Abstract
Nutritional and genetic risk factors for intestinal tumors are additive on mouse tumor phenotype, establishing that diet and genetic factors impact risk by distinct combinatorial mechanisms. In a mouse model of dietary-induced sporadic small and large intestinal cancer in WT mice in which tumor etiology, lag, incidence, and frequency reflect >90% of intestinal cancer in Western societies, dietary-induced risk altered gene expression profiles predominantly in villus cells of the histologically normal mucosa, in contrast to targeting of crypt cells by inheritance of an Apc1638N allele or homozygous inactivation of p21Waf1/cip1, and profiles induced by each risk factor were distinct at the gene or functional group level. The dietary-induced changes in villus cells encompassed ectopic expression of Paneth cell markers (a lineage normally confined to the bottom of small intestinal crypts), elevated expression of the Wnt receptor Fzd5 and of EphB2 (genes necessary for Paneth cell differentiation and localization to the crypt bottom), and increased Wnt signaling in villus cells. Ectopic elevation of these markers was also present in the colon crypts, which are also sites of sporadic tumors in the nutritional model. Elevating dietary vitamin D3 and calcium, which prevents tumor development, abrogated these changes in the villus and colon cells. Thus, common intestinal cancer driven by diet involves mechanisms of tumor development distinct from those mechanisms that cause tumors induced by the rare inheritance of a mutant adenomatous polyposis coli (Apc) allele. This is fundamental for understanding how common sporadic tumors arise and in evaluating relative risk in the population.
Keywords: sporadic, colon cancer, Western diet, microarray
Both large and small intestinal tumors develop in WT C57BL/6 mice after long-term (1.5–2 y) feeding of a fully defined Western-style diet that combines a number of risk factors for human colon cancer (NWD1: higher fat, lower calcium, vitamin D3, donors to the single carbon pool, and fiber, and carbohydrate supplied by sucrose, a refined sugar commonly consumed in developed but not undeveloped countries) (1, 2). These dietary-induced tumors arise with an incidence, frequency, and lag (i.e., two-thirds of the host lifespan) similar to those of human colon tumors, making them a useful model of sporadic intestinal cancer, the form of the disease responsible for >90% of intestinal cancer in the human (1, 2).
In contrast, inheritance of a mutant Apc allele causes much more rapid development of mouse intestinal tumors (3–5), similar to tumor development in familial adenomatous polyposis (FAP) patients who inherit mutations in Apc, a syndrome that accounts for 1–2% of colon tumors in humans (6). Western-style diets increase and accelerate the tumor phenotype of Apc+/− mice (7, 8), establishing that dietary and genetic stimulation of tumorigenesis involve at least partially distinct and interactive effects. Indeed, although there are overlaps in the expression signatures of intestinal epithelial cells of mice fed the Western-style diet and Apc1638N/+ mice, there are also many differences (2). Furthermore, inactivation of the cdk inhibitors p21Waf1/cip1 or p27Kip1 in Apc1638N/+ mice also produced a more aggressive tumor phenotype than in Apc1638N/+ mice, but even in these compound mutant mice, a Western-style diet amplified and accelerated tumor development, establishing that multiple distinct pathways interact in altering probability of intestinal tumorigenesis (9–11).
Therefore, we determined how the Western-style diet, as a model of sporadic intestinal cancer, altered programming in either crypt or villus cells of the histologically normal mucosa before tumors developed compared with reprogramming in these cells caused by inheritance of an Apc mutation or inactivation of p21WAF1/cip1. Altered expression profiles and functional gene groups in either crypt or villus cells in the histologically normal mucosa were distinct for the three models: Apc1638N/+ mice, p21−/− mice, and WT mice fed the Western-style diet. Moreover, the Western-style diet induced alterations primarily in villus cells in contrast to the principal effects of either genetic mutation in crypt cells. Importantly, this encompassed induction by diet but not genotype of ectopic expression of Paneth cell markers in villus cells, elevated expression of the Wnt receptor Fzd5 and the EphB2 receptor (both necessary for normal differentiation and localization of Paneth cells) (12–15), and elevated Wnt signaling in villus cells. Similar changes were also seen in the colon of mice fed the Western diet (also a site of sporadic tumor development linked to diet), and the changes in both the small intestinal and colonic mucosa were abrogated by elevating vitamin D3 and calcium in the Western-style diet, which eliminates eventual tumor development (2).
These alterations in the histologically normal mucosa have important implications for how common, sporadic tumors develop compared with rare tumors that arise because of inheritance of genetic mutation, and they may provide markers for evaluation of relative probability for tumor development in the general population.
Results
Altered Gene Expression Along the Crypt-Villus Axis (CVA) by Dietary and Genetic Factors.
