Abstract
Jellyfish blooms occur in many estuarine and coastal regions and may be increasing in their magnitude and extent worldwide. Voracious jellyfish predation impacts food webs by converting large quantities of carbon (C), fixed by primary producers and consumed by secondary producers, into gelatinous biomass, which restricts C transfer to higher trophic levels because jellyfish are not readily consumed by other predators. In addition, jellyfish release colloidal and dissolved organic matter (jelly-DOM), and could further influence the functioning of coastal systems by altering microbial nutrient and DOM pathways, yet the links between jellyfish and bacterioplankton metabolism and community structure are unknown. Here we report that jellyfish released substantial quantities of extremely labile C-rich DOM, relative to nitrogen (25.6 ± 31.6 C:1N), which was quickly metabolized by bacterioplankton at uptake rates two to six times that of bulk DOM pools. When jelly-DOM was consumed it was shunted toward bacterial respiration rather than production, significantly reducing bacterial growth efficiencies by 10% to 15%. Jelly-DOM also favored the rapid growth and dominance of specific bacterial phylogenetic groups (primarily γ-proteobacteria) that were rare in ambient waters, implying that jelly-DOM was channeled through a small component of the in situ microbial assemblage and thus induced large changes in community composition. Our findings suggest major shifts in microbial structure and function associated with jellyfish blooms, and a large detour of C toward bacterial CO2 production and away from higher trophic levels. These results further suggest fundamental transformations in the biogeochemical functioning and biological structure of food webs associated with jellyfish blooms.
Keywords: biogeochemical cycling, jelly carbon shunt, fisheries production
Dramatic spatial increases and temporal shifts in gelatinous organisms (ctenophores and medusae, hereafter referred to as “jellyfish”) are thought to have occurred in many estuarine, coastal, and open-ocean ecosystems worldwide over the past several decades (1–3). The proximate causes for these changes are unknown, but likely include a combination of factors, including eutrophication, overharvesting of fish, climate variations, accidental introductions or translocations, increased polyp abundances, and habitat modifications (2, 4). Given current and projected global increases in ocean temperature, combined with anthropogenic influences, these trends are likely to continue into the near future (5) with unknown consequences at the ecosystem level.
Voracious predation by coastal jellyfish assimilates large quantities of carbon (C) [as well as nitrogen (N) and phosphorus (P)] fixed by primary producers and consumed by secondary producers (e.g., copepods) into gelatinous biomass, which is not readily consumed by higher trophic-level predators (4). As a result, jellyfish may alter trophic C transfer by limiting C availability and potentially representing a trophic “dead-end” for C to higher trophic levels (e.g., planktivorous fish) (5). However, jellyfish generate large amounts of colloidal and dissolved organic matter (jelly-DOM) (6, 7), which is released to the water and which may be used by the ambient bacterioplankton (4, 8). Bacterial use of jelly-DOM could lead to the repackaging of jelly C and its reincorporation into the food web during planktonic jellyfish blooms, as opposed to this C being assimilated into jellyfish biomass. The magnitude of this trophic link between jellyfish and the microbial food web is poorly understood, because little is known about the large-scale effects of DOM release by jellyfish blooms on the biomass production and respiration by bacterioplankton (8).
Here we assess the role of jellyfish on water-column C pathways, focusing on potential alterations to bacterial community structure and to bacterial C metabolism at the ecosystem level. We have explored these questions in the York River estuary, a southern Chesapeake Bay tributary, using a combination of experimental determinations of jellyfish and bacteria metabolism and C cycling, and of field surveys of jellyfish biomass. Chesapeake Bay and its tributaries have consistently high jellyfish biomass of different species (Fig. S1). There is also abundant background information on this system, making it ideal to explore the influence of jellyfish on estuarine and coastal biogeochemical processes.
Our study focuses on blooms of two native species, Mnemiopsis leidyi ctenophores and Chrysaora quinquecirrha scyphomedusae, which dominate throughout the estuary during late spring (May) and summer (June–August), leading to significant “top down” changes to the zooplankton community (4). Blooms of both Mnemiopsis and Chrysaora (along with other members of the Pelagiidae) have been recorded worldwide as causing major problems for marine food webs and human activities (e.g., fisheries and tourism), and their increased abundance in both eutrophic (e.g., the Baltic Sea) and oligotrophic (e.g., Pelagia and Mnemiopsis in the Mediterranean) exotic regions suggests that these jellyfish have considerable invasive capabilities (9–12). The inclusion of these jellyfish species in this study is therefore relevant from a global perspective and serves as a relevant model to represent possible jellyfish-mediated changes to coastal biogeochemical pathways.
Results and Discussion
Jellyfish DOM Production.
