Abstract
The enzyme guanylyltransferase (GTase) plays a central role in the three-step catalytic process of adding an m7GpppN cap cotranscriptionally to nascent mRNA (pre-mRNAs). The 5′-mRNA capping process is functionally and evolutionarily conserved from unicellular organisms to human. However, the GTases from viruses and yeast have low amino acid sequence identity (∼25%) with GTases from mammals that, in contrast, are highly conserved (∼98%). We have defined by limited proteolysis of human capping enzyme residues 229–567 as comprising the minimum enzymatically active human GTase (hGTase) domain and have determined the structure by X-ray crystallography. Seven related conformational states of hGTase exist in the crystal. The GTP-binding site is evolutionarily and structurally conserved. The positional variations of the oligonucleotide/oligosaccharide binding fold lid domain over the GTP-binding site provide snapshots of the opening and closing of the active site cleft through a swivel motion. The pattern of conserved surface residues in mammals, but not in yeast, supports the finding that the recognition of the capping apparatus by RNA polymerase II and associated transcription factors is highly conserved in mammals, and the mechanism may differ somewhat from that in yeast. The hGTase structure should help in the design of biochemical and molecular biology experiments to explore the protein∶protein and protein∶RNA interactions that ensure regulated transcription of genes in humans and other mammals.
Keywords: gene expression, 5′ modification, RNA initiation
Addition of a 5′ terminal m7GpppN cap structure to nascent transcripts is a critical step in the formation of RNA polymerase II (PolII) functional products, notably mRNAs. This modification is one of the earliest events in gene expression and a characteristic feature of eukaryotic transcription (1, 2). Capping is cotranscriptional and occurs when the chain length of the mRNAs (pre-mRNAs) reaches approximately 25 nucleotides (3–5). PolII with its associated factors then facilitate the binding of capping enzyme (CE) to the complex and the pre-mRNA initiated 5′ end becomes accessible to, and modified by, the PolII-bound CE.
In addition to marking transcription start sites on genes and blocking a potential exonucleolytic attack of pre-mRNAs/mRNAs (6), the cap facilitates splicing (7, 8) and is recognized by a nuclear cap-binding protein complex for nucleocytoplasmic transport of processed mRNAs (9). Subsequently, a different, cytoplasmic cap-binding complex promotes ribosome attachment and translation initiation as well as its regulation (10–13). In light of these effects of cap on key stages in gene expression, it is not surprising that capping is essential for viability from yeast to human (14–19).
Capping of nascent RNA 5′ ends is accomplished in eukaryotic cells and for most viruses in three sequential catalytic steps: removal of the gamma phosphate by RNA triphosphatase (RTase), addition of GMP from GTP by guanylyltransferase (GTase) via a phosphoamide linked GMP–enzyme intermediate, and N7 methylation of the added GMP by RNA (guanine-N7) methyltransferase (MTase) (20). The first two steps are catalyzed in metazoans by a bifunctional CE consisting of N-terminal RTase and C-terminal GTase domains. However, in yeast species these activities are contained in separate but necessarily interacting enzymes (21). In addition, yeast RTase is cation-dependent (22) whereas mammalian and Caenorhabditis elegans RTases use a cation-independent protein tyrosine phosphatase catalytic mechanism involving formation of a phosphocysteine intermediate at an active site motif, (I/V)HCXXGXXR(S/T)G (23–25) that is absent in the yeast enzyme. Although differing in organization, sequence, and—in the case of RTase—catalytic mechanism, capping enzymes are functionally conserved from single-cell to multicellular organisms as demonstrated by growth complementation of yeast deletion mutants by mammalian enzymes (15) as well as the viability of yeast in which the endogenous capping system has been completely replaced by mammalian enzymes (26).