Dietary groups were WT C57BL/6 mice fed one of the following three diets for 55 mo (∼1 y) from weaning: control AIN76A diet, NWD1 (based on AIN76A but higher in fat and phosphate and lower in calcium, vitamin D3, methyl donors, and fiber), reflecting consumption levels of these nutrients by large segments of the population (1, 16) and causing ∼25% of the mice to develop one to two tumors in the small and large intestine between 1.5 and 2 y of age (1, 2), and NWD2, in which calcium and vitamin D3 in the NWD1 is elevated, preventing this tumor formation (2). Tumor development caused by the NWD1 reflects the etiology, lag, incidence, and frequency of most intestinal cancer that develops in the United States, and it is a model of sporadic intestinal cancer (1, 2). Genetic groups used were Apc1638N/+ mice, in which three to five intestinal tumors develop from 6 to 12 mo of age, and p21Waf1/cip1−/− mice, which do not develop intestinal tumors but introduction of this mutation into Apc1638N/+ mice accelerates and increases intestinal tumor phenotype (9). All genetic strains were on a C57BL/6 background, maintained ad libitum on control AIN76A-defined diet, and compared with WT littermates fed the same diet for the same period (3 mo).
RNA of intestinal epithelial cells from the top of the villi (F1) or bottom of the crypts (F10) of these six groups (four mice per group except for the NWD2-fed mice, for which villus cells were analyzed from three mice) was used for expression profiling with Affymetrix 430 2.0 mouse arrays. Unsupervised clustering of the data for the 35,112 probe sets that passed initial filtering for all analyses yielded two main branches with 22 of 23 villus samples (F1) in branch A and 24 of 24 crypt samples (F10) in branch B (Fig. S1); this observation was consistent with the expectation and our previously reported data (17, 18) that villus cells are highly reprogrammed relative to crypt cells and that the method of cell isolation (Methods and Fig. S7) yields cell fractions highly enriched for each compartment.
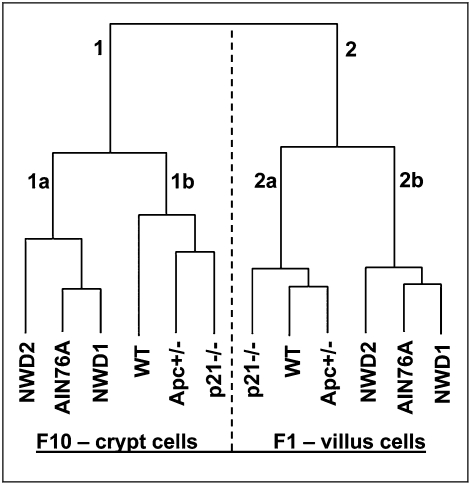
Clustering of the mean value for each probe set for mice in each of the six different genetic/dietary groups again clearly separated villus and crypt cell samples (Fig. 1, branches 1 and 2). Within each branch, the genetic groups (Apc1638N/+, p21Waf1/cip1−/−, and respective WT controls) clustered together (Fig. 1, branches 1b and 2a) and separated from the dietary groups (Fig. 1, branches 1a and 2b). Because the control groups in each case are congenic C57BL/6 mice fed the same standardized control AIN76A diet but separated along these lines, this likely reflects the different ages of the genetic and dietary groups, which were chosen in each case to be ∼6 mo before tumors are detected (9 or 18 mo, respectively, for the genetic and dietary-induced tumors).
Fig. 1.
Unsupervised clustering of Affymetrix expression data for crypt and villus cells based on the mean of each probe for the four mice of each group. Expression profiles of villus and crypt cells are distinctly independent of dietary/genetic risk. Fig. S1 has details of data generation and analyses.
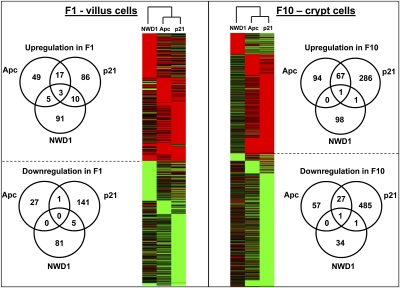
Pair-wise comparison among all groups was based on both magnitude and significance of change [greater than twofold and P < 0.05 (Student's t test), respectively], and also, it included sequences present in all samples of one group but not detectably above background in the second group. Differentially expressed genes for all comparisons, with overlaps among groups, are presented in Table S1 (available at augenlichtlab.com) and summarized in Fig. 2. As reported, inheritance of the Apc1638N mutant allele had more pronounced effects on altering gene expression in the crypt (18), the site where Wnt signaling, which it regulates, is active in maintaining a crypt progenitor cell population and in lineage allocation of cells (19); for Apc1638N/+ mice, 162 and 85 genes were up- and down-regulated in the crypt, respectively, which was more than two times the 74 and 28 up- and down-regulated genes in the villus (Fig. 2), respectively. Similarly, for p21−/− mice, genes that increased and decreased in expression in the crypt exceeded those in the villus by three- and fivefold, respectively. These genotype-generated alterations contrasted with dietary-induced risk; for the NWD1, there was a bias to increased expression changes in villus cells, especially for down-regulated genes (36 in the crypt but 86 in the villus). There was very limited overlap among up- and down-regulated genes in genetic risk (the mucosa of Apc1638N/+ mice fed AIN76A) compared with dietary risk (C57BL/6 mice fed NWD1) (Fig. 2), and there was little overlap among all three groups (Fig. 2), with distinct gene subsets unique to each of the three risk factors. Distinction among the effects of these risk factors extended to functional group analysis (Fig. S2 and Table S2). Therefore, both limited overlap among effects of these risk factors on expression patterns in the mucosa as well as differential targeting in relation to crypt–villus architecture explain why the three factors are additive in generating tumor phenotype.