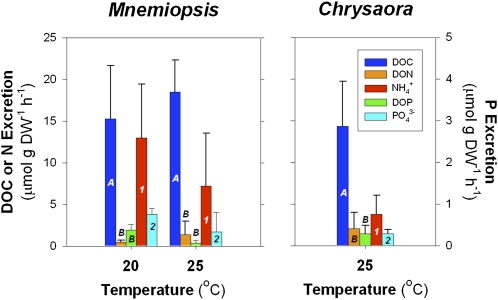
We measured the simultaneous release of DOM (C, N, and P), ammonium (NH4+), and phosphate (PO43−) by M. leidyi ctenophores and C. quinquecirrha medusae during short laboratory incubations. Jellyfish directly release assimilated material as DOM and inorganic nutrients via excretion and mucus production. Because organic and inorganic constituents are produced via different processes but originate from assimilated material, we use the terms “jelly-C” or “jelly-DOM” to describe material directly released by jellyfish. DOM may also be released indirectly through leaching from fecal material and sloppy feeding. Jelly-DOM release rates were normalized to jellyfish dry weight to allow comparison between jellyfish species. Both jellyfish species released consistently high amounts of dissolved organic C (DOC) (Fig. 1), with variable stoichiometry relative to dissolved organic N (DON) release. M. leidyi ctenophores, in particular, produced more C-rich DOM, as reflected in the high molar DOC:DON ratios of their excretia (25.6 ± 31.6C:1N), compared with the canonical Redfield ratio of 6.6C:1N (Fig. 1). The fact that DOM release per unit jellyfish biomass in our experiments appeared to be a function of temperature tends to confirm that C and nutrient release were not caused by the experimental manipulation, but rather physiological mechanisms (6, 7, 13).
Fig. 1.
Jellyfish release of DOM and inorganic nutrients. Weight-specific release rates of DOC, DON, phosphorus (DOP), and inorganic N (ammonium, NH4+) and P (phosphate, PO43−) by (Left) M. leidyi ctenophores and (Right) C. quinquecirrha medusae at 20 °C (M. leidyi only) and 25 °C. Dissolved organic and inorganic excretion rates denoted by the same letter or number are not significantly different (ANOVA, P > 0.05). Excretion rates denoted by different letters or numbers are significantly different (ANOVA, P < 0.05). Error bars are ± 1 SEM.
Bacterial Metabolism of Jelly-DOM.
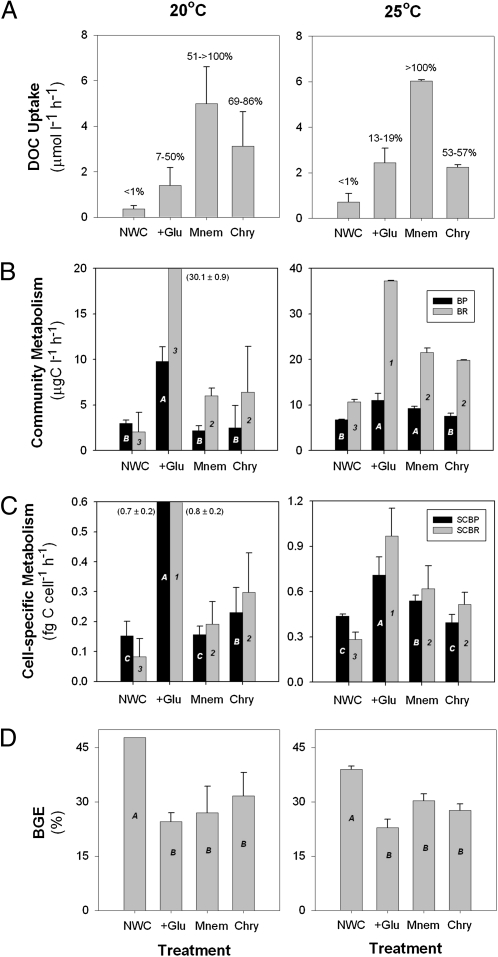
The biochemical nature and biological reactivity of the DOM released by jellyfish is unknown, although it is likely to contain a combination of polysaccharides and other C-rich compounds (14, 15). Our results show that this organic matter is highly bioavailable to heterotrophic bacteria. We assessed the bacterial consumption of jelly-DOM and its subsequent utilization by bacteria, by measuring bacterial production and respiration on natural, free-living microbial assemblages in closed incubations with and without jelly-DOM, and compared the results to control incubations with natural water and with natural water plus glucose as an added labile source of C-rich DOM. Glucose was chosen as a labile DOC control because it is the most abundant neutral sugar in the ocean, and it is preferentially used by bacteria relative to other monosaccharides (16). Our experimental results show that a large proportion of jelly-DOM was consumed by free-living bacterial communities, with an estimated 51% to >100% and 52% to 86% of jelly-C produced by Mnemiopsis and Chrysaora jellyfish disappearing within 6 to 8 h, respectively (Fig. 2A and Fig. S2). In turn, this rapid utilization of jelly-DOM greatly enhanced bacterial C metabolism and altered C flows through bacterial communities. Compared with the natural seawater control, the presence of jelly-DOM had pronounced effects on microbial C consumption, reflected in significant increases in both respiration and production, in absolute terms and also on a per cell basis (ANOVA, P < 0.05) (Fig. 2 B and C and Fig. S2).