In mammalian cells, as in yeast, CE binds to the largest subunit of PolII, Rpb1, via the GTase domain (15, 27, 28). Binding occurs early in transcription and depends on transcription factor IIH (TFIIH)-catalyzed specific phosphorylation of serine residues in the YSPTSPS heptad repeats comprising the Rpb1 C-terminal domain (CTD) (29). This protein∶protein interaction stimulates capping (30, 31). CE activity is also increased by binding to transcription factor Spt5 (31), a subunit of the 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole sensitivity-inducing factor (DSIF). CE binding to DSIF relieved transcription repression by negative elongation factor, consistent with a possible role of CE in elongation checkpoint control during promoter clearance (29). These and other results point to a functional connection between capping, transcription, and protein targeting in a multifaceted cellular network of interacting pathways required for viability.
To decipher how protein∶protein interactions involving CE, in particular the GTase domain, may regulate gene expression in human cells, it is important to know the structure of the human enzyme at atomic resolution. Structures have been determined for yeast monofunctional CE (32, 33) and several viruses that replicate in the cytoplasm and often assemble viral genome-encoded RNA polymerases and capping enzymes into infectious virions. Examples include the DNA-containing Paramecium bursaria Chlorella virus 1 (PBCV-1) that encodes a monofunctional GTase (34), and vaccinia virus that contains a trifunctional capping protein (35, 36). The dsRNA reoviruses also apparently contain a multifunctional virus encoded capping protein with GTase and MTase activities (37, 38). Other variations on the capping theme have evolved among animal viruses, e.g., influenza virus “cap snatching” from host transcripts in the nucleus as primers of viral transcription (39). Although the structure of the RTase domain of human capping enzyme (hCE) has been determined (PDB ID code 2C46), no structures of mammalian full-length CE or the GTase domain are available. Here we report the crystal structure of the GTase domain of hCE and compare its salient features with those of GTases from PBCV-1 virus and two yeast species, Saccharomyces cerevisiae and Candida albicans. The structure of human GTase (hGTase) should provide insight into a deeper understanding of the mechanisms of regulated transcription of human and other mammalian genes.
Results and Discussion
Structure Solution.
We have determined the crystal structure of the hGTase domain of hCE at 3.0 Å resolution (Table S1). Numerous attempts to crystallize the full-length hCE (amino acids 1–597) over many years failed to produce crystals. Amino acid sequence analysis of hCE indicated that the N-terminal human RNA triphosphatase (hRTase) segment and C-terminal hGTase segments are connected by a linker of approximately 25 amino acid residues that are predicted to form a flexible loop structure. Additionally, previous studies have shown that the two segments may not have any stable interaction (40). We turned to a systematic proteolytic digestion experiment to generate stable, active hGTase fragments from the full-length hCE. Bovine plasmin cleaved hCE into two main, stable fragments that were resistant to further digestion. Mass spectrometry and SDS-PAGE analysis of the digest demonstrated products of approximately 25.6 and 40.5 kDa, suggesting that they correspond to the N-terminal hRTase and C-terminal hGTase domains, respectively (Fig. 1A and data not shown). Several hGTase constructs were then generated based on the mass of the plasmin-cleaved putative hGTase domain and protein structure predictions; fragments 222–574, 224–567, 229–567, and 229–569 exhibited GTase activities comparable to that of full-length hCE as measured by the formation of hGTase–GMP intermediate (Fig. 1 B and C, Table 1) and transfer of the P32-labeled GMP to RTase-processed, 5′-diphosphate-ended single-stranded RNA (31, and data not shown).
Fig. 1.
Plasmin digestion of CE and guanylylation of GT truncations. Plasmin-digested hCE was analyzed by MALDI–TOF mass spectrometry (A). Fragments corresponding in mass to RTase (∼25.6 kDa) and to GTase (∼40.5 kDa) are apparent. (B and C) hGTase activity was measured by α[32P]GTP binding and labeling of full-length CE and GTase truncations; α[32P]GTP (10 μCi, 3,000 Ci/mmol) was incubated with 20 ng (B) or 100 ng (C) of the indicated constructs, and the GMP-labeled enzyme capping intermediates were resolved by SDS-PAGE and exposed to storage phosphor image plates.
Table 1.