Fig. 2.
Limited overlap of gene expression changes for each dietary and genetic risk group in both villus and crypt cells.
Dietary-Induced Alteration of Paneth Cell Markers in Villus Cells.
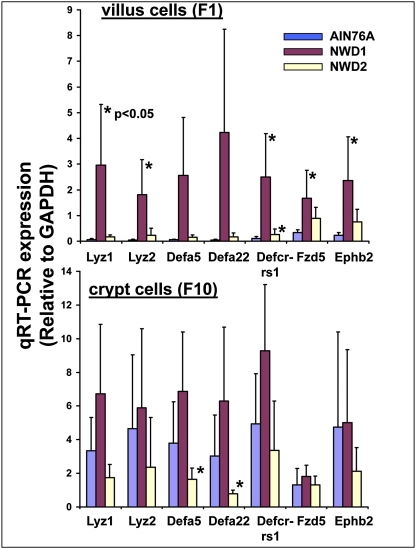
Functional group analysis identified the category “defense response to bacterium” uniquely enriched in villi of WT mice fed NWD1 (P < 0.0001) (Table S2). These markers are normally confined to Paneth cells at the bottom of the small intestinal crypt. We, therefore, analyzed a panel of five Paneth cell markers (lysozyme 1 and 2 and three defensins) by quantitative RT-PCR (qRT-PCR) in four mice per group. In WT mice fed control AIN76A diet (Fig. 3, blue bars), expression of each Paneth cell marker in villus cells was very low (Fig. 3 Upper) at <5% the level detected in crypt cells (Fig. 3 Lower), consistent with normal localization of Paneth cells to the lower crypt. However, feeding the high-risk NWD1 elevated expression of each marker 10- to 50-fold in villus cells (Fig. 3 Upper). There were similar, although much smaller, increases in these markers in crypt cells (Fig. 3 Lower), but these were not statistically significant. Similar changes were seen in the array data (Fig. S3), showing that these changes were not because of changes in GAPDH used as the control for the qRT-PCR analyses. Furthermore, as suggested by the array data (www.ncbi.nlm.nih.gov/geo/) and functional group analysis (Fig. S2 and Table S2), qRT-PCR assay confirmed that neither inheritance of the Apc1638N allele nor homozygous inactivation of p21 had detectable effects on Paneth cell markers in either villus or crypt cells (available at augenlichtlab.com). This contrasts with reports that reduction of the inherited Apc mutation to homozygosity and thus very high levels of Wnt signaling can drive elevated Paneth cell differentiation, but only in cells of the crypt (12–14).
Fig. 3.
Level of expression of Paneth cell markers, Fzd5, and Ephb2 in villus and crypt cells of mice maintained on AIN76A, NWD1, and NWD2 from weaning for 1 y. Expression levels determined by qRT-PCR and normalized to GAPDH expression, determined similarly.
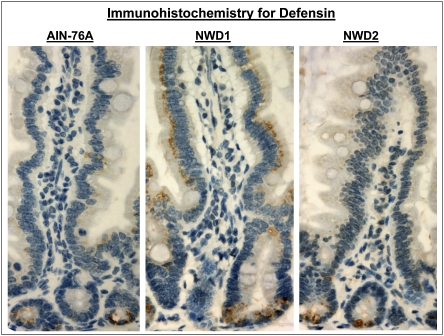
The NWD1-induced elevation in defensin and lysozyme in villus cells was also detected by Western blot analysis (Fig. S4) and pursued by immunohistochemistry. Defensin expression was detected only in crypt cells in mice fed AIN76A, but in mice fed NWD1, positive staining was all along the villus (Fig. 4). Villus cells that stained positive do not have the appearance of Paneth cells; staining is intracellular in the apical portion of enterocytes. Therefore, it is likely that this represents ectopic expression of the gene in inappropriate epithelial cell types rather than altered allocation of cells to the mature Paneth cell lineage. Lysozyme expression was also expressed ectopically in villus cells in mice fed NWD1 compared with AIN76A (Fig. S5), although the pattern of villus staining was more focal than for defensin, and there was some staining of goblet cell vacuoles. The differences in staining pattern between defensin and lysozyme may be related to a strong Tcf binding site (Wnt signaling response element) in the defensin promoter but not in the lysozyme promoter (14). Hence, as suggested, the Wnt signaling regulation of the two genes may be through different mechanisms and/or involve different threshold levels for activation (13).