Fig. 2.
Bacterial DOC uptake, metabolism, and growth efficiency. Results from 6-h closed bacterial incubations conducted during July at 20 °C (Left) and 25 °C (Right) measuring (A) DOC uptake, changes in (B) community BP and BR, changes in (C) cell-specific production (SCBP) and respiration (SCBR), and (D) BGE of free-living communities between M. leidyi (Mnem) and C. quinquecirrha (Chry) excretia treatments, and glucose addition (+Glu) and natural water controls (NWC). Mnem and Chry treatments were prepared by incubating M. leidyi ctenophores or C. quinquecirrha medusae with microbial communities before incubations for 6 h. Results from treatments were compared with controls by ANOVA. (A) Percentage values equal the proportion of DOC consumed in the NWC, or glucose, Mnem, and Chry DOC pools used by bacterial communities after 6 h. (D) BGE (%) = BP/(BP + BR) × 100 following del Giorgio and Cole (17). Treatment and controls connected by the same letter or numbers are not significantly different (P > 0.05). Treatment and controls denoted by different letters or numbers are significantly different (ANOVA, P < 0.05). Error bars are ± 1 SD.
The impact and fate of the jelly-DOM consumed by bacteria in jellyfish-dominated systems depends greatly on how much of this C bacteria channel into respiration versus biomass. This result will be reflected in changes in bacterial growth efficiency (BGE), which is the ratio of biomass produced (bacterial production, BP) to substrate assimilated (BP + bacterial respiration, BR) (17). An increase in BGE would imply a more efficient functioning of the microbial loop with a higher proportion of jellyfish-derived C incorporated into microbial biomass and available to other consumers. In contrast, a decrease in BGE would imply a greater shunt of jelly-DOM to CO2. Our simultaneous measurements of BP and BR suggest a disproportionate increase in respiration in the presence of jelly-DOM. This pattern was stronger at summer temperatures (20 and 25 °C) when BR increased 82% to 159% compared with natural water controls (NWCs), whereas BP increased 92% to 128% within jellyfish treatments (Fig. 2B and Fig. S2). At lower temperatures (14 °C), increases in BP and BR were similar, perhaps because DOC release rates in M. leidyi ctenophores are linked to metabolic activity and therefore were reduced at lower temperatures (Fig. S2). Overall, the jellyfish treatments resulted in increased BR and decreased BGE of between 10% and 15% relative to the control incubations at the two higher temperatures (Fig. 2D).
Collectively, these results suggest that jelly-C is rapidly used by bacteria (on the order of minutes to hours), and that it has the potential to enhance bacterial metabolism. This increased total bacterial metabolism, however, is accompanied by significant declines in BGE compared with NWCs (ANOVA, P < 0.05), which suggests a shunt to respiration and an overall decline in the efficiency of C transfer through the microbial loop during jellyfish blooms. Interestingly, similar trends between BP, BR, and BGE were observed in the glucose-addition control (Fig. 2 B–D), suggesting that the response in BGE might be related to an increase in the availability of C relative to other nutrients (18), and thus reflect the stoichiometry of C and nutrient release from jellyfish. Bacteria grown in energy and substrate-rich media (e.g., glucose) often display an uncoupling of catabolism (respiration) from anabolism (growth), resulting in nongrowth energy dissipation (i.e., overflow metabolism) and decreased BGE (18). Although our results show that jellyfish release relatively high amounts of inorganic N and P together with DOC (Fig. 1), it would appear that the organic C:N and C:P stoichiometry and biochemical quality of jellyfish material released into labile DOM pools might induce significant declines in BGE.
We further evaluated the impact of jellyfish blooms on total bacterial metabolism at the ecosystem level by calculating daily amounts of Mnemiopsis- and Chrysaora-derived DOC production, and the potential for jelly-DOM to fuel respiration relative to biomass production by bacterial communities in the York River estuary (Table 1). Our estimates of the potential bacterial C respiration and production directly linked to utilization of jellyfish DOM indicate that jellyfish blooms in the York River could influence the overall microbial metabolism in the system, and ultimately the fate of this C. The BR potentially associated with peaks in jellyfish biomass was high, with large increases in BR relative to bacterial production for M. leidyi during June to July (up to 96 mg C m−3·day−1) and for C. quinquecirrha during July to August (up to 60 mg C m−3·day−1) (Table 1). This relative increase in respiration versus production was substantial compared with seasonal background BP rates. Our estimates suggest that microbes were potentially respiring up to 45% and 73% (respectively) of DOC released during M. leidyi and C. quinquecirrha blooms, rather than using this C for BP (Fig. S1 and Tables S1–S3). This high proportion of respired material has consequences for C transfer within jellyfish-dominated food webs and implies that jellyfish repackage and reroute planktonic organic matter into a respiratory sink, rather then being potentially reincorporated into and transferred up the food web.