Mutation analysis of GTase constructs of hCE
| Mutants | GTP binding |
| GT222-574 | +++ |
| GT229-569 | +++ |
| GT240-569 | insoluble |
| GT224-468 | + |
| GT224-475 | + |
| GT224-551 | +/− |
| GT224-567 | +++ |
| GT229-468 | + |
| GT229-475 | + |
| GT229-551 | + |
| GT229-567 | +++ |
| GT229-567 (M335I/M437V/RM476KV/M543V/M667I) | +++ |
| GT229-569 (M396A/M437A/M527A) | +/− |
| (M335A/M437A/M527A/M543A) | − |
| (M335A/M396A/M437A/M527A/M543A) | − |
| GT229-569 E234A | +++ |
| GT229-569 K458A | − |
| GT229-569 K460A | − |
| GT229-569 R528A | + |
| GT229-569 R530A | − |
| GT229-569 D532A | − |
| GT229-569 K533A | − |
GT229-567, GT229-569, and the listed methionine mutants containing Se-Met had activities comparable to the corresponding unsubstituted proteins. Activity levels: +++ > 85% of WT; + 10–22% of WT; +/- < 10% of WT; - undetectable.
The limited proteolysis results followed by cloning and expression of hGTase segments revealed amino acid residues 229–567 as the minimum segment that has activity comparable to hCE. The C-terminal 30-residue stretch (568–597) that contains a patch of five consecutive prolines and five basic residues (KRKHH) apparently is not required for GTase activity and hindered crystallization. Fragments 229–569 and 229–567 exhibited similar enzymatic activity and produced crystals using ammonium sulfate as precipitant that diffracted to 3.5 and 3.0 Å, respectively, at high-flux synchrotron beamlines. These hGTase constructs were monomeric and monodisperse in solution as revealed by dynamic light scattering; however, aggregation occurred rapidly with increasing concentration and decreasing temperature. There are eight cysteine residues in hGTase, and the refined structure later confirmed that it contained no disulfide bonds.
No mammalian GTase X-ray structure is available, and the sequence identity of hGTase with the GTases from PBCV-1 virus (41) and C. albicans (32) is only 22 and 25%, respectively. Despite the low sequence identity, hGTase is assumed to have a structure related to the structures of the PBCV-1 and C. albicans GTases as all three use GTP catalytically to form a stable enzyme intermediate by covalent phosphoamide linkage with GMP which is then transferred to the 5′-end of pre-mRNA, forming a GpppN premethylation cap structure. However, our attempts to solve hGTase structures using the monomeric structures of GTases from PBCV-1 or C. albicans failed to generate an unambiguous molecular replacement (MR) solution. The self-rotation function in Patterson space indicated the presence of a twofold noncrystallographic symmetry (NCS) axis, and in consideration of the twofold NCS, the unit cell content analysis predicted that the number of hGTase molecules in an asymmetric unit could be four, six, or eight. Our hGTase construct contains 12 methionine residues, and although selenomethionine (Se-Met)-substituted hGTase yielded active protein (Table 1), various Se-Met hGTase constructs readily aggregated and did not produce crystals. For example, hGTase constructs containing all 12 methionine residues replaced by Se-Met or only seven Se-Met residues, produced by mutating the remaining five methionines to isoleucine or valine based on homology analysis, were also active (Table 1) but failed to produce crystals. Finally, we took a tedious trial-and-error approach in which MR solutions were generated, followed by NCS averaging among the molecules in an asymmetric unit. The map correlation and map quality were checked for each solution.
After several failed attempts, a promising solution was obtained that contained two trimers per asymmetric unit using the domain 1 of the PBCV-1 GTase (PDB ID code 1CNK; amino acids 1–236). A sixfold NCS-averaged map guided the modeling of the hGTase protein residues 229–460. Refinement of the partial structure and further MR search in the presence of the six partial hGTase molecules found the existence of a seventh hGTase molecule in the asymmetric unit which does not obey the twofold NCS. Sevenfold NCS averaging was employed to improve the quality of electron density maps and to overcome model bias. The averaged electron density maps were of good quality that guided the building of the seven hGTase molecules and their side chains, even at a moderate resolution of 3.0 Å. Cycles of model building and NCS averaging improved and extended the model (Fig. 2) that refined to a final R and R-free of 0.258 and 0.296, respectively (Table S1).