Fig. 4.
Immunohistochemical detection of the Paneth cell marker defensin in the intestinal mucosa of mice maintained on AIN76A, NWD1, or NWD2 for 1 y from weaning. Positive staining is brown and localized to Paneth cells in the crypt for mice fed AIN76A or NWD2, but also, it is present in the apical region of normal-appearing enterocytes in mice fed NWD1.
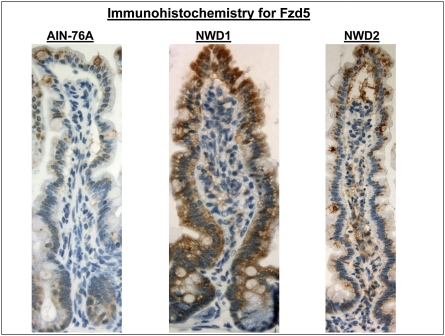
Regulation of Paneth cell differentiation by Wnt signaling (12–14) is through the Wnt receptor Fzd 5 (14), which is normally confined in expression to crypt cells (20). Indeed, Fzd5 expression was undetectable in the villus cell array data of control mice fed AIN76A (available at augenlichtlab.com) and was very low in these cells by qRT-PCR assay, but it was highly elevated in mice fed NWD1, which coincides with elevation in Paneth cell markers (Fig. 3 Upper). This was also confirmed by immunohistochemistry (Fig. 5). There was also a significant increase in the NWD1-fed mice of EphB2, a gene necessary for migration of Paneth cells to their normal crypt location (15) (Fig. 3 Upper). These changes were not seen in villus cells of Apc1638N/+ or p21−/− mice (available at augenlichtlab.com).
Fig. 5.
Immunohistochemical detection of the Wnt receptor Fzd 5 in mice fed AIN76A, NWD1, or NWD2 for 1 y from weaning.
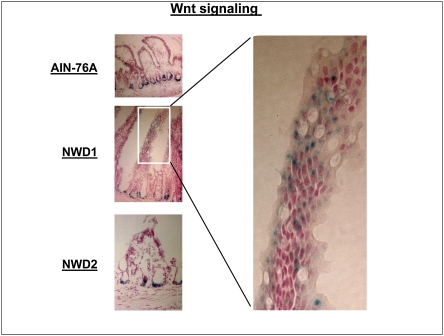
The Wnt ligands 2, 4, and 5a are expressed by mesenchymal cells along the villi, especially at the villus top (20). Wnt 5a is a ligand for Fzd5 (21), raising the possibility that elevated Fzd5 may increase Wnt signaling in villus cells. This is suggested by preferential enrichment of expression changes in the functional group “Wnt signaling” in villus cells of mice fed NWD1 (P = 0.01) (Table S2). To investigate this possibility, a mouse strain in which expression of a lacZ transgene is a marker of Wnt signaling (22) was backcrossed for 10 generations onto the C57BL/6 background. These mice were then fed different diets for 3 mo from weaning, and frozen sections were analyzed for β-galactosidase activity, the product encoded by lacZ. In mice fed AIN76A, β-galactosidase activity was confined to crypt cells, but when fed NWD1, activity expanded into the villus region (Fig. 6).
Fig. 6.
Detection of Wnt signaling by assay of β-galactosidase activity (blue) in frozen sections of the mucosa of C57BL/6 mice that harbor a lacZ transgene driven by a minimal fos promoter under regulation of three tandem Tcf4 binding sites. In this case, mice were fed the three diets from weaning for 3 mo.
Colonic tissue, also a site of sporadic tumor formation in mice fed the NWD1, was also investigated. Defensin, lysozyme, and Fzd5 were all expressed at a very low level in the colon of mice fed the control AIN76A diet, consistent with the absence of Paneth cells in this tissue (Fig. 7). However, each was markedly elevated in the colon when mice were fed NWD1, with lysozyme expression again more focal than defensin like in the small intestine. Furthermore, as in the small intestine, this was associated with increased Wnt signaling throughout the colonic crypts detected by immunostaining for β-galactosidase (Fig. 7).
Fig. 7.
Immunohistochemical detection of defensin, lysozyme, Fzd5, and β-galactosidase in the colon of mice fed AIN76A, NWD1, or NWD2 for 1 y from weaning (3 mo for beta-galactosidase).