Table 1.
Bacterial respiration of jelly-DOM
| Bacterial respiration increases associated with jellyfish DOC production (BRJ) |
||||
|
Mnemiopsis blooms |
Chrysaora blooms |
|||
| Month | BRJ (mg C m−3·day−1) | % BRJ of BPb | BRJ (mg C m−3·day−1) | % BRJ of BPb |
| May | 1.2 ± 1.3 (3.0) | 9.2 ± 12.5 (32.1) | 0 | 0 |
| June | 13.0 ± 21.2 (96.3) | 12.7 ± 14.1 (45.2) | 0.1 ± 0.3 (1.2) | 0.1 ± 0.5 (2.4) |
| July | 4.9 ± 9.5 (30.7) | 3.7 ± 6.7 (18.7) | 10.1 ± 19.1 (60.1) | 7.2 ± 13.1 (34.2) |
| August | 0.8 ± 1.8 (7.1) | 0.5 ± 0.7 (2.7) | 7.3 ± 15.1 (54.6) | 9.4 ± 19.8 (73.1) |
Increased respiration by York River bacterial populations because of uptake of DOC released by M. leidyi and C. quinquecirrha jellyfish blooms. BRJ is increased bacterial respiration (BR) on jelly-C, and BPb is bulk bacterial community production of unfiltered water (Table S1). Bacterial metabolism data presented as monthly mean values ± 1 SD. Maximum values are in parentheses. DOC production by jellyfish blooms (Table S2) was determined by multiplying daily weight-specific DOC release rates (Fig. 1) by biomass of M. leidyi and C. quinquecirrha populations (Fig. S1). BRJ was based on differences in cell-specific BR between NWC and both jellyfish treatments normalized to DOC released by jellyfish in the experiments (Fig. 2 and Fig. S2). Final calculations of BRJ were determined by multiplying jelly-DOC–normalized single-cell BR by in situ bacterial abundances (cells m−3) and then by total daily amounts of jelly-C released by M. leidyi and C. quinquecirrha populations (Tables S1–S3). % BRJ of BPb = (BRJ/BPb) × 100.
Shifts in Bacterial Community Composition.
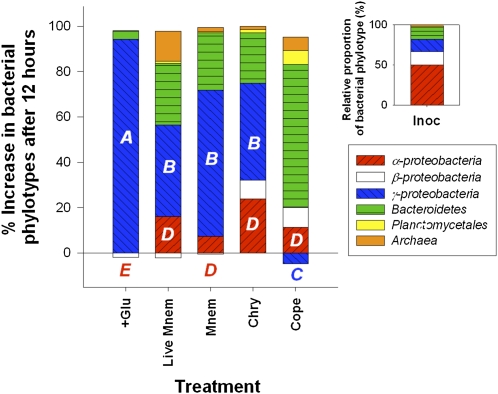
Finally, we analyzed to what extent these dramatic changes in bacterial metabolism induced by jellyfish were accompanied by shifts in bacterial community composition, by examining prokaryotic community structure from a summer incubation experiment using FISH (Tables S4 and S5). Comparisons between broad bacterial phylogenetic groupings have shown differences in the effectiveness of their uptake of specific types of organic substrates (19, 20), so shifts in bacterial community structure that underlie changes in metabolism at the community and single-cell level may reflect major changes in the dominant organic substrates in labile DOM pools (21, 22). Differences in prokaryote community structure were characterized by comparing changes after 12 h between the initial inoculum (i.e., ambient free-living microbial assemblage), a glucose addition, and jelly-DOM treatments (Fig. 3). A copepod DOM excretia treatment was included as an additional zooplankton control. Compared with untreated control containers that exhibited a decrease in all bacterial cells, we observed strong and consistent shifts in composition in the various treatments, with growth and increased abundance of resource-specific phylotypes that were not abundant in the ambient microbial assemblage. In particular, we observed high net growth and a 54% to >100% increase in γ-proteobacteria cells (based on changes in γ-proteobacteria relative to total bacteria after 12 h) versus low or no growth of the dominant in situ phylotype, α-proteobacteria, in all jellyfish and glucose addition containers (Fig. 3). Our ability to detect these changes in the microbial assemblage using FISH was also high with our probe set (Table S5), accounting for on average 92.5% ± 17.5% of total bacterial cells (hybridization efficiencies were 82.6% ± 15.1%; calculated as the total eubacteria counts to total bacteria counts stained with DAPI). Furthermore, these changes were not the result of bottle effects or nutrient enrichment as there was a decrease in γ-proteobacteria density and different resource-specific phylotypes (e.g., Bacteroidetes) emerged in copepod excretia (Fig. 3).
Fig. 3.