Fig. 2.
Structure of the hGTase domain of hCE. hGTase has an ATP-grasp fold domain with two subdomains (green and blue) and an OB-fold domain (orange). The secondary structural elements (1–15 β-strands and A–G α-helices) are labeled. The GTP sits between the blue and green subdomains with the phosphates pointing to the cleft. The conserved residue K294 forms a phosphoamide-linked GMP intermediate with the removal of PPi, followed by transfer of the GMP to the diphosphate-ended pre-mRNA.
Structure of hGTase.
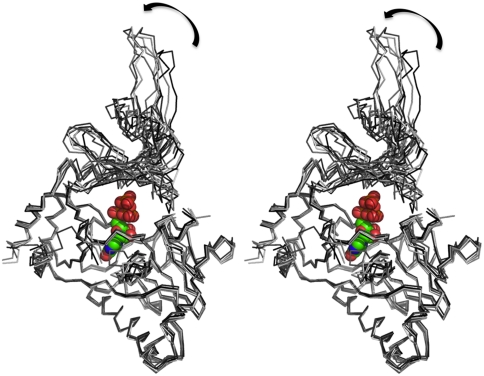
The hGTase structure is composed of seven helices and 15 β-strands distributed into three antiparallel β-sheets composed of seven, five, and three strands, and the structure has an overall GTase/DNA ligase fold (Fig. 2) (42, 43). The N-terminal nucleotide transferase domain has an ATP-grasp fold. In the context of hGTase structure, we termed the two subdomains of the N-terminal domain as the base (amino acids 271–415) and hinge (amino acids 229–270, 416–461, and 553–567). The domain composed of amino acid residues 462–552 has an oligonucleotide/oligosaccharide binding fold (OB-fold) that is positioned as a lid over the base subdomain (Fig. 2). The GTP-binding site is between the base and the hinge and highly conserved in GTases from virus to human. A comparison of the crystal structure of hGTase with the structures of GTases from PBCV-1 virus and C. albicans reveals that the structures of the guanine-binding pocket and the hinge region are most conserved, and the structural differences are larger as one moves away from this region (Fig. S1). The lid part shows the highest deviations; relative positioning of the base and lid domains defines the open/closed conformations of the cleft that (i) accommodates the triphosphate moiety of GTP, (ii) contains Lys294, which forms a covalent link with GMP as GTase cleaves off the β,γ-phosphates, (iii) accommodates the RTase-processed 5′ diphosphate end of pre-mRNA, and (iv) transfers GMP to the 5′-end. The C. albicans GTase has a wide-open cleft and the PBCV-1 GTase structure has an open and a closed conformation (41, 42). The seven monomers of hGTase molecules in an asymmetric unit define seven conformations of the cleft. The superposition of the seven hGTase molecules (Fig. 3) showed large variations in the positioning of the lid domain and some variations in the positioning of the β6–β7 loop.
Fig. 3.
Structural variations among the hGTase molecules. A stereoview showing the superposition of the seven hGTase molecules in the asymmetric unit. The large variations in the positioning of the OB-domain with respect to the nucleotide transferase domain reveals the mode of interdomain movements that is essential for the activity of the enzyme. In the DNA ligase domain, the GTP-binding pocket is highly superimposable; however, the distant structural motif β6–β7 hairpin shows structural variation. The GTP molecule is positioned based on structural superposition of the Chlorella PBCV-1 virus GTase (PDB ID code 1CKN-A) on hGTase.
hGTase vs. Other GTase Structures.
