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and Power in Genetic Association Studies. Nature Genetics. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- De La Vega FM, Lazaruk KD, Rhodes MD, Wenz MH. Assessment of two flexible and compatible SNP genotyping platforms: Taqman SNP genotyping assays and the SNPlex genotyping system. Mutation Research. 2005;573:111–135. doi: 10.1016/j.mrfmmm.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Schug J, Overton GC. TESS: Transcription Element Search Software on the WWW. Technical Report CBIL-TR-1997-1001-v0.0. Computational Biology and Informatics Laboratory School of Medicine University of Pennsylvania. 1997. http://www.cbil.upenn.edu/tess
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiard S, Charlier C, Coppieters W, Georges M, Baurain D. Patrocles: a database of polymorphic miRNA-mediated gene regulation in vertebrates. Nucleic Acids Research. 2010. pp. D640–D651. [DOI] [PMC free article] [PubMed]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Menashe I, Rosenberg PS, Chen BE. PGA: Power calculator for case-control genetic association analyses. BMC Genetics. 2008;9:36. doi: 10.1186/1471-2156-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of ld and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. Plink: A tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury I, Mo Y, Gao L, Kazi A, Fisher AB, Feinstein SI. Oxidant Stress Stimulates Expression of the Human Peroxiredoxin 6 (Prdx6) Gene by a Transcriptional Mechanism Involving an Antioxidant Response Element. Free Radical Biology and Medicine. 2009;46:146–153. doi: 10.1016/j.freeradbiomed.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Feinstein SI, Fisher AB. H2O2-dependent Hyperoxidation of Peroxiredoxin 6 (Prdx6) Plays a Role in Cellular Toxicity via Up-regulation of iPLA2. Journal of Cellular Biochemistry. 2008;104:1274–1285. doi: 10.1002/jcb.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukosz M, Jakob S, Büchner N, Zschauer T, Altschmied T, Haendeler J. Antioxidants & Redox Signaling. 2010. pp. 713–742. [DOI] [PubMed]
- Laird PW. Principles and Challenges of Genome-wide DNA Methylation Analysis. Nature Reviews Genetics. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA Genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Ho JN, Lee SB, Lee SS, Yoon SH, Kang GY, Hwang SG, Um HD. Phospholipase A2 activity of peroxiredoxin 6 promotes invasion and metastasis of lung cancer cells. Molecular Cancer Therapuetics. 2010;9(4):825–32. doi: 10.1158/1535-7163.MCT-09-0904. [DOI] [PubMed] [Google Scholar]
- Moscow JA, Fairchild CR, Madden MJ, Ransom DT, Wieand HS, O'Brien EE, Poplack DG, Cossman J, Myers CE, Cowan KH. Expression of anionic glutathione-S-transferase and P-glycoprotein in human tissues and tumors. Cancer Research. 1989;49(6):1422–1428. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR primers and cycling conditions.
Detectable relative risk vs. disease allele frequency.
Sequencing variants separated by race and case-control status.
Confidence score for sequencing genotypes with only one variant.
Hardy-Weinberg equilibrium values.
Dominant and recessive models in African Americans and European Americans.