Effects of jelly-C uptake on bacterial community composition. Percent composition of bacterial phylotypes and the domain archaea in the natural water inoculum (Inoc, Insert), and proportional changes in these phylotypes after 12 h between the live Mnemiopsis (Live Mnem) and ctenophore (Mnem), Chrysaora medusa (Chry), and copepod (Cope) excretia treatments and glucose addition control (+Glu) in the bacterial dilution experiment (25 °C). The copepod treatment was included as an additional control. Phylotypes were detected with FISH using oligonucleotide probes labeled with CY3 (Table S5). FISH results from NWCs are not included because there was a decrease in all bacterial phylotypes in these containers. The emergence of Bacteroidetes rather than γ-proteobacteria in the copepod treatment suggests that changes in community structure to uptake of jelly-C were not the result of bottle or nutrient enrichment effects. Treatments and controls with different letters are significantly different (P < 0.05) with respect to increases in cell counts of α>- and γ-proteobacteria.
It is unknown what compounds are primarily used by γ-proteobacteria for their growth (19, 20). However, it is clear that when exposed to jelly-DOM, bacteria belonging to this group outcompeted the dominant α-proteobacteria in terms of increases in cell abundance and specific growth rates, and we hypothesize that this was because of a higher affinity for that type of organic matter (Fig. 3). This result suggests that by changing the quantity and presumably composition of labile DOM pools (Fig. 1), jellyfish can greatly influence not only the balance of microbial community metabolism (Fig. 2), but also the structure of bacterioplankton communities by favoring the development of specific groups and thus possibly altering not only the microbial C pathways, but also trophic interactions within the microbial loop.
Jelly-Carbon Shunt.
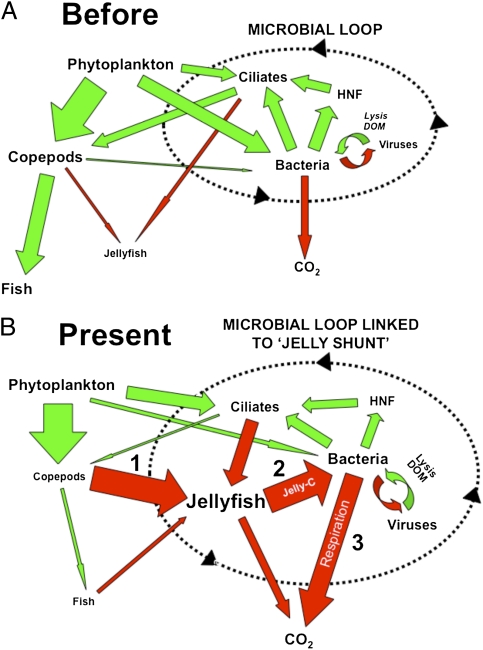
Our results show that the jellyfish–bacteria link resulted in an alternative C pathway for resource-specific bacterioplankton metabolism during jellyfish blooms in coastal and estuarine systems. Although we have demonstrated that jelly-DOM was mostly respired by bacteria cells, a small portion of jelly-DOM was incorporated as new bacterial biomass (Fig. 2, Fig. S2, and Table S3). Whether this jellyfish-derived bacterial C can be reincorporated into the planktonic food web will initially be a trade-off between the response of viruses and heterotrophic nanoflagellates (23, 24) (Fig. 4), with flagellate bacterivory representing the primary mechanism to reintroduce jellyfish C into the planktonic food web (25). In turn, potential increased flagellate biomass may have a “bottom-up” effect, resulting in increases in predators of flagellates (e.g., ciliates) and their predators (copepods). As jellyfish also consume ciliates (26, 27) and maintain copepod populations at low levels during blooms (4), flagellate C would potentially be assimilated back into gelatinous biomass. This process creates a hypothetical positive-feedback “jelly loop” that is controlled centrally by jellyfish predation and the pumping of jelly-DOM through the microbial loop: an ecosystem-scale process that we have defined as the “jelly-C shunt” (Fig. 4). In terms of C flows, the importance of microbial respiration in this cycle and strong links between jellyfish and bacterial metabolism are further emphasized, because BR releases C from the loop in a form (i.e., CO2) that reduces uptake and direct reincorporation into food webs by jellyfish, heterotrophic microbes, and other heterotrophic processes.
Fig. 4.
The “jelly carbon shunt.” Hypothetical changes in C pathways within the planktonic food web (A) before and (B) after (present) increases in jellyfish blooms. Green arrows indicate flows reincorporating C into the planktonic food web and potential transfer to higher trophic levels. Red arrows signify C pathways impacted by jellyfish. The size of the arrow indicates relative magnitude of C flow, and the size of the text indicates relative size of C pool. The direct link and increased influence of jellyfish and microbial pathways are emphasized by (i) the shunting of C away from fish production, (ii) the conversion of C into jellyfish biomass and subsequent release in excretia jelly-C, and (iii) utilization of jellyfish material for bacterial metabolism, especially respiration. The “jelly loop” involves the cycling of C between jellyfish, bacteria, heterotrophic nanoflagellates (HNF), and ciliates.