A Dali search against one of the hGTase monomers revealed Chlorella PBCV-1 GTase (PDB ID codes 1CKM and 1CKN) as the structurally closest neighbor with a rmsd of approximately 3 Å, whereas the C. albicans (PDB ID code 1P16) and S. cerevisiae (PDB ID code 3KYH) structures had an rmsd of approximately 5 Å or higher. hGTase has only 22, 25, and 28% amino acid sequence identity with PBCV-1, C. albicans, and S. cerevisiae GTases, respectively. The conserved amino acid sequences of the four structurally characterized GTase molecules (Fig. S1B) are primarily located at the GTP-binding site or form the hydrophobic cores of GTases; consequently, the overall fold of the enzyme is evolutionarily conserved. Sequence analysis shows high conservation (∼98%) among mammalian GTases, including hGTase. Comparison of the electrostatic potential surfaces of the four structurally characterized GTases revealed different shape and surface charge distributions (Fig. 4). Viral GTases would be expected to work in association with a viral polymerase or a part of a viral polymerase complex, and in either case, the GTase would function in the cytoplasm of the infected host cell. In contrast, during transcription in the nucleus of yeast and mammalian cells, GTase is recognized by YSPTSPS heptad repeat sequences at the CTD of the Rpb1 subunit of PolII and transcription factors such as Spt5 (31). In the light of the conserved cellular mechanism, it is surprising that the amino acid sequence and structural features such as shape and surface charge distribution are distinct (Fig. 4), possibly reflecting differences in the pre-mRNA capping process in mammalian vs. unicellular eukaryotic systems.
Fig. 4.
Molecular surfaces of GTases. Electrostatic potential surface of four structurally characterized GTase shows significant variations in shape and surface electrostatic potential. hGTase has relatively close resemblance to Chlorella PBCV-1 virus GTase but significantly different from C. albicans and S. cerevisiae GTases.
Structural Conservation Among the GTases.
Despite high variability on the surfaces of GTases, the GTP-binding sites, the phosphoamide linking lysine (K294 in hGTase) and surrounding residues, and the electropositivity of the active site clefts are conserved in all structures. However, the shapes of the cleft, which are primarily determined by the shape and position of the lid domain, are significantly different in the different GTase structures (Fig. 4). In C. albicans and S. cerevisiae GTase structures, the active site cleft is predominantly open. The Chlorella PBCV-1 virus GTase structure revealed two distinct, open and closed, conformations of the active site cleft (41, 42). Comparison of the two conformations suggests that the lid domain may open and close like a jaw to facilitate GTP binding and addition of GMP to the 5′-ppN end of pre-mRNA to form the unmethylated cap. The hGTase structure revealed different conformations of the cleft from closed to half-open (Fig. 3). A superposition of the seven hGTase molecules in the asymmetric unit of the crystal indicates that the lid domain opens in a swivel motion relative to the base.
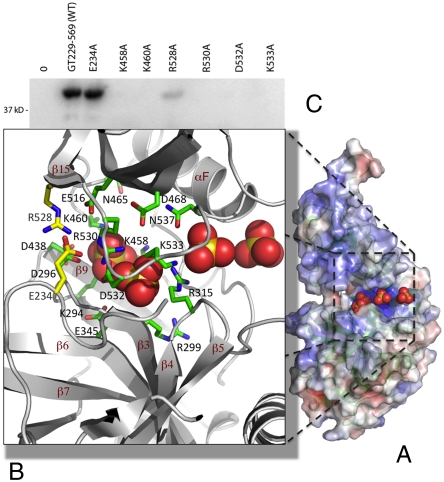
The crystals were grown in presence of ammonium sulfate, and one of the hGTase molecules has four sulfate ions bound at the active site cleft (Fig. 5). We predict that the sulfate binding reflects the binding of phosphates of GTP/RNA to hGTase. The first sulfate ion is positioned near K294 of the conserved KXDG active site sequence, analogous to the phosphate of GMP, and the remaining three sulfates define a track from the first sulfate ion to the solvent region through the positively charged cleft. The S–S distance between the consecutive sulfate ions is 6.5–7.5 Å, which is intriguingly similar to the distance between consecutive phosphates in single-stranded RNA. The sulfate-binding track is highly conserved, and mutation of residues interacting with the sulfates (motif V) has been shown to be lethal in mouse CE (mCE) tested in a yeast complementation assay (44). The study identified seven highly conserved regions in GTases including motifs III and IIIa that are primarily responsible for the fold of the enzyme. Unlike the yeast open-cleft GTase structures, all seven conserved motifs surround the sulfate-binding track in the hGTase structure, and motifs V (hGTase amino acids 456–468) and VI (amino acids 524–537) are most involved. K458 and K460 in motif V are likely to interact with the phosphate of GMP, and motif VI in the loop connecting β15 and αF of hGTase is closed down on the base domain. We made a set of individual mutations in motifs V and VI and assayed them for enzymatic activity. K458A, K460A, R528A, R530A, D532A, and K533A were inactive or, in the case of R528A, formed significantly less phosphoamide-linked GMP-enzyme intermediate which suggests that these residues are responsible for GTP binding/formation of GMP-linked GTase (Table 1). By contrast, alanine mutation of position E234 located away from the sulfate-binding track did not inhibit the formation of the phosphoamide intermediate (Fig. 5). The GMP-enzyme intermediate state would be expected to interact with the 5′-end of RTase-processed pre-mRNA, and the conserved sulfate-bound track (Fig. 5A) may facilitate binding of the 5′diphosphate end of pre-mRNA to the GMP-linked GTase intermediate. Alternatively, the RNA may enter the cleft from the opposite side (Fig. 4).