Elevating calcium and vitamin D3 in the NWD1 (termed NWD2) prevents development of dietary-induced tumors in both the small and large intestine (1, 2) or the increase in tumors in Apc1638/N+ mice caused by a related Western-style diet also higher in fat and lower in calcium and vitamin D3 (8). We identified a subset of sequences in the mucosa for which expression tracked with dietary calcium and vitamin D3 levels in NWD1 and NWD2 (2). In this regard, Paneth cell marker elevation, up-regulation of Fzd5 and EphB2, and elevated Wnt signaling in villus cells were all prevented by elevating dietary calcium and vitamin D3 in NWD2 (Figs. 3 Upper and 4–6 and Figs. S3 and S4), and elevated expression of Paneth cell markers, up-regulation of Fzd5, and elevation in Wnt signaling were also prevented in the colon (Fig. 7). The correspondence between the molecular and immunohistochemical data is noteworthy; both methods show almost complete prevention of defensin expression in villus cells but an incomplete prevention of Fzd5 expression by feeding of NWD2 (Figs. 3 Upper, 4, and 5).
Alteration of Other Lineage Markers.
Inheritance of the Apc1638N allele perturbs the dynamics of Wnt and Notch signaling along the crypt–villus axis in the histologically normal mucosa, which is associated with dampening of the reprogramming of intestinal epithelial cells and altered expression of goblet, enteroendocrine, and enterocyte markers in cells of the villi (18). Markers of these lineages were similarly altered by the NWD1 and inactivation of p21; there was a general decrease in secretory cell markers (goblet and enteroendocine) and increase in enterocyte markers (Fig. S6). In each case, the sole exception to this pattern was the enteroendocrine marker Gcg. However, these differences were much more modest than for dietary induction of Paneth cell markers, and they do not satisfy the criteria for alterations described in Fig. 2 (twofold change and P < 0.05). Nevertheless, they seem to be consistent, if subtle, alterations of the mucosa at risk, regardless of etiology.
Discussion
Intestinal carcinoma develops in a classic multistep manner (6, 23). High-risk factors, such as genetic mutations in oncogenes or tumor suppressor genes, and habitual preference for a diet higher in fat but lower in calcium, vitamin D, methyl donors, and fiber establish predisposition for intestinal tumorigenesis. Although subsequent genetic hits, including mutations or epigenetic changes in Apc, p53, and ras and perhaps in a large number of other genes and pathways, are required for tumor formation and progression (24, 25), dysregulation of cellular functions preexists in the histologically normal intestinal mucosa at elevated probability of tumor formation long before detectable alterations in histopathology or frank tumor formation (2, 18, 26). Here, we show that the programming of crypt and villus cells in mucosa at higher probability for tumor development in relation to nutritional factors differs distinctly from that of either Apc1638N/+ or p21Waf1/cip1-/- mice. Moreover, in the small intestine, the dietary effects are more prominent in villus cells in contrast to the genetically induced effects that are concentrated in crypt cells.
Among the dietary-induced changes in the villi is elevation of Paneth cell markers accompanied by elevated expression of Fzd5, a Wnt receptor fundamental for specification of this lineage (14). Importantly, several Wnt ligands, including Wnt 2b, 4, and 5a, are expressed by mesenchymal cells along the length of the villi, particularly at the villus tips, and therefore, especially Wnt 5a (21), could function as cognate ligands for this dietary-induced expression of Fzd5. Indeed, Wnt signaling, restricted to crypt cells in mice fed the control diet, is elevated in villus cells in mice fed high-risk NWD1. Paneth cell localization also depends on EphB guidance receptor interaction with EphrinB expressed on other cell types, causing repulsion of Paneth cells and thus, their migration to the bottom of the crypt (15). Thus, the up-regulation of EphB2 along with Fzd5 in the villus of mice fed the NWD1 may drive dietary expression and mislocalization of cells expressing Paneth cell markers. The dietary-induced increases in Paneth cell markers, Fzd5, and Wnt signaling are also seen throughout the crypt of the colon, also a site for sporadic tumor development in mice fed the Western-style diet (1, 2).
Although complete loss of Apc function and thus, high Wnt signaling that initiates tumor development induces Paneth cell differentiation in crypts (12–14), we did not detect increased expression of Paneth cell markers in either crypt or villus cells of the Apc1638N/+ or p21−/− mice. However, loss of a single Apc allele would likely alter Wnt signaling much more modestly than loss of both alleles, and it has been emphasized that expression of Paneth cell markers may be highly sensitive to levels of Wnt signaling (13).
Elevating calcium and vitamin D3 in the NWD2 prevents the increases in the Paneth cell markers, Fzd5, and EphB2 and elevated Wnt signaling in villus cells and the colon as well as dietary-induced small and large intestinal tumor formation in the mice (1, 2). This provides a strong link among these changes and probability of tumor development. This may involve several mechanisms. It has recently been reported that Paneth cells likely signal to Lgr5+ stem cells at the bottom of the crypt, providing a niche necessary for survival and stem cell functions of the Lgr5+ cells (27), a cell population that is the necessary target for transformation by Apc mutation (28). How the dietary-induced ectopic expression of Paneth cell markers may affect this Lgr5+ population or other aspects of mucosal function and physiology, such as modulation of the microfloral populations in the intestine or host response to these commensal bacteria, may be subtle: only a few, at most, of the ∼1012 cell divisions that occur in the mucosa over the lifetime of these mice lead to tumors, which is also true of human sporadic intestinal tumors.