Ultimately, the strength of the jelly-C shunt will depend upon the interplay between the amount and activity of jellyfish biomass, local primary production, and the proportion of primary production that is assimilated initially by copepod populations and subsequently by jellyfish blooms (Fig. 4). In coastal and estuarine systems, the potential for primary production to be shunted into gelatinous biomass is high, with C-based estimates of copepod grazing on primary production ranging from 12% to 103% d−1, and jellyfish predation on copepod production ranging from 27% to 242% d−1 (4, 28). In addition, this C pathway may be accentuated because of other environmental forcing factors, such as climate change, which can alter the phenology of plankton communities (29). For example, many coastal and estuarine systems (e.g., Chesapeake Bay) have experienced major increases in spring-water temperatures over the past 40 y, resulting in the early appearance and prolonged seasonal persistence of Mnemiopsis blooms (4, 30, 31). These temporal shifts have major implications for food webs and in the functioning of the jelly-C shunt because ctenophores are then able to process and assimilate larger quantities of primary and secondary production into gelatinous biomass, which increases residence times for C assimilated in jellyfish (4). On seasonal time scales, these jellyfish-mediated processes may further impact coastal fisheries production by shunting C flows away from higher trophic levels.
Future Considerations.
Although we show how jellyfish blooms may affect C biogeochemistry at the ecosystem level by altering microbial community structure, metabolism, and trophic pathways in coastal and estuarine food webs, many of the processes controlling this jelly-C shunt may also apply in open-ocean systems. For example, blooms of pelagic tunicates (salps and doliolids) consume high amounts of picoautotrophic biomass (32, 33), thereby providing a more efficient and direct assimilation pathway for converting primary production into gelatinous biomass. However, we know very little about the causes, magnitude, and extent of jellyfish blooms in the open ocean. Furthermore, although we have demonstrated that the jelly-C shunt may be a major jellyfish-mediated process in estuaries and coastal oceans, the fate and biochemical composition of the C assimilated in gelatinous biomass, but not released as jelly-DOM, is still unclear. Is the fate of C in jellyfish blooms channeled through higher trophic level predators, such as turtles and pelagic fish, which consume jellyfish, or do massive die-out events occur whereby carcasses sink as “jelly-flux” to the benthos (34, 35), or are they decomposed on the way down by planktonic microbial communities (36)? With anticipated further increase in the magnitude and extent of jellyfish populations in the coming decades, our results suggest fundamental shifts in the biological structure and biogeochemical functioning of the marine systems affected, with potentially significant environmental, societal, and economic implications.
Methods
Jelly-DOM Release.
For each experiment, individual jellyfish were incubated in the dark at 20 or 25 °C for 4 to 12 h in 1.2-L (for M. leidyi) or 4-L (for C. quinquecirrha) acid-cleaned polycarbonate containers filled with 0.2 μm filtered (Nucleopore polycarbonate) low-nutrient Sargasso seawater diluted with Nanopure Diamond (Barnstead) water to in situ York River salinity [20–22 practical salinity units (psu)]. At the start of the experiment, one jellyfish was randomly added to each experimental container (treatment) and the release of DOM and inorganic N and P determined through measured changes in concentrations in the water. Experimental controls consisted of chambers absent of jellyfish, although the addition of animals was mimicked. The collection techniques and handling of animals did not damage or harm these jellyfish; however, all animals were rigorously inspected regularly before, during, and after experiments. In addition, animals were suspended in the containers by gently swirling the containers regularly during the incubation.
Bacterial Growth Efficiency.
Experiments consisted of measuring differences in BP (anabolism), BR (catabolism), and BGE in free-living bacterial assemblages exposed to jellyfish excretia (treatment), natural conditions (York River control), or added glucose (labile DOC control). York River surface water (0–2 m, 20–22 psu) was filtered through 1.2-μm AP15 glass-fiber filters using peristaltic pumps and acid-washed tubing, and 10 L of filtrate (York River control) was added to individual acid-washed buckets. Jellyfish treatments were prepared by incubating either 10 M. leidyi ctenophores (Mnemiopsis treatment) or one C. quinquecirrha medusa (Chrysaora treatment, summer only) in control water. After preparation, jellyfish were removed from treatments, and experimental water was screened through acid-washed 100-μm Nitex sieves before being added to triplicate closed, airtight 4-L Erlenmeyer experimental incubation setups (see SI Methods for detailed description of setup). Samples for bacterial abundance (BA), BP, BR, DOM, and inorganic nutrients were taken at the start of treatment preparation and during incubations at the initial (t0), mid- (t3, bacteria only), and final (t6) time points.
Bacterial Metabolism.
For bacterial counts (BA), fixed cells were stained with SYTO-13 and enumerated using flow cytometry. BP was measured using the 3H-leucine incorporation technique (37). For field samples, BA and BP were determined on whole (bulk) and filtered (< 1.2 μm) bacterial fractions. BR was determined by quantifying the decline in oxygen (O2) (38), as determined using membrane-inlet mass spectrometry (39). O2 uptake was transformed to C respired assuming a respiratory quotient of 1. BGE was determined as: BGE = BP/(BP + BR) (17).