Fig. 5.
Active site cleft of hGTase. (A) Four sulfate ions [in Corey–Pauling–Koltun (CPK) model] are bound at the active site cleft of one of the hGTase molecules; the molecule is viewed at approximately 70° clockwise rotation about the y axis with respect to the view in Fig. 4. The amino acid residues surrounding the sulfate ions are highly conserved and, if mutated, impair or eliminate GTase activity in human (Table 1) and mCE (44). (C) hGTase containing E234A and R528A mutations (positions shown as yellow side-chains in B) retained full and 10% activity, respectively.
The GTase enzyme fold and the architecture of the active site region are evolutionarily conserved; however, the overall shape and the molecular surface of hGTase, that are expected to be conserved in mammalian GTases based on amino acid sequence conservation, are different in unicellular eukaryotic organisms. The structural information of the hGTase enzyme, together with the available structures of human RTase and mouse MTase (45) will guide structural and molecular studies to elucidate further the mRNA capping events in transcription of mammalian genes.
Materials and Methods
His-tagged full-length hCE, expressed and purified as described (31), was digested at 5 mg/mL with plasmin (0.035 mg/mL), and the resulting protease-resistant core fragments were analyzed by SDS-PAGE and mass spectrometry. Regions corresponding to the C-terminal GTase domain were PCR amplified, sequenced, cloned, expressed, purified, and characterized. GTase construct 229–567 was crystallized, and the structure was determined by X-ray crystallography. Details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Dr. J. Marcotrigiano for helpful comments on the manuscript, A.D. Clark, Jr. and C. Dharia for their help with experiments, and Drs. J. Marcotrigiano and S. Gurla for discussion and suggestions. We acknowledge Brookhaven National Laboratory and Cornell High Energy Synchrotron Source for their generous help with data collection at X25 and F1 beamlines, respectively. This work was supported in part by National Institute of Allergy and Infectious Diseases R21 award AI 087201 (to K.D.), National Institutes of Health R37 MERIT Award AI 27690 (to E.A.), and National Institute of General Medical Sciences Protein Structure Initiative Grant U54 GM094597 (to G.T.M.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The crystallography, atomic coordinates, and structure factors for hGTase have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3S24).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106610108/-/DCSupplemental.