Finally, our data showing that about two-thirds of the expression changes in Apc1638N/+ mice are localized to the crypt are consistent with the fact that, in mice that inherit a mutant Apc allele, somatic reduction to homozygosity at this locus causes hyperproliferation and budding of new glands from the crypt region, with the outgrowths expanding to form adenomas (28–30). However, in mice fed the Western-style diet, which is a model of human sporadic colon cancer (similar incidence, frequency, and lag in regard to lifespan and dietary risk factors), the majority of the changes were in villus cells in the small intestine, and changes in Paneth cell markers were distributed throughout the colonic crypt. In this regard, it is interesting that genetic analysis of early-stage human colon tumors, specifically excluding tumors in FAP patients, suggested that human sporadic colon tumors may form in a top-down manner, with transformed cells and dysplastic crypts farther up along the crypt–luminal axis expanding down into the crypt (31). Thus, although APC regulation of Wnt signaling is fundamental to both genetic and dietary-induced tumorigenesis, the vast majority of human intestinal tumors that are sporadic may develop with a distinctly different morphogenesis than mouse genetic models that target genes active in the proliferating/progenitor cell compartment. It has been discussed that this pattern of top-down morphogenesis does not require that initiation (e.g., gene mutation) take place in cells outside the crypt, only that such initiated cells eventually expand from extracrypt regions (32, 33).
In summary, reprogramming of crypt and villus cells in a dietary mouse model of common human sporadic intestinal cancer and the concentration of changes along the crypt–villus axis differ from that caused by mutations in either the Apc or the p21WAF1/cip1 genes, which initiate and accelerate intestinal tumorigenesis, respectively. These findings explain why the three risk factors act combinatorially in causing intestinal tumors. Changes in the mucosa initiated by these factors impinge on a number of aspects of the normal lineage-specific reprogramming of cells along the crypt–luminal axis, especially the distribution of Paneth cell markers throughout the small and large intestine, with important implications for mechanisms that drive elevated probability of tumor development. The changes in differentiation markers are also of great significance for two additional reasons. First, if early changes in the mucosa at risk involved only alterations in cell proliferation, this might give rise to hyperplastic lesions that have limited malignant potential rather than malignant tumors that are characterized by abnormal cell differentiation and gland morphology. Second, analyses of these early changes provide approaches to evaluate relative risk for intestinal tumors in individuals in the general population.
Methods
Mice.
Generation and genotyping of the Apc1638N/+ and p21Waf1/cip1−/− mice, both on a C57BL/6 background, were reported (4, 9). WT C57BL/6 mice were from Jackson Laboratory. Genotypic comparisons were among Apc1638N/+, p21WAF1/cip1−/−, or WT C57BL/6 mice fed AIN76A control diet from weaning for 3 mo before being euthanized. For analysis of dietary effects, C57BL/6 mice were randomized at weaning to maintenance on AIN76A, NWD1, or NWD2 for 55 wk before being euthanized. The diets were supplied by Research Diets and have been described in detail (1, 2) (note that, although vitamin D3 and calcium are altered in the Western-style diets from levels in AIN76A, the ratio to phosphorous is not kept constant). The exceptions were the Tcf4-lacZ mice backcrossed for 10 generations to a C57BL/6 background and maintained on diets from weaning for 3 mo before being used for analysis of Wnt signaling.
Fractionation of Intestinal Epithelium.
Ten fractions of epithelial cells were sequentially isolated from the top of the villus (F1) to the bottom of the crypt (F10) of histologically normal mouse small intestinal mucosa as reported (34, 35), and as modified and extensively validated (17, 18, 36, 37). For example, there are gradients of expression in these fractions of c-myc, which drive proliferation of the cells in the progenitor cell compartment at the bottom of the crypt, and Math1, a component of the Notch signaling pathway that specifies secretory cell differentiation in the intestinal mucosa and is elevated when Notch signaling decreases as cells leave the progenitor cell compartment (details of fractionation provided in Fig. S7).
Expression Analysis.
RNA preparation, microarray hybridization, and data analysis were as described in Fig. S1.
qRT-PCR.
Steady-state levels of mRNA were determined by qRT-PCR (Prism 7900HT Sequence Detector; Applied Biosystems); 5 μg total RNA were reverse-transcribed into cDNA (Superscript II Reverse Transcriptase; Invitrogen). Optimal primer sequences were designed using Primer-BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) or were obtained from Primer Bank (http://pga.mgh.harvard.edu/primerbank/; primer pairs listed in Table S3). Specificity of each primer pair was confirmed by observation of a single amplified product by melting curve analysis (7900HT; Applied Biosystems) and agarose electrophoresis. PCR was performed with SYBR-Green PCR-Master Mix (Applied Biosystems). A standard curve method quantified relative target gene expression, with normalization to expression of the Gapdh gene.