Phylogenetic Diversity.
To analyze the responses of various prokaryote phylotypes to jellyfish DOM, we conducted a regrowth experiment in the dark at 25 °C (summer) in which an inoculum of natural, free-living bacteria (1.2 μm filtered York River water) was added to acid-washed 8-L polycarbonate carboys containing 0.2-μm filtered jelly- or copepod-DOM water (zooplankton treatments), York River water (control), or York River water with added glucose (labile DOC control). Bacterial community composition was determined by FISH using oligonucleotide probes conjugated with CY3 (21, 24) (see Table S5 for list of probes), and changes in community composition based on differences in phylotypes between the initial (t0) and 12-h time points.
See SI Methods for a full description of methods.
Supplementary Material
Acknowledgments
We thank C. Beauchemin, J. Cope, E. Condon, Q. Roberts, M. Erickson, J. Apple, R. I. E. Newell, D. Stoecker, B. Crump, S. Goldthwait, M. Sanderson, and H. Quinby for field and laboratory assistance; and K. Tang, E. Condon, J. Peloquin, M. Lomas, J. M. Gasol, G. Jackson, and two anonymous reviewers, whose comments improved the manuscript. This study was supported by National Science Foundation Grant OCE 0221825 (to D.K.S., D.B., and W. Smith).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015782108/-/DCSupplemental.
References
- 1.Mills CE. Jellyfish blooms: Are populations increasing globally in response to changing ocean conditions? Hydrobiologia. 2001;451:55–68. [Google Scholar]
- 2.Purcell JE, Uye S-I, Lo W-T. Anthropogenic causes of jellyfish blooms and their direct consequences for humans: A review. Mar Ecol Prog Ser. 2007;350:153–174. [Google Scholar]
- 3.Richardson AJ, Bakun A, Hays GC, Gibbons MJ. The jellyfish joyride: Causes, consequences and management responses to a more gelatinous future. Trends Ecol Evol. 2009;24:312–322. doi: 10.1016/j.tree.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Condon RH, Steinberg DK. Development, biological regulation, and fate of ctenophore blooms in the York River estuary, Chesapeake Bay. Mar Ecol Prog Ser. 2008;369:153–168. [Google Scholar]
- 5.Jackson JBC, et al. Historical overfishing and the recent collapse of coastal ecosystems. Science. 2001;293:629–637. doi: 10.1126/science.1059199. [DOI] [PubMed] [Google Scholar]
- 6.Hansson LJ, Norrman B. Release of dissolved organic carbon (DOC) by the scyhozoan jellyfish Aurelia aurita and its potential influence on the production of planktic bacteria. Mar Biol. 1995;121:527–532. [Google Scholar]
- 7.Kremer P. Respiration and excretion by the ctenophore Mnemiopsis leidyi. Mar Biol. 1977;44:43–50. [Google Scholar]
- 8.Carlson CA. Production and removal processes. In: Hansell DA, Carlson CA, editors. Biochemistry of Marine Dissolved Organic Matter. San Diego: Academic Press; 2002. pp. 91–151. [Google Scholar]
- 9.Faasse MA, Bayha KM. The ctenophore Mnemiopsis leidyi A. Agassiz 1865 in coastal waters of the Netherlands: An unrecognized invasion? Aquat Inv. 2006;1:270–277. [Google Scholar]
- 10.Licandro P, et al. A blooming jellyfish in the northeast Atlantic and Mediterranean. Biol Lett. 2010;6:688–691. doi: 10.1098/rsbl.2010.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuentes VL, Atienza D, Gili J-M, Purcell JE. First records of Mnemiopsis leidyi A. Agassiz 1865 off the NW Mediterranean coast of Spain. Aquat Inv. 2009;4:671–674. [Google Scholar]
- 12.Purcell JE, Shiganova TA, Decker MB, Houde ED. The ctenophore Mnemiopsis in native and exotic habitats: U.S. estuaries versus the Black Sea basin. Hydrobiologia. 2001;451:145–176. [Google Scholar]
- 13.Condon RH, Steinberg DK, Bronk DA. Production of dissolved organic matter and inorganic nutrients by gelatinous zooplankton in the York River estuary, Chesapeake Bay. J Plankton Res. 2010;32:153–170. [Google Scholar]
- 14.Ducklow HW, Mitchell R. Composition of mucus released by coral reef coelenterates. Limnol Oceanogr. 1979;24:706–714. [Google Scholar]
- 15.Wild C, Woyt H, Huettel M. Influence of coral mucus on nutrient fluxes incarbonate sands. Mar Ecol Prog Ser. 2005;287:87–98. [Google Scholar]
- 16.Rich JH, Ducklow HW, Kirchman DL. Concentrations and uptake of neutral monosaccharides along 14OdegW in the equatorial Pacific: Contribution of glucose to heterotrophic bacterial activity and the DOM flux. Limnol Oceanogr. 1996;41:595–604. [Google Scholar]