References
- 1.Shatkin AJ. Capping of eukaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 2.Furuichi Y, et al. Methylated, blocked 5 termini in HeLa cell mRNA. Proc Natl Acad Sci USA. 1975;72:1904–1908. doi: 10.1073/pnas.72.5.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagler J, Shuman S. A freeze-frame view of eukaryotic transcription during elongation and capping of nascent mRNA. Science. 1992;255:983–986. doi: 10.1126/science.1546295. [DOI] [PubMed] [Google Scholar]
- 4.Coppola JA, Field AS, Luse DS. Promoter-proximal pausing by RNA polymerase II in vitro: Transcripts shorter than 20 nucleotides are not capped. Proc Natl Acad Sci USA. 1983;80:1251–1255. doi: 10.1073/pnas.80.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci USA. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furuichi Y, LaFiandra A, Shatkin AJ. 5′-terminal structure and mRNA stability. Nature. 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 7.Konarska MM, Padgett RA, Sharp PA. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 8.Edery I, Sonenberg N. Cap-dependent RNA splicing in a HeLa nuclear extract. Proc Natl Acad Sci USA. 1985;82:7590–7594. doi: 10.1073/pnas.82.22.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izaurralde E, Stepinski J, Darzynkiewicz E, Mattaj IW. A cap binding protein that may mediate nuclear export of RNA polymerase II-transcribed RNAs. J Cell Biol. 1992;118:1287–1295. doi: 10.1083/jcb.118.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filipowicz W, et al. A protein binding the methylated 5′-terminal sequence, m7GpppN, of eukaryotic messenger RNA. Proc Natl Acad Sci USA. 1976;73:1559–1563. doi: 10.1073/pnas.73.5.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonenberg N, Rupprecht KM, Hecht SM, Shatkin AJ. Eukaryotic mRNA cap binding protein: Purification by affinity chromatography on sepharose-coupled m7GDP. Proc Natl Acad Sci USA. 1979;76:4345–4349. doi: 10.1073/pnas.76.9.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonenberg N, Trachsel H, Hecht S, Shatkin AJ. Differential stimulation of capped mRNA translation in vitro by cap binding protein. Nature. 1980;285:331–333. doi: 10.1038/285331a0. [DOI] [PubMed] [Google Scholar]
- 13.Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA. 2011;2:277–298. doi: 10.1002/wrna.52. [DOI] [PubMed] [Google Scholar]
- 14.Mao X, Schwer B, Shuman S. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol Cell Biol. 1995;15:4167–4174. doi: 10.1128/mcb.15.8.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yue Z, et al. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukamoto T, et al. Isolation and characterization of the yeast mRNA capping enzyme beta subunit gene encoding RNA 5′-triphosphatase, which is essential for cell viability. Biochem Biophys Res Commun. 1997;239:116–122. doi: 10.1006/bbrc.1997.7439. [DOI] [PubMed] [Google Scholar]
- 17.Srinivasan P, Piano F, Shatkin AJ. mRNA capping enzyme requirement for Caenorhabditis elegans viability. J Biol Chem. 2003;278:14168–14173. doi: 10.1074/jbc.M212102200. [DOI] [PubMed] [Google Scholar]
- 18.Shafer B, Chu C, Shatkin AJ. Human mRNA cap methyltransferase: Alternative nuclear localization signal motifs ensure nuclear localization required for viability. Mol Cell Biol. 2005;25:2644–2649. doi: 10.1128/MCB.25.7.2644-2649.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu C, Shatkin AJ. Apoptosis and autophagy induction in mammalian cells by small interfering RNA knockdown of mRNA capping enzymes. Mol Cell Biol. 2008;28:5829–5836. doi: 10.1128/MCB.00021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furuichi Y, Muthukrishnan S, Tomasz J, Shatkin AJ. Mechanism of formation of reovirus mRNA 5′-terminal blocked and methylated sequence, m7GpppGmpC. J Biol Chem. 1976;251:5043–5053. [PubMed] [Google Scholar]
- 21.Ho CK, Lehman K, Shuman S. An essential surface motif (WAQKW) of yeast RNA triphosphatase mediates formation of the mRNA capping enzyme complex with RNA guanylyltransferase. Nucleic Acids Res. 1999;27:4671–4678. doi: 10.1093/nar/27.24.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho CK, Pei Y, Shuman S. Yeast and viral RNA 5′ triphosphatases comprise a new nucleoside triphosphatase family. J Biol Chem. 1998;273:34151–34156. doi: 10.1074/jbc.273.51.34151. [DOI] [PubMed] [Google Scholar]