Histochemistry.
Mouse intestinal tissues were fixed with 10% formalin and embedded in paraffin, and sections were deparaffinized in Histo-Clear (National Diagnostics) and rehydrated in graded ethanol. Endogenous peroxidase was blocked by incubation in 3% H2O2 solution in 50% methanol two times for 15 min. Antigen retrieval was performed by boiling sections for 10 min in 10 mM sodium citrate buffer (pH 6.0) in a microwave oven; the exception was lysozyme detection, for which sections were incubated in 200 μg/mL proteinase K in 50 mM Tris (pH 7.4) at room temperature for 5 min. Nonspecific binding was blocked for 45 min with 10% goat serum in PBS, with sections then incubated at 4 °C overnight with rabbit antidefensin Ab (diluted 1:500; gift from Andre Ouellette, University of Southern California, Los Angeles, CA), rabbit anti-Fzd5 Ab (1:25 0, ab75234; Abcam), rabbit antilysozyme Ab (1:1,000, A0099; Dako), or rabbit anti–β-galactosidase Ab (1:500, A-11132; Invitrogen) followed by biotinylated goat anti-rabbit IgG (1:500, BA-1000; Vector Lab,) for 30 min. The reaction was visualized using the Vectastain Elite ABC Kit (Vector Lab) and DAB Quanto kit (Thermo Fisher Scientific). Sections were counterstained with hematoxylin, mounted in Permount (Thermo Fisher Scientific), and examined under a light microscope. For enzymatic analysis of β-galactosidase activity in tissue from lacZ mice, dissected tissue was embedded in optimal cutting temperature (OCT) compound (Sakura), frozen on dry ice, and sectioned by cryostat. Activity was detected using the LacZ Tissue staining kit (Invivogen), which was modified: sections were fixed 5′ at room temperature (2% paraformaldehyde, 0.125% glutaraldehyde in PBS), washed five times in PBS, and incubated with staining buffer (6 mM potassium ferricyanide, 6 mM potassium ferrocyanide, 2 mM MgCl2, 0.02% Igepal, 0.01% Na deoxycholate in PBS) without X-gal for 5 min at room temperature followed by using staining solution (1 mg/mL X-Gal in staining buffer) for 14 h at 37 °C, washing with PBS five times, and counterstaining with fast red.
Statistical Analysis.
BRB-ArrayTools was used for microarray group comparisons. Student's t test was performed in group-wise comparison of gene expression. A P value < 0.05 combined with a greater than twofold change were considered significant in accordance with the recommendations of the MicroArray Quality Control project group, which were shown to achieve a balance of sensitivity, specificity, and reproducibility (38, 39).
Supplementary Material
Acknowledgments
This work was supported in part by National Cancer Institute Grants U54 CA100926, CA114265, CA135561, and P30-CA13330 and American Institute of Cancer Research Grant AICR 09A020.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data (MIAMI compliant) reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE29538).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017668108/-/DCSupplemental.
References
- 1.Newmark HL, et al. A Western-style diet induces benign and malignant neoplasms in the colon of normal C57Bl/6 mice. Carcinogenesis. 2001;22:1871–1875. doi: 10.1093/carcin/22.11.1871. [DOI] [PubMed] [Google Scholar]
- 2.Yang K, et al. Dietary induction of colonic tumors in a mouse model of sporadic colon cancer. Cancer Res. 2008;68:7803–7810. doi: 10.1158/0008-5472.CAN-08-1209. [DOI] [PubMed] [Google Scholar]
- 3.Su L-K, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 4.Fodde R, et al. A targeted chain-termination mutation in the mouse Apc gene results in multiple intestinal tumors. Proc Natl Acad Sci USA. 1994;91:8969–8973. doi: 10.1073/pnas.91.19.8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oshima M, et al. Loss of Apc heterozygosity and abnormal tissue building in nascent intestinal polyps in mice carrying a truncated Apc gene. Proc Natl Acad Sci USA. 1995;92:4482–4486. doi: 10.1073/pnas.92.10.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 7.Wasan HS, Novelli M, Bee J, Bodmer WF. Dietary fat influences on polyp phenotype in multiple intestinal neoplasia mice. Proc Natl Acad Sci USA. 1997;94:3308–3313. doi: 10.1073/pnas.94.7.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang K, et al. Dietary modulation of carcinoma development in a mouse model for human familial adenomatous polyposis. Cancer Res. 1998;58:5713–5717. [PubMed] [Google Scholar]
- 9.Yang WC, et al. Targeted inactivation of the p21(WAF1/cip1) gene enhances Apc-initiated tumor formation and the tumor-promoting activity of a Western-style high-risk diet by altering cell maturation in the intestinal mucosal. Cancer Res. 2001;61:565–569. [PubMed] [Google Scholar]