- 17.del Giorgio PA, Cole JJ. Bacterial growth efficiency in natural aquatic systems. Annu Rev Ecol Syst. 1998;29:503–541. [Google Scholar]
- 18.Carlson CA, del Giorgio PA, Herndl GJ. Microbes and the dissipation of energy and respiration: From cells to ecosystems. Oceanography (Wash DC) 2007;20(2):89–100. [Google Scholar]
- 19.Alonso-Sáez L, Gasol JM. Seasonal variations in the contributions of different bacterial groups to the uptake of low-molecular-weight compounds in northwestern Mediterranean coastal waters. Appl Environ Microbiol. 2007;73:3528–3535. doi: 10.1128/AEM.02627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cottrell MT, Kirchman DL. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol. 2000;66:1692–1697. doi: 10.1128/aem.66.4.1692-1697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouvier TC, del Giorgio PA. Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol Oceanogr. 2002;47:453–470. [Google Scholar]
- 22.del Giorgio PA, Bouvier TC. Linking the physiologic and phylogenetic sucessions in free-living bacterial communities along an estuarine salinity gradient. Limnol Oceanogr. 2002;47:471–486. [Google Scholar]
- 23.Fuhrman JA, Suttle CA. Viruses in marine planktonic systems. Oceanography. 1993;6:51–63. [Google Scholar]
- 24.Bouvier TC, del Giorgio PA. Key role of selective viral-induced mortality in determining marine bacterial community composition. Environ Microbiol. 2007;9:287–297. doi: 10.1111/j.1462-2920.2006.01137.x. [DOI] [PubMed] [Google Scholar]
- 25.Kirchman DL. Microbial Ecology of the Oceans. New York: Wiley-Liss; 2000. p. 542. [Google Scholar]
- 26.Stoecker DK, Michaels AE, Davis LH. Grazing by the jellyfish, Aurelia aurita, on microzooplankton. J Plankton Res. 1987;9:901–915. [Google Scholar]
- 27.Stoecker DK, Verity PG, Michaels AE, Davis LH. Feeding by larval and post-larval ctenophores on microzooplankton. J Plankton Res. 1987;9:667–683. [Google Scholar]
- 28.White JR, Roman MR. Seasonal study of grazing by metazoan zooplankton in the mesohaline Chesapeake Bay. Mar Ecol Prog Ser. 1992;86:251–261. [Google Scholar]
- 29.Edwards M, Richardson AJ. Impact of climate change on marine pelagic phenology and trophic mismatch. Nature. 2004;430:881–884. doi: 10.1038/nature02808. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan BL, van Keuren D, Clancy M. Timing and size of blooms of the ctenophore Mnemiopsis leidyi in relation to temperature in Narragansett Bay, RI. Hydrobiologia. 2001;451:113–120. [Google Scholar]
- 31.Costello JH, Sullivan BK, Gifford DJ, Van Keuren D, Sullivan LJ. Seasonal refugia, shoreward thermal amplification and metapopulation dynamics of the ctenophore Mnemiopsis leidyi in Narragansett Bay, Rhode Island. Limnol Oceanogr. 2006;51:1819–1831. [Google Scholar]
- 32.Madin LP, et al. Periodic swarms of the salp Salpa aspera in the Slope Water off the NE United States: Biovolume, vertical migration, grazing, and vertical flux. Deep Sea Res Part I Oceanogr Res Pap. 2006;53:804–819. [Google Scholar]
- 33.Sutherland KR, Madin LP, Stocker R. Filtration of submicrometer particles by pelagic tunicates. Proc Natl Acad Sci USA. 2010;107:15129–15134. doi: 10.1073/pnas.1003599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Billett DSM, Bett BJ, Jacobs CL, Rouse IP, Wigham BD. Mass deposition of jellyfish in the deep Arabian Sea. Limnol Oceanogr. 2006;51:2077–2083. [Google Scholar]
- 35.Lebrato M, Jones DOB. Mass deposition event of Pyrosoma atlanticum carcasses off Ivory Coast (West Africa) Limnol Oceanogr. 2009;54:1197–1209. [Google Scholar]
- 36.Titelman J, et al. Turnover of dead jellyfish: Stimulation and retardation of microbial activity. Mar Ecol Prog Ser. 2006;325:43–58. [Google Scholar]
- 37.Smith DC, Azam F. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar Microb Food Webs. 1992;6:107–114. [Google Scholar]
- 38.Apple JK, Del Giorgio PA, Kemp WM. Temperature regulation of bacterial production, respiration, and growth efficiency in a temperate salt-marsh estuary. Aquat Microb Ecol. 2006;43:243–254. [Google Scholar]
- 39.Kana TM, et al. Membrane inlet mass spectrometer for rapid high-precision determination of N2, O2, and Ar in environmental water samples. Anal Chem. 1994;66:4166–4170. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.