- 23.Takagi T, Moore CR, Diehn F, Buratowski S. An RNA5′-triphosphatase related to the protein tyrosine phosphatases. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 24.Wen Y, Yue Z, Shatkin AJ. Mammalian capping enzyme binds RNA and uses protein tyrosine phosphatase mechanism. Proc Natl Acad Sci USA. 1998;95:12226–12231. doi: 10.1073/pnas.95.21.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lima CD, Wang LK, Shuman S. Structure and mechanism of yeast RNA triphosphatase: An essential component of the mRNA capping apparatus. Cell. 1999;99:533–543. doi: 10.1016/s0092-8674(00)81541-x. [DOI] [PubMed] [Google Scholar]
- 26.Saha N, Schwer B, Shuman S. Characterization of human, Schizosaccharomyces pombe, and Candida albicans mRNA cap methyltransferases and complete replacement of the yeast capping apparatus by mammalian enzymes. J Biol Chem. 1999;274:16553–16562. doi: 10.1074/jbc.274.23.16553. [DOI] [PubMed] [Google Scholar]
- 27.McCracken S, et al. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandal SS, et al. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc Natl Acad Sci USA. 2004;101:7572–7577. doi: 10.1073/pnas.0401493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho CK, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 31.Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 1999;13:1774–1779. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabrega C, Shen V, Shuman S, Lima CD. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol Cell. 2003;11:1549–1561. doi: 10.1016/s1097-2765(03)00187-4. [DOI] [PubMed] [Google Scholar]
- 33.Gu M, Rajashankar KR, Lima CD. Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA capping apparatus. Structure. 2010;18:216–227. doi: 10.1016/j.str.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho CK, Van Etten JL, Shuman S. Expression and characterization of an RNA capping enzyme encoded by Chlorella virus PBCV-1. J Virol. 1996;70:6658–6664. doi: 10.1128/jvi.70.10.6658-6664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venkatesan S, Gershowitz A, Moss B. Modification of the 5′ end of mRNA. Association of RNA triphosphatase with the RNA guanylyltransferase-RNA (guanine-7-) methyltransferase complex from vaccinia virus. J Biol Chem. 1980;255:903–908. [PubMed] [Google Scholar]
- 36.Wang SP, Deng L, Ho CK, Shuman S. Phylogeny of mRNA capping enzymes. Proc Natl Acad Sci USA. 1997;94:9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luongo CL, Contreras CM, Farsetta DL, Nibert ML. Binding site for S-adenosyl-L-methionine in a central region of mammalian reovirus lambda2 protein. Evidence for activities in mRNA cap methylation. J Biol Chem. 1998;273:23773–23780. doi: 10.1074/jbc.273.37.23773. [DOI] [PubMed] [Google Scholar]
- 38.Luongo CL, Reinisch KM, Harrison SC, Nibert ML. Identification of the guanylyltransferase region and active site in reovirus mRNA capping protein lambda2. J Biol Chem. 2000;275:2804–2810. doi: 10.1074/jbc.275.4.2804. [DOI] [PubMed] [Google Scholar]
- 39.Plotch SJ, Bouloy M, Ulmanen I, Krug RM. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 40.Yagi Y, Mizumoto K, Kaziro Y. Limited tryptic digestion of messenger RNA capping enzyme from Artemia salina. Isolation of domains for guanylyltransferase and RNA 5′-triphosphatase. J Biol Chem. 1984;259:4695–4698. [PubMed] [Google Scholar]
- 41.Hakansson K, Doherty AJ, Shuman S, Wigley DB. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 42.Gu M, Lima CD. Processing the message: Structural insights into capping and decapping mRNA. Curr Opin Struct Biol. 2005;15:99–106. doi: 10.1016/j.sbi.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 43.Doherty AJ. Conversion of a DNA ligase into an RNA capping enzyme. Nucleic Acids Res. 1999;27:3253–3258. doi: 10.1093/nar/27.16.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sawaya R, Shuman S. Mutational analysis of the guanylyltransferase component of mammalian mRNA capping enzyme. Biochemistry. 2003;42:8240–8249. doi: 10.1021/bi034396d. [DOI] [PubMed] [Google Scholar]
- 45.Fabrega C, Hausmann S, Shen V, Shuman S, Lima CD. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol Cell. 2004;13:77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.