- 10.Yang W, Bancroft L, Nicholas C, Lozonschi I, Augenlicht LH. Targeted inactivation of p27kip1 is sufficient for large and small intestinal tumorigenesis in the mouse, which can be augmented by a Western-style high-risk diet. Cancer Res. 2003;63:4990–4996. [PubMed] [Google Scholar]
- 11.Yang W, Bancroft L, Liang J, Zhuang M, Augenlicht LH. p27kip1 in intestinal tumorigenesis and chemoprevention in the mouse. Cancer Res. 2005;65:9363–9368. doi: 10.1158/0008-5472.CAN-05-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreu P, et al. Crypt-restricted proliferation and commitment to the Paneth cell lineage following Apc loss in the mouse intestine. Development. 2005;132:1443–1451. doi: 10.1242/dev.01700. [DOI] [PubMed] [Google Scholar]
- 13.Andreu P, et al. A genetic study of the role of the Wnt/beta-catenin signalling in Paneth cell differentiation. Dev Biol. 2008;324:288–296. doi: 10.1016/j.ydbio.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 14.van Es JH, et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- 15.Batlle E, et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- 16.Newmark HL. Nutrient density: An important and useful tool for laboratory animal studies. Carcinogenesis. 1987;8:871–873. doi: 10.1093/carcin/8.7.871. [DOI] [PubMed] [Google Scholar]
- 17.Mariadason JM, et al. Gene expression profiling of intestinal epithelial cell maturation along the crypt-villus axis. Gastroenterology. 2005;128:1081–1088. doi: 10.1053/j.gastro.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 18.Wang D, et al. Altered dynamics of intestinal cell maturation in Apc1638N/+ mice. Cancer Res. 2010;70:5348–5357. doi: 10.1158/0008-5472.CAN-09-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van de Wetering M, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 20.Gregorieff A, et al. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 21.He X, et al. A member of the Frizzled protein family mediating axis induction by Wnt-5A. Science. 1997;275:1652–1654. doi: 10.1126/science.275.5306.1652. [DOI] [PubMed] [Google Scholar]
- 22.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 23.Kinzler KW, Vogelstein B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature. 1997;386:761–763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 24.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 25.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 26.Yeung AT, et al. One-hit effects in cancer: Altered proteome of morphologically normal colon crypts in familial adenomatous polyposis. Cancer Res. 2008;68:7579–7586. doi: 10.1158/0008-5472.CAN-08-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 29.Oshima H, Oshima M, Kobayashi M, Tsutsumi M, Taketo MM. Morphological and molecular processes of polyp formation in Apc(delta716) knockout mice. Cancer Res. 1997;57:1644–1649. [PubMed] [Google Scholar]
- 30.Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: From endoderm to cancer. Genes Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- 31.Shih I-M, et al. Top-down morphogenesis of colorectal tumors. Proc Natl Acad Sci USA. 2001;98:2640–2645. doi: 10.1073/pnas.051629398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preston SL, et al. Bottom-up histogenesis of colorectal adenomas: Origin in the monocryptal adenoma and initial expansion by crypt fission. Cancer Res. 2003;63:3819–3825. [PubMed] [Google Scholar]
- 33.Thirlwell C, et al. Clonality assessment and clonal ordering of individual neoplastic crypts shows polyclonality of colorectal adenomas. Gastroenterology. 2010;138:1441–1454. doi: 10.1053/j.gastro.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 34.Weiser MM. Intestinal epithelial cell surface membrane glycoprotein synthesis. I. An indicator of cellular differentiation. J Biol Chem. 1973;248:2536–2541. [PubMed] [Google Scholar]
- 35.Weiser MM. Intestinal epithelial cell surface membrane glycoprotein synthesis. II. Glycosyltransferases and endogenous acceptors of the undifferentiated cell surface membrane. J Biol Chem. 1973;248:2542–2548. [PubMed] [Google Scholar]
- 36.Smartt HJ, et al. p27kip1 Regulates cdk2 activity in the proliferating zone of the mouse intestinal epithelium: Potential role in neoplasia. Gastroenterology. 2007;133:232–243. doi: 10.1053/j.gastro.2007.04.043. [DOI] [PubMed] [Google Scholar]
- 37.Flandez M, Guilmeau S, Blache P, Augenlicht LH. KLF4 regulation in intestinal epithelial cell maturation. Exp Cell Res. 2008;314:3712–3723. doi: 10.1016/j.yexcr.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi L, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi L, et al. The balance of reproducibility, sensitivity, and specificity of lists of differentially expressed genes in microarray studies. BMC Bioinformatics. 2008;9(Suppl 9):S10. doi: 10.1186/1471-2105-9-S9